Abstract
The purpose of the present work was to determine the identity of the enzymes that synthesize N-acetylaspartylglutamate (NAAG), the most abundant dipeptide present in vertebrate central nervous system (CNS), and β-citrylglutamate, a structural analogue of NAAG present in testis and immature brain. Previous evidence suggests that NAAG is not synthesized on ribosomes but presumably is synthesized by a ligase. As attempts to detect this ligase in brain extracts failed, we searched the mammalian genomes for putative enzymes that could catalyze this type of reaction. Mammalian genomes were found to encode two putative ligases homologous to Escherichia coli RIMK, which ligates glutamates to the C terminus of ribosomal protein S6. One of them, named RIMKLA, is almost exclusively expressed in the CNS, whereas RIMKLB, which shares 65% sequence identity with RIMKLA, is expressed in CNS and testis. Both proteins were expressed in bacteria or HEK293T cells and purified. RIMKLA catalyzed the ATP-dependent synthesis of N-acetylaspartylglutamate from N-acetylaspartate and l-glutamate. RIMKLB catalyzed this reaction as well as the synthesis of β-citrylglutamate. The nature of the reaction products was confirmed by mass spectrometry and NMR. RIMKLA was shown to produce stoichiometric amounts of NAAG and ADP, in agreement with its belonging to the ATP-grasp family of ligases. The molecular identification of these two enzymes will facilitate progress in the understanding of the function of NAAG and β-citrylglutamate.
Keywords: Brain Metabolism, Development, Enzymes, Neuropeptide, Spermatogenesis, ATP-grasp, Ligase
Introduction
N-Acetylaspartylglutamate (NAAG),3 the most abundant dipeptide in the CNS, is present in brain and in the spinal cord, most particularly in neurons of the anterior horn (1). NAAG can be released from neurons upon calcium depolarization (reviewed in Ref. 1) and is a substrate for two glial peptidases, glutamate carboxypeptidase-II (2) and, to a lower extent, glutamate carboxypeptidase-III (3), which are anchored to the plasma membrane with their catalytic site oriented toward the outside of the cell. NAAG long has been thought to be a neurotransmitter able to bind to the metabotropic glutamate receptors mGluR3 (4). Two recent reports suggest, however, that the effects of NAAG as a neurotransmitter are due to a 0.5% glutamate contamination present in commercial NAAG (5, 6). The function of NAAG is, therefore, presently unknown.
β-Citrylglutamate (BCG), which is structurally close to NAAG, is less well characterized. It was first identified in newborn rat brain, where its concentration reaches 0.5–1 μmol/g at birth and then decreases with age (7). BCG also was detected in kidneys and heart and to a much lower extent in intestine, spinal cord, and lung of newborn rats. The content of BCG in all organs decreased with age to the noticeable exception of testes, where its concentration increases during sexual maturation and remains constant in adulthood (8, 9). BCG is present not only in the testes of mammals but also in those of amphibians and fishes. There is evidence in germinal cells for a role in spermatogenesis (9). BCG recently has been proposed to be an iron chelator (10).
Little is known regarding the synthesis of NAAG and BCG. NAAG is synthesized in cells even in the presence of protein synthesis inhibitors, from N-acetylaspartate (NAA), suggesting that it is formed by a ligase and not on ribosomes (11, 12). The finding that no NAAG is synthesized in a patient with a defect in aspartate N-acetyltransferase (NAT8L) indicates that NAAG is indeed synthesized from NAA and glutamate (13, 14). The synthesis of β-citrylglutamate by tissues has not been studied. Progression in our understanding of the function of NAAG and BCG would be greatly facilitated by molecular identification of the enzyme(s) that make(s) these two compounds. Our recent success in the identification of aspartate and cysteinyl-S-conjugate N-acetyltransferases by a database search approach led us to attempt the identification of these (this) enzyme(s) with a similar strategy (14, 15).
MATERIALS AND METHODS
Cloning and Preparation of Expression Vectors
The DNA sequences of RIMKLA and RIMKLB (GenBankTM accession nos. NM_177572 and NM_027664) were PCR-amplified from mouse brain cDNA using 5′ primers (RIMKLA, ttg agg tac cat atg tgc gcg cag gtc tgg; RIMKLB, tt cgg tac cat atg tgt agc tca gtg act gg) containing KpnI and NdeI restriction sites (in boldface type) and 3′ primers (RIMKLA, gcg gat cct taa tgc tgt aac cag gct tgg g; RIMKLB, gcg gat cct cac tcc acc agg agt ttg att tc) containing a BamHI site and inserted in the pEF6Myc-His eukaryotic expression vector at the KpnI-BamHI sites. To create fusion proteins with the polyhistidine tag at the C terminus, the stop codons were removed by mutagenesis. The resulting pEF6Myc-His constructs were used to transfect HEK293T cells (results not shown). To express the His-tagged RIMKLA/B proteins in bacteria, the DNA fragment corresponding to the RIMKLA/B open reading frames in fusion with the C terminus polyhistidine tag were excised from the pEF6MycHis constructs by restriction with NdeI and MssE (blunt) and inserted into a (5′)NdeI-(3′)blunt pET3a vector. A BL21(DE3)pLysS strain was used for the expression of His-tagged RIMKLA, and a BL21 GroE strain was used for His-tagged RIMKLB. Bacterial expression of the non-His-tagged RIMKLA and RIMKLB was performed using the pET3a expression plasmid. PCR-amplified RIMKLA and RIMKLB were inserted in the pET3a expression vector at the NdeI-BamHI sites. A BL21(DE3)pLysS strain was used for the expression of RIMKLA, and a BL21 GroE strain was used for RIMKLB. All expression vectors were checked by sequencing.
Expression in Bacteria
1 liter of LB medium was inoculated with 25 ml of preculture in the presence of 200 μg/l ampicillin (BL21(DE3)pLysS and BL21 GroE strains) and 25 μg/l chloramphenicol (BL21(DE3)pLysS strain only) and incubated at 37 °C under vigorous shaking until A600 reached 0.6. The flasks were then cooled on ice for 15 min. Isopropyl 1-thio-β-d-galactopyranoside was added to a final concentration of 0.4 mm, and the culture was pursued for 24 h at 16 °C. Bacterial extracts were prepared as described previously and centrifuged for 40 min at 20,000 × g (16).
Expression in HEK293T Cells
HEK293T cells were cultured and transfected essentially as described by Ref. 17 using the jetPEITM procedure. After 48 h at 37 °C, the cells were washed once with 5 ml of PBS. They were scraped into 0.8 ml of buffer A (25 mm Hepes, pH 7.1, 1 mm α-toluenesulfonyl fluoride, 1 μg/ml leupeptin and antipain), frozen in liquid nitrogen, thawed, and lysed by vortex-mixing.
Purification
RIMKLA was purified on DEAE-Sepharose, Q-Sepharose, and gel filtration. 50 ml of a bacterial extract supernatant (corresponding to 2 liters of culture) were diluted in 150 ml of buffer A (25 mm Hepes, pH 7.1, 1 mm TSF, 1 μg/ml leupeptin and antipain) and loaded onto a 25-ml DEAE column (GE Healthcare) in a Bio-Rad FPLC. The column was washed with 75 ml buffer A, a linear 250 ml gradient (0 to 0.5 m NaCl in buffer A) was applied, and fractions were collected. NAAG synthase activity was assayed, and active fractions were pooled, diluted with 4 volumes of buffer B (25 mm Tris, pH 8.0, 1 mm DTT, 1 μg/ml leupeptin and antipain), and loaded onto a 20-ml Q-Sepharose column. The column was washed with 50 ml of buffer B, a linear 250-ml gradient (0 to 0.5 m NaCl in buffer B) was applied, and fractions were collected. Fractions containing NAAG synthase activity were pooled, concentrated to 2 ml on a Vivaspin 15 concentration unit (Sartorius), and applied onto a S-200 gel filtration column (GE Healthcare) equilibrated with buffer C (25 mm Hepes, pH 7.1, 200 mm NaCl, 1 mm DTT, 1 μg/ml leupeptin and antipain), and fractions were collected. The purification of His-tagged RIMKLA and RIMKLB was performed as for RIMKLA, except that the Q-Sepharose purification step was replaced by a purification on a HisTrap column (5-ml, GE Healthcare) performed as described in Ref. 16.
RIMKLB was purified starting from transfected HEK293T cells (30 dishes of 60 cm2). Cells were collected and resuspended in buffer A, thawed, lysed by vortex-mixing, and centrifuged for 30 min at 20,000 × g. The supernatant contained the β-citrylglutamate synthase activity (>90%) and was used for the purification. The purification procedure was identical to the purification of RIMKLA (see above).
Western Blots
Western blots were performed as described previously (17) with serum of rabbits immunized against mouse RIMKLA or RIMKLB peptides coupled to KLH (Eurogentec) diluted 1:2000 in PBS containing 1% (w/v) BSA. After washing, membranes were probed for 1 h at room temperature with peroxidase-conjugated anti-rabbit Ig antibody (Sigma-Aldrich). Detection was performed using the ECL Western blot analysis system (GE Healthcare) according to the manufacturer's protocol. Peptide sequences used for RIMKLA antibodies production were CGLQLSQKPLTTFPD and ILPGLASPREKNEPNGC, and sequences for RIMKLB were CDPESTTEREMLTKLP and AGRLTRRMSLLS (see Fig. 1).
FIGURE 1.
Sequence alignment of mouse RIMKLA and RIMKLB with E. coli RIMK and E. coli glutathione synthase. The following sequences are shown: mouse RIMKLA (MmusA, NP_808240), mouse RIMKLB (MmusB, NP_081940), E. coli RIMK (EcRIM, NP_415373), E. coli glutathione synthetase (EcGlS, NP_417422, Protein Data Bank code 1GSH). Strictly conserved residues are indicated in boldface type. Asterisks indicate residues of the catalytic site involved in the binding of glutathione synthase substrates (see “Discussion”). The peptides used to prepare anti-RIMKLA and anti-RIMKLB antibodies are underlined.
Enzymatic Assays
RIMKLA and RIMKLB were assayed radiochemically in a mixture (final volume, 200 μl) comprising, unless otherwise stated, 25 mm Tris, pH 8.0, 5 mm DTT, 5 mm MgATP, 5 mm MgCl2, 1 mm EGTA, 50,000 cpm l-[U-14C]glutamate (GE Healthcare), l-glutamate (in a concentration ranging from 0.05 to 1 mm), 1 mg/ml BSA, and 5 mm NAA or citrate. After 30 min at 30 °C, the reaction was stopped by a 5-min incubation at 80 °C and centrifuged for 20 min at 15,000 × g, and the supernatant was diluted in 0.8 ml of 5 mm Hepes, pH 7.1. The sample was applied onto a 1 ml Dowex AG1-X8 column (Cl− form, 100–200 mesh, Acros Organics) prepared in a Pasteur capillary pipette. The latter was washed with 2 ml of 5 mm Hepes, followed by 5 ml of 150 mm NaCl in the same buffer to elute unreacted glutamate and 2 × 2 ml of 500 mm NaCl to elute NAAG or β-citrylglutamate. Radioactivity was counted in the presence of Ultima Gold (PerkinElmer Life Sciences) in a liquid scintillation counter.
NMR and MS Characterization of NAAG Formed Enzymatically by RIMKLA
50 milliunits of purified His-tagged RIMKLA was added to a 100-ml solution containing 25 mm Tris, pH 8,0, 5 mm NAA, 5 mm MgATP, 5 mm MgCl2, 10 mm DTT, 0.2 mg/ml bovine serum albumin, 5 mm glutamic acid, and 500,000 cpm l-[U-14C]glutamate. The reaction mixture was incubated overnight at 30 °C under stirring and then heated for 10 min at 80 °C. The mixture was centrifuged 30 min at 18,000 × g to remove proteins, and the supernatant was treated with 2% (w/v) activated charcoal to remove nucleotides. The charcoal was filtered, and the filtrate was loaded onto a 25-ml AG1-X8 Dowex column (Cl− form). The column was washed with 100 ml of water, a linear gradient of NaCl was applied (0 to 1 m NaCl in 300 ml), and fractions (5 ml) were collected. Fractions containing radioactivity corresponding to NAAG were pooled, concentrated to 2 ml in a lyophilizer, and loaded onto a Bio-Gel P2 column (Bio-Rad; 50 cm × 1.0 cm) equilibrated with water to separate NaCl from NAAG. Desalted fractions containing NAAG were evaporated and analyzed by NMR and MS. MS analysis was performed on a LCQ Deca XP ion-trap spectrometer equipped with an electrospray ionization source (ThermoFinnigan, San Jose, CA). The LCQ was operated in positive mode under manual control in the Tune Plus view with default parameters and active automatic gain control. MS/MS analysis was done to confirm the structure of the precursor ions using low energy collision-induced dissociation with a relative collision energy of 25%. For NMR analysis, the sample was dissolved in 500 μl of H2O/D2O (9:1) and transferred to a 5-mm NMR tube. Spectra were recorded on a Bruker Avance 400 MHz UltrashieldTM spectrometer.
NMR and MS Characterization of β-Citrylglutamic Formed Enzymatically by RIMKLB
β-Citrylglutamic acid was enzymatically prepared using His-tagged RIMKLB (50 mU), and the same reaction mixture as described above except that NAA was replaced by citrate. For the synthesis of 13C-citrate-labeled citrylglutamate, 13C-citrate (CortecNet) was used, and the final volume was reduced to 2 ml. Purification and MS analysis of β-citrylglutamate was performed as for NAAG. NMR analysis was performed on purified 13C citrate-labeled citrylglutamate. The sample was dissolved in 500 μl of H2O/D2O (9:1) and transferred to a 5-mm NMR tube for spectroscopic analyses. All spectra were acquired on a Bruker AVANCE III 800 spectrometer (Bruker, Rheinstetten, Germany) working at a proton operating frequency of 800.33 MHz, equipped with a three channel 5-mm inverse detection probe head with pulse field gradients along the Z axis. Spectra were run at 25 °C using standard Bruker pulse programs. 1H and 13C chemical shifts are referenced to 3-(trimethylsilyl)propane sulfonic acid. The 1H-13C heteronuclear multiple bond connectivity spectrum (HMBC) was modified to include a water presaturation pulse during the relaxation delay and a carbon decoupling GARP4 sequence (18) during the acquisition time. A delay of 71 ms was used for the evolution of the JH,C long range couplings.
RESULTS
Identification of Putative Mammalian NAAG Synthetases
As attempts to detect the activity of the enzyme synthesizing NAAG in mouse brain extracts failed, we decided to use a database search approach to identify this enzyme. Previous reports indicated that NAAG is synthesized from N-acetylaspartate and glutamate in the presence of ATP (12, 19, 20). This suggests that the synthesis of NAAG is catalyzed by a ligase using ATP. Many ligases catalyzing amide bond formation belong to the “ATP-grasp” clan. Searching in the Pfam database (21) for mouse or human enzymes of unknown function and belonging to this clan indicated that two of them, RIMKLA and RIMKLB, belonged to a family (the RIMK family) that comprises three enzymes ligating the α-amino group of glutamate to the carboxylic group of an acceptor. The prototypic enzyme RIMK adds a glutamate at the C terminus of ribosomal protein S6 (22), whereas two other enzymes participate in the synthesis of glutamate containing cofactors, namely coenzyme F420 and tetrahydrosarcinapterin (23). These findings suggested that RIMKLA and RIMKLB also served to ligate free glutamate to (an) acceptor(s).
RIMKLA and RIMKLB are ≈ 390 amino acid proteins that are predicted to be soluble. They share ∼85% sequence identity among each other, 30% identity with E. coli RIMK, and 25% identity with E. coli glutathione synthase (Fig. 1). Compared with the latter two sequences, RIMKLA and RIMKLB present C-terminal extensions of ∼70 residues that show a much lower degree of conservation. BioGPS expression data indicate that RIMKLA is mostly expressed in the CNS, most particularly in the spinal cord and in dorsal root ganglions, but also in the retina (GeneAtlas MOE430; probe 1555378). RIMKLB is expressed in testis, placenta, and various regions of the CNS (probe 1435532). These findings suggested that RIMKLA and RIMKLB could be involved in the synthesis of NAAG and β-citrylglutamate, respectively.
Characterization of RIMKLA
To check the function of RIMKLA, we produced it in Escherichia coli either unmodified or as a fusion protein with a poly-His tag at the C terminus. Extracts of cells expressing these recombinant proteins displayed NAAG synthase activity, as indicated by the conversion of radiolabeled glutamate to a more anionic product in the presence of NAA and ATP. No conversion was observed in the absence of either of these compounds or when a control bacterial extract was used. Western blotting using an anti-RIMKLA peptide antibody and an anti-His tag antibody indicated that the His-tagged protein was partially (>50%) proteolysed, whereas this was not the case for the nontagged protein. Both the tagged and the nontagged proteins were purified as described in “Materials and Methods,” to purities of ∼60 and 30% as estimated from Coomassie Blue stained SDS-PAGE gels (data not shown). Both preparations were active and showed similar kinetic properties (see below), despite the fact that the purified His-tagged protein was partially proteolysed (data not shown). Fig. 2A shows the elution profile of the nontagged protein from a Sephacryl S-200 column. The activity was present in the flow-through fractions and coeluted with a band with the expected size (43 kDa) that reacted with antipeptide antibodies in Western blotting. A comparison of its elution profile with that of molecular mass standards indicated that the apparent molecular mass of the holoenzyme was >250 kDa, i.e. that the enzyme comprises at least six monomers. Similar results were obtained with the His-tagged protein.
FIGURE 2.
Purification of recombinant untagged RIMKLA (A) and RIMKLB (B) by gel filtration on Sepharose S-200. Mouse RIMKLA was produced in E. coli and purified by chromatography on DEAE-Sepharose and Q-Sepharose. The most active fractions of the latter column were applied on a Sepharose S-200 column. NAAG synthase activity (●) was measured in the fractions. RIMKLA was detected by Western blotting using anti-RIMKLA peptide antibodies. Mouse RIMKLB was produced in HEK cells and purified as RIMKLA before being loaded on the gel filtration column. The β-citrylglutamate synthase activity (●) of this enzyme was measured in fractions. mU, milliunits absorbance at 280 nm.
To confirm the identity of the product made by RIMKLA from NAA and glutamate in the presence of ATP, His-tagged RIMKLA was allowed to synthesize ∼1 μmol of product, which was purified by anion exchange chromatography and gel filtration. Mass spectrometry analysis of this product in the positive mode indicated the presence of a major peak of m/z 305 corresponding to NAAG. Fragmentation revealed the appearance of a major fragment with m/z 148, corresponding to protonated glutamic acid, indicating the loss of the NAA moiety. This information indicated that the synthesized molecule contained NAA and glutamate but did not disclose whether the amide bond involved the α- or β-carboxylic group of NAA. To determine the structure of the synthesized compound, its 13C NMR spectrum was recorded and compared with those of commercial α- and β-NAAG. The spectrum of the enzymatically synthesized NAAG matched that of α-NAAG and was distinctly different from that of β-NAAG (supplemental Table S1). These data led to the conclusion that RIMKLA indeed synthesizes α-NAAG, the physiological form of NAAG that is present in vertebrate CNS.
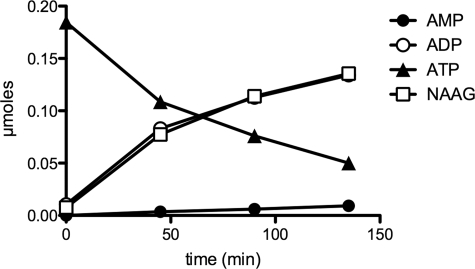
As enzymes of the ATP-grasp family catalyze the synthesis of covalent bonds with concomitant hydrolysis of ATP to ADP and inorganic phosphate, we checked the identity of the nucleotide released when RIMKLA synthesizes NAAG. The enzyme was found to convert ATP to ADP in stoichiometric amounts with respect to the formation of NAAG. No AMP was formed (Fig. 3).
FIGURE 3.
Stoichiometry of the formation of NAAG and ADP by RIMKLA. Purified RIMKLA (10.5 μg) was incubated at 30 °C in a final volume of 1 ml of 25 mm Tris, pH 8.0, 5 mm DTT, 1 mm MgATP, 5 mm MgCl2, 1 mm EGTA, and 5 mm glutamate as well as 250,000 cpm of l-[U-14C]glutamate, 1 mg/ml BSA, and 5 mm NAA. At the indicated times, an aliquot (200 μl) was taken; 100 μl of the reaction mixture were loaded onto a 1-ml Dowex AG1-X8 (Cl− form) to assay radiochemically glutamate consumption and NAAG formation (see “Material and Methods”), and 100 μl were used to measure nucleotides by high pressure liquid chromatography as described in Ref. 34.
The partially purified untagged enzyme was used to study the kinetic properties of RIMKLA. As expected, the activity of this enzyme was dependent on the simultaneous presence of NAA, glutamate, and ATP. No activity was observed when NAA was replaced by aspartate. As shown in Fig. 4, the pH curve showed an optimum at ∼8. The enzyme was markedly stimulated by dithiothreitol, which increased the activity by ∼5-fold (data not shown). Dithiothreitol was therefore added to all enzymatic assays. The Km for NAA, glutamate and MgATP were 1.48 mm, 0.88 mm, and 0.065 mm, respectively (Table 1). The Vmax amounted to 1.09 μmol/min/mg protein, corresponding to a kcat of ≈ 2.6/s assuming that the purity of the enzyme was ≈ 30%. The enzyme also catalyzed the synthesis of β-citrylglutamate, with an activity that was ∼75-fold lower than its NAAG synthase activity (Table 1). Similar Km values and pH activity dependence were observed for the His-tagged (partially proteolysed) enzyme (data not shown).
FIGURE 4.
pH curve of the NAAG and citrylglutamate synthase activities of RIMKLA (A) and RIMKLB (B). Purified RIMKLA and RIMKLB activities were measured as described under “Materials and Methods” using NAA (open symbols) or citrate (closed symbols) as substrate at the indicated pH values. The buffers used were MES (○), Hepes (□), and Tris (▵).
TABLE 1.
Kinetic parameters of RIMKLA and RIMKLB
Values are means of three measurements using two and three independent preparations of RIMKLA and RIMKLB, respectively. Activities were measured as described under “Materials and Methods” at pH 8.0. The Vmax calculation for RIMKLA takes into consideration the 30% purity of the preparation. In the case of RIMKLB, the exact purity of the enzyme could not be estimated precisely enough, and it was therefore not integrated in the calculation of the Vmax.
| RIMKLA |
RIMKLB |
|||
|---|---|---|---|---|
| Km | Vmax | Km | Vmax | |
| mm | μmol/min/mg | mm | nmol/min/mg | |
| N-Acetyl aspartate | 1.48 | 1.088 | 4.59 | 2.35 |
| Citrate | 0.87 | 0.014 | 1.24 | 2.83 |
| Glutamate | 0.88 | 1.014 | 0.73 | 2.50 |
| ATP | 0.065 | 0.778 | 0.0096 | 2.30 |
Characterization of RIMKLB
Recombinant RIMKLB was much more difficult to produce in soluble form than RIMKLA. Unmodified RIMKLB was exclusively present in pellets of centrifuged (at 12,000 × g for 45 min) bacterial extracts, as indicated by Western blotting with antipeptide antibodies. The situation was somewhat better with His-tagged RIMKLB, ≈ 10% of which was recovered in the supernatant. This form of enzyme could be purified to ≈ 20% purity by chromatography on DEAE-Sepharose and His-trap columns (data not shown). As for RIMKLA, this enzyme preparation was partially proteolysed. Nontagged RIMKLB produced by transfection of HEK cells was soluble. It was partially purified by chromatography on DEAE-Sepharose, Q-Sepharose, and gel filtration on Sephacryl S-200. Western blotting indicated that it had the expected size (42 kDa) and was not proteolysed (Fig. 2B). It was, however, much less pure than the His-tagged protein prepared from bacterial extracts.
The nonproteolysed preparation of RIMKLB produced in HEK cells was used to study the kinetic properties. Remarkably, this enzyme catalyzed the synthesis of both NAAG and β-citrylglutamate with similar Vmax (Table 1), the Km being ∼3-fold lower for citrate than for NAA. Similar to RIMKLA, the activity of RIMKLB was markedly stimulated (up to 5-fold) by DTT (data not shown), which was therefore included in all assays and purification buffers. The pH curves indicated that the rates for the two reactions were similar between pH 7 and 8, but that the β-citrylglutamate synthetase was severalfold higher than the NAAG synthetase activity at more alkaline pH (Fig. 4B). The more purified, but proteolysed preparation also catalyzed the synthesis of NAAG and β-citrylglutamate at nearly equal rates. From the Vmax (1.4 and 0.7 μmol/min/mg protein), kcat values of 3 and 1.5/s were calculated for the two activities, respectively.
To ascertain its identity, the product made by RIMKLB from citrate and glutamate was purified by anion exchange chromatography and gel filtration. Its MS analysis disclosed a major cation with m/z 322, as expected for β-citrylglutamate (data not shown). NMR analysis was performed on a compound formed from uniformly labeled 13C-citrate (Fig. 5). The proton spectrum showed six major signals, five corresponding to the resonances of the glutamate moiety and a sixth one with a complex coupling pattern corresponding to the proton frequencies of the CH2 groups of citrate, which appear as an AB system split by various JH,C couplings arising from the uniform 13C labeling of this moiety (Fig. 5). The carbon spectrum presents four major signals, which were assigned based on the carbon-carbon couplings and chemical shifts. This assignment was further confirmed by the acquisition of a carbon-carbon correlation spectroscopy (COSY) NMR spectrum. The 1H/13C-HMBC (Fig. 5) allowed us to firmly establish the amide bond between the NH group of glutamate and the carboxyl group at position 4 of citrate. In fact, there are clear connectivities between the NH group and carbons at position 4 and 3 of citrate, but not to positions 1 and 2. Also, the proton at position 2 of glutamate correlates to carbons 4 and 3 of citrate. This spectrum also allowed us to unequivocally assign the carbon frequencies of the glutamate moiety. The spectroscopic data are summarized in supplemental Table S2.
FIGURE 5.
1H/13C-HMBC spectrum of β-citrylglutamate formed by RIMKLB. 13C-Citrate-labeled citrylglutamate was synthesized enzymatically with RIMKLB and purified as described under “Materials and Methods.” All spectra were acquired on a Bruker AVANCE III 800 spectrometer.
Species Distribution and Evolutionary Tree
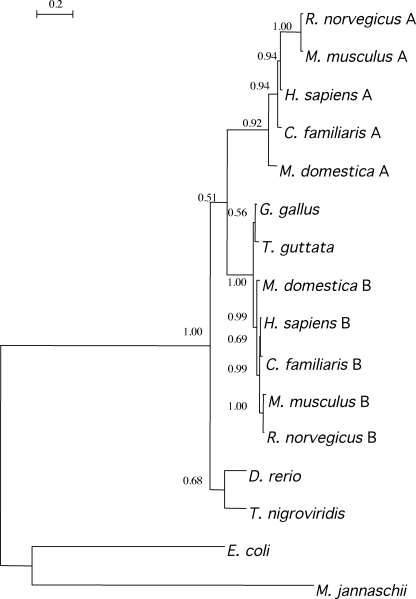
Blast searches indicated that mammalian genomes encode both RIMKLA and RIMKLB. Only one gene is found in the genomes of the fishes Tetraodon nigroviridis and Danio rerio and in the two birds Gallus gallus and Taeniopygia guttata. In birds, it encodes a product closer to RIMKLB than to RIMKLA. A phylogenetic tree is shown in Fig. 6. As is indicated by the tree, a gene duplication has occurred probably after the fish radiation but before the separation of mammals and birds. However, the precise order of events could not be determined because of the low support values for the branching points in this part of the tree. A striking feature of this tree is that the branches leading to RIMKLA are longer than those leading to RIMKLB, suggesting a faster rate of evolution.
FIGURE 6.
Phylogenetic tree of RIMKLA and RIMKLB. Protein sequences were obtained form the translated GenBankTM nucleotide database (http://www.ncbi.nlm.nih.gov/guide) and aligned with each other using ClustalX (version 2.0) (35). MrBayes (version 3.1.2) (36) was used for the inference of a phylogenetic tree. The best results were obtained with the WAG (Whelan and Goldman) model of protein evolution, with a discrete γ rate variation model with four rate categories and a proportion of invariant residues equal to 0. The program was run for 50,000 generations, with a burnin proportion of 0.4. Support values for the nodes are indicated at the respective branching points. The horizontal bar represents 20 accepted mutations per 100 residues. The E. coli and Methanococcus jannaschii sequences were used as the outgroup. Homo sapiens A (RIMKLA), NP_775913; H. sapiens (RIMKLB), NP_065785; Rattus norvegicus A, XP_002729492; R. norvegicus B, XP_342750; Mus musculus, A NP_808240; M. musculus B, NP_081940; Canis familiaris A, XP_539562, C. familiaris B, XP_543825; Monodelphis domestica A, XP_001367054; M. domestica B, XP_001368145; Gallus gallus, XP_416481; T. guttata, XP_002192984; D. rerio, NP_001004554; T. nigroviridis, CAF96644; E. coli, NP_415373; M. jannaschii, NP_247604.
DISCUSSION
Identification of the Reaction Catalyzed by RIMKLA and RIMKLB
We report in this study the identification of two enzymes that ligate the α-amino group of l-glutamate to the α-carboxylic group of N-acetylaspartate or the β-carboxylic group of citrate. The identity of these two products was established by NMR analysis. Synthesized NAAG showed a 1H NMR spectrum superimposable with that of synthetic α-NAAG but distinctly different from β-NAAG. HMBC analysis disclosed that the bond made by RIMKLB between glutamate and citrate involved the β-carboxylic group of citrate, as in the compound isolated from tissues (7). The finding that RIMKLB was able to ligate both NAA and citrate to l-glutamate at similar rates is not surprising if one considers that NAA and citrate are almost isosteric, with the β-carboxylic group of citrate being in equivalent position as the α-carboxylic group of NAA.
Agreement with Previous Results Suggesting That NAAG Is Synthesized by a Ligase
The identification of ligases permitting the synthesis of NAAG from NAA and glutamate is consistent with previous observations pointing to the existence of a ligase catalyzing this reaction from NAA and glutamate rather than to a ribosome-dependent process. NAAG synthesis from radiolabeled amino acids is insensitive to inhibition of protein synthesis as demonstrated both in spinal sensory ganglia in vivo (11) or in isolated spinal cords (12). Furthermore, labeling studies in vivo (24), in spinal cords (12) and in SH-SY5Y human neuroblastoma cells (25) all indicate that NAA is a better precursor for NAAG than free aspartate. Finally, almost four decades ago Reichelt and Kvamme (19) demonstrated the existence of NAAG synthesis in homogenates of mouse brain and showed it to be absolutely dependent on the presence of ATP. Further attempts to purify this enzyme, have, however, not met with success. We now have the proof that NAAG is indeed synthesized by a ligase reaction. This is true also for β-citrylglutamate, for which this work represents the first report on its synthesis.
Agreement between Localization of Products of Reaction and RIMKLA/B
Because of the reaction they catalyze in vitro, RIMKLA and RIMKLB are most likely responsible for the synthesis of NAAG and β-citrylglutamate in vivo. This is in keeping with the tissue distribution of these enzymes. RIMKLA is indeed mostly present in the CNS, particularly in the spinal cord, in dorsal root ganglions and in the deep nuclei of the cerebellum (Bio-GPS; Allen Atlas). In situ hybridization images of mouse brain (Allen Atlas) indicate a marked RIMKLA signal in motor neurons of the anterior horn and in the dentate nucleus of the cerebellum, which is in agreement with the localization of NAAG (26). The RIMKLA signal is restricted to a more limited number of cells than the aspartate N-acetyltranferase (NAT8L) signal, consistent with the much wider distribution of NAA than NAAG in neurons (26). It is likely, therefore, that RIMKLA is expressed only in cells that express NAT8L. Whether RIMKLA is restricted to neurons or is also present in other brain cell types is not known at present. Its high Km for NAA makes, however, that it can function at a high rate only in cells that have a sufficiently high concentration of NAA.
RIMKLB is also present in the CNS, but it seems to be expressed at the highest level in testis, consistent with the presence of elevated levels of β-citrylglutamate in this organ. As NAA and NAT8L are absent from this organ, RIMKLB is only involved in the synthesis of β-citrylglutamate in testis. Interestingly, RIMKLB appears to be up-regulated by 3–7-fold upon differentiation of spermatogonia to leptotene/zygotene spermatocytes (27), suggesting that RIMKLB could be involved in spermatogenesis.
The presence of both RIMKLB and NAT8L in Purkinje cells in the cerebellum (Allen Atlas) suggests, however, that RIMKLB may act to synthesize both NAAG and β-citrylglutamate in some cell types. RIMKLB is most likely responsible for the synthesis of both compounds in the brain of birds, as there is no RIMKLA in these species. Finally, the presence of RIMKLB in placenta (where NAT8L is absent) suggests that β-citrylglutamate may have a role in this organ. Remarkably, the RIMKLB gene has recently been shown to be 3-fold more expressed in the placenta of female mice than of male mice (28). The significance of this observation is at present unknown.
Evolution of RIMKLA and RIMKLB
Orthologues of both RIMKLA and RIMKLB are found in all mammals and in Xenopus, whereas the fishes and birds genomes comprise only one homologous gene, which is closer to RIMKLB than to RIMKLA. These findings suggest that a gene duplication event occurred during or after fish radiation and that the second copy was lost in the bird lineage. This is consistent with the finding that the genes that surround the chicken RIMKL gene are homologous to those that surround the mammalian RIMKLB gene, whereas the genetic environment of the RIMKLA gene is completely different (not shown). The ancestral form of RIMKLA and RIMKLB had presumably both activities, consistent with the presence of both NAAG and β-citrylglutamate in fishes (29). The finding that the NAAG/NAA ratio in fish CNS is much lower than in mammals or amphibians (26, 29) suggests, however, that the fish enzyme is not very good at synthesizing NAAG.
After its separation from RIMKLB, RIMKLA specialized to become a more specific NAAG synthase, consistent with the higher rate of evolution of this enzyme compared with RIMKLB. The literature describes other examples of gene duplication events that were followed by a more rapid evolution of one of the two copies, corresponding to a specialization of the function of this copy (30, 31). In the present case, this specialization corresponds to a loss of the β-citrylglutamate synthase activity. This may be advantageous to reach elevated levels of NAAG without diverting too much of an essential metabolic intermediate (citrate) or because β-citrylglutamate exerts toxic effects in some cellular environments or competes with NAAG for some essential function.
NAA and NAAG have been described in the crayfish giant axon (20), and it is therefore likely that these molecules are synthesized in invertebrates by homologues of mammalian NAT8L and RIMKLA/B. Unfortunately, no nuclear genomic sequence is available for the crayfish or any other crustacean.
RIMKLA and RIMKLB as Typical Members of the ATP-Grasp Family
Sequence comparisons indicate that RIMKLA and RIMKLB both belong to the ATP-grasp superfamily of proteins. Enzymes of this superfamily catalyze the synthesis of a bond between a carboxylic group and a nucleophile, often an amine, at the expense of ATP, which is converted to ADP and Pi (32). Accordingly, RIMKLA was found to form stoichiometric amounts of ADP and NAAG. Comparison with E. coli glutathione synthase indicates the conservation of several amino acid residues of the catalytic site. This enzyme has been crystallized with glutathione, ADP, sulfate (which presumably occupies the position of the γ-phosphoryl group of ATP) and two Mg2+ ions that bind to the phosphates of ADP and the sulfate (33). The conserved residues (see Fig. 1) include Gln198 (numbering for glutathione synthase), whose side chains bind the amino group of adenine; Asp208, whose side chain binds O3′ of the ribose moiety of ADP; Lys160, which binds the α-phosphoryl group of ADP; Lys125 and Asn235, which bind the β-phosphoryl group; the Mg2+ binding residues Asp273, Glu281, and Asn283; Arg210, which binds the nucleophilic carboxylic group of glutamyl cysteine, as well as the sulfate replacing the γ-phosphoryl group; and Arg225, which binds the carboxylic group of glycine. It is likely that the conserved residues in RIMKLA and RIMKLB play equivalent roles. These findings indicate that RIMKLA and RIMKLB presumably have a similar reaction mechanism as glutathione synthase, involving the transient formation of an acylphosphate intermediate. The structural comparison indicates also that the C-terminal extension of RIMKLA and RIMKLB are distant from the catalytic domain and have no equivalent in glutathione synthase. This agrees with our observation that their removal by proteolytic breakdown does not affect significantly the kinetic properties of the two ligases. Their function is presently unknown.
Conclusion
In conclusion, our work led to the identification of the enzymes that synthesize NAAG and β-citrylglutamate. This should help create cellular models to understand the physiological function of these molecules, and in particular, the mechanisms by which they are (at least in the case of NAAG) excreted from cells as well as their physiological role in vivo. Our work illustrates also the opportunity offered by the availability of extensive protein family and expression databases to identify enzymes that, for various reasons, are very difficult to purify.
Supplementary Material
Acknowledgments
We thank Geneviève Connerotte and Gaëtane Noël for excellent technical assistance and Helena Santos for contribution to NMR analysis.
This work was supported by grants from the Interuniversity Attraction Poles Program-Belgian Science Policy (Networks P6/05 and P6/28), by the Désordres Inflammatoires dans les Affections Neurologiques center of excellence program of the Région Wallonne, and by a grant from Asco Industries (to E. V. S.). The NMR spectrometers are part of the National NMR Network (REDE/1517/RMN/2005), supported by “Programa Operacional Ciência e Inovação 2010” and Fundação para a Ciência e a Tecnologia, Portugal.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
- NAAG
- N-acetylaspartylglutamate
- BCG
- β-citrylglutamate
- HMBC
- heteronuclear multiple bond connectivity spectrum.
REFERENCES
- 1.Neale J. H., Bzdega T., Wroblewska B. (2000) J. Neurochem. 75, 443–452 [DOI] [PubMed] [Google Scholar]
- 2.Bzdega T., Turi T., Wroblewska B., She D., Chung H. S., Kim H., Neale J. H. (1997) J. Neurochem. 69, 2270–2277 [DOI] [PubMed] [Google Scholar]
- 3.Bzdega T., Crowe S. L., Ramadan E. R., Sciarretta K. H., Olszewski R. T., Ojeifo O. A., Rafalski V. A., Wroblewska B., Neale J. H. (2004) J. Neurochem. 89, 627–635 [DOI] [PubMed] [Google Scholar]
- 4.Wroblewska B., Wroblewski J. T., Pshenichkin S., Surin A., Sullivan S. E., Neale J. H. (1997) J. Neurochem. 69, 174–181 [DOI] [PubMed] [Google Scholar]
- 5.Chopra M., Yao Y., Blake T. J., Hampson D. R., Johnson E. C. (2009) J. Pharmacol. Exp. Ther. 330, 212–219 [DOI] [PubMed] [Google Scholar]
- 6.Fricker A. C., Mok M. H., de la Flor R., Shah A. J., Woolley M., Dawson L. A., Kew J. N. (2009) Neuropharmacology 56, 1060–1067 [DOI] [PubMed] [Google Scholar]
- 7.Miyake M., Kakimoto Y., Sorimachi M. (1978) Biochim. Biophys. Acta 544, 656–666 [DOI] [PubMed] [Google Scholar]
- 8.Miyake M., Kakimoto Y. (1981) J. Neurochem. 37, 1064–1067 [DOI] [PubMed] [Google Scholar]
- 9.Miyake M., Kume S., Kakimoto Y. (1982) Biochim. Biophys. Acta 719, 495–500 [DOI] [PubMed] [Google Scholar]
- 10.Hamada-Kanazawa M., Kouda M., Odani A., Matsuyama K., Kanazawa K., Hasegawa T., Narahara M., Miyake M. (2010) Biol. Pharm. Bull. 33, 729–737 [DOI] [PubMed] [Google Scholar]
- 11.Cangro C. B., Namboodiri M. A., Sklar L. A., Corigliano-Murphy A., Neale J. H. (1987) J. Neurochem. 49, 1579–1588 [DOI] [PubMed] [Google Scholar]
- 12.Gehl L. M., Saab O. H., Bzdega T., Wroblewska B., Neale J. H. (2004) J. Neurochem. 90, 989–997 [DOI] [PubMed] [Google Scholar]
- 13.Burlina A. P., Schmitt B., Engelke U., Wevers R. A., Burlina A. B., Boltshauser E. (2006) Adv. Exp. Med. Biol. 576, 283–287; discussion 361–283 [DOI] [PubMed] [Google Scholar]
- 14.Wiame E., Tyteca D., Pierrot N., Collard F., Amyere M., Noel G., Desmedt J., Nassogne M. C., Vikkula M., Octave J. N., Vincent M. F., Courtoy P. J., Boltshauser E., Van Schaftingen E. (2010) Biochem. J. 425, 127–136 [DOI] [PubMed] [Google Scholar]
- 15.Veiga-da-Cunha M., Tyteca D., Stroobant V., Courtoy P. J., Opperdoes F. R., Van Schaftingen E. (2010) J. Biol. Chem. 285, 18888–18898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maliekal P., Vertommen D., Delpierre G., Van Schaftingen E. (2006) Glycobiology 16, 165–172 [DOI] [PubMed] [Google Scholar]
- 17.Rzem R., Veiga-da-Cunha M., Noël G., Goffette S., Nassogne M. C., Tabarki B., Schöller C., Marquardt T., Vikkula M., Van Schaftingen E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16849–16854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaka A. J., Barker P. B., Freeman R. (1985) J. Magn. Reson. 64, 547–552 [Google Scholar]
- 19.Reichelt K. L., Kvamme E. (1973) J. Neurochem. 21, 849–859 [DOI] [PubMed] [Google Scholar]
- 20.Lieberman E. M., Achreja M., Urazaev A. K. (2006) Adv. Exp. Med. Biol. 576, 303–315; discussion 361–363 [DOI] [PubMed] [Google Scholar]
- 21.Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., Gavin O. L., Gunasekaran P., Ceric G., Forslund K., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A. (2010) Nucleic Acids Res. 38, D211–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang W. K., Icho T., Isono S., Kitakawa M., Isono K. (1989) Mol. Gen. Genet. 217, 281–288 [DOI] [PubMed] [Google Scholar]
- 23.Li H., Xu H., Graham D. E., White R. H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9785–9790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyson R. L., Sutherland G. R. (1998) Neurosci. Lett. 251, 181–184 [DOI] [PubMed] [Google Scholar]
- 25.Arun P., Madhavarao C. N., Moffett J. R., Namboodiri M. A. (2006) J. Neurochem. 98, 2034–2042 [DOI] [PubMed] [Google Scholar]
- 26.Moffett J. R., Namboodiri A. M. (2006) Adv. Exp. Med. Biol. 576, 7–26; discussion 361–363 [DOI] [PubMed] [Google Scholar]
- 27.Kogo H., Kowa-Sugiyama H., Yamada K., Bolor H., Tsutsumi M., Ohye T., Inagaki H., Taniguchi M., Toda T., Kurahashi H. (2010) J. Hum. Genet. 55, 293–299 [DOI] [PubMed] [Google Scholar]
- 28.Mao J., Zhang X., Sieli P. T., Falduto M. T., Torres K. E., Rosenfeld C. S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 5557–5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyake M., Kakimoto Y., Sorimachi M. (1981) J Neurochem 36, 804–810 [DOI] [PubMed] [Google Scholar]
- 30.Delplanque J., Delpierre G., Opperdoes F. R., Van Schaftingen E. (2004) J. Biol. Chem. 279, 46606–46613 [DOI] [PubMed] [Google Scholar]
- 31.Kellis M., Birren B. W., Lander E. S. (2004) Nature 428, 617–624 [DOI] [PubMed] [Google Scholar]
- 32.Galperin M. Y., Koonin E. V. (1997) Protein Sci 6, 2639–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hara T., Kato H., Katsube Y., Oda J. (1996) Biochemistry 35, 11967–11974 [DOI] [PubMed] [Google Scholar]
- 34.Drozak J., Veiga-da-Cunha M., Vertommen D., Stroobant V., Van Schaftingen E. (2010) J. Biol. Chem. 285, 9346–9356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huelsenbeck J. P., Ronquist F. (2001) Bioinformatics 17, 754–755 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.