Abstract
Epigenetic mechanisms, in particular the enzymatic modification of histones, are a crucial element of cell differentiation, a regulated process that allows a precursor cell basically to turn into a different cell type while maintaining the same genetic equipment. We have previously described that the promoters of adipogenic genes display significant levels of dimethylation at the Lys4 of histone H3 (H3K4) in preadipocytes, where these genes are still silenced, thus maintaining the chromatin of the precursor cell in a receptive state. Here, we show that the expression of several histone demethylases and methyltransferases increases during adipogenesis, suggesting an important role for these proteins in this process. Knockdown of the H3K4/K9 demethylase LSD1 results in markedly decreased differentiation of 3T3-L1 preadipocytes. This outcome is associated with decreased H3K4 dimethylation and increased H3K9 dimethylation at the promoter of transcription factor cebpa, whose expression must be induced >200-fold upon stimulation of differentiation. Thus, our data suggest that LSD1 acts to maintain a permissive state of the chromatin in this promoter by opposing the action of a H3K9 methyltransferase. Knockdown of H3K9 methyltransferase SETDB1 produced the opposite results, by decreasing H3K9 dimethylation and increasing H3K4 dimethylation levels at the cebpa promoter and favoring differentiation. These findings indicate that the histone methylation status of adipogenic genes as well as the expression and function of the proteins involved in its maintenance play a crucial role in adipogenesis.
Keywords: Adipocyte, Chromatin, Differentiation, Gene Regulation, Histone Methylation
Introduction
It has been extensively shown that the posttranslational modifications of histones participate in the regulation of transcription, both by altering chromatin structure locally and through the recruitment of regulatory complexes that recognize and bind to the modified histone tails (1). Among these modifications, the role of lysine acetylation is the best established, usually associating with transcriptional activation (2, 3). Unlike acetylation, histone lysine methylation has been linked with either transcriptional activation or repression, depending on the modified residue. Thus, methylation of the residue Lys4 of histone H3 (H3K4) correlates with gene activation (4, 5), whereas H3 Lys9 (H3K9) methylation is associated with transcriptional repression and the establishment and maintenance of silent heterochromatin regions (6). Moreover, lysine residues may be mono-, di-, or trimethylated (for review, see Ref. 1), thus resulting in a vast potential for functional responses.
Histone lysine methylation was until recently considered to be a permanent mark because no demethylases had been identified (7). However, the recent discovery of two classes of histone lysine demethylases (HDMs)4 (8–12) established the dynamic nature of this modification. The first HDM described, lysine-specific histone demethylase 1, LSD1 (also known as AOF2, BHC110, and KDM1), is an amine oxidase that mediates histone demethylation via a FAD-dependent oxidative reaction and has been found to be associated with a number of corepressor complexes such as those nucleated by REST corepressor (CoREST) or C-terminal-binding protein (CtBP) (13, 14). In agreement with these data, LSD1 has been shown specifically to demethylate in vitro mono- and dimethylated H3K4 and has been suggested to mediate gene repression in vivo by maintaining an unmethylated H3K4 status on a set of target promoters (8, 13). However, the substrate specificity of LSD1 seems to be modulated by the proteins with which it is associated (13, 15) and by other histone marks displayed on the histone tail (16, 17). In this regard, LSD1 has also been connected to gene activation through demethylation of H3K9 as a component of the MLL1 activator complex (14, 18) or when associated with the androgen or estrogen receptors (19, 20). LSD1 is widely recruited to active promoters in estrogen-stimulated cells and opposes the silencing function of H3K9 methyltransferases (HMTs) such as RIZ1 or SETDB1, which bind to the estrogen receptor in the absence of ligand, thus maintaining high levels of methylated H3K9 at its target promoters (20). Interestingly, H3K9 methyltransferase SETDB1 is also able to repress transcription of nuclear receptor PPARγ target genes in mesenchymal stem cells, consequently influencing cell fate by reducing adipogenesis while favoring osteoblastogenesis (21, 22).
Adipogenesis is a complex process tightly regulated by a well established cascade of sequence-specific transcription factors that concludes with increased expression and activation of CCAAT/enhancer-binding protein α (C/EBPα) and peroxisome proliferator activator receptor γ (PPARγ) (23). These two factors are between them responsible for the transcription of the largest part of adipocyte-specific genes, including the hormone adiponectin (apm1) or the transporters fabp4 and glut4. A number of recent studies have demonstrated the crucial role of epigenetic mechanisms in adipogenesis (for review, see 24). In our laboratory, we described a role for histone H3K4 dimethylation in labeling specific adipocyte genes such as apm1 as “poised” for transcription in undifferentiated fibroblasts that do not yet express them (25). Recently, the essential role of histone methylation in adipogenesis has been confirmed by two independent studies. Targeted deletion in mice of the H3K4 methyltransferase MLL3 results in drastically decreased adipose tissue, even under a high fat diet (26), whereas disruption of the gene encoding the H3K9 demethylase JMJD1A results in obesity (27). These data emphasize the active role of histone methylation in the development of adipocytes from undifferentiated cells.
In the present study, we evaluate how the expression levels of different HDMs and HMTs are regulated during adipocyte differentiation. Furthermore, we show that expression of HDM LSD1 is necessary for adipogenesis to proceed adequately. LSD1 participates in the regulation of adipogenic transcription factor cebpa by decreasing H3K9 dimethylation and maintaining H3K4 dimethylation at its promoter, opposing the function of H3K9 methyltransferase SETDB1. Taken together, these results indicate that histone methylation plays a key role in the regulation of the transcription of key adipogenic genes during adipogenesis and consequently in the differentiation process itself.
EXPERIMENTAL PROCEDURES
Cell Culture and Differentiation
Mouse 3T3-L1 preadipocytes were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% calf serum (Sigma) and 10T1/2 pluripotent fibroblasts were grown in Basal Medium Eagle supplemented with 10% fetal bovine serum (FBS). Differentiation of 3T3-L1 and 10T1/2 cells to adipocytes was induced by treatment of confluent cells (designated day 0, D0) with an adipogenic mixture consisting of 850 nm insulin, 1 μm dexamethasone, and 0.5 mm isobutylmethylxanthine in the presence of 10% FBS (all reagents from Sigma). The differentiation medium was replaced 2 days later (D2) with medium supplemented with 10% FBS and 850 nm insulin. After 3 more days in insulin-containing medium (D5), the cells were cultured in DMEM containing 10% FBS without further supplements. Isolation of mouse preadipocytes, in vitro differentiation, and transfection of D5 adipocytes are discussed in supplemental Methods.
siRNA Transfection
For RNA analysis and Oil Red O staining, 3T3-L1 or 10T1/2 fibroblasts were trypsinized 1 h before transfection and distributed into 24-well plates (7–8 × 104 cells/well). For ChIP assays or nuclear extract preparation, 2 × 106 cells were plated in a 100-mm dish 1 h prior to transfection. Cells were transfected with 60 nm corresponding double-stranded siRNAs using Metafectene Pro (Biontex, Martinsried, Germany) at a 1:3 (w:v) ratio with the siRNA. Two different siRNAs for each target were used. Sequence details are presented in supplemental Table 1. When required, cell differentiation was started when the transfected cells reached confluence, usually 48–72 h after transfection. Otherwise, RNA was extracted at 24, 48, and 72 h after transfection. For Western blotting and ChIP assays, cells were used 72 h after transfection.
Oil Red O Staining
Intracellular triglyceride was stained with Oil Red O (Sigma). Briefly, cells were fixed overnight with 4% paraformaldehyde, washed with 60% isopropyl alcohol, and stained with Oil Red O solution (0.21% Oil Red O in 60% isopropyl alcohol) for 1 h at room temperature. Excess stain was removed with 60% isopropyl alcohol, and cells were washed extensively with tap water before being photographed under a light microscope (magnification, ×400).
Nuclear Extract Preparation and Western Blotting
Nuclear extracts from 3T3-L1 cells were prepared as described (28). Equal amounts of protein were resolved by 9% SDS-PAGE and transferred to a 0.45-μm nitrocellulose membrane (Schleicher & Schuell). The primary antibodies were diluted 1/1,000 in TBS (20 mm Tris, pH 7.5, 137 mm NaCl) supplemented with 5% nonfat milk and visualized by blotting with HRP-conjugated secondary antibodies. Chemiluminescence was detected using the ECL Plus reagents (GE Healthcare), in a LAS3000 Lumi-Imager (Fuji Photo Film Inc., Valhalla, NY). Antibodies were from Upstate Biotechnology.
RNA Isolation and Real Time RT-PCR
Total RNA was extracted using TRI reagent (Sigma) according to the instructions of the manufacturer. Random-primed cDNA synthesis was performed at 37 °C starting with 0.5 μg of RNA, using the High Capacity cDNA Archive kit (Applied Biosystems). Gene expression was measured by real time PCR in an ABI Prism 7900HT Real Time PCR system using TaqMan FAM-labeled specific probes (Applied Biosystems). A list of the probes used is presented in supplemental Table 2. Results were normalized to actb expression. Statistical differences were assessed by two-tailed Student's t test using the mean ± S.D. calculated from three independent experiments performed in duplicate.
Chromatin Immunoprecipitation (ChIP) Assays
Dimethyl and trimethyl H3K4 ChIP assays were performed as described (25, 29). Dimethyl H3K9, LSD1, and SETDB1 ChIP assays were performed using the EpiQuik chromatin immunoprecipitation kit (Epigentek, Brooklyn, NY) following the instructions of the manufacturer. All antibodies used were from Upstate Biotechnology. ChIP assays were analyzed by real time PCR using TaqMan specific primers and FAM-labeled probes designed to amplify segments located in a region of approximately 500 bp around the transcription initiation site of selected genes for analysis of promoter regions (see supplemental Table 3). Results shown are the mean ± S.D. calculated from three independent ChIP experiments performed in duplicate. Statistical differences were assessed by two-tailed Student's t test.
RESULTS
H3K4 and H3K9 Dimethylation at Adipogenic Promoters Display a Tightly Regulated and Complementary Pattern of Enrichment throughout Adipogenesis
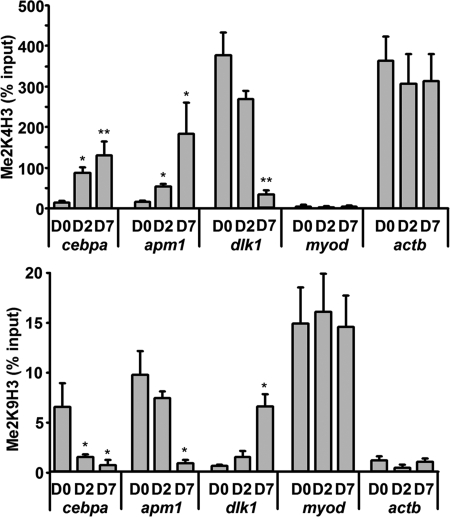
On a previous work (25) we showed that adipogenic genes such as the hormone adiponectin (apm1) display significant levels of H3K4 dimethylation on their promoter regions in preadipocytes. We proposed that this epigenetic signal, usually associated with actively transcribed genes (3, 4), labels those still silenced genes as poised to be expressed once the cells are induced to differentiate. To build on this information, we used ChIP to analyze those same promoters for the presence of H3K9 dimethylation, a mark usually associated with silent chromatin, in 3T3-L1 fibroblasts (D0), cells in the early stages of the differentiation process (D2), and mature adipocytes (D7). Our data show that, in addition to H3K4 dimethylation (Fig. 1, upper panel), adipogenic genes such as apm1 or cebpa, encoding transcription factor C/EBPα, one of the master regulators of adipogenesis, display significant levels of H3K9 dimethylation on their promoters in D0 preadipocytes (Fig. 1, lower panel). In contrast, the promoter of the silent muscle-specific gene myod1 displays high levels of H3K9 dimethylation throughout adipogenesis, but no detectable H3K4 dimethylation was observed. On the other hand, high H3K4 dimethylation levels, but not H3K9 dimethylation, were easily detected on the promoter of the constitutively active and heavily expressed actb gene during differentiation. Thus, adipogenic genes in preadipocytes exhibit characteristics shared with both inactive (H3K9 dimethylation) and active (H3K4 dimethylation) genes. Moreover, H3K4 and H3K9 dimethylation follow inverse patterns throughout differentiation on those promoters, with H3K4 dimethylation significantly increasing at the time that the expression of the genes increases (Fig. 1 and supplemental Fig. 1), and H3K9 dimethylation decreasing, reaching background levels in mature adipocytes (D7), where those genes are highly expressed. Conversely, the promoter of dlk1, a gene that follows an inverse pattern of expression to apm1 or cebpa, being highly expressed in preadipocytes and progressively silenced throughout adipogenesis, displays high levels of H3K4 dimethylation but undetectable H3K9 dimethylation in 3T3-L1 D0 fibroblasts. During adipogenesis, mirroring the silencing of the gene, H3K4 dimethylation gradually decreases whereas H3K9 dimethylation increases at the dlk1 promoter, thus leaving a bivalent H3K4/K9 dimethylation mark on the silenced promoter in mature adipocytes (Fig. 1 and supplemental Fig. 1).
FIGURE 1.
H3K4 and H3K9 dimethylation at adipogenic promoters follow a tightly regulated and complementary pattern of enrichment throughout adipogenesis. ChIP analysis shows that the promoters of adipogenic genes such as cebpa or apm1 display significant levels of both H3K4 and H3K9 dimethylation in 3T3-L1 D0 fibroblasts. Throughout differentiation, H3K4 dimethylation steadily increases (upper panel), whereas H3K9 dimethylation decreases (lower panel), paralleling the induction on gene expression (see supplemental Fig. 1). On the other hand, the promoter of dlk1, which follows an inverted pattern of expression, displays high levels of H3K4 but not H3K9 dimethylation in preadipocytes. During adipogenesis, H3K4 dimethylation decreases and H3K9 dimethylation increases, paralleling transcriptional silencing of the gene. Finally, silenced genes such as myod display only H3K9 dimethylation in the absence of H3K4 dimethylation, whereas constitutively active genes such as actb present only high levels of H3K4 dimethylation that remain stable during differentiation. *, p < 0.05; **, p < 0.005. Error bars, S.D.
Expression of Several HMTs and HDMs Increases during Adipogenesis
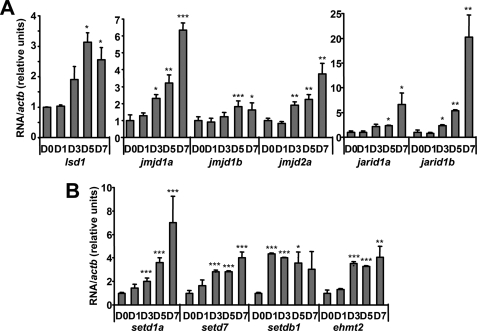
The observed regulated pattern of histone modifications in correlation with gene expression changes of key adipogenic genes suggests an important role for histone methylation on the regulation of adipogenesis. Thus, we set out to study the expression of some of the enzymes involved in the regulation of histone methylation turnover in preadipocytes and differentiating cells. Gene expression levels measured by real time RT-PCR in 3T3-L1 cells at different days during the differentiation process show a significant increase of a number of HDMs (Fig. 2A) and HMTs (Fig. 2B). A similar pattern of gene expression was observed in mouse primary preadipocytes at different days during in vitro differentiation (supplemental Fig. 2).
FIGURE 2.
Expression of histone methyltransferases and demethylases increases throughout adipogenesis. A and B, mRNA levels of different HDMs (A) and HMTs (B) during 3T3-L1 cell differentiation were assessed by real-time RT-PCR using specific TaqMan primers and probes. Values presented are the ratio of the specific RNAs corrected by housekeeping actb at different days during differentiation. *, p < 0.05; **, p < 0.005; ***, p < 0.001. Error bars, S.D.
Knockdown of lsd1 Blocks Adipogenesis
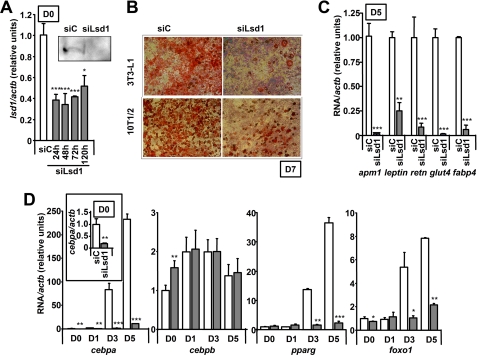
LSD1 was initially described as a H3K4 demethylase, therefore functioning as a transcriptional repressor (8), but it was later linked to gene activation depending on the chromatin context and its associated partners (19, 20), thus establishing its dual nature as a H3K4/K9 demethylase. Because we were interested in studying the impact of modifying the levels of H3K4 and H3K9 methylation during adipogenesis, we knocked down the expression of lsd1 in 3T3-L1 fibroblasts by transient transfection of a specific siRNA duplex. Two different siRNAs were used to confirm the results. Lsd1 gene expression was blocked with high efficiency until at least 120 h after transfection, as measured by real time RT-PCR (Fig. 3A). LSD1 protein expression was almost undetectable in siLsd1-transfected cells by Western blotting 72 h after transfection (Fig. 3A, inset).
FIGURE 3.
Knockdown of lsd1 decreases adipogenesis. A, transfection of a siRNA directed against lsd1 in 3T3-L1 D0 fibroblasts results in significantly decreased lsd1 mRNA and protein levels. B, lsd1 knockdown blocks adipogenesis as measured by Oil Red O lipid staining. 3T3-L1 and 10T1/2 cells were transfected and differentiated for 7 days as usual. The images shown are representative of three experiments performed in duplicate. C, lsd1 knockdown blocks adipogenesis on 3T3-L1 cells as measured by the expression of characteristic adipogenic genes. 3T3-L1 fibroblasts were transfected with siC or siLsd1 and differentiated for 5 days. RNA was extracted and analyzed by real time RT-PCR. D, lsd1 knockdown in 3T3-L1 fibroblasts results in decreased induction of adipogenesis. 3T3-L1 preadipocytes were transfected and induced to differentiate 72 h after transfection. Transcription factors cebpa, pparg, and foxo1 were readily induced several orders of magnitude in the siC-transfected cells but remained at basal levels in siLsd1-transfected cells. Expression of cebpa was already significantly decreased in undifferentiated D0 cells (inset). Values are mean ± S.D. (error bars) of three experiments performed in duplicate. *, p < 0.05; **, p < 0.005; ***, p < 0.001.
To investigate the role of LSD1 in adipogenesis, 3T3-L1 preadipocytes transfected with either siLsd1 or a control scrambled siRNA (siC) were stimulated to differentiate under normal conditions. Differentiation was allowed to proceed until D7, and mature adipocytes were identified under light microscopy by Oil Red O staining, which specifically dyes lipid droplets. Lipid accumulation, a distinctive feature of adipocyte differentiation, was effectively decreased in lsd1-knocked down cells with respect to controls (Fig. 3B). To confirm this decrease in adipocyte differentiation, expression of genes specific of mature adipocytes was also analyzed. 3T3-L1 preadipocytes were transfected and differentiated as usual. Total RNA was extracted at D5, and expression of adipogenic genes was measured by real time RT-PCR. A significant decrease in the hormones apm1, leptin, and retn, as well as the lipid transporter fabp4 and glucose transporter glut4 was observed in D5 siLsd1 cells compared with controls (Fig. 3C).
To check whether the effect of knocking down lsd1 on adipogenesis was restricted to the 3T3-L1 cell line or was on the contrary a more general phenomenon, we performed the same experiments on the pluripotent mouse cell line C3H 10T1/2. These cells are pluripotent fibroblasts that can be differentiated into adipocytes, chondrocytes, or myocytes (30). Indeed, low passage C3H 10T/2 cells could be easily differentiated into mature adipocytes under standard conditions, but adipogenesis was significantly decreased after siLsd1 transfection (Fig. 3B). Thus, our data indicate that lsd1 expression is needed for adipogenesis both in already committed 3T3-L1 preadipocytes and in pluripotent C3H 10T1/2 fibroblasts.
To identify possible LSD1 target genes, we used real time RT-PCR to analyze the expression of a number of genes involved in the regulation of adipogenesis in 3T3-L1 cells transfected with either siC or siLsd1 and stimulated to differentiate. Interestingly, expression of some of the leading adipogenic transcription factors was altered in D0 preadipocytes (Fig. 3D). However, this effect was not seen when we transfected already mature adipocytes at D5 (supplemental Fig. 3). This lack of an effect in differentiated cells is not due to deficient transfection of mature adipocytes because lsd1 mRNA levels were decreased to an extent similar to that observed in D0 siLsd1 cells (supplemental Fig. 3B). In D0 lsd1-knocked down preadipocytes, expression of cebpa was significantly decreased (Fig. 3D, inset), whereas expression of cebpb was increased. Expression of pparg and foxo1, on the other hand, was not significantly modified at 48 h after transfection, although transient significant changes could be detected at other time points (supplemental Fig. 3D). However, the studied transcription factors failed to be induced upon stimulation of adipogenesis in lsd1-knocked down cells, so that in mature cells at D3 or D5, the expression of cebpa, pparg, and foxO1 was much decreased compared with control siC-transfected cells (Fig. 3D). The expression of cebpb, on the other hand, is only limitedly induced during adipogenesis and was not affected by lsd1 knockdown (Fig. 3D).
LSD1 Regulates H3K4 and H3K9 Dimethylation at the cebpa Promoter in Preadipocytes
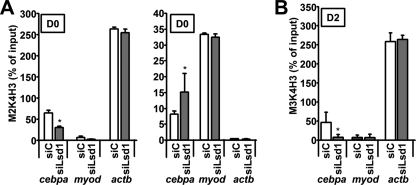
The dramatic decrease of cebpa expression observed in siLsd1-transfected cells at D0 may explain at least in part the observed effect on adipogenesis because C/EBPα is one of the master regulators of this process, and cebpa−/− mice lack white adipose tissue in many depots (31). Thus, we studied the histone methylation status of the cebpa promoter in control and siLsd1-transfected 3T3-L1 preadipocytes. Interestingly, ChIP assays demonstrate that H3K4 dimethylation was significantly decreased at the cebpa promoter 72 h after siLsd1 transfection with respect to the control. In contrast, H3K9 dimethylation in the same region was increased (Fig. 4A). As a control, H3K4 and H3K9 dimethylation at the actb and myod promoters, respectively, did not show significant changes (Fig. 4A). The increase of H3K9 dimethylation in absence of LSD1 would indicate a more repressive status of the chromatin and correlates with the observed silencing of the gene.
FIGURE 4.
LSD1 regulates H3K4 and H3K9 methylation at the cebpa promoter. A, ChIP assays show that lsd1 knockdown results in decreased H3K4 dimethylation (left panel) and increased H3K9 dimethylation (right panel) at the cebpa promoter in 3T3-L1 preadipocytes, without affecting the levels of those modifications at the promoters of control genes myod or actb. B, lsd1 knockdown decreases the induction of H3K4 trimethylation at the cebpa promoter in 3T3-L1 cells at D2. Values are the mean ± S.D. (error bars) of three experiments performed in duplicate. *, p < 0.05.
Similar to what we had previously described for other adipogenic genes (25), the cebpa promoter does not display detectable H3K4 trimethylation in D0 preadipocytes (data not shown). However, this epigenetic signal, usually associated with gene activity, increases steadily throughout differentiation at adipogenic promoters, reaching maximum levels in mature cells, when expression of the genes is maximal (25). To study whether this programmed enrichment of H3K4 trimethylation was affected by knocking down lsd1, we transfected 3T3-L1 preadipocytes and proceeded to differentiation as usual. At D2, cells were harvested and used for ChIP assays. Our data show that in siLsd1 cells, H3K4 trimethylation at the promoter of cebpa remains undetectable (Fig. 4B). In contrast, siC cells at D2 display significant levels of H3K4. These results are in accordance with the observed lack of induction of the gene (Fig. 3D). Taken together, our data suggest that in 3T3-L1 preadipocytes LSD1 is acting as an activator on the cepba promoter by maintaining low levels of dimethylated H3K9, probably opposing the function of a H3K9 methyltransferase. Maintenance of H3K9 dimethylation at this basal level is a prerequisite for the fast and robust induction of the gene that takes place after adipogenesis is induced.
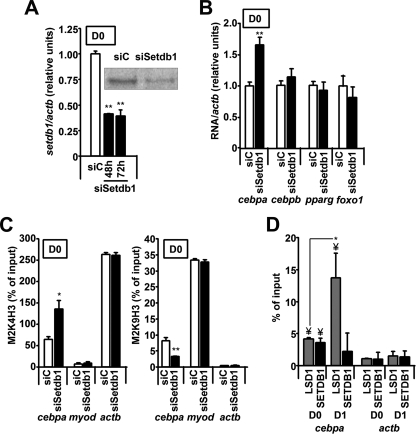
H3K9 Methyltransferase SETDB1 Regulates cebpa Expression and Histone Methylation
We were next interested in identifying the H3K9 HMT involved in the regulation of the cebpa promoter. Recently, SETDB1 was shown to play a crucial role in pluripotent mesenchymal cells as an inhibitor of adipogenesis (21). Moreover, an opposite action of LSD1 and SETDB1 has been reported in the regulation of the estrogen receptor α target promoters (20). Thus, we considered SETDB1 to be a promising candidate and knocked down its expression in 3T3-L1 preadipocytes. Setdb1 mRNA and protein levels were effectively decreased by siRNA transfection (Fig. 5A). Interestingly, in contrast with the situation in siLsd1 cells, D0 siSetdb1 cells displayed significantly increased cebpa expression compared with controls at 48 h after transfection (Fig. 5B). Moreover, when we studied by ChIP assays the promoter region of cebpa in siC and siSetdb1 preadipocytes, we observed a significant decrease in H3K9 dimethylation accompanied by an increase of H3K4 dimethylation in setdb1-knocked down cells (Fig. 5C). Thus, in the early stages of adipogenesis, SETDB1 helps to maintain high levels of H3K9 dimethylation at the cebpa promoter to repress gene activation. In absence of the factor, decreased H3K9 methylation at the cebpa promoter correlates with enhanced expression of the gene.
FIGURE 5.
SETDB1 regulates cebpa expression and histone methylation. A, transfection of a siSetdb1 siRNA into 3T3-L1 preadipocytes effectively decreases setdb1 expression as measured by real time RT-PCR and Western blotting. B, knockdown of setdb1 results in increased cebpa expression in 3T3-L1 preadipocytes. RNA was extracted from transfected cells 48 h after transfection and analyzed by real time RT-PCR. C, knockdown of setdb1 results in increased H3K4 dimethylation (left panel) and decreased H3K9 dimethylation (right panel) at the cebpa promoter in D0 preadipocytes as analyzed by ChIP. D, both SETDB1 and LSD1 can be detected at the cebpa promoter on D0 fibroblasts by ChIP. On D1, following induction of adipogenesis, recruitment of LSD1 significantly increases, whereas recruitment of SETDB1 remains unchanged. ¥, p < 0.05 with respect to actb gene promoter;*, p < 0.05; **, p < 0.005.
These data suggest a direct effect of LSD1 and SETDB1 over the cebpa promoter. Thus, we studied the recruitment of both proteins by means of ChIP assays. On D0 cells, we observe detectable levels of both LSD1 and SETDB1 bound to the cebpa promoter (Fig. 5D). After induction of adipogenesis (D1 cells), we detect a significant increase in LSD1 recruitment, whereas the levels of SETDB1 remain unchanged.
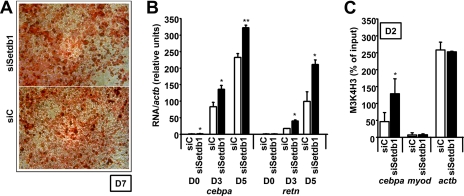
Knockdown of setdb1 Favors Adipogenesis
Because knockdown of setdb1 resulted in increased basal levels of cebpa and a more open status of its promoter in preadipocytes, we set out to study how absence of the factor affects adipogenesis. We transfected 3T3-L1 preadipoctes with siC and siSetdb1 siRNAs and proceeded to differentiation as usual. Interestingly, setdb1-knocked down cells displayed even higher levels of differentiation than control cells (Fig. 6, A and B). Oil Red O staining of D7 adipocytes shows that lipid storage is not affected in siSetdb1-transfected cells (Fig. 6A). Moreover, expression of adipogenic genes such as cebpa itself or the hormone retn were more strongly induced at D3 and D5 in siSetdb1-transfected cells than in controls as analyzed by real time RT-PCR (Fig. 6B). In addition, this stronger induction was accompanied by increased levels of H3K4 trimethylation at the cebpa promoter on D2 cells (Fig. 6C). Taken together, all of these data indicate that SETDB1 acts in preadipocytes to preserve the repressed status of cebpa by maintaining basal H3K9 dimethylation levels. In absence of the factor, H3K9 dimethylation decreases at the cebpa promoter, resulting in a more open status of the chromatin and consequently favoring adipogenesis. Histone methylation at the promoters of adipogenic genes thus emerges as an important element in the regulation of adipogenesis.
FIGURE 6.
Knockdown of setdb1 favors adipogenesis. A, setdb1 knockdown does not affect lipid accumulation of 3T3-L1 cells as measured by Oil Red O staining. The images shown are representative of three experiments performed in duplicate. B, setdb1 knockdown favors adipogenesis. Total RNA was extracted at D3 and D5 of differentiation from siC- and siSetdb1-transfected cells and analyzed by real time RT-PCR. Values presented are the ratio of the specific RNAs corrected by housekeeping actb. C, siSetdb1 transfection significantly increases the induction of H3K4 trimethylation at the cebpa promoter in D2 cells. *, p < 0.05; **, p < 0.005. Error bars, S.D.
DISCUSSION
A number of recent works have established the key role of histone modifications in cell differentiation in general and in adipogenesis in particular (25–27, 32–36). In our laboratory, we have previously shown that H3K4 dimethylation labels the promoters of adipogenic genes as potentially active in undifferentiated 3T3-L1 fibroblasts (25). Here, we extend those findings by showing that these promoters also display significant levels of H3K9 dimethylation, a well established epigenetic silencing mark. Moreover, both signals follow inverse patterns of enrichment throughout adipogenesis, with H3K4 dimethylation steadily increasing in parallel with gene activation, and H3K9 dimethylation decreasing. The presence of a similar pattern of bivalent signals has been observed in the promoters of developmentally important genes in stem cells (34, 35, 37, 38), and its alteration results in aberrant gene expression and/or spontaneous differentiation (39, 40). Our data suggest that the presence of this bivalent mark (H3K4/K9 dimethylation) in preadipocytes labels those promoters as silent but primed for transcription. H3K9 dimethylation prevents gene activation, whereas H3K4 dimethylation maintains the chromatin in a receptive state, allowing the fast and robust induction that these genes undergo when differentiation begins.
In agreement with this crucial role of histone methylation in adipogenesis, we observe that several HMTs and HDMs significantly increase throughout adipogenesis. Interestingly, a general decrease of histone deacetylases during adipogenesis has been described earlier (36), and inhibition of histone deacetylase activity has been long known to favor adipogenesis (36, 41). Thus, a striking change in the pattern of the most abundant histone-modifying proteins takes place during adipogenesis, suggesting that histone methylation plays an important role in both adipocyte differentiation and the regulation of homeostasis in mature cells. Indeed, as the cells differentiate and acquire their definitive gene expression patterns, a considerable number of genes must either be induced or silenced. It is thus reasonable to speculate that the expression and activity of the enzymes involved in histone methylation turnover must be tightly controlled during differentiation, as each of them actively participates in setting up the expression levels of a broad number of genes. Accordingly, alteration of the expression of several HMTs and HDMs has been recently shown to exert a powerful effect over adipogenesis (26, 27, 32, 33).
In this regard, here we demonstrate for the first time an association between the H3K4/K9 histone demethylase LSD1 and adipogenesis. By knocking down lsd1 gene expression in 3T3-L1 preadipocytes and pluripotent fibroblasts C3H 10T1/2, a more undifferentiated model of adipogenesis, we observe a significant decrease in lipid accumulation and adipogenic marker gene expression. This effect over adipocyte differentiation seems to be exerted at least in part by the regulatory action of LSD1 over cebpa gene expression. Transcription factor C/EBPα is one of the master regulators of adipogenesis, and its expression has been shown to be necessary for the development of mature adipocytes both in vivo and in vitro (31, 42). It is important to remark that this effect of LSD1 over cebpa expression is not observed in mature adipocytes, once the gene is heavily expressed. These data indicate that LSD1 is required to exert its function at a precise time window, corresponding to the early stages of adipogenesis, when the definitive expression levels of key adipogenic genes are being established. It should be noted that we transfected cells 48–72 h prior to adipogenesis induction. Thus, it is probable that the knockdown effect is restricted to the first 2–3 days of the differentiation process. In fact, in transfected cells differentiated up to D5/D7, lsd1 levels in siLsd1-transfected cells are comparable with those in control cells (data not shown). Again, these data suggest that the presence of LSD1 is essential for the first stages of adipogenesis but more dispensable for later steps.
Intriguingly, the expression of other adipogenic transcription factors such as pparg or cebpb is unchanged or actually increased in the siLsd1-transfected preadipocytes. PPARγ is to date the only factor considered to be both necessary and sufficient to direct adipogenesis (23, 42). However, increased pparg expression accompanied by decreased cebpa expression and adipogenesis has been observed in MLL3−/− mice (26), which display decreased adipose tissue even when subjected to a high fat diet. The H3K4 methyltransferase MLL3 has been shown to play a key role in PPARγ-dependent adipogenesis (26), by being recruited to PPARγ target promoters and increasing H3K4 methylation, thus favoring gene expression. In a separate study, knock out of Pax transactivation domain-interacting protein, a component of the MLL3 complex, also results in impaired adipogenesis by blocking the H3K4 trimethylation enrichment that takes place in the promoter regions of cebpa and pparg itself throughout adipogenesis (32).
We observe that siLsd1-transfected cells display increased levels of H3K9 dimethylation at the cebpa promoter, correlating with decreased expression of the gene already in D0 undifferentiated 3T3-L1 fibroblasts, before adipogenesis is induced, thus turning the cells less responsive. Cebpa−/− mouse embryonic fibroblasts are unable to undergo adipogenesis under normal conditions. However, reintroduction of cebpa activity in these cells rescues pparg expression and adipogenesis, whereas replenishment of pparg in the same cells also rescues adipogenesis (43). These and other data indicate that the main role of C/EBPα in the differentiation process is to maintain pparg expression (42, 43). In agreement with these data, when lsd1-knocked down preadipocytes, with severely decreased cebpa expression, are stimulated to differentiate with the adipogenic mixture, pparg induction is blunted, although expression of the factor was not affected in D0 fibroblasts prior to addition of the adipogenic mix (Fig. 3D). Similarly, cebpa expression is not induced, in contrast with the situation in control cells, where by D3 of the differentiation process, cebpa expression has been induced by almost 100-fold. Moreover, this defect in gene induction in siLsd1 cells correlates with a defect in enrichment in H3K4 trimethylation at the cebpa promoter, similar to the situation described for mouse embryonic fibroblasts lacking Pax transactivation domain-interacting protein (43). Interestingly, expression of cebpb, a gene that is already highly expressed in preadipocytes and does not need to be induced as strongly as cebpa or pparg during adipogenesis, is not affected in either our model or in the absence of Pax transactivation domain-interacting protein (44). Thus, our data suggest that LSD1 is required in the early stages of adipogenesis to allow H3K9 demethylation in adipogenic promoters and thus favor H3K4 trimethylation enrichment and gene expression. Although initially identified as a repressor (8, 13, 16), LSD1 was shown to participate in the transactivating activity of nuclear receptors such as those for androgens or estrogens (19, 20). A recent report has shown that α-herpesvirus recruits the host cell LSD1 to its promoter to erase repressive H3K9 dimethylation, allowing transcription of viral immediate early genes (18). In the particular case of the estrogen receptor α, LSD1 was described to facilitate gene activation by opposing the silencing function of the H3K9 HMT SETDB1 (20). Interestingly, SETDB1 was also shown to repress PPARγ-dependent gene transcription in mesenchymal stem cells by increasing H3K9 methylation on PPARγ target promoters and thus blocking adipogenesis in favor of osteogenesis (21).
Here, we show that LSD1 and SETDB1 exhibit opposite effects in adipogenesis. SETDB1 knockdown in preadipocytes results in significantly increased cebpa gene expression associated with decreased H3K9 dimethylation and increased H3K4 dimethylation at the gene promoter. Moreover, in accordance with results published recently by another group (33), adipogenesis is enhanced in 3T3-L1 cells by the absence of SETDB1.
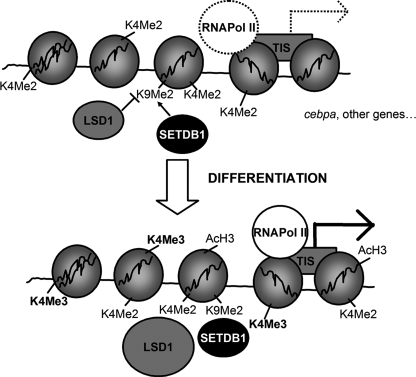
We propose the following model (Fig. 7): in fibroblasts that are committed to an adipogenic lineage such as preadipocytes 3T3-L1, the promoter regions of key adipogenic genes display detectable levels of both H3K4 dimethylation and H3K9 dimethylation. This bivalent signal maintains the chromatin in a silent but receptive state. In the cebpa promoter, H3K9 dimethylation is maintained by the combined action of the H3K9 methyltransferase SETDB1 and demethylase LSD1. When adipogenesis is induced, enhanced recruitment of LSD1 to the promoter results in decreased H3K9 methylation. This in turn allows increased H3K4 dimethylation and trimethylation and subsequent activation of the gene. Alteration of the basal levels of any of these signals in preadipocytes deeply disturbs adipogenesis, thus highlighting the important role that epigenetic mechanisms play in cell differentiation.
FIGURE 7.
Model showing the proposed interplay between LSD1 and SETDB1 on the regulation of the histone methylation status of the cebpa promoter in 3T3-L1 preadipocytes. The cebpa promoter displays significant levels of both the activation signal H3K4 dimethylation and the silencing mark H3K9 dimethylation in 3T3-L1 preadipocytes. H3K9 dimethylation levels are maintained at the basal state by the interplay of HDM LSD1 and HMT SETDB1. When adipogenesis is induced, decreased H3K9 demethylation driven by enhanced recruitment of LSD1 favors increased H3K4 dimethylation and subsequent H3K4 trimethylation at the time that transcription of the gene is robustly induced.
Supplementary Material
Acknowledgment
We thank Dr. Rosa Gasa for a critical reading of the manuscript.
This work was supported by Spanish Ministry of Science and Innovation (MICINN) Grants BFU2009-09988/BMC (to M. P.), SAF2007-63353 (to P. K.), and SAF2006-07382 (to R. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods, Figs. 1–3, and Tables 1–3.
- HDM
- histone demethylase
- C/EBP
- CCAAT/enhancer-binding protein
- HMT
- histone methyltransferase
- LSD
- lysine-specific histone demethylase
- PPAR
- peroxisome proliferator-associated receptor
- RT
- reverse transcription.
REFERENCES
- 1.Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 2.Kurdistani S. K., Tavazoie S., Grunstein M. (2004) Cell 117, 721–733 [DOI] [PubMed] [Google Scholar]
- 3.Pokholok D. K., Harbison C. T., Levine S., Cole M., Hannett N. M., Lee T. I., Bell G. W., Walker K., Rolfe P. A., Herbolsheimer E., Zeitlinger J., Lewitter F., Gifford D. K., Young R. A. (2005) Cell 122, 517–527 [DOI] [PubMed] [Google Scholar]
- 4.Bernstein B. E., Humphrey E. L., Erlich R. L., Schneider R., Bouman P., Liu J. S., Kouzarides T., Schreiber S. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8695–8700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santos-Rosa H., Schneider R., Bannister A. J., Sherriff J., Bernstein B. E., Emre N. C., Schreiber S. L., Mellor J., Kouzarides T. (2002) Nature 419, 407–411 [DOI] [PubMed] [Google Scholar]
- 6.Lachner M., O'Carroll D., Rea S., Mechtler K., Jenuwein T. (2001) Nature 410, 116–120 [DOI] [PubMed] [Google Scholar]
- 7.Kubicek S., Jenuwein T. (2004) Cell 119, 903–906 [DOI] [PubMed] [Google Scholar]
- 8.Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., Casero R. A., Shi Y. (2004) Cell 119, 941–953 [DOI] [PubMed] [Google Scholar]
- 9.Klose R. J., Yamane K., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. (2006) Nature 442, 312–316 [DOI] [PubMed] [Google Scholar]
- 10.Tsukada Y., Fang J., Erdjument-Bromage H., Warren M. E., Borchers C. H., Tempst P., Zhang Y. (2006) Nature 439, 811–816 [DOI] [PubMed] [Google Scholar]
- 11.Whetstine J. R., Nottke A., Lan F., Huarte M., Smolikov S., Chen Z., Spooner E., Li E., Zhang G., Colaiacovo M., Shi Y. (2006) Cell 125, 467–481 [DOI] [PubMed] [Google Scholar]
- 12.Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. (2006) Cell 125, 483–495 [DOI] [PubMed] [Google Scholar]
- 13.Shi Y. J., Matson C., Lan F., Iwase S., Baba T., Shi Y. (2005) Mol. Cell 19, 857–864 [DOI] [PubMed] [Google Scholar]
- 14.Wang J., Scully K., Zhu X., Cai L., Zhang J., Prefontaine G. G., Krones A., Ohgi K. A., Zhu P., Garcia-Bassets I., Liu F., Taylor H., Lozach J., Jayes F. L., Korach K. S., Glass C. K., Fu X. D., Rosenfeld M. G. (2007) Nature 446, 882–887 [DOI] [PubMed] [Google Scholar]
- 15.Lee M. G., Wynder C., Cooch N., Shiekhattar R. (2005) Nature 437, 432–435 [DOI] [PubMed] [Google Scholar]
- 16.Forneris F., Binda C., Vanoni M. A., Battaglioli E., Mattevi A. (2005) J. Biol. Chem. 280, 41360–41365 [DOI] [PubMed] [Google Scholar]
- 17.Forneris F., Binda C., Dall'Aglio A., Fraaije M. W., Battaglioli E., Mattevi A. (2006) J. Biol. Chem. 281, 35289–35295 [DOI] [PubMed] [Google Scholar]
- 18.Liang Y., Vogel J. L., Narayanan A., Peng H., Kristie T. M. (2009) Nat. Med. 15, 1312–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metzger E., Wissmann M., Yin N., Müller J. M., Schneider R., Peters A. H., Günther T., Buettner R., Schüle R. (2005) Nature 437, 436–439 [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Bassets I., Kwon Y. S., Telese F., Prefontaine G. G., Hutt K. R., Cheng C. S., Ju B. G., Ohgi K. A., Wang J., Escoubet-Lozach L., Rose D. W., Glass C. K., Fu X. D., Rosenfeld M. G. (2007) Cell 128, 505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takada I., Mihara M., Suzawa M., Ohtake F., Kobayashi S., Igarashi M., Youn M. Y., Takeyama K., Nakamura T., Mezaki Y., Takezawa S., Yogiashi Y., Kitagawa H., Yamada G., Takada S., Minami Y., Shibuya H., Matsumoto K., Kato S. (2007) Nat. Cell Biol. 9, 1273–1285 [DOI] [PubMed] [Google Scholar]
- 22.Takada I., Suzawa M., Matsumoto K., Kato S. (2007) Ann. N.Y. Acad. Sci. 1116, 182–195 [DOI] [PubMed] [Google Scholar]
- 23.Rosen E. D., MacDougald O. A. (2006) Nat. Rev. Mol. Cell Biol. 7, 885–896 [DOI] [PubMed] [Google Scholar]
- 24.Musri M. M., Gomis R., Párrizas M. (2010) Organogenesis 6, 14–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Musri M. M., Corominola H., Casamitjana R., Gomis R., Párrizas M. (2006) J. Biol. Chem. 281, 17180–17188 [DOI] [PubMed] [Google Scholar]
- 26.Lee J., Saha P. K., Yang Q. H., Lee S., Park J. Y., Suh Y., Lee S. K., Chan L., Roeder R. G., Lee J. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19229–19234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tateishi K., Okada Y., Kallin E. M., Zhang Y. (2009) Nature 458, 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schreiber E., Matthias P., Müller M. M., Schaffner W. (1989) Nucleic Acids Res. 17, 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Musri M. M., Gomis R., Párrizas M. (2008) Methods Mol. Biol. 456, 231–247 [DOI] [PubMed] [Google Scholar]
- 30.Taylor S. M., Jones P. A. (1979) Cell 17, 771–779 [DOI] [PubMed] [Google Scholar]
- 31.Linhart H. G., Ishimura-Oka K., DeMayo F., Kibe T., Repka D., Poindexter B., Bick R. J., Darlington G. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12532–12537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho Y. W., Hong S., Jin Q., Wang L., Lee J. E., Gavrilova O., Ge K. (2009) Cell Metab. 10, 27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wakabayashi K., Okamura M., Tsutsumi S., Nishikawa N. S., Tanaka T., Sakakibara I., Kitakami J., Ihara S., Hashimoto Y., Hamakubo T., Kodama T., Aburatani H., Sakai J. (2009) Mol. Cell. Biol. 29, 3544–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyer L. A., Plath K., Zeitlinger J., Brambrink T., Medeiros L. A., Lee T. I., Levine S. S., Wernig M., Tajonar A., Ray M. K., Bell G. W., Otte A. P., Vidal M., Gifford D. K., Young R. A., Jaenisch R. (2006) Nature 441, 349–353 [DOI] [PubMed] [Google Scholar]
- 35.Bracken A. P., Dietrich N., Pasini D., Hansen K. H., Helin K. (2006) Genes Dev. 20, 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoo E. J., Chung J. J., Choe S. S., Kim K. H., Kim J. B. (2006) J. Biol. Chem. 281, 6608–6615 [DOI] [PubMed] [Google Scholar]
- 37.Atkinson S. P., Koch C. M., Clelland G. K., Willcox S., Fowler J. C., Stewart R., Lako M., Dunham I., Armstrong L. (2008) Stem Cells 26, 1174–1185 [DOI] [PubMed] [Google Scholar]
- 38.Bilodeau S., Kagey M. H., Frampton G. M., Rahl P. B., Young R. A. (2009) Genes Dev. 23, 2484–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Azuara V., Perry P., Sauer S., Spivakov M., Jørgensen H. F., John R. M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Fisher A. G. (2006) Nat. Cell Biol. 8, 532–538 [DOI] [PubMed] [Google Scholar]
- 40.Kim D., Patel S. R., Xiao H., Dressler G. R. (2009) Stem Cells 27, 1516–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fajas L., Egler V., Reiter R., Hansen J., Kristiansen K., Debril M. B., Miard S., Auwerx J. (2002) Dev. Cell 3, 903–910 [DOI] [PubMed] [Google Scholar]
- 42.Rosen E. D., Hsu C. H., Wang X., Sakai S., Freeman M. W., Gonzalez F. J., Spiegelman B. M. (2002) Genes Dev. 16, 22–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu Z., Rosen E. D., Brun R., Hauser S., Adelmant G., Troy A. E., McKeon C., Darlington G. J., Spiegelman B. M. (1999) Mol. Cell 3, 151–158 [DOI] [PubMed] [Google Scholar]
- 44.Cho Y. W., Hong T., Hong S., Guo H., Yu H., Kim D., Guszczynski T., Dressler G. R., Copeland T. D., Kalkum M., Ge K. (2007) J. Biol. Chem. 282, 20395–20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.