Abstract
Neuronally enriched RGS4 plays a critical role attenuating G protein signaling in brain, although the mechanisms regulating RGS4 expression are unknown. Here we describe a novel mechanism for transcriptional activation of RGS4 in neuron-like PC6 cells, where RGS4 is markedly induced during confluence-induced growth arrest. Transcriptional activation of RGS4 in confluent PC6 cells was accompanied by impaired Gi/o-dependent MAPK activation. In the human RGS4 gene promoter, we identified three phylogenetically conserved cis-elements: an inverted CCAAT box element (ICE), a cAMP response element, and a B-cell lymphoma 6 (Bcl6)-binding site. The ICE and the cAMP response element mediate activation, and the Bcl6 site mediates repression of RGS4 transcription. Activation of RGS4 transcription in confluent PC6 cells is accompanied by increases in NF-YA and C/EBPβ and decreases in Bcl6 levels in the nucleus. Increases in NF-YA and C/EBPβ lead to their increased binding to the RGS4 promoter in vivo, and dominant negative forms of these proteins repressed RGS4 promoter activity. Acetylation of NF-YA and Bcl6 were increased in postconfluent cells. Trichostatin A stimulation of RGS4 promoter activity, accompanied by increased binding of NF-YA and decreased binding of Bcl6 to the promoter, was abolished by mutation of the ICE and enhanced by mutation of the Bcl6 site. These findings demonstrate a dynamic and coordinated regulation of nuclear levels and acetylation status of trans-acting factors critical in determining the off/on state of the RGS4 promoter.
Keywords: G Proteins, Gene Transcription, Signal Transduction, Transcription Factors, Transcription Regulation, Acetylation, Bcl6, NF-YA, RGS4 Gene
Introduction
Regulator of G protein signaling 4 (RGS4)2 is a member of the mammalian RGS family of proteins of which 30 members exist in humans. RGS proteins were discovered as essential negative regulators of heterotrimeric G protein signaling by genetic studies in yeast and Caenorhabditis elegans (1, 2). These proteins act as GTPase-activating proteins for heterotrimeric Gα subunits (3), thereby accelerating the shut-off mechanism for G protein signaling. Some RGS proteins, including RGS4, can also act as effector antagonists (4, 5) or can directly or indirectly interact with G protein-coupled receptors (6–8), actions that contribute to their negative regulatory effects on G protein signaling in cells.
Although G protein signaling is involved in virtually every known physiological process and RGS proteins are an important component of this signaling, the mechanisms regulating expression of RGS genes are largely unknown. Studies have shown that several RGS genes are induced under certain physiological conditions, for example RGS1 during mitogenic activation of lymphocytes (9), RGS2 in the early stage of 3T3-L1 differentiation to adipocytes (10), RGS16 during genotoxic stress (11), and the yeast RGS gene SST2 by pheromone (12). Regulation of RGS4 expression has also been noted in facial motoneuronal precursors during embryonic development in mice (13), in epithelial and endothelial cells during tubulogenesis (14), and in rat PC12 cells during cell confluence (15). In the study by Grillet et al. (13), in situ hybridization was used to demonstrate that RGS4 transcription is increased in differentiating and post-mitotic neurons in the developing mouse nervous system. Here we found that neuronal-like PC6 cells exhibit a similar activation of RGS4 transcription during confluence-induced mitotic exit. These cells thus present an attractive experimental system to study the mechanism of RGS4 gene activation observed during neuronal differentiation.
We undertook studies to identify and characterize the human RGS4 promoter and to gain insights into the molecular mechanisms regulating the expression of the RGS4 gene. Here we provide the first comprehensive analysis and functional evaluation of a promoter for an RGS gene, identifying critical cis- and trans-acting elements and a dynamic and coordinated regulatory mechanism to control the on/off status of the RGS4 gene. We also assessed possible roles of up-regulated RGS4 in G protein signaling.
EXPERIMENTAL PROCEDURES
Materials
Marathon®-ready cDNA was purchased from Clontech Laboratories Inc. Elongase was from Invitrogen. cDNAs encoding various transcription factors used in this study were generously provided by other investigators: NF-YA from Dr. Hiroyoshi Ariga; NF-YA29, the dominant negative mutant of NF-YA that lacks DNA binding activity (16), from Dr. Roberto Mantovani; and A-C/EBP, the dominant negative C/EBPβ that lacks DNA binding and trans-activation domains (17), from Dr. Charles Vinson. Human histone deacetylase 1 cDNA was amplified from human brain cDNA (Clontech) and cloned into the pCMV-Tag mammalian expression vector (Stratagene). The LAP isoform of rat C/EBPβ was amplified from rat genomic DNA and subcloned in pcDNA3 vector (Invitrogen). RGS4-specific antibody (U1079) (15) was generously provided by Dr. Susan Mumby. Oligonucleotide primers and other molecular biological reagents were obtained from the University of Iowa DNA Core Facility.
Identification and Cloning of the Human RGS4 Gene Promoter
The transcription start site of RGS4 was identified using 5′-RACE with Marathon®-ready cDNA and RGS4-specific primers essentially as we described previously (18). The transcription start site of RGS4 was designated as +1 with the translation start site as +124. The promoter region of human RGS4 (bp −5874 to +124) was then cloned into the luciferase reporter plasmid pGL3-basic using PCR. The cloned RGS4 promoter sequence was validated by sequencing at the University of Iowa DNA Core Facility. Base substitution mutants of RGS4 promoter were constructed using a site-directed mutagenesis kit (Stratagene). The ICE was mutated from ATTGG (wild type) to an ACCGG (mutant), the CRE was mutated from TCGTCA (wild type) to an TAATCA (mutant), and the Bcl6 binding site was mutated from CTAGA (wild type) to an GGAGT (mutant). Mutations were confirmed by sequencing at the University of Iowa DNA Core Facility.
Cell Culture, Transfection, and Luciferase Reporter Assay
Rat PC6 cells were cultured in RPMI 1640 medium supplemented with 10% horse serum, 5% fetal bovine serum, and 0.1% gentamicin. Subconfluent and confluent cultures were defined as cultures covering 35–45 and 100% areas of the culture dish, respectively. Postconfluent cultures were allowed to grow 1 more day after reaching confluence. Transfection was performed in 24-well plates 24 h after plating cells, using FuGENE 6 (Roche Applied Science) according to the manufacturer's procedure. Wild type and mutants of the RGS4 promoter in pGL3 were co-transfected with the pRL-SV40 plasmid, which was used as a control for transfection efficiency. 24–36 h after transfection, the cells were harvested and assayed for firefly and Renilla luciferase activities using the dual luciferase assay kit, according to the manufacturer's protocol (Promega). The final results were expressed as relative luciferase units, with firefly luciferase activity normalized to Renilla luciferase activity.
Real Time PCR
PC6 cells were plated in 6-well plates at various densities so that 1 day later they were subconfluent, confluent, and postconfluent. Total RNA was isolated from these cells using a Qiagen RNeasy kit according to the manufacturer's recommendations. The first strand of cDNA was synthesized from 2 μg of total RNA using a SuperScriptIII first strand synthesis system (Invitrogen). Real time PCR was carried out using iQTM SYBR® Green Supermix (Bio-Rad) by following the manufacturer's protocol. The primers were the following: RGS4 forward, GCA AAG GAT ATG AAA CAT CGG CTG GGA; RGS4 reverse, TTC TTG GCT CAC CCT CTG GCA AGT T; 18 S rRNA forward, CAA AGA TTA AGC CAT GCA TGT CTA AGT ACG C; 18 S rRNA reverse, GGC ATG TAT TAG CTC TAG AAT TAC CAC AGT TAT CC; ChIP primer 1, GTA GTT TCT TCC CCC TTT CTA A; ChIP primer 2, GGT CTC TTT TAT AGC CCA GCC AC; ChIP primer 3, GCG CTG ATT TTT GTA CCT AGT; and ChIP primer 4, TTA GAA AGG GGG AAG AAA CTA C.
Nuclear Extracts and Fluorescent EMSA
Nuclear extracts were prepared using a modified protocol of Yang et al. (19). Briefly, PC6 cells were collected and resuspended in ice-cold buffer A and then lysed with 0.5% of Nonidet P-40. The nuclei were pelleted by centrifugation at 5,000 × g for 1 min and then lysed in 200 μl of lysis buffer (50 mm Hepes/potassium hydroxide, pH 7.9, 150 mm KCl, 0.5% Triton X-100, 10% glycerol, 1 mm EDTA, and 1× protease inhibitors (Roche Applied Science)). After centrifugation at 12,000 × g for 10 min at 4 °C, the resulting supernatants were saved as nuclear extracts. Fluorescent EMSA were performed with a protocol described previously (20). Briefly, 2 μg of nuclear extracts were mixed with 1 pmol of DNA oligonucleotide in a reaction containing 0.6 μg of poly(dI-dC)·(dI-dC), 50 mm Hepes/potassium hydroxide, pH 7.9, 20% glycerol, 2.5 mm MgCl2, 2.5 mm DTT, 100 mm KCl, and 1.25 mm EDTA. For binding of endogenous Bcl6, 4 μg of nuclear extracts from PC6 cells were used. One μg of nuclear extract from Cos-7 cells, which alone had no binding to Bcl6 element, was added to each reaction to stabilize Bcl6 binding. A similar strategy was used to stabilize the binding of SREBP-1c to its binding site on PEPCK-C gene promoter (20). For the Bcl6 positive control reaction, 0.2 μg of nuclear extract from Cos-7 cells overexpressing Bcl6 was used. DNA fragments were generated by annealing two complementary oligonucleotides, one of which was labeled with IRDye700 at the 5′-end (LI-COR). The labeled oligonucleotides were 5′-/IRDye700/TAC GAC CGT CCA GCC AAT CAG ACG ACC GCT CTG-3′ for wild type ICE sequence and 5′-/IRDye700/CAG AGG TTT AGA ATT TCT AGA AAA GGA AAA AAA GTG-3′ for wild type Bcl6 element.
Biotin-labeled DNA Pulldown Assay
PC6 cells at subconfluence and postconfluence were lysed with radioimmune precipitation assay lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS). A biotin-labeled human RGS4 promoter DNA fragment was mixed with 500 μg of the above cell lysates and 100 μl of 4% streptavidin-agarose beads. After overnight incubation at 4 °C, the beads were washed three times with cold radioimmune precipitation assay lysis buffer, and precipitated complexes were eluted with 50 μl of SDS-PAGE sample buffer by boiling tubes for 5 min. The eluents were analyzed by SDS-PAGE and immunoblotting for NF-YA. Biotin-labeled DNA fragments were generated using PCR from wild type and ICE mutant RGS4 promoters using following primers: forwarding primer, 5′-/5Bio/CTG CAG GGC GGT CGT CTG ATT GGC TGG ACG GTC GTA GCT GGG TAT AAA AGA GA-3′; and reverse primer, 5′-TCT CTT TTA TAC CCA GCT ACG ACC GTC CAG CCA ATC AGA CGA CCG CCC TGC AG-3′.
Immunoblotting and Immunoprecipitation
SDS-PAGE and immunoblotting was performed as we described (21), using antibodies against RGS4 (U1079, 1:5000 dilution), NF-YA (SC-10779X, 1:10000 dilution), C/EBPβ (SC-150X, 1:10000 dilution), Bcl6 (SC-368X, 1:10000 dilution) (Santa Cruz Biotechnology Inc.), and LPA1r (10005280, 1:200 dilution) (Cayman Chemical Company, MI). For measurements of Erk1/2 phosphorylation, subconfluent and confluent PC6 cells were incubated in serum-free medium, with and without 100 ng/ml pertussis toxin (Sigma) for 6 h prior to stimulation with vehicle, EGF, or LPA for 5 min. The cells were harvested in radioimmune precipitation assay lysis buffer, and lysates were subjected to immunoblotting using antibodies against phosphorylated (SC-7383, 1:500 dilution) (Santa Cruz Biotechnology Inc.) or total (catalog number 9102, 1:6000 dilution) (Cell Signaling Technology) Erk1/2 MAPK. For immunoprecipitation studies, 100 μg of nuclear extracts were incubated at 4 °C for 2 h with 1 μg of antibody against NF-YA (BD Biosciences) or antibody against Bcl6 (Santa Cruz Biotechnology Inc.), followed by an additional 2-h incubation with 30 μl of protein G-agarose beads (Santa Cruz Biotechnology Inc.) at 4 °C. At the end of the incubation, the beads were collected by centrifugation at 1,000 × g for 5 min at 4 °C and washed three times with 1× TTBS (1 mm Tris-HCl, pH 7.5, 0.9% NaCl, 0.05% Tween 20). After the final wash, immunoprecipitates were eluted from the beads with 20 μl of 2× SDS-PAGE sample buffer by heating the tube at 95 °C for 5 min and were immunoblotted with antibody against acetyl-lysine (catalog number 9441S, 1:5000 dilution) (Cell Signaling Technology).
ChIP Assay
ChIP assays were carried out as described (22). Briefly, the cells were cross-linked with 0.5% formaldehyde at room temperature for 10 min and then lysed with lysis buffer (20 mm Tris-HCl, pH 8.0, 140 mm NaCl, 1% Triton X-100, 1% SDS, 1% deoxycholic acid, 2 mm EDTA, and freshly added protease inhibitors). The resultant chromatin was sheared using a probe sonicator (Sonics & Materials, Inc.), power output settings at 3, and 25 cycles of 1-s on and 5-s off. Sheared chromatin was precleared with protein G-agarose beads, salmon sperm DNA, and preimmune serum. Approximately 25 μg of chromatin DNA was used for one immunoprecipitation. The antibodies used were against NF-YA, C/EBPβ (Santa Cruz Biotechnology Inc.), or water (negative control). Immunoprecipitates were subjected to sequential washes beginning with lysis buffer, then lysis-500 buffer (20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 1% Triton X-100, 0.1% SDS, 1% deoxycholic acid, and protease inhibitor mixture), then LiCl detergent solution (0.5% deoxycholic acid, 1 mm EDTA, 250 mm LiCl, 0.5% Nonidet P-40, 20 mm Tris-HCl, pH 8.0, and protease inhibitor mixture), and ending with 1× TBS. Precipitated chromatin was eluted with 200 μl of elution buffer (1% SDS, 1 mm Tris-HCl, pH 8.0, and 1 mm EDTA) at 65 °C for 10 min. The protein was degraded with proteinase K, and DNA was purified using a QIAquick PCR purification kit (Qiagen). The DNA isolated from the immunoprecipitates was amplified using PCR and resolved using 1% agarose gels stained with ethidium bromide. The PCR primers were 5′-CTG GGA GGC AAG TTC TTT AGA CAT TGA TTT TAC A-3′ and 5′-GGA GCA ACG GTG GTT TCT ATT TTG AGC A-3′ for the target region and 5′-ATC TAC AAT GAG TTC ATC TCT GTG CAG GC-3′ and 5′-TAG AAT CGA GAC TTG AGG AAA CGA CGG TA-3′ for the nontarget region control.
Statistics
The experimental data were expressed as the means ± S.E. The significance of differences was determined by the unpaired Student's t test using SigmaPlot (Systat Software, CA); a p < 0.05 was considered to be statistically significant.
RESULTS
Cell Density-dependent Transcriptional Activation of RGS4 Is Associated with Inhibition of G Protein-coupled Receptor Signaling
RGS4, a tightly regulated GTPase-activating protein (13–15, 23), plays a critical role as a negative modulator of G protein-coupled receptor signaling via Gαi or Gαq (3, 4, 49, 50) in brain and other tissues. Fig. 1A shows that RGS4 protein levels were markedly higher in confluent and postconfluent PC6 cells compared with subconfluent cells. Thus, PC6 cells provide an excellent experimental system to understand how RGS4 expression and actions are regulated in a neuron-like cell. To ascertain whether the increase in RGS4 expression under these conditions is associated with altered G protein signaling, we evaluated LPA receptor-mediated activation of Erk1/2 in PC6 cultures of varying densities. LPA receptors are known to promote activation of Gαi and Gαq in a variety of cells (24–26). EGF, which activates Erk1/2 independently of heterotrimeric G proteins, was used as a control. Phosphorylation of Erk1/2 induced by EGF was found to be unaffected by cell density (Fig. 1B). However, LPA-stimulated phosphorylation of Erk1/2 was attenuated in confluent cells relative to subconfluent cells (Fig. 1B), although LPA1 receptor expression was the same or slightly higher in confluent PC6 cells (not shown). Treatment of cells with pertussis toxin eliminated the cell density-dependent changes in LPA-stimulated Erk1/2 phosphorylation, suggesting that the observed reduction in Erk1/2 activation with increasing cell density reflected an attenuation of Gαi/o signaling as would be predicted to occur with increased RGS4 protein levels.
FIGURE 1.
Cellular density-dependent transcriptional activation of RGS4 in PC6 cells is associated with inhibited G protein-coupled receptor signaling. A, RGS4 levels in subconfluent (SubC), confluent (C), and postconfluent (PostC) PC6 cells. The inset shows a representative Western blot. The intensities of RGS4 bands were quantified, normalized to α-tubulin level, and then plotted. The results are expressed as the means ± S.E. of four experiments. *, p < 0.03 as compared with values from subconfluent cells. B, loss of pertussis toxin-sensitive activation of MAPK by LPA receptor activation in confluent PC6 cells. Subconfluent (SubC) or confluent (C) PC6 cells, treated with and without pertussis toxin (PTX, 100 ng/ml) in serum-free medium for 6 h, were stimulated with vehicle, 100 ng/ml of EGF, or 10 μm LPA for 5 min. The levels of total and phosphorylated Erk1/2 were determined by Western blotting. The inset shows a representative Western blot. The intensities of bands from Western blotting were quantified. The levels of phosphorylated Erk1/2 were normalized to total Erk1/2 levels and expressed as fold stimulation over vehicle-treated control cells. The results shown are the means ± S.E. of three independent experiments (*, p < 0.05). C, quantification of RGS4 mRNA in PC6 cells using real time PCR. The data are expressed as the means ± S.E. of three experiments (*, p < 0.006). D, the human RGS4 promoter (bp −435 to +124) in pGL3 basic vector was transfected into subconfluent (SubC), confluent (C), and postconfluent (PostC) PC6 cells. Luciferase activity was determined 48 h after transfection as described under “Experimental Procedures.” The results represent the means ± S.E. of three independent experiments (*, p < 0.01).
We undertook studies to determine whether transcriptional activation of the RGS4 gene contributed to RGS4 up-regulation in PC6 cells. Real time PCR results showed that RGS4 mRNA in confluent PC6 cells was 2.5-fold that in subconfluent cells (Fig. 1C), which indicated that increases in RGS4 protein were partly, if not completely, contributed by transcriptional activation of this gene promoter. Using 5′-RACE, we identified the transcription start site for the human RGS4 gene at 124 bp upstream of the translation start site. To investigate transcriptional activation of the RGS4 promoter in PC6 cells, a segment of the human RGS4 promoter (bp −5874/+124) was subcloned into the pGL3-Basic luciferase reporter plasmid. Transient transfection of a reporter plasmid with a segment of human RGS4 promoter (bp −435/+124) into PC6 cells showed a 2.5- and 3.4-fold increase in luciferase activity in confluent and postconfluent cells over subconfluent cultures (Fig. 1D), which indicated that the transiently transfected promoter behaved similarly to its endogenous counterpart. Indeed, PC6 cells stably expressing a segment of the RGS4 promoter (bp −195/+124) fused to luciferase exhibited similar increases in luciferase activity when cells became confluent (not shown). PC6 cells thus represent a suitable model to study the mechanism of RGS4 gene regulation, which is critical to a complete understanding of the biological functions of RGS4.
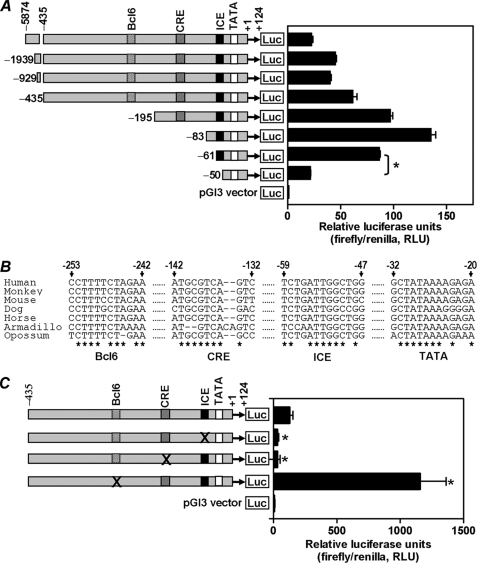
An ICE Is Required for Transcriptional Activation of the RGS4 Gene Promoter
To delineate the region(s) of the RGS4 promoter responsible for its transcriptional activation in confluent PC6 cells, sequentially truncated RGS4 promoters were subcloned into the pGL3-Basic vector to generate luciferase reporter plasmids (Fig. 2A, left panel). Transient transfection of these reporter constructs showed the presence of both positive and negative regulatory elements in the RGS4 promoter. Basal transcription increased when the promoter sequence between bp −435 and −83 was deleted (Fig. 2A, right panel), indicating possible repressor elements in this segment of the RGS4 promoter. When the sequence between bp −83 and −50 was deleted, there was a marked reduction in transcription of RGS4 gene promoter. It appeared that the sequence located between bp −61 and −50 was critical for transcriptional activation of the RGS4 promoter (Fig. 2A). By aligning RGS4 promoter sequences from seven mammals, namely human, monkey, mouse, dog, horse, armadillo, and opossum, we were able to identify four phylogenetically conserved transcription factor-binding sites (Fig. 2B), namely the Bcl6 element, CRE, ICE, and TATA box. Particularly noteworthy is that the conserved ICE site at bp −59 to −47 almost coincides with the sequence (bp −61 to −50) identified by truncation analyses to play a critical role in RGS4 gene transcription (Fig. 2A). To assess the function of these conserved sites, each was mutated by replacing key nucleotides with ones that would lead to a loss of binding to its transcription factor(s). Mutants of ICE and CRE significantly reduced the basal activity of RGS4 promoter, whereas mutation of Bcl6 markedly increased RGS4 basal transcription (Fig. 2C). Indeed, truncation analyses of the RGS4 promoter indicated the existence of binding site(s) for transcriptional repressors in the segment between bp −435 and −83 of RGS4 promoter (Fig. 2A). The Bcl6 site is likely one of them, because transcription factor Bcl6 is known to be a strong repressor for many gene promoters. Together, three conserved binding sites were critical for transcriptional regulation of RGS4.
FIGURE 2.
Identification of transcription factor binding element(s) critical for transcriptional activation of the RGS4 gene promoter. A, sequential 5′-truncated human RGS4 promoters were linked with a luciferase reporter gene and transfected into confluent PC6 cells. The drawing of the RGS4 promoter is in scale. Firefly and Renilla luciferase activities were determined as described under “Experimental Procedures.” The results are the means ± S.E. of three independent experiments (*, p < 0.001). B, phylogenetic analysis of RGS4 promoters from seven mammals revealed four evolutionarily conserved transcription factor-binding sites, namely TATA box, ICE, CRE, and Bcl6. C, RGS4 promoters, WT or mutants at each of ICE, CRE, and Bcl6 sites, were linked with a luciferase gene and transfected into confluent PC6 cells. Promoter activity was determined as described under “Experimental Procedures.” The results are the means ± S.E. of three experiments (*, p < 0.02). × indicates the base substitution mutation for each of ICE, CRE, and Bcl6 sites.
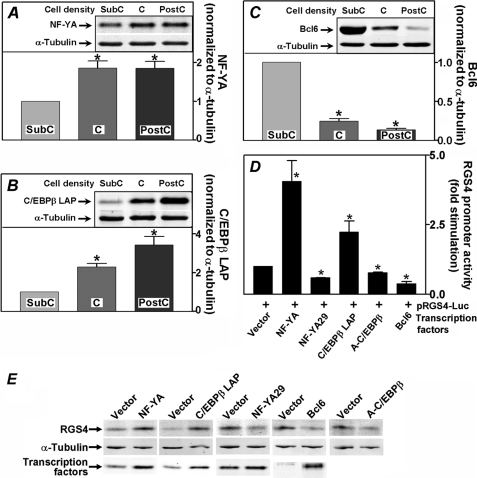
NF-YA and C/EBPβ Stimulate Whereas Bcl6 Inhibits Transcription of the RGS4 Gene Promoter
A number of transcription factors including NF-Y, C/EBP, ICB90, and YB-1 are known to interact with ICE sequences in genes to regulate their activity (27–30). NF-Y is comprised of three heterologous subunits (31), of which NF-YA functions as a regulatory subunit, whereas levels of NF-YB and -YC remain constant (32). C/EBPβ, a basic leucine zipper transcription factor known to interact with ICE, is involved in regulating a number of genes (33–36). The transcription repressor Bcl6 has been shown to inhibit gene promoter activity by interacting with complexes containing histone deacetylases and co-repressors including SMRT, SIN3A, and BCoR (37–42). When we measured endogenous levels of NF-YA, we found that NF-YA was ∼85% higher in confluent and postconfluent PC6 cells compared with subconfluent cells (Fig. 3A). Similarly, endogenous levels of C/EBPβ LAP isoform, the activator form of C/EBPβ, were increased in confluent and postconfluent cells compared with subconfluent cells by 2.3- and 3.4-fold, respectively (Fig. 3B). In contrast, transcription repressor Bcl6 levels were markedly lower in confluent and postconfluent cells, averaging 25 and 14%, respectively, of the level of Bcl6 found in subconfluent PC6 cells (Fig. 3C). To elucidate effects of these three transcription factors on transcriptional regulation of the RGS4 promoter, NF-YA and C/EBPβ LAP were overexpressed in subconfluent PC6 cells, where endogenous levels of these proteins were low. NF-YA and C/EBPβ LAP stimulated RGS4 promoter activity by 4.1- and 2.2-fold, respectively (Fig. 3D), consistent with the observed increases in endogenous RGS4 levels in response to their expression (Fig. 3E). When dominant negative forms of these two transcription factors, NF-YA29 (16), A-C/EBP (17), or wild type Bcl6 were overexpressed in confluent PC6 cells, RGS4 promoter activity was reduced by 41, 22, and 62%, respectively. Endogenous RGS4 levels were also decreased accordingly (Fig. 3E). Thus, transcription factors NF-YA and C/EBPβ transactivate, whereas Bcl6 represses the RGS4 promoter.
FIGURE 3.
Confluence-induced changes in levels of NF-YA, C/EBPβ LAP, and Bcl6. A–C, endogenous levels of NF-YA (A), C/EBPβ LAP (B), and Bcl6 (C) in subconfluent (SubC), confluent (C), and postconfluent (PostC) PC6 cell lysates were analyzed using Western blotting. The insets show representative Western blots. The intensities of bands were quantified, normalized to that of α-tubulin, and plotted. The results shown are the means ± S.E. of seven experiments (*, p < 0.006). D, 200 ng of luciferase reporter plasmid containing the human RGS4 promoter (bp −435 to +124) was co-transfected with 50 ng of constructs expressing NF-YA, C/EBPβ LAP, NF-YA29, A-C/EBPβ, Bcl6, or the empty vector. Luciferase activity was determined as described under “Experimental Procedures.” The results are expressed as fold stimulation of cells transfected with a transcriptional factor over cells transfected with empty vector DNA. The values are the means ± S.E. of six experiments for NF-YA and C/EBPβ LAP and of three experiments for NF-YA29, A-C/EBPβ, and Bcl6. *, significant difference (p < 0.003) from the empty vector-transfected cells. E, transfection of transcription factors or their dominant negative forms in PC6 cells altered endogenous RGS4 expression. Subconfluent PC6 cells were transfected with 200 ng of constructs expressing NF-YA, C/EBPβ LAP, or the empty vector, and confluent PC6 cells were transfected with 200 ng of constructs expressing NF-YA29, A-C/EBPβ, Bcl6, or the empty vector. Endogenous levels of RGS4 and transfected transcription factor levels were determined using Western blotting (A-C/EBPβ levels are not shown because of its migration at the dye front caused by its small size of ∼10 kDa).
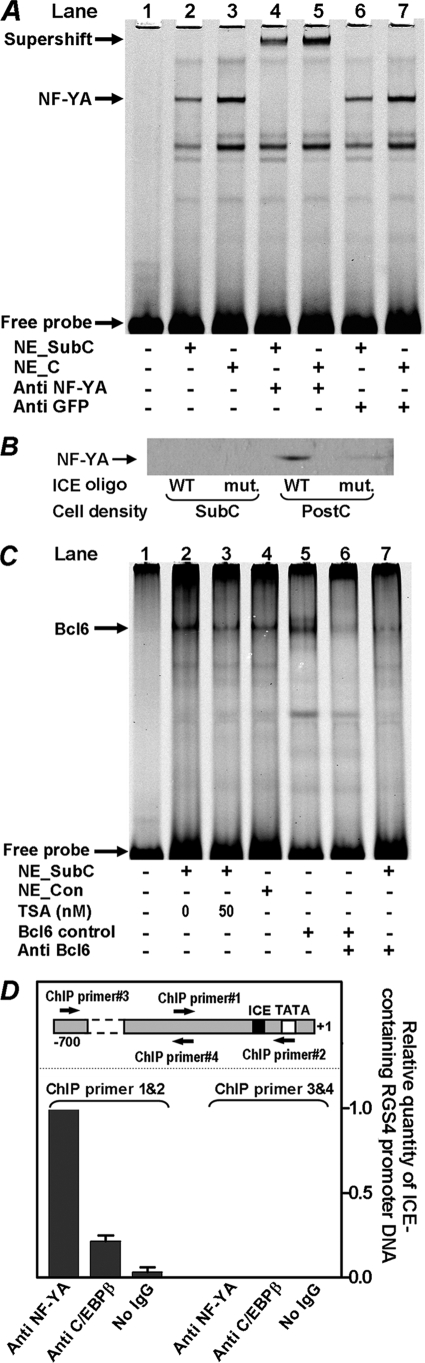
To determine whether interaction of NF-YA with the ICE element in the RGS4 promoter is altered when cell density changes, fluorescent EMSA were performed using nuclear extracts isolated from subconfluent and confluent cells. Binding of NF-YA to its target DNA element was markedly augmented in reactions using nuclear extracts from confluent cells relative to those from subconfluent cells (Fig. 4A, lanes 2 and 3). Binding of NF-YA was authenticated by a supershift assay with antisera against NF-YA or GFP (control) (Fig. 4A, lanes 4–7). The DNA binding activity of NF-YA is impaired by its phosphorylation by Cdk2 (43). To assess whether phosphorylation regulates the binding of NF-YA to ICE, nuclear extracts from both subconfluent and confluent cells were treated with alkaline phosphatase. However, binding of NF-YA to the target oligonucleotide did not change in EMSA assays (not shown). We speculate that dephosphorylation of NF-YA is not essential for its binding to RGS4 promoter in PC6 cells or that NF-YA is not phosphorylated in confluent PC6 cells. To further prove that NF-YA binds to the ICE on the RGS4 promoter in confluent cells, we also performed a biotin-labeled DNA pull-down assay. A DNA fragment containing the wild type ICE sequence was able to pull down NF-YA from cell lysates isolated from postconfluent but not subconfluent PC6 cells. No appreciable pull-down was observed using a DNA fragment in which the ICE was mutated (Fig. 4B). It is therefore clear that NF-YA interacts with the ICE of the RGS4 promoter.
FIGURE 4.
Interaction of NF-YA, Bcl6, and C/EBPβ with RGS4 promoter. A, binding of NF-YA to the ICE element. Nuclear extracts isolated from subconfluent (NE_SubC) or confluent (NE_C) PC6 cells were mixed with 1 pmol of DNA oligonucleotide containing the ICE sequence for EMSA assays. Supershifting was done by adding 1.6 μg of antisera for NF-YA or for GFP (control) to the EMSA reactions. B, biotin-labeled DNA fragments (8 μg) containing WT or mutant ICE (mut.) were mixed with 500 μg of total protein extract from subconfluent (SubC) or postconfluent (PostC) PC6 cells and 100 μl of 4% streptavidin-agarose gel beads. Isolated complexes were resolved using SDS-PAGE. NF-YA protein in the isolated complexes was determined using Western blotting. C, binding of Bcl6 to the Bcl6 element in RGS4 promoter. The nuclear extracts were isolated from the following cells: subconfluent (NE_SubC) PC6 cells that were treated with or without 50 nm TSA, confluent (NE_C) PC6 cells, Cos-7 cells that overexpressed Bcl6 (Bcl6 positive control), and Cos-7 cells. EMSA were performed as described under “Experimental Procedures.” Supershifting was done by adding 2 μg of antisera for Bcl6 to the EMSA reactions. D, in vivo binding of NF-YA and C/EBPβ to the ICE. Real time PCR was used to determine in vivo binding of NF-YA and C/EBPβ to the ICE element. The inset shows relative locations of ChIP primers 1 and 2 that are specific for amplifying ICE-containing RGS4 promoter sequence and relative locations of ChIP primers 3 and 4 that are upstream of ICE-containing region. DNA templates were generated from chromatin IP assay. The value of PCR amplification from the NF-YA bound ICE was set as 1.
We further determined interactions of Bcl6 with the RGS4 promoter using an EMSA assay. The binding of Bcl6 to the Bcl6 element was lower in confluent than subconfluent PC6 cells (Fig. 4C, lane 4 versus lane 2). Nuclear extract from Cos-7 cells in which Bcl6 was overexpressed was used as positive control (Fig. 4C, lane 5), and the binding of Bcl6 was also authenticated by the disappearance of Bcl6 shift bands in supershift assays with antisera against Bcl6 (Fig. 4C, lanes 5–7). C/EBPβ is known to interact with ICE in a number of gene promoters (33–36). However, we failed to detect the binding of C/EBPβ to ICE in vitro using EMSA assay, presumably because the RGS4 promoter ICE element is not an optimal binding site for C/EBPβ, which prefers a consensus sequence of TKNNGYAAK. To determine whether C/EBPβ binds to the ICE of the RGS4 promoter in vivo, ChIP assay was performed using subconfluent PC6 cells. The ICE-containing DNA fragment was precipitated using antibodies against C/EBPβ or NF-YA versus no IgG control and quantified using real time PCR. ChIP primers 1 and 2 are specific for amplification of the ICE region, whereas ChIP primer 3 is located ∼500 bp upstream of ChIP primer 1 (Fig. 4D, inset). Fig. 4D demonstrates that both NF-YA and C/EBPβ bind to the ICE of the RGS4 promoter. The higher pull down of NF-YA versus C/EBPβ might be due to the ICE being an optimal and suboptimal binding sequence for NF-YA and C/EBPβ, respectively, or may reflect different pull-down efficiencies of the two antibodies. Consistently, there were no detectable PCR amplifications when ChIP primers 3 and 4 were used, demonstrating that NF-YA and C/EBPβ binding is specific to the ICE-containing region of the RGS4 promoter.
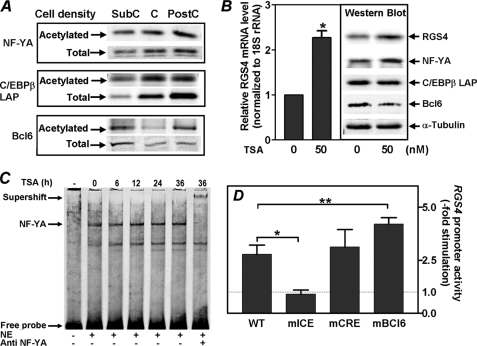
Acetylation of NF-YA and Bcl6 Regulates RGS4 Promoter Activity
In view of recent evidence that acetylation of NF-YA (32), C/EBPβ (44, 45), and Bcl6 (46) can modulate their transcriptional activities, we undertook studies to determine whether acetylation of these transcription factors was associated with activation of the RGS4 promoter at cellular confluency. Acetylation was assessed by immunoprecipitation of these proteins followed by anti-acetyl-lysine immunoblotting. Fig. 5A (upper panel) shows that increases in NF-YA protein levels in confluent and postconfluent PC6 cells were accompanied by comparable increases in NF-YA acetylation. Recently, Manni et al. (32) demonstrated that acetylation of NF-YA increased both its stability and transactivation activity. Thus, it is likely that NF-YA acetylation plays a similar role in activation of the RGS4 promoter during confluent growth of PC6 cells. As we found for NF-YA, increases in C/EBPβ protein were accompanied by increases in its acetylation (Fig. 5A, middle panel) in confluent and postconfluent PC6 cells. However, the observed increases in C/EBPβ protein levels (2–4-fold) under these conditions were larger than the increases in its acetylation (1-fold or less). Ceseña et al. (45) recently demonstrated that acetylation of C/EBPβ enhances its transcriptional activity. Therefore, it is likely that increases in both acetylated and total C/EBPβ protein are involved in its activation of the RGS4 promoter, with our subsequent studies (Fig. 6) suggesting a major contribution from the latter. In contrast to NF-YA and C/EBPβ, Bcl6 is known to be inactivated by acetylation (46). Interestingly, acetylation of Bcl6 first decreased in confluent PC6 cells but then increased markedly in postconfluent cells, whereas the total Bcl6 level was markedly low in both growth conditions as compared with subconfluent cells (Fig. 5A, bottom panel). These findings suggest that relief of Bcl6 repression activity on the RGS4 promoter may be achieved in two steps: first by a reduction in Bcl6 protein level and then by acetylation of the residual Bcl6. Taken together, our results demonstrate a coordinated acetylation of NF-YA, C/EBPβ, and Bcl6 during activation of the RGS4 promoter, with acetylation playing a positive role in transactivation by NF-YA and C/EBPβ and a negative role in the repressive actions of Bcl6.
FIGURE 5.
Acetylation of NF-YA, C/EBPβ, and Bcl6 and regulation of transcription from the RGS4 gene promoter. A, acetylation status of NF-YA, C/EBPβ LAP, and Bcl6 were determined in PC6 cells at subconfluent (SubC), confluent (C), and postconfluent (PostC) densities. These transcription factors were first immunoprecipitated and then analyzed for their acetylation as described under “Experimental Procedures.” The total levels of NF-YA, C/EBPβ LAP, and Bcl6 were also measured at three cellular densities using Western blotting. B, TSA treatment increased endogenous levels of RGS4 mRNA and protein in PC6 cells. Subconfluent PC6 cells were treated with 50 nm TSA or vehicle for 18 h, and the endogenous RGS4 mRNA level was determined using real time PCR protocol as described under “Experimental Procedures.” The data are the means ± S.E. of three experiments (*, p < 0.001). Endogenous levels of RGS4, NF-YA, C/EBPβ LAP, and Bcl6 were also determined by Western blotting. The α-tubulin level was used as loading control. C, TSA treatment enhanced NF-YA binding to ICE element. PC6 cells were treated with 50 nm of TSA for the time periods indicated at the top of the panel. Binding of NF-YA to ICE was measured using EMSA assay as described in Fig. 4A. NE, nuclear extract. D, effects of RGS4 promoter mutations on TSA-stimulated activation of RGS4 promoter. Luciferase reporter plasmids containing the wild type RGS4 promoter (bp −435 to +124) or with mutations in ICE (mICE), CRE (mCRE), or Bcl6 (mBCL6) were transfected into confluent PC6 cells. The cells were then treated with 25 nm TSA or vehicle for 24 h. Fold stimulation of RGS4 promoter activity was calculated by division of luciferase activities of TSA-treated cells over vehicle-treated ones. The results shown are the means ± S.E. of three experiments (*, p < 0.005; **, p < 0.03).
FIGURE 6.
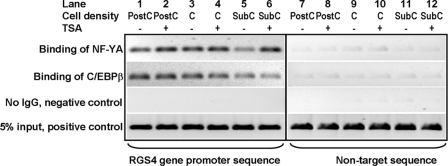
In vivo binding of NF-YA and C/EBPβ to the RGS4 gene promoter. PC6 cells at subconfluent (SubC), confluent (C), and postconfluent (PostC) densities were treated with vehicle or 50 nm TSA for 24 h. ChIP assays were performed using chromatin isolated from these cells. Immunoprecipitated RGS4 promoter DNA and nontarget sequence DNA (located in the coding sequence of RGS4) were determined using semi-quantitative PCR. Five percent of total chromatin used for one ChIP assay (5% input) was used as a positive control.
Protein acetylation is controlled by both acetyltransferase and deacetylase activities. To determine whether perturbing cellular overall acetylation status alters RGS4 promoter activity, we treated PC6 cells with trichostatin A (TSA), a known histone deacetylase inhibitor. Both endogenous RGS4 mRNA and protein increased in TSA-treated PC6 cells (Fig. 5B), which can be explained by the combined increase in NF-YA and decrease in Bcl6 levels (Fig. 5B, Western blot). It appeared that TSA treatment had little effect upon the level of C/EBPβ LAP (Fig. 5B, Western blot). Thus, TSA treatment of PC6 cells promoted RGS4 promoter activation and alterations in NF-YA and Bcl6 protein levels like those observed during confluent growth of these cells. The question that remains is whether TSA produces increases in NF-YA binding and decreases in Bcl6 binding to the RGS4 promoter like those we observed in response to confluent growth (Fig. 4, A and C). Fig. 5C shows that binding of NF-YA to the ICE element of the RGS4 promoter was enhanced in TSA-treated cells in a time-dependent manner and reached steady state 24 h after treatment; binding of Bcl6 to its element was attenuated in TSA-treated cells (Fig. 4C, lane 3 versus lane 2). Moreover, mutation of the ICE abolished TSA-induced activation of the RGS4 promoter (Fig. 5D, second bar); mutation at CRE had no effect (Fig. 5D, third bar), whereas mutation at Bcl6 site led to a 4-fold increase in TSA-induced activation of the RGS4 promoter (Fig. 5D, fourth bar). These findings suggest that acetylation of NF-YA and Bcl6 is involved in regulation of RGS4 promoter by coordinately enhancing the binding of NF-YA and attenuating the binding of Bcl6 to their respective elements.
NF-YA and C/EBPβ Interact with the RGS4 Promoter in Vivo
ChIP assays were used to determine interaction of NF-Y and C/EBPβ with the RGS4 promoter in vivo in PC6 cells during confluent growth and treatment with TSA. PC6 cells were grown at subconfluent, confluent, and postconfluent conditions and stimulated with and without TSA. Binding of NF-YA and C/EBPβ to the RGS4 promoter was increased when cell densities increased from subconfluent to postconfluent (Fig. 6, lane 5 versus lanes 1 and 3). However, treating cells with 50 nm TSA had differential effects on the binding of these two transcription factors to the RGS4 promoter. TSA treatment augmented interaction of NF-YA with the RGS4 promoter but had little effect on interaction of C/EBPβ (Fig. 6). These findings demonstrate that two transcription factors that transactivate the RGS4 promoter and whose levels increase during cellular confluence (Fig. 3, A, B, and D) bind to the RGS4 promoter in vivo and that this binding increases at confluence. In addition, the ability of TSA to stimulate binding of NF-YA suggests that the observed increase in acetylated NF-YA levels in confluent PC6 cells plays a key role in transactivation of the RGS4 promoter. Unlike NF-YA, binding of C/EBPβ to the RGS4 promoter in PC6 cells was not regulated by acetylation.
DISCUSSION
Expression of RGS proteins is tightly regulated in a number of biological processes (9–15, 23, 47). However, the molecular mechanisms underlying transcriptional regulation of these genes remain largely unknown. This study is the first to systematically identify and provide functional evaluation of a promoter for a member of the RGS gene family, of which 30 genes exist in humans. We focused on the RGS4 gene in view of its association with a number of pathologies and the important role of neurally enriched RGS4 in G protein signaling. Acute increases in RGS4 transcripts in confluent PC6 cells (15) presented an excellent experimental system to elucidate how the RGS4 gene is activated in a neuronal-like cell. First, we identified and cloned the human RGS4 promoter. We determined that there are four key cis-elements in the RGS4 promoter controlling its transcription. In addition to the TATA box, which is obviously critical for controlling gene transcription, a Bcl6 site, CRE, and ICE are highly conserved in the RGS4 promoter among seven mammals, indicating that these sites are important in gene regulation. Transcription factors that interact with Bcl6 and ICE elements appear to be coordinately regulated to control the overall activation of the RGS4 promoter. That is, cellular levels of transcriptional activators NF-YA and C/EBPβ increased, whereas those of the transcriptional repressor Bcl6 decreased markedly at cellular confluence. Interestingly, increases in RGS4 transcripts in PC6 cells were associated with even greater increases in RGS4 protein levels (Fig. 1, A and B). Therefore, it is possible that RGS4 expression may also be regulated post-transcriptionally. Indeed, Xie et al. (48) recently reported that the stability of RGS4 is regulated by the proteasome in MDA-MB-231 cells.
Acetylation/deacetylation of transcription factors/co-regulators represents an important layer of regulation of gene promoter activities (49–51). Manni et al. (32) recently showed that the function of the CCAAT box-binding transcription factor NF-Y complex is regulated by acetylation of the NF-YA subunit. Because the NF-YA subunit is regulatory in this complex, acetylation of NF-YA and an attendant increase in its stability and transactivation activity is of obvious importance in regulating transcription from CCAAT box bearing gene promoters. We found that the level of acetylated NF-YA was higher in confluent PC6 cells, paralleling the increase in NF-YA protein levels and possibly reflecting stabilization of NF-YA by acetylation. This is the first evidence that NF-YA acetylation, reported previously in response to expression of p300 or cellular treatment with TSA, is biologically regulated in cells. Binding of NF-YA to the RGS4 promoter was increased in PC6 cells at confluence and in cells treated with TSA, providing new evidence that NF-YA acetylation increases its binding to a promoter. The ICE of the RGS4 promoter, for which we documented binding of NF-YA and C/EBPβ, is essential for TSA-induced transcriptional activation of RGS4. The finding that dominant negative NF-YA29 and A-C/EBPβ significantly decreases RGS4 promoter activity in confluent cells supports this model. Acetylation of C/EBPβ has also been implicated in enhancing its transcriptional activity in adipose tissue (45). However, we found that TSA treatment had little effect on the binding of C/EBPβ to the RGS4 promoter. We speculate that acetylation/deacetylation of C/EBPβ plays a minor role in activation of the RGS4 promoter in confluent PC6 cells. Indeed, increases in total C/EBPβ protein levels were much larger than increases in its acetylation during confluent growth of PC6 cells. These results suggest that transactivation of the RGS4 promoter by C/EBPβ may depend more upon increases in its cellular concentration than its acetylation status.
We also found that Bcl6 (or zinc finger protein 51) plays a critical role in RGS4 gene activation. Bcl6 is a BTB/POZ zinc finger transcription factor, known by its involvement in B-cell-derived non-Hodgkin lymphoma resulting from chromosomal translocations of its gene (52) (reviewed in Ref. 53). We found that the Bcl6 protein level was markedly decreased in confluent and postconfluent PC6 cells compared with subconfluent cells. Mutation of the RGS4 promoter at the Bcl6 site dramatically increased transcription, attesting to the strong repressor activity of Bcl6 reported in prior studies. Bereshchenko et al. (46) showed that acetylation of Bcl6 markedly decreases its function as a transcriptional repressor. Thus, it is noteworthy that acetylation of Bcl6 markedly increased in postconfluent PC6 cells. Our results suggest that relief of Bcl6 repression on RGS4 promoter activity might occur first by marked decreases in Bcl6 protein levels and then by acetylation of the residual Bcl6. This would then allow full activation of the RGS4 promoter by transcriptional activators NF-YA and C/EBPβ. Our results suggest that RGS4 promoter activity is controlled by coordinated changes in the steady state level and/or acetylation status of three critical transcription factors, NF-YA, C/EBPβ, and Bcl6, that determine the on/off state of the RGS4 promoter (Fig. 7).
FIGURE 7.
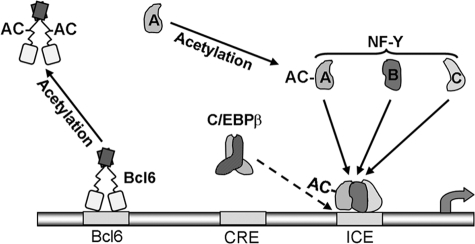
Model for transcriptional activation of the RGS4 gene promoter. The transcriptional state of the RGS4 promoter is controlled by interactions of three transcription factors with two phylogenetically conserved cis-elements in the RGS4 promoter, i.e. binding of transcriptional activators NF-Y and C/EBPβ (with a low affinity, dotted arrow) to the ICE and binding of transcriptional repressor Bcl6 to the Bcl6 element. Activation of RGS4 transcription in confluent PC6 cells is mediated by increases in levels of NF-YA and C/EBPβ LAP and their binding to the ICE and decreases in the level of Bcl6 and its binding to the Bcl6 element. Under these same conditions, acetylation of NF-YA and Bcl6 occurs, leading to increases in NF-YA stability and binding to the ICE and a reduction in binding of Bcl6 to the Bcl6 element.
RGS4 is linked to various pathologies, including Parkinson disease (54) and schizophrenia (55). It might be assumed that the function of RGS4 in such pathological processes would be attributed to its attenuation of Gαi/o- or Gαq-mediated G protein-coupled receptor signaling, functioning as a GTPase-activating protein (3) or effector antagonist (4), respectively. Indeed, we observed a cell density-dependent loss of Gi/o-mediated activation of Erk1/2 by stimulation of LPA receptors in PC6 cells, as would be predicted with increased RGS4 levels in confluent PC6 cells. As expected, confluent PC6 cells exhibited cell cycle arrest as measured by key regulators of cell cycle progression (cyclin D1 and pRb) and cell counts (not shown). Thus, it is intriguing that Grillet et al. (13) showed that RGS4 expression is increased in post-mitotic neurons. In that study, RGS4 transcripts were expressed only transiently in facial motoneuronal precursors during development, in contrast to constitutive expression of RGS4 in adult brain. RGS4 expression in the neuronal precursors occurred concomitant with or subsequent to cell cycle exit, with RGS4 expression switched on again later in adult stages. This link between RGS4 gene activation in post-mitotic neuronal precursors or differentiated neurons is of considerable interest in view of our evidence that RGS4 promoter activity is low in proliferating PC6 cells and increases dramatically at confluence-induced growth arrest. The obvious implication of RGS4 gene activation in such neurons is provision of feedback modulation of G protein signaling by RGS4 for the multitude of neurotransmitter receptors that signal by activating Gi, Go, or Gq.
RGS proteins were discovered in yeast in the form of the SST2 gene product whose loss led to supersensitivity to the α factor pheromone. SST2 is pheromone-inducible, and SST2 mRNA and protein increase during prolonged Ste2p receptor stimulation, suggesting built-in feedback for modulating G protein signaling at the transcriptional level (12). These early studies pointed to an important role for transcriptional activation of RGS genes in modulating RGS protein function. Despite the universal importance of mammalian RGS proteins in regulating G protein signaling, the mechanisms regulating expression of RGS genes are largely unknown. Here, we provide new insights into the cis- and trans-acting factors involved in activation of the prototypic member of the mammalian RGS protein gene family. Our findings support a role for promoter activation as an important mechanism of control of RGS4 expression and signaling, suggesting conservation of transcriptional control of RGS genes as a regulatory mechanism in organisms ranging from yeast to human. Hopefully this work will serve as a model of inquiry to provide further understanding of how RGS4 and other RGS genes are regulated in various physiological and pathophysiological situations.
This work was supported, in whole or in part, by National Institutes of Health Grants GM075033 and GM075033-03s1 (to R. A. F.).
- RGS
- regulator of G protein signaling
- bp
- base pair(s)
- Bcl6
- B-cell lymphoma 6
- C/EBP
- CCAAT/enhancer binding protein
- LAP
- liver-enriched transcriptional activator protein
- ChIP
- chromatin immunoprecipitation
- CRE
- cyclic AMP response element
- ICE
- inverted CCAAT element
- LPA
- lysophosphatidic acid
- NF-Y
- nuclear factor Y
- PC6 cells
- pheochromocytoma-6 cells
- RACE
- rapid amplification of cDNA ends
- TSA
- trichostatin A.
REFERENCES
- 1.Dohlman H. G., Apaniesk D., Chen Y., Song J., Nusskern D. (1995) Mol. Cell. Biol. 15, 3635–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koelle M. R., Horvitz H. R. (1996) Cell 84, 115–125 [DOI] [PubMed] [Google Scholar]
- 3.Berman D. M., Wilkie T. M., Gilman A. G. (1996) Cell 86, 445–452 [DOI] [PubMed] [Google Scholar]
- 4.Hepler J. R., Berman D. M., Gilman A. G., Kozasa T. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinnarajah S., Dessauer C. W., Srikumar D., Chen J., Yuen J., Yilma S., Dennis J. C., Morrison E. E., Vodyanoy V., Kehrl J. H. (2001) Nature 409, 1051–1055 [DOI] [PubMed] [Google Scholar]
- 6.Wang Q., Liu M., Mullah B., Siderovski D. P., Neubig R. R. (2002) J. Biol. Chem. 277, 24949–24958 [DOI] [PubMed] [Google Scholar]
- 7.Xu X., Zeng W., Popov S., Berman D. M., Davignon I., Yu K., Yowe D., Offermanns S., Muallem S., Wilkie T. M. (1999) J. Biol. Chem. 274, 3549–3556 [DOI] [PubMed] [Google Scholar]
- 8.Leontiadis L. J., Papakonstantinou M. P., Georgoussi Z. (2009) Cell Signal. 21, 1218–1228 [DOI] [PubMed] [Google Scholar]
- 9.Hong J. X., Wilson G. L., Fox C. H., Kehrl J. H. (1993) J. Immunol. 150, 3895–3904 [PubMed] [Google Scholar]
- 10.Cheng Y. S., Lee T. S., Hsu H. C., Kou Y. R., Wu Y. L. (2008) J. Cell. Biochem. 105, 922–930 [DOI] [PubMed] [Google Scholar]
- 11.Buckbinder L., Velasco-Miguel S., Chen Y., Xu N., Talbott R., Gelbert L., Gao J., Seizinger B. R., Gutkind J. S., Kley N. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 7868–7872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dohlman H. G., Thorner J. W. (2001) Annu. Rev. Biochem. 70, 703–754 [DOI] [PubMed] [Google Scholar]
- 13.Grillet N., Dubreuil V., Dufour H. D., Brunet J. F. (2003) J. Neurosci. 23, 10613–10621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albig A. R., Schiemann W. P. (2005) Mol. Biol. Cell 16, 609–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krumins A. M., Barker S. A., Huang C., Sunahara R. K., Yu K., Wilkie T. M., Gold S. J., Mumby S. M. (2004) J. Biol. Chem. 279, 2593–2599 [DOI] [PubMed] [Google Scholar]
- 16.Mantovani R., Li X. Y., Pessara U., Hooft van Huisjduijnen R., Benoist C., Mathis D. (1994) J. Biol. Chem. 269, 20340–20346 [PubMed] [Google Scholar]
- 17.Zhang J. W., Tang Q. Q., Vinson C., Lane M. D. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 43–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chatterjee T. K., Liu Z., Fisher R. A. (2003) J. Biol. Chem. 278, 30261–30271 [DOI] [PubMed] [Google Scholar]
- 19.Yang J., Kawai Y., Hanson R. W., Arinze I. J. (2001) J. Biol. Chem. 276, 25742–25752 [DOI] [PubMed] [Google Scholar]
- 20.Chakravarty K., Wu S. Y., Chiang C. M., Samols D., Hanson R. W. (2004) J. Biol. Chem. 279, 15385–15395 [DOI] [PubMed] [Google Scholar]
- 21.Liu Z., Fisher R. A. (2004) J. Biol. Chem. 279, 14120–14128 [DOI] [PubMed] [Google Scholar]
- 22.Aparicio O., Geisberg J. V., Sekinger E., Yang A., Moqtaderi Z., Struhl K. (2005) in Current Protocols in Molecular Biology (Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. eds) pp. 21.23.21–21.23.33, John Wiley & Sons, Inc., New York: [DOI] [PubMed] [Google Scholar]
- 23.Hu W., Li F., Mahavadi S., Murthy K. S. (2009) Am. J. Physiol. Cell Physiol. 296, C1310–C1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radeff-Huang J., Seasholtz T. M., Matteo R. G., Brown J. H. (2004) J. Cell. Biochem. 92, 949–966 [DOI] [PubMed] [Google Scholar]
- 25.Kranenburg O., Moolenaar W. H. (2001) Oncogene 20, 1540–1546 [DOI] [PubMed] [Google Scholar]
- 26.Sorensen S. D., Nicole O., Peavy R. D., Montoya L. M., Lee C. J., Murphy T. J., Traynelis S. F., Hepler J. R. (2003) Mol. Pharmacol. 64, 1199–1209 [DOI] [PubMed] [Google Scholar]
- 27.Mantovani R. (1998) Nucleic Acids Res. 26, 1135–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Q. S., Qian B., Levy D. (2004) J. Biol. Chem. 279, 29902–29910 [DOI] [PubMed] [Google Scholar]
- 29.Hopfner R., Mousli M., Jeltsch J. M., Voulgaris A., Lutz Y., Marin C., Bellocq J. P., Oudet P., Bronner C. (2000) Cancer Res. 60, 121–128 [PubMed] [Google Scholar]
- 30.Jurchott K., Bergmann S., Stein U., Walther W., Janz M., Manni I., Piaggio G., Fietze E., Dietel M., Royer H. D. (2003) J. Biol. Chem. 278, 27988–27996 [DOI] [PubMed] [Google Scholar]
- 31.Sinha S., Maity S. N., Lu J., de Crombrugghe B. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 1624–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manni I., Caretti G., Artuso S., Gurtner A., Emiliozzi V., Sacchi A., Mantovani R., Piaggio G. (2008) Mol. Biol. Cell 19, 5203–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu L., Wu Q., Yang C. P., Horwitz S. B. (1995) Cell Growth & Differ. 6, 1505–1512 [PubMed] [Google Scholar]
- 34.Jump D. B., Badin M. V., Thelen A. (1997) J. Biol. Chem. 272, 27778–27786 [DOI] [PubMed] [Google Scholar]
- 35.Chen G. K., Sale S., Tan T., Ermoian R. P., Sikic B. I. (2004) Mol. Pharmacol. 65, 906–916 [DOI] [PubMed] [Google Scholar]
- 36.Sikder H., Zhao Y., Balato A., Chapoval A., Fishelevich R., Gade P., Singh I. S., Kalvakolanu D. V., Johnson P. F., Gaspari A. A. (2009) J. Immunol. 183, 1657–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polo J. M., Ci W., Licht J. D., Melnick A. (2008) Blood 112, 644–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahmad K. F., Melnick A., Lax S., Bouchard D., Liu J., Kiang C. L., Mayer S., Takahashi S., Licht J. D., Privé G. G. (2003) Mol. Cell 12, 1551–1564 [DOI] [PubMed] [Google Scholar]
- 39.Huynh K. D., Fischle W., Verdin E., Bardwell V. J. (2000) Genes Dev. 14, 1810–1823 [PMC free article] [PubMed] [Google Scholar]
- 40.Dhordain P., Lin R. J., Quief S., Lantoine D., Kerckaert J. P., Evans R. M., Albagli O. (1998) Nucleic Acids Res. 26, 4645–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huynh K. D., Bardwell V. J. (1998) Oncogene 17, 2473–2484 [DOI] [PubMed] [Google Scholar]
- 42.Wong C. W., Privalsky M. L. (1998) Mol. Cell. Biol. 18, 5500–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yun J., Chae H. D., Choi T. S., Kim E. H., Bang Y. J., Chung J., Choi K. S., Mantovani R., Shin D. Y. (2003) J. Biol. Chem. 278, 36966–36972 [DOI] [PubMed] [Google Scholar]
- 44.Ceseña T. I., Cardinaux J. R., Kwok R., Schwartz J. (2007) J. Biol. Chem. 282, 956–967 [DOI] [PubMed] [Google Scholar]
- 45.Ceseña T. I., Cui T. X., Subramanian L., Fulton C. T., Iñiguez-Lluhí J. A., Kwok R. P., Schwartz J. (2008) Mol. Cell. Endocrinol. 289, 94–101 [DOI] [PubMed] [Google Scholar]
- 46.Bereshchenko O. R., Gu W., Dalla-Favera R. (2002) Nat. Genet. 32, 606–613 [DOI] [PubMed] [Google Scholar]
- 47.Lin T. C., Huang L. T., Huang Y. N., Chen G. S., Wang J. Y. (2009) Epilepsy Behav. 14, 316–323 [DOI] [PubMed] [Google Scholar]
- 48.Xie Y., Wolff D. W., Wei T., Wang B., Deng C., Kirui J. K., Jiang H., Qin J., Abel P. W., Tu Y. (2009) Cancer Res. 69, 5743–5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Calao M., Burny A., Quivy V., Dekoninck A., Van Lint C. (2008) Trends Biochem. Sci. 33, 339–349 [DOI] [PubMed] [Google Scholar]
- 50.Ellis L., Hammers H., Pili R. (2009) Cancer Lett. 280, 145–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buchwald M., Krämer O. H., Heinzel T. (2009) Cancer Lett. 280, 160–167 [DOI] [PubMed] [Google Scholar]
- 52.Ye B. H., Lista F., Lo Coco F., Knowles D. M., Offit K., Chaganti R. S., Dalla-Favera R. (1993) Science 262, 747–750 [DOI] [PubMed] [Google Scholar]
- 53.Ci W., Polo J. M., Melnick A. (2008) Curr. Opin. Hematol. 15, 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding J., Guzman J. N., Tkatch T., Chen S., Goldberg J. A., Ebert P. J., Levitt P., Wilson C. J., Hamm H. E., Surmeier D. J. (2006) Nat. Neurosci. 9, 832–842 [DOI] [PubMed] [Google Scholar]
- 55.Buckholtz J. W., Meyer-Lindenberg A., Honea R. A., Straub R. E., Pezawas L., Egan M. F., Vakkalanka R., Kolachana B., Verchinski B. A., Sust S., Mattay V. S., Weinberger D. R., Callicott J. H. (2007) J. Neurosci. 27, 1584–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]