Abstract
The first committed step in triterpenoid biosynthesis is the cyclization of oxidosqualene to polycyclic alcohols or ketones C30H50O. It is catalyzed by single oxidosqualene cyclase (OSC) enzymes that can carry out varying numbers of carbocation rearrangements and, thus, generate triterpenoids with diverse carbon skeletons. OSCs from diverse plant species have been cloned and characterized, the large majority of them catalyzing relatively few rearrangement steps. It was recently predicted that special OSCs must exist that can form friedelin, the pentacyclic triterpenoid whose formation involves the maximum possible number of rearrangement steps. The goal of the present study, therefore, was to clone a friedelin synthase from Kalanchoe daigremontiana, a plant species known to accumulate this triterpenoid in its leaf surface waxes. Five OSC cDNAs were isolated, encoding proteins with 761–779 amino acids and sharing between 57.4 and 94.3% nucleotide sequence identity. Heterologous expression in yeast and GC-MS analyses showed that one of the OSCs generated the steroid cycloartenol together with minor side products, whereas the other four enzymes produced mixtures of pentacyclic triterpenoids dominated by lupeol (93%), taraxerol (60%), glutinol (66%), and friedelin (71%), respectively. The cycloartenol synthase was found expressed in all leaf tissues, whereas the lupeol, taraxerol, glutinol, and friedelin synthases were expressed only in the epidermis layers lining the upper and lower surfaces of the leaf blade. It is concluded that the function of these enzymes is to form respective triterpenoid aglycones destined to coat the leaf exterior, probably as defense compounds against pathogens or herbivores.
Keywords: Enzymes, Lipid Synthesis, Mass Spectrometry (MS), Plant, Sterol, Cyclase, Enzyme specificity, Plant Leaf Wax, Secondary Metabolites, Triterpenoids
Introduction
Triterpenoids are a very diverse group of natural products with complex alicyclic structures containing C30 backbones or derivatives formed from C30 parent structures by side chain addition/elimination (1). They are found in virtually all organisms, serving various important functions as membrane constituents (steroids in eukaryotes, hopanoids in prokaryotes), as hormones (steroids in plants and animals), and as defense compounds (pentacyclic triterpenoids in plants). The greatest structural diversity of triterpenoids is found in higher plants, where hundreds of different backbone structures have been reported, with even higher diversity arising from oxidative modification to form various aglycones and their glycosylation to saponins (2).
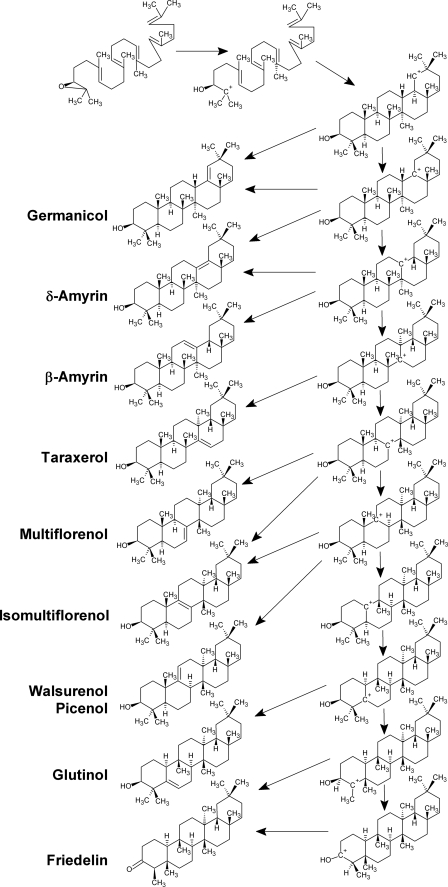
Triterpenoid biosynthesis in eukaryotes occurs by cyclization of epoxysqualene, leading in a single enzyme-catalyzed reaction to an initial product C30H50O that can contain 1–5 rings (3). Although many organisms have single product-specific oxidosqualene cyclase (OSC)4 enzymes, plants can contain arrays of cyclases competing for the same substrate to form diverse steroids and pentacyclic triterpenoids (2, 4, 5). This cyclization is, thus, a branch point between primary metabolism and secondary metabolism. Triterpenoid cyclization reactions comprise a sequence of four steps, including an initial protonation, a polyene addition cascade plus ring widening to form cycles A–E, 1,2-shifts of hydride and/or methyl groups, and a final deprotonation (Scheme 1) (3). Substantial variation in carbon backbone structures is due to differences in the numbers of rearrangement steps in the third stage of the reaction.
SCHEME 1.
Mechanism for the first committed step during biosynthesis of pentacyclic triterpenoids. The reaction starts with the protonation of oxidosqualene (top) and then involves a series of carbocationic intermediates that first undergo cyclization and then various rearrangements (right) and finally ends by deprotonation into various natural products (left).
The sequence of rearrangements can be predicted based on the occurrence of tertiary carbocations (Scheme 1) and has, for the early steps along the mechanistic pathway, been confirmed by experimental evidence using inhibitor studies and enzyme domain swapping/site-directed mutagenesis (6–15). The sequence of rearrangements shifts the position of the carbocation from the E ring, where it resides at the end of the polyene reaction, toward the A ring. The cyclization products may, accordingly, be classified depending on the C=C double bond position and the number of rearrangement steps and carbocationic intermediates occurring en route. Thus, germanicane derivatives can be considered as the structures with five six-membered rings with the minimum number of rearrangements, whereas β-amyrin formation involves two more intermediates, isomultiflorenol involves another two, and glutinol involves three more. Friedelin is of special interest because it is the triterpenoid product with the maximum number of rearrangements possible. Once the entire series of rearrangements is completed and the positive charge has migrated back to the A ring, then deprotonation from the hydroxyl group rather than from a methylene unit becomes possible. Due to abstraction of the same proton that was added in the initial step of the reaction, friedelin is the only triterpenoid ketone formed directly by cyclization, in contrast to all of the other C30H50O isomers that are synthesized as unsaturated alcohols. It seems likely that the same amino acid that is donating the proton initially will accept it back after the ring formation and rearrangement steps have occurred. The residue responsible for substrate protonation is known to be an aspartate in the DCTAE motif well conserved among all OSC enzymes (16).
OSCs from many plant species have been cloned and characterized by heterologous expression in yeast. Most of these enzymes were found to be product-specific lupeol or β-amyrin synthases, with only a few enzymes also capable of synthesizing triterpenoid products further along the rearrangement sequence (2). The currently known OSC catalyzing the highest number of rearrangement steps is a product-specific enzyme from Luffa cylindrica that forms isomultiflorenol (17). However, corresponding OSCs synthesizing friedelin either as the sole product or in a mixture with other triterpenoid products have not been described to date, leaving the possibility that this triterpenoid might be generated by modification of preformed pentacyclic triterpenols through rearrangements (i.e. in a two-enzyme process) rather than by direct cyclization of oxidosqualene catalyzed by a single enzyme. Corey and co-workers (18) have recently calculated free energies of various carbocations involved in the rearrangement series and found that the later rearrangement reactions are slightly endergonic. Therefore, it was concluded that the late rearrangement reactions must be coupled with earlier ones in a single enzymatic reaction in order to occur. Based on this finding, friedelin synthases are likely to be true OSC enzymes and should be cloned and characterized as such.
The goal of the current investigation was to identify and characterize an enzyme forming friedelin. To that end, we had recently screened a number of Crassulaceae species for the accumulation of friedelin because multiple occurrences of this compound had previously been reported for this plant family. Special focus was on plant surface waxes, because they are known to be principal lipophilic pools containing triterpenoid aglycones. Kalanchoe daigremontiana leaf waxes were found to be particularly rich in friedelin (19) and were chosen for further studies because this plant species is easy to cultivate and to use for molecular genetic experiments. K. daigremontiana is of further interest because its leaf surface waxes contain three other triterpenoids besides friedelin, raising the question whether these different products are formed by one promiscuous or several specific OSC enzymes. Triterpenoids accumulate mainly during the first 30 days of leaf development, indicating that activities of relevant triterpenoid synthases and, thus, expression of corresponding genes peak early during leaf development (20). Based on these findings, the current study aimed at the isolation of mRNA coding for enzymes involved in the formation of triterpenoids in K. daigremontiana, followed by cloning of corresponding cDNAs into yeast and finally the biochemical characterization of the enzymes overexpressed there.
EXPERIMENTAL PROCEDURES
Plant Growth and Triterpenoid Analysis
K. daigremontiana plants were grown in standard soil under temperate greenhouse conditions. Leaves were harvested, surface-extracted with CHCl3 to remove cuticular waxes, and then ground to powder in liquid nitrogen. The powder was extracted with CHCl3, and the lipid mixture was fractionated by TLC (20 × 20 cm, silica gel, 0.5 mm; Merck) using CHCl3 as the mobile phase. Resulting bands were stained with primuline, viewed under UV light, scratched from the plates, extracted with CHCl3, and analyzed by GC-MS and GC-flame ionization detection for identification and quantification, respectively, of the triterpenoids contained in the internal leaf tissues. GC conditions were used as described below for yeast extract analyses.
OSC Cloning
K. daigremontiana leaves were harvested, immediately placed in liquid nitrogen, and ground thoroughly with a mortar and pestle. Total RNA was extracted from the powder using the TRIzol reagent (Invitrogen) and used for cDNA synthesis by SuperScript II reverse transcriptase (Invitrogen) following standard protocols. The resulting cDNA mixture served directly as template for the following PCRs. Degenerate oligonucleotide primers OGL3S (5′-TTYGGNWGYCARNHRTGGGAT-3′), OGC1S (5′-TAYAATGGMAGYCARYTVTGGGA-3′), and OGN3A (5′-GGTGGDTGGRGDGARAGYYA-3′), previously designed to target conserved regions in plant OSCs (21), were used here for the amplification of the core fragments of OSCs from K. daigremontiana. PCR was carried out with recombinant TaqDNA polymerase (Fermentas) and the following cycling conditions: 2 min at 94 °C (45 s at 94 °C, 60 s at 50 °C, and 90 s at 72 °C) × 30 cycles, with a final extension of 10 min at 72 °C. The resulting PCR products were separated by gel electrophoresis (1% agarose) and extracted using the QIAquick gel extraction kit (Qiagen). A DNA product of the desired length was cloned into the pGEM-T vector (Promega) and transformed into Top10 competent cells (Invitrogen). Plasmid DNA was purified from transformed cells using the QIAprep spin miniprep kit (Qiagen) and sequenced. All further PCR products mentioned below were subcloned and sequenced by the same procedure.
The core fragment was extended in a second PCR (conditions as above) using the primer BWN1S (5′-CARWAYRDDGAYGGNGGNTGGG-3′) and gene-specific primers synthesized according to the obtained core sequence (supplemental Table 1). For 3′-end amplification of the open reading frame (ORF), first strand synthesis was carried out for 1 h at 42 °C using 5 μg of total RNA, AP-primer (5′-GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTT-3′), and reverse transcriptase in 20 μl. The product served as template in a PCR (conditions as above except annealing temperature 55 °C) with the adapter primer AUAP (5′-GGCCACGCGTCGACTAGTAC-3′) and a specific primer (supplemental Table 1). To obtain the 5′-end of the cDNA, the 5′-rapid amplification of cDNA ends system of Invitrogen was employed with the two nested gene-specific antisense primers (supplemental Table 1).
The corresponding full-length cDNAs were amplified using K. daigremontiana cDNA as a template and specific N- and C-terminal primers (supplemental Table 1). The PCRs were conducted with Phusion high fidelity DNA polymerase (New England Biolabs) in a final volume of 50 μl (1 μm each primer and 1 μl of cDNA) under the following conditions: 30 s at 98 °C (10 s at 98 °C, 30 s at 55 °C, and 75 s at 72 °C) × 28 cycles, 5 min at 72 °C. The resulting 2.3-kb PCR products were purified by gel electrophoresis. Sequences were analyzed by NCBI BLAST (available from the National Institutes of Health Web site) and DNAstar software (available on the World Wide Web). Sequence information for the putative triterpenoid synthases has been deposited in GenBankTM (accession numbers KdTAS HM623868, KdGLS HM623869, KdFRS HM623870, KdLUS HM623871, KdCAS HM623872).
Functional Expression of OSC cDNAs in Yeast
The full-length cDNAs of the putative triterpenoid synthases were digested with appropriate restriction enzymes and ligated into the yeast expression vector pYES2 (Invitrogen) behind the GAL1 promoter. The resulting constructs were transformed into the mutant yeast strain GIL77 by the LiAc/SS-DNA/PEG method (22), and yeast expression experiments were carried out as described previously (21). In brief, cells were induced with galactose, incubated in 0.1 m potassium phosphate-containing glucose and hemin for 24 h, collected, refluxed for 5 min in 20% KOH, 50% EtOH, and extracted twice with 5 ml of hexane. Both hexane solutions were combined, the solvent was removed under a gentle stream of N2, and the residue was redissolved in 0.2 ml of CHCl3 for chemical analyses.
Chemical Analysis of Yeast Expression Products
Yeast lipid extracts were either analyzed directly by GC-MS or first purified by TLC and then analyzed by GC-MS and NMR. For direct GC-MS analyses, extracts were derivatized using bis-N,O-trimethylsilyltrifluoroacetamide (in pyridine at 70 °C for 60 min) or Ac2O (in pyridine at 70 °C for 4 h), dried under N2, and taken up in CHCl3. The qualitative composition was studied using a 6890N Network GC system (Agilent) equipped with a mass spectrometric detector (Agilent 5973N) and an HP-1 capillary column (Agilent, length 30 m, inner diameter 320 μm, 1-μm film thickness). 1 μl of each sample was injected on column into a flow of helium gas held constant at 1.4 ml/min. The oven temperature was programmed for 2 min at 50 °C, followed by a 40 °C/min ramp to 200 °C, held at 200 °C for 2 min, increased by 3 °C/min to 320 °C, and held at 320 °C for 30 min. Triterpenoids were identified by comparison with literature data and with authentic compounds. Standards of friedelin, β-amyrin, and lupeol were purchased from Roth. A standard of glutinol had previously been isolated from K. daigremontiana wax, purified by TLC as well as HPLC, and authenticated using GC-MS and NMR (20). Finally, taraxerol was generated semisynthetically by lithium aluminum hydride reduction of taraxerone isolated as previously described from the surface wax mixture on Macaranga hypoleuca leaves and was purified by TLC and characterized by GC-MS as acetate and TMS derivative.
Further structure confirmation was obtained from NMR analyses. To this end, compounds were purified from yeast extracts using TLC as described above for plant lipid separation. 13C NMR spectra of the resulting fractions were recorded at 400 MHz in CDCl3 solution. Chemical shifts, expressed as values relative to the solvent signals δ 77.0 (t) for 13C were recorded for two triterpenoids isolated from transgenic yeast: compound 3 δ 16.41 (C-25), 18.43 (C-1), 18.63 (C-26), 19.83 (C-27), 23.86 (C-7), 25.66 (C-23), 28.04 (C-2), 28.46 (C-20), 29.17 (C-24), 30.31 (C-17), 30.57 (C-12), 32.25 (C-28), 32.30 (C-21), 32.61 (C-30), 33.34 (C-11), 34.74 (C-29), 34.83 (C-15), 35.06 (C-9), 35.30 (C-16), 36.24 (C-19), 38.06 (C-14), 39.17 (C-22), 39.52 (C-13), 41.03 (C-4), 43.29 (C-18), 47.65 (C-8), 49.91 (C-10), 76.54 (C-3), 122.26 (C-6), 141.84 (C-5); compound 4 δ 7.04 (C-23), 14.88 (C-24),18.16 (C-25), 18.46 (C-7), 18.88 (C-27), 20.48 (C-30), 22.50 (C-1), 28.39 (C-20), 30.22 (C-17), 30.73 (C-21), 32.00 (C-26), 32.31 (C-28), 32.65 (C-12), 33.00 (C-15), 35.24 (C-29), 35.57 (C-11), 35.85 (C-16), 36.24 (C-19), 37.67 (C-9), 38.52 (C-13), 39.47 (C-22), 39.92 (C-14), 41.52 (C-6), 41.75 (C-2), 42.36 (C-5), 43.03 (C-8), 53.33 (C-18), 58.45 (C-4), 59.71 (C-10), 213.40 (C-3).
The quantitative composition of the yeast lipid mixtures was studied using capillary GC with a flame ionization detector under the same GC conditions as above but H2 carrier gas at 2.0 ml/min. Five independent analyses were carried out for each transgenic yeast line.
Phylogenetic Analyses
Sequence alignments and phylogenetic analyses based on a neighbor-joining method were carried out with the ClustalX program Version 1.83 (23), using the amino acid sequences of cloned and characterized plant OSCs. A phylogenetic tree was created based on 1,000 bootstrap replications with the MEGA3.1 program. The GenBankTM data base accession numbers of the sequences used in the analysis are shown in supplemental Table 2.
RT-PCR Analysis
The epidermal cell layers were peeled from the upper and lower surfaces of K. daigremontiana leaves, and the remaining inner tissue as well as the epidermis preparations were immediately frozen in liquid nitrogen, ground into powders using a mortar and pestle, and used to extract RNA with TRIzol reagent (Invitrogen). The RNA samples were used for cDNA synthesis by SuperScript II reverse transcriptase (Invitrogen) following standard protocols. Gene-specific primers were designed to amplify fragments of the five different OSCs (listed in supplemental Table 1). Additionally, a 287-bp fragment of the actin gene was amplified as a positive control using the primers 5′-CGTTCTCTCCTTGTATGCCAGTGGTC-3′ and 5′-GAGCTGCTCTTGGCAGTCTCAAGTTC-3′. PCR cycle numbers and template amounts were optimized to yield products in the linear range of the reaction. PCR conditions for comparison of transcription levels for the five OCSs were as follows: 25 ng of template cDNA denatured at 94 °C for 2 min, followed by 23 cycles of 94 °C for 30 s, 55 °C for 45 s, and 72 °C for 60 s. Reactions were maintained at 72 °C for 10 min before separation of PCR products by electrophoresis in a 1% agarose gel.
Quantitative RT-PCR Analysis
RNA samples from leaf epidermal cell layers were used for cDNA synthesis by SuperScript II Reverse Transcriptase (Invitrogen) following standard protocols. Gene-specific primers were designed to amplify fragments of the four different OSCs, and the specificity of these primers was confirmed by a series of PCRs with plasmid DNA as a template. Quantitative PCR was carried out using SYBR GreenER qPCR SuperMix Universal kit (Invitrogen). The quantitative PCRs were programmed at 95 °C for 9 min, followed by 40 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s on an MJ MiniOpticon real-time PCR system (Bio-Rad).
RESULTS AND DISCUSSION
The goal of the current investigation was to identify and characterize enzymes forming pentacyclic triterpenoids through skeletal rearrangement series much longer than those occurring in previously known OSCs. In particular, the synthesis of friedelin had been predicted to involve the maximum number of 1,2-methyl and 1,2-hydride shifts after initial cyclization and thus requires an OSC capable of stabilizing more high energy carbocation intermediates than any other comparable enzymes (18). K. daigremontiana leaf surface lipids accumulate friedelin, together with a number of other pentacyclic triterpenoids, suggesting that this species must contain a friedelin synthase (19). It was therefore chosen as a model for the current study.
OSC Cloning and Sequence Analysis
Previous analyses had shown that K. daigremontiana leaves accumulate triterpenoids during the first stages of expansive growth, for ∼40 days after onset of leaf development (20). Here, it was further found that the major portion of these triterpenoids is accumulating in the cuticular waxes coating the exterior walls of epidermal cells, with only 2% of the total amounts of glutinol and of friedelin being present in the internal tissues of K. daigremontiana leaves. Therefore, it was assumed that OSC expression should be largely restricted to the epidermal cell layers in this tissue, and they were peeled from both sides of young K. daigremontiana leaves (<20 days old) to harvest mRNA enriched in target OSC sequences. The resulting mRNA population was transcribed into cDNA, which served as a template for the following PCR-based cloning. In a first PCR experiment, degenerate primers targeting the highly conserved consensus sequences of published OSCs were employed to amplify the core segment of potential OSC sequences. The PCR product mixture was separated by gel electrophoresis, yielding one major band corresponding to the expected size of 750 bp that was isolated, cloned, and amplified in E. coli. Based on restriction and PCR analyses, 32 bacterial colonies containing plasmids with inserts of the expected length were selected for sequencing. Inserts with five different sequences were found, each of them represented between three and six times. Extended core sequences were obtained by a set of PCRs. Finally, 5′- and 3′-rapid amplification of cDNA end reactions with specific primers yielded five full-length sequences named KdTAS, KdGLS, KdFRS, KdLUS, and KdCAS, each containing one of the core segments (Fig. 1).
FIGURE 1.
Amino acid sequences of five OSCs from K. daigremontiana. The conserved QW motifs thought to stabilize OSC structure are highlighted by single underlines, whereas the highly conserved DCTAE motif containing the aspartate residue involved in substrate protonation is marked by a double underline.
All of the cloned K. daigremontiana OCSs possess the amino acid motif DCTAE implicated in substrate binding (16) and the four QW motifs characteristic of the OSC superfamily (24). The ORFs of the K. daigremontiana OSCs range from 2,286 to 2,340 nucleotides in length, sharing between 57.4 and 94.3% with each other (Table 1). The amino acid sequences of these four genes show relatively high levels of identity with known β-amyrin synthases from other plant species, whereas the KdCAS ORF has substantially higher similarity with corresponding cycloartenol synthases (Table 2).
TABLE 1.
DNA sequence identities between OSCs cloned from K. daigremontiana
| KdGLS | KdFRS | KdTAS | KdLUS | |
|---|---|---|---|---|
| % | % | % | % | |
| KdFRS | 94.3 | |||
| KdTAS | 72.5 | 75.0 | ||
| KdLUS | 94.3 | 92.8 | 75.1 | |
| KdCAS | 57.4 | 61.3 | 59.4 | 60.4 |
TABLE 2.
Sequence identities between OSCs cloned from K. daigremontiana and selected OSCs previously described in the literature
| Plant species | Major product | Accession number | Amino acid sequence similarity with K. daigremontiana OSCs (amino acid number in parentheses) |
||||
|---|---|---|---|---|---|---|---|
| KdGLS (767) | KdFRS (761) | KdTAS (779) | KdLUS (765) | KdCAS (764) | |||
| % | % | % | % | % | |||
| B. platyphylla | β-Amyrin | AB055512 | 76.5 | 76.7 | 84.7 | 78.6 | 57.5 |
| P. ginseng | β-Amyrin | AB014057 | 74.6 | 74.9 | 82.3 | 75.8 | 57.3 |
| P. ginseng | β-Amyrin | AB009030 | 75.8 | 75.9 | 80.6 | 77.5 | 57.4 |
| Rhizophora stylosa | Multifunctional | AB263203 | 75.2 | 75.9 | 80.6 | 77.1 | 56.4 |
| Euphorbia tirucalli | β-Amyrin | AB206469 | 75.3 | 75.7 | 80.7 | 77.0 | 57.6 |
| Lathyrus japonicus | Cycloartenol | AB181246 | 58.0 | 58.1 | 58.7 | 59.7 | 79.8 |
| P. ginseng | Cycloartenol | AB009029 | 57.0 | 58.2 | 59.2 | 58.6 | 79.2 |
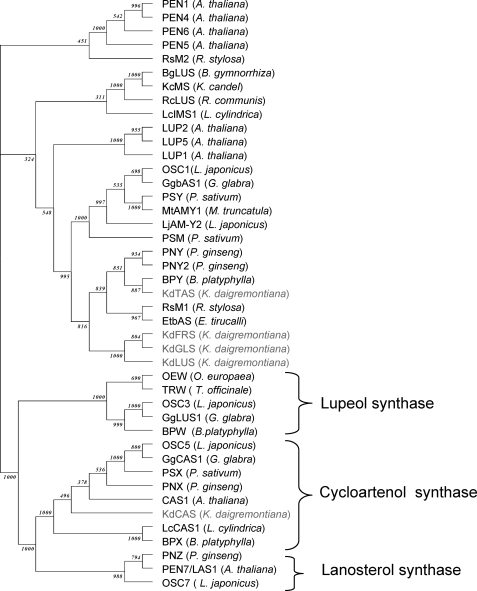
Phylogenetic analyses using the neighbor-joining method (ClustalX) (23) showed that KdFRS, KdGLS, KdLUS, and KdTAS all cluster very closely together within a clade of OSC enzymes previously characterized as β-amyrin synthases (Fig. 3). Although the first three enzymes from K. daigremontiana form a new branch within this cluster of OSC enzymes, KdTAS is more closely related to the β-amyrin synthases BPY, PNY, and PNY2 from Betula platyphylla and Panax ginseng. Each of the four OSCs from K. daigremontiana shares at least 76% sequence identity with one or more OSCs reported in the literature to date (Table 2). Finally, KdCAS was found to group together with the cycloartenol synthases from various other species (Fig. 2).
FIGURE 3.
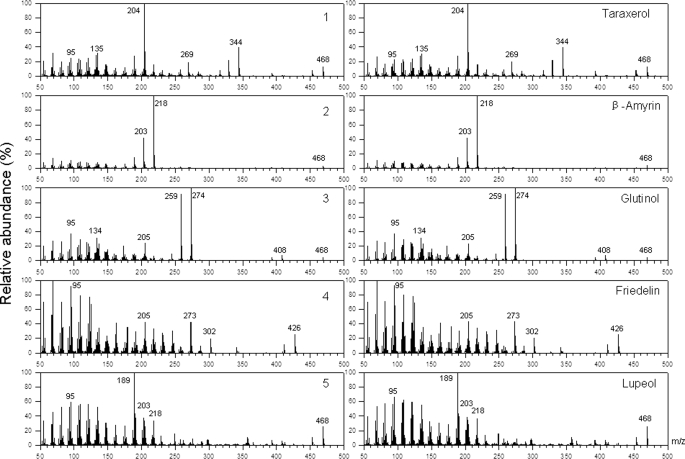
Mass spectra of pentacyclic triterpenoids. Compounds 1–5, formed by yeast strains heterologously expressing various OSCs from K. daigremontiana (left), have fragmentation patterns identical with those of authentic standards (right). For the triterpenoid alcohols 1–3 and 5 as well as the corresponding standards, spectra of acetates are shown.
FIGURE 2.
Phylogenetic analysis comparing the new OSC cDNAs cloned from K. daigremontiana with previously known OSCs from other plant species.
Functional cDNA Expression in Yeast
In order to biochemically characterize the putative OSCs from K. daigremontiana, their full-length ORFs were heterologously expressed in Saccharomyces cerevisiae. The yeast mutant GIL77 (gal2 hem3-6 erg7 ura3-167) was used for this purpose because it is deficient in lanosterol synthase and therefore accumulates high concentrations of 2,3-oxidosqualene that can serve as substrate for OSC-catalyzed cyclization (25). However, growth of this strain requires exogenous ergosterol, which must be added to the medium and is consequently detected in triterpenoid analyses together with the products of the transgenic OSCs. All five K. daigremontiana cDNAs were subcloned into the yeast expression vector pYES2, and the resulting constructs were transformed into GIL77. In parallel, the empty vector was transformed into the yeast mutant to serve as a negative control. Three independent lines were selected from each transformation experiment and were further grown in the presence of ergosterol, induced with galactose, and extracted. The extracts were acetylated and analyzed by GC-MS in order to identify triterpenoids, whereas GC-flame ionization detection was used to assess relative quantities of the compounds. No differences between parallel lines were found, and therefore results are shown here for only one line harboring each OSC construct.
All six yeast transformants were found to contain two compounds in common, which were identified as ergosterol and 2,3-oxidosqualene. In addition, the extracts from yeast harboring K. daigremontiana cDNAs contained several compounds not present in the control, most of them also differing between the lines expressing different OSC candidate cDNAs. The structures of all major compounds were elucidated by comparison of mass spectral characteristics with those of authentic standards (Fig. 3) and of GC elution behavior (Fig. 4). Most of the triterpenoids in the yeast extracts were found to contain hydroxyl functions and could therefore be derivatized in order to improve chromatographic resolution and spectral quality. Acetate esters and trimethylsilyl ethers were generated for all these triterpenoid alcohols, and both derivatives together were used to assign structures. Lupeol and β-amyrin were identified by comparison with commercial standards, whereas glutinol was identified by comparison with a standard that had been extracted from K. daigremontiana leaf wax and authenticated by 1H NMR and 13C NMR analysis (20). The structure of taraxerol was assigned by comparing MS data for the acetate ester and TMS ether with those reported in the literature and found for the corresponding triterpenoid isolated from M. hypoleuca leaf wax (26). Finally, one compound in the yeast extracts was identified as the triterpenoid ketone friedelin based on direct comparison with a commercial standard of this compound and 1H NMR and 13C NMR analysis.
FIGURE 4.
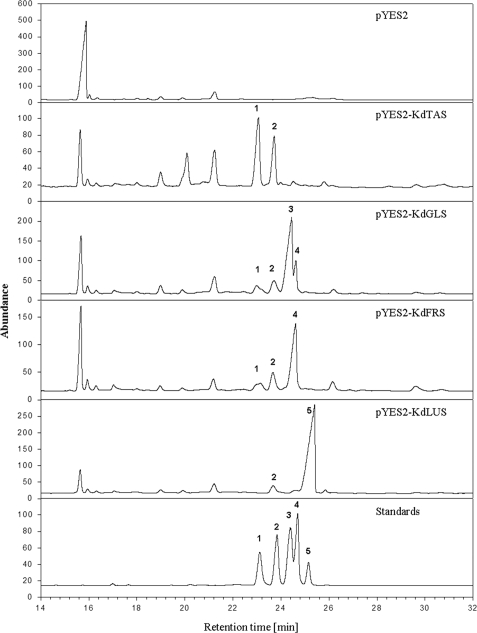
Gas chromatographic analysis of triterpenoids in yeast strains harboring the vectors indicated in the various panels. Triterpenoid alcohols were transformed into acetates prior to GC analysis. A and B were present in all yeast samples and were identified as epoxysqualene and ergosterol, respectively. Compounds 1–5 showed retention behavior similar to those of authentic standards of taraxerol, β-amyrin, glutinol, friedelin, and lupeol, respectively.
Quantitative analyses were carried out on five independent parallels of each transgenic yeast line. They showed that all of the lines expressing putative OSC cDNAs contained 2,3-oxidosqualene at markedly lower concentrations than the empty vector control (Fig. 4). Yeast heterologously expressing KdTAS cDNA produced a mixture of taraxerol (60 ± 1%; mean ± S.D., n = 5) and β-amyrin (40% ± 1%), whereas expression of KdGLS cDNA prompted the formation of glutinol (67 ± 2%) together with friedelin (14 ± 1%), β-amyrin (11 ± 0.3%), and taraxerol (8 ± 0.2%). Expression of the plasmid pYES2-KdFRS resulted in the formation of friedelin (72 ± 1%), accompanied by smaller amounts of β-amyrin (18 ± 0.1%) and taraxerol (10 ± 1%). Lines harboring KdLUS cDNA produced a mixture of lupeol (93 ± 2%) and β-amyrin (7 ± 2%). The products formed by the four OSCs together account for all of the major triterpenoids found in the cuticular wax on K. daigremontiana leaves (19, 20).
The relative activity of the four OSC genes characterized here can be estimated based on our GC analyses of the transgenic yeast extracts (Fig. 4). Because exactly the same amounts of ergosterol had been added to all cultures and samples had been taken and worked up in the same way, the steroid can serve as an internal standard to estimate the amounts of triterpenoid products formed. Hence, it can be concluded that KdLUS was the most active enzyme when tested in yeast, followed by KdFRS and KdGLS, whereas KdTAS was found to be a much less active OSC. Finally, extracts of GIL77 yeast cells functionally expressing KdCAS cDNA were found to contain cycloartenol together with some unidentified cyclization products (data not shown).
OSC Expression Patterns in the K. daigremontiana Leaf
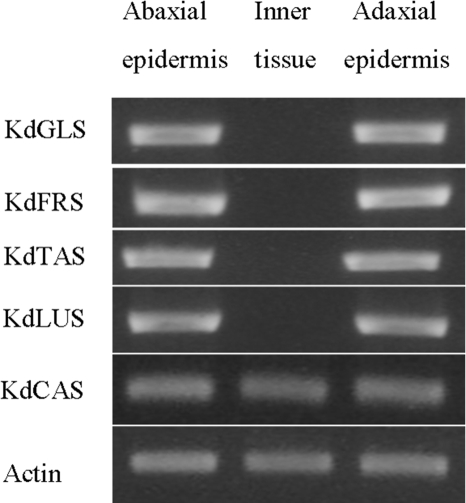
To investigate whether OSC transcription is tissue-specific, RT-PCR analyses were carried out. Five pairs of PCR primers were designed based on the OSC sequences and first tested for gene specificity using plasmids harboring full-length cDNAs of the five OSCs. It was found that each primer pair allowed amplification of its target OSC sequence fragment but not of the other four OSCs (data not shown). Total RNA was isolated from leaf inner tissue as well as the abaxial and adaxial epidermal layers of K. daigremontiana leaves, and the RT-PCR analysis using these RNA samples showed that the KdCAS enzyme is expressed equally in all tissues within the leaf (Fig. 5), in accordance with its likely role in membrane steroid production of all cells. In contrast, the KdTAS, KdFRS, KdGLS, and KdLUS enzymes are expressed only in the epidermal cells on both sides of the leaf and not in internal leaf tissues (Fig. 5). These results are in agreement with the accumulation of the triterpenoid products to high concentrations in the cuticular waxes on both leaf surfaces. Our results show that the major biological function of all four genes is in formation of triterpenoids targeted to the plant surface, where they probably play important roles in protecting the tissue from herbivores and pathogens.
FIGURE 5.
RT-PCR analysis of the expression patterns of five OSC genes in K. daigremontiana leaves. The four enzymes generating pentacyclic triterpenoids were found expressed only in the epidermis layers of the leaves, whereas the cycloartenol synthase was equally expressed in all tissues within the leaf.
OSC Expression Levels in the K. daigremontiana Leaf
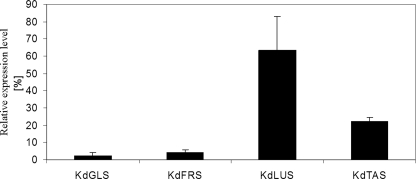
Although the above results show that the four OSCs are involved in the formation of cuticular triterpenoids in the K. daigremontiana leaf, it remains to be elucidated how much each enzyme contributes to the product mixture. However, the enzyme activities within the tissue cannot be determined directly due to the low solubility of substrates and products. In order to assess the relative contribution of the four OSCs, the relative expression levels of the corresponding genes were assessed using quantitative RT-PCR. It was found that the expression levels were highest for KdLUS, intermediate for KdTAS, and lowest for KdGLS and KdFRS (Fig. 6).
FIGURE 6.
Quantitative RT-PCR analysis of the four OSC genes in K. daigremontiana leaves generating pentacyclic triterpenoids. The relative expression levels were determined in whole, still growing leaves and normalized (n = 3, S.D. (error bars)). Lupeol synthase was found to be expressed most actively, taraxerol synthase at an intermediate level, and glutinol as well as friedelin synthase least of the genes.
Taking the gene expression levels together with the relative activities of the OSCs (determined in yeast; see above), it seems likely that KdLUS should have the highest absolute activity in the K. daigremontiana leaf. The other three OSC activities are predicted to be lower than KdLUS and similar to each other because higher relative activities of KdGLS and KdFRS might be compensated for by the higher expression level of KdTAS. These predicted activity levels of the four OSCs do not match the triterpenoid profile in the cuticular waxes of K. daigremontiana leaves, which had been reported to contain ∼62% glutinol (and its derivatives), 20% friedelin, 13% taraxerol, and 5% β-amyrin, whereas lupeol had not been detected at all. It should be noted that germanicol had been described for the leaf wax, but further analyses using authentic standard as described here have now confirmed that the true structure of the compound is taraxerol both in the wax and in the yeast product.
The discrepancy between our biochemical data and the wax triterpenoid profile might be due to one or more of the following reasons. 1) The apparent OSC activities in yeast might be misleading because of artifacts in the heterologous environment, 2) large portions of the triterpenoid products formed by the OSCs in the plant might be transformed and/or sequestered in inner leaf tissues and therefore might not be reflected in previous surface wax analyses, or 3) other OSCs might be present in K. daigremontiana that contribute further to the product profile.
Predictions for OSC Residues Determining Product Specificities for Friedelin and Glutinol
Friedelin is a very late quench product of the 1,2-hydride and 1,2-methyl rearrangement series, and the OSC forming this product can thus be expected to have catalytic site features stabilizing the maximum possible cationic intermediates. Interestingly, a second OSC from K. daigremontiana was found to form predominantly glutinol, the triterpenoid product generated immediately before friedelin along the rearrangement cascade. Both enzymes share very high sequence identity, and thus the small differences in amino acid structures between them can now be used to identify amino acids that may be involved in determining product specificities for either friedelin or glutinol.
Residues Ala79, Ser127, Gln172, His278, Lys350, and Phe423 in the sequence of KdFRS stand out because they differ between the friedelin synthase and most other OSCs and because they involve aromatic amino acids either in KdFRS or in the other enzymes. Aromatic residues are thought to be important for the cyclization mechanism because they can stabilize the cationic intermediates. Further amino acids of interest in KdFRS are Ile323 and Thr733 because they differ in this enzyme from all other OSCs, including KdGLS. Finally, amino acids His103, Leu249, Gln297, Phe497, Thr367, and Thr370 not only differ between KdFRS and other OSCs, but they are also predicted to line the active site or to be located near it, according to three-dimensional structure modeling (data not shown) based on the crystal structure available for human lanosterol synthase (27).
The role of the amino acids listed above in OSC product specificity is presently being investigated using site-directed mutagenesis experiments. To identify further residues possibly involved in the cyclization mechanism, domain-swapping experiments are also currently being carried out.
CONCLUSION
Altogether, five different OSCs from one plant species have been cloned and fully characterized by heterologous expression in yeast. Very diverse triterpenoids were identified as products of the enzymes, ranging from taraxerol over glutinol to friedelin. This is the first report on the sequence and biochemical characteristics of such a friedelin synthase, confirming the prediction that this late triterpenoid product is indeed formed in one sequence of catalytic steps. In comparison with friedelin and glutinol, taraxerol is a relatively early product of the rearrangement series, and the OSC forming this product must be stabilizing fewer cationic intermediates than the others. This is also the first report describing a taraxerol synthase, now providing the opportunity to perform further comparative studies of the amino acid structures determining earlier rearrangement steps in the sequence.
Supplementary Material
Acknowledgments
We thank Bangjun Wang and Ortwin Guhling for technical help and fruitful discussions and Dr. Y. Ebizuka (Tokyo) for providing the yeast strain GIL77.
This work was supported by a Natural Sciences and Engineering Research Council of Canada Special Research Opportunity Grant, by the Canada Foundation for Innovation, and by the Canada Research Chairs Program.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2.
- OSC
- oxidosqualene cyclase.
REFERENCES
- 1.Xu R., Fazio G. C., Matsuda S. P. T. (2004) Phytochemistry 65, 261–291 [DOI] [PubMed] [Google Scholar]
- 2.Phillips D. R., Rasbery J. M., Bartel B., Matsuda S. P. T. (2006) Curr. Opin. Plant Biol. 9, 305–314 [DOI] [PubMed] [Google Scholar]
- 3.Abe I., Rohmer M., Prestwich G. D. (1993) Chem. Rev. 93, 2189–2206 [Google Scholar]
- 4.Fazio G. C., Xu R., Matsuda S. P. T. (2004) J. Am. Chem. Soc. 126, 5678–5679 [DOI] [PubMed] [Google Scholar]
- 5.Lodeiro S., Xiong Q., Wilson W. K., Kolesnikova M. D., Onak C. S., Matsuda S. P. T. (2007) J. Am. Chem. Soc. 129, 11213–11222 [DOI] [PubMed] [Google Scholar]
- 6.Cattel L., Ceruti M. (1998) Crit. Rev. Biochem. Mol. Biol. 33, 353–373 [DOI] [PubMed] [Google Scholar]
- 7.Segura M. J., Jackson B. E., Matsuda S. P. (2003) Nat. Prod. Rep. 20, 304–317 [DOI] [PubMed] [Google Scholar]
- 8.Morikubo N., Fukuda Y., Ohtake K., Shinya N., Kiga D., Sakamoto K., Asanuma M., Hirota H., Yokoyama S., Hoshino T. (2006) J. Am. Chem. Soc. 128, 13184–13194 [DOI] [PubMed] [Google Scholar]
- 9.Abe I., Liu W., Oehlschlager A. C., Prestwich G. D. (1996) J. Am. Chem. Soc. 118, 9180–9181 [Google Scholar]
- 10.Corey E. J., Cheng H., Baker C. H., Matsuda S. P., Li D., Song H. (1997) J. Am. Chem. Soc. 119, 1277–1288 [Google Scholar]
- 11.Corey E. J., Cheng H., Baker C. H., Matsuda S. P., Li D., Song X. (1997) J. Am. Chem. Soc. 119, 1289–1296 [Google Scholar]
- 12.Hart E. A., Hua L., Darr L. B., Wilson W. K., Pang J., Matsuda S. P. T. (1999) J. Am. Chem. Soc. 121, 9887–9888 [Google Scholar]
- 13.Herrera J. B., Wilson W. K., Matsuda S. P. (2000) J. Am. Chem. Soc. 122, 6765–6766 [Google Scholar]
- 14.Kushiro T., Shibuya M., Masuda K., Ebizuka Y. (2000) J. Am. Chem. Soc. 122, 6816–6824 [Google Scholar]
- 15.Lodeiro S., Schulz-Gasch T., Matsuda S. P. (2005) J. Am. Chem. Soc. 127, 14132–14133 [DOI] [PubMed] [Google Scholar]
- 16.Abe I., Prestwich G. D. (1995) Lipids 30, 231–234 [DOI] [PubMed] [Google Scholar]
- 17.Hayashi H., Huang P., Inoue K., Hiraoka N., Ikeshiro Y., Yazaki K., Tanaka S., Kushiro T., Shibuya M., Ebizuka Y. (2001) Eur. J. Biochem. 268, 6311–6317 [DOI] [PubMed] [Google Scholar]
- 18.Kürti L., Chein R. J., Corey E. J. (2008) J. Am. Chem. Soc. 130, 9031–9036 [DOI] [PubMed] [Google Scholar]
- 19.van Maarseveen C., Jetter R. (2009) Phytochemistry 70, 899–906 [DOI] [PubMed] [Google Scholar]
- 20.Van Maarseveen C., Han H., Jetter R. (2009) Plant. Cell Environ. 32, 73–81 [DOI] [PubMed] [Google Scholar]
- 21.Guhling O., Hobl B., Yeats T., Jetter R. (2006) Arch. Biochem. Biophys. 448, 60–72 [DOI] [PubMed] [Google Scholar]
- 22.Gietz R. D., Woods R. A. (2002) Methods Enzymol. 350, 87–96 [DOI] [PubMed] [Google Scholar]
- 23.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poralla K., Hewelt A., Prestwich G. D., Abe I., Reipen I. G., Sprenger G. A. (1994) Trends Biochem. Sci. 19, 157–158 [DOI] [PubMed] [Google Scholar]
- 25.Kushiro T., Shibuya M., Ebizuka Y. (1998) Eur. J. Biochem. 256, 238–244 [DOI] [PubMed] [Google Scholar]
- 26.Markstädter C., Federle W., Jetter R., Riederer M., Hölldobler B. (2000) Chemoecology 10, 33–40 [Google Scholar]
- 27.Thoma R., Schulz-Gasch T., D'Arcy B., Benz J., Aebi J., Dehmlow H., Hennig M., Stihle M., Ruf A. (2004) Nature 432, 118–122 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.