Abstract
Mutations in the C terminus of titin, situated at the M-band of the striated muscle sarcomere, cause tibial muscular dystrophy (TMD) and limb-girdle muscular dystrophy (LGMD) type 2J. Mutations in the protease calpain 3 (CAPN3), in turn, lead to LGMD2A, and secondary CAPN3 deficiency in LGMD2J suggests that the pathomechanisms of the diseases are linked. Yeast two-hybrid screens carried out to elucidate the molecular pathways of TMD/LGMD2J and LGMD2A resulted in the identification of myospryn (CMYA5, cardiomyopathy-associated 5) as a binding partner for both M-band titin and CAPN3. Additional yeast two-hybrid and coimmunoprecipitation studies confirmed both interactions. The interaction of myospryn and M-band titin was supported by localization of endogenous and transfected myospryn at the M-band level. Coexpression studies showed that myospryn is a proteolytic substrate for CAPN3 and suggested that myospryn may protect CAPN3 from autolysis. Myospryn is a muscle-specific protein of the tripartite motif superfamily, reported to function in vesicular trafficking and protein kinase A signaling and implicated in the pathogenesis of Duchenne muscular dystrophy. The novel interactions indicate a role for myospryn in the sarcomeric M-band and may be relevant for the molecular pathomechanisms of TMD/LGMD2J and LGMD2A.
Keywords: Calpain, Muscular Dystrophy, Protein Kinase A (PKA), Protein-Protein Interactions, Skeletal Muscle, Sarcomere, Titin
Introduction
Titin forms a continuous filament system in the myofibrils of striated muscle, with single molecules spanning from the sarcomeric Z-disc to the M-band (1). Titin guides myofibrillogenesis, provides myofibrils with elasticity, distributes forces across sarcomeres, and maintains the sarcomeric structure during muscle contraction (reviewed in Ref. 2). The titin regions located at the sarcomeric Z-disc, N2-line, and M-band sense the mechanical status of the sarcomere and convey information for signaling pathways regulating muscle function (3–5).
Mutations in the extreme C terminus of titin, situated in the periphery of the M-band, underlie two muscle diseases. When present on one allele, mutations lead to tibial muscular dystrophy (TMD,2 MIM #600334), a late-adult-onset distal myopathy typically restricted to the anterior muscles of the lower leg (6, 7). The same mutations, when present on both alleles, result in limb-girdle muscular dystrophy type 2J (LGMD2J, MIM #608807), in which most skeletal muscle groups are affected, leading to early disability (6–8).
The known TMD mutations are missense or truncating changes affecting the unique sequence region is7 or the Ig-like domain M10 of titin (Fig. 1A) (6, 9, 10). The Finnish founder mutation, FINmaj, changes four amino acids in M10 (EVTW → VKEK), presumably leading to domain misfolding (6). The downstream mechanisms leading to muscular dystrophy remain unknown, but in the absence of major ultrastructural defect in the sarcomere (6, 11), the pathogenesis is likely to depend on altered regulatory functions of M-band titin. Immunofluorescence microscopy of LGMD2J muscles reveals an absence of titin epitopes from the region M9-is7-M10 (6), suggesting that the mutations lead to a grossly altered conformation or proteolytic cleavage of the entire mutant C terminus. In either case, loss of protein interactions of the affected titin region is a likely consequence.
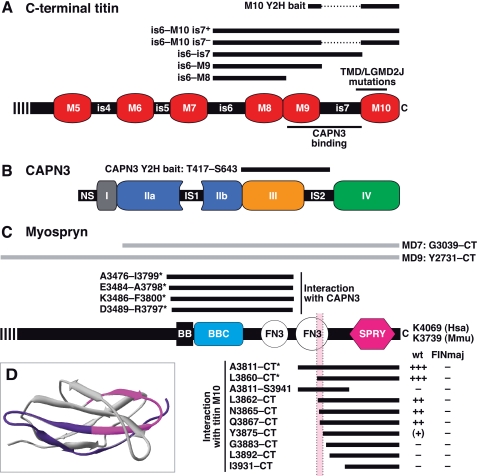
FIGURE 1.
Schematic structures of M-band titin, CAPN3, and myospryn and regions covered by the protein constructs. A, C-terminal part of M-band titin, showing the Ig-like domains M5–M10 and intervening sequence regions is4–is7. Thick bars indicate the regions covered by the Y2H and mammalian expression constructs used. Thin bars show the CAPN3-binding site and the region harboring the known TMD/LGMD2J mutations. B, CAPN3 is composed of domains I, II (split into IIa and IIb), III, and IV, common to all calpains, and CAPN3-specific sequence regions NS, IS1, and IS2. The bar indicates the bait region used in the Y2H screen. C, the C-terminal part of myospryn contains a BBox' zinc finger domain (designated BB), a BBC (BBox C-terminal coiled coil) domain, two FN3 (fibronectin type 3-like) domains, and a SPRY (SPlA and RYanodine receptor) domain. Numbers at the C terminus indicate the C-terminal amino acid residues in human (Hsa) and mouse (Mmu) myospryn. Gray bars show the MD7 and MD9 constructs used in coimmunoprecipitation and cardiomyocyte transfection studies; numbers indicate the amino acid range covered in mouse myospryn. Black bars, specified by the amino acid ranges covered in human myospryn, represent regions tested in Y2H for interaction with titin M10 or CAPN3. Asterisks indicate the interacting prey clones originally identified in the Y2H screens. For titin interactions, the strength of Y2H reporter signals indicating the interaction with wild-type (wt) and FINmaj mutant M10 bait is scored from − (no signal) to +++ (moderate signal). Pink shading indicates a region that may directly participate in titin M10 binding, corresponding to the pink highlighting in D. D, structural model of the C-terminal FN3 domain of myospryn. According to Y2H, the C-terminal half of the domain (colored pink and blue) is required for the interaction with titin M10. Amino acid residues directly participating in titin binding may be situated in the region Leu3862–Tyr3875, shown in pink (compare with Figs. 1C and 2B).
The skeletal muscle-specific protease calpain 3 (CAPN3) (Fig. 1B) binds M-band titin at is7 (12, 13), within the region affected by the TMD/LGMD2J mutations. Accordingly, a secondary deficiency of CAPN3 in LGMD2J muscle is evident in Western blotting (14) and in immunofluorescence microscopy.3
Calpains function in irreversible modulation of protein function through defined proteolytic events (15). The identified substrates of CAPN3 are sarcomeric or cytoskeletal components, and CAPN3 has been proposed to participate in sarcomere maintenance and remodeling (16, 17), to regulate the dysferlin-dependent membrane repair system (18), and to act in the titin-based signaling complexes (19).
Mutations in CAPN3 lead to another form of recessive limb-girdle muscular dystrophy, LGMD2A (MIM #253600) (20), suggesting that the pathogenetic mechanisms underlying TMD/LGMD2J and LGMD2A may be partly shared. Interactions with titin may protect CAPN3 from autolytic activation (12), and their disruption in titinopathies could result in CAPN3 dysregulation.
To elucidate the pathogenetic processes of TMD/LGMD2J and LGMD2A, we searched for proteins interacting with M-band titin and calpain 3 and identified myospryn (CMYA5, cardiomyopathy-associated 5) as a ligand for both proteins. Myospryn is a large protein (449 kDa; 4069 amino acids in human), comprising a repetitive, acidic N-terminal part and a C-terminal domain structure related to the tripartite motif proteins (Fig. 1C) (21). Myospryn is specifically expressed in striated muscle (21, 22), where it has been suggested to associate with sarcoplasmic reticulum (SR) and costameres (23, 24).
Myospryn seems to play a dual role in protein kinase A (PKA) signaling and vesicular trafficking (reviewed in Ref. 25). It binds the regulatory RIIα subunit of PKA and functions as an protein kinase A-anchoring protein regulating the spatial specificity of cAMP-PKA signaling (26). A role in vesicular trafficking or protein sorting is implied by the interaction of myospryn with dysbindin-1, a subunit of BLOC-1 (biogenesis of lysosome-related organelles complex 1) (21), and evidence exists for myospryn participating in lysosomal biogenesis and positioning (24). Recently, myospryn was implicated in the pathogenesis of Duchenne muscular dystrophy; in the dystrophin-deficient mdx mouse, disrupted interaction of myospryn with dystrophin leads to mislocalization of myospryn and RIIα and to impaired PKA signaling (27).
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid Constructs
The titin bait constructs pGBKT7-M10 WT and FINmaj were produced by cloning the corresponding cDNA sequences to the pGBKT7 vector of the Matchmaker 3 system (Clontech). The baits spanned the 132 C-terminal amino acids of the human titin is7− isoform, thus covering the M10 domain preceded by the last 34 amino acids of M9 (Fig. 1A). To generate the bait construct for the CAPN3 interaction screen, the cDNA sequence encoding Thr417–Ser643 of human CAPN3 isoform a (Fig. 1B) was cloned into the pB27 plasmid as a LexA C-terminal fusion (28).
To generate the myospryn prey constructs pGADT7-CMYA53811–CT and pGADT7-CMYA53860–CT used in further Y2H studies, coding regions of the corresponding myospryn inserts were PCR-amplified from the original pACT2 prey constructs, cloned into pCR-Blunt II-TOPO (Invitrogen), and transferred to pGADT7 (Clontech). The same procedure was used for producing the myospryn deletion constructs covering the regions Ala3811–Ser3941, Leu3862–CT (C terminus), Asn3865–CT, Gln3867–CT, Tyr3875–CT, Gly3883–CT, Leu3892–CT, and Ile3931–CT (Fig. 1C).
Mammalian Expression Constructs
M-band titin constructs pEF6-is6-M8-V5, -is6-M9-V5, -is6-is7-V5, and -is6-M10-V5 (is7+/is7−, WT/FINmaj) were generated by cloning the corresponding wild-type (WT) or FINmaj mutant titin cDNA sequences into pEF6/V5-His-TOPO (Invitrogen). These constructs encoded different fragments of is7+ and is7− titin isoforms (see Fig. 1A), fused to a C-terminal 45-residue tag containing V5 and His6 sequences. An untagged construct pEF6-is6-M10 is7+ WT was produced in a similar fashion.
To produce pAHC-is6-M10 is7+ WT, encoding the is6-M10 region N-terminally HA-tagged (HA-is6-M10), the insert was PCR-amplified, cloned into pCR-Blunt II-TOPO, and then subcloned into pAHC (modified pCI-Neo (29)). The construct pEGFP-M10 WT, encoding the wild-type human titin M10 domain with an N-terminal GFP tag (GFP-M10), has been described previously (30).
The construct pCMV-Myc-MD7, encoding the 701 C-terminal amino acid residues of mouse myospryn (corresponding to 707 C-terminal amino acids of the human protein) with an N-terminal Myc tag (Myc-MD7), has been described previously (21). The myospryn insert MD9, encoding the 1009 C-terminal amino acids of mouse myospryn (1018 amino acids in human), was PCR-amplified from pCI-Neo-MD9 (21) and cloned into pCR-Blunt II-TOPO. N-terminally Myc-tagged myospryn constructs pAMC-MD9 (encoding Myc-MD9) and pAMC-CMYA53811–CT (Myc-CMYA53811–CT) were produced by cloning the respective inserts to pAMC (modified pCI-Neo, (29)). Corresponding constructs with N-terminal GFP tags, pEGFP-MD9 (GFP-MD9) and pEGFP-CMYA53811–CT (GFP-CMYA53811–CT), were produced by cloning the inserts to pEGFP-C1 (Clontech). For an overview of myospryn constructs, see Fig. 1C.
The utilized calpain 3 constructs have been described previously. The constructs pTOM-rCAPN3 and pTOM-rCAPN3C129S encoded wild type and proteolytically inactive C129S versions of rat CAPN3, tagged N-terminally with YFP and C-terminally with CFP (31). pSRD-rCAPN3C129S encoded inactive rat CAPN3 without tags (32).
Interaction Screens
The yeast two-hybrid screen for identifying ligands of the titin M10 domain was performed at the two-hybrid core facility of Biocentrum Helsinki, University of Helsinki. The bait pGBKT7-M10 WT was screened against a human skeletal muscle prey library in pACT2 (Clontech). Yeast clones positive for interaction were selected by culturing on SD-LWHA + 2.5 mm 3-amino-1,2,4-triazole + X-gal. The prey plasmids from positive colonies were isolated and screened by AluI digestion, and inserts encoding putative interacting proteins were identified by sequencing.
The yeast two-hybrid screen for identifying CAPN3 ligands was performed at Hybrigenics S.A. (Paris, France), using a mating method and following previously described protocols for prey library construction, large scale screening, and identification of interacting fragments (28). The prey library was constructed from adult (catalogue no. AM7983 Ambion; Applied Biosystems, Austin, TX) and fetal (catalogue no. 778020, Stratagene, La Jolla, CA) human skeletal muscle poly(A) RNAs. Random-primed cDNA fragments were isolated from the two RNA pools, cloned into pB6 as C-terminal fusions of the GAL4 transcription-activating domain, and separately amplified in Escherichia coli (50–100 million independent bacterial clones). Equimolar fractions of the two cDNA libraries were pooled and used to transform the Y187 yeast. The CAPN3Thr-417–Ser-643 bait was screened against the prey library, and the growth ability of 106 million diploid clones (equivalent to 10-fold coverage of the library) was tested on appropriate medium. Prey fragments from all positive clones were PCR-amplified and identified by sequencing.
Further Yeast Two-hybrid Studies
To verify the results of the titin interaction screen, selected putative ligands of M10 were studied in pairwise Y2H experiments using the Matchmaker 3 system. The pGBKT7-M10 WT and FINmaj baits were tested against various pGADT7 prey constructs. As negative controls, appropriate empty vectors were tested against the different bait and prey constructs. The pair pGBKT7–53/pGADT7-T served as a positive control. The experiments were carried out with the mating strategy as described in the Clontech Yeast Protocols Handbook, with the bait constructs in AH109 and prey constructs in the Y187 strain. Activity of the nutritional reporter genes was assayed by culturing on different selection plates (SD-LWH, SD-LWHA, and SD-LWHA + 2.5 mm 3-amino-1,2,4-triazole) for up to 11 days. Activity of the β-galactosidase reporter was assayed with the Herskowitz laboratory X-gal overlay method. Same procedures were used for testing the myospryn deletion constructs against the pGBKT7-M10 WT and FINmaj baits.
Antibodies
The following previously described primary antibodies (ab) were used in Western blotting (WB), immunofluorescence (IF), and proximity ligation assay (PLA) studies: rabbit polyclonal ab M10-1 against a peptide epitope from the titin M10 domain (10) at 1:1000 (WB); rabbit polyclonal ab Tm8ra against the titin M8 domain (33) at 1:50 (IF); mouse monoclonal ab T51 against the titin M9 domain (33) at 1:20 (IF, PLA); mouse monoclonal ab T41 against the titin M-is4 region (33) at 1:30 (PLA); rabbit polyclonal ab 653 against sarcomeric α-actinin (34) at 1:200 (IF); and rabbit polyclonal ab Des122 against myospryn (21) at 1:1000 (WB)/1:50 (IF, PLA).
In addition, the following commercial primary antibodies were used: mouse monoclonal Myc ab 9E10 for IF at 1:100 (Roche Applied Science) and for WB at 1:1000 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA); mouse monoclonal anti-Myc ab R950-CUS (Invitrogen) at 1:5000 (WB); mouse monoclonal V5 ab SV5-P-k (Invitrogen) at 1:5000 (WB); rat monoclonal HA ab 3F10 (Roche Applied Science) at 1:100 (IF) and mouse monoclonal sarcomeric α-actinin ab EA-53 (Sigma) at 1:500–1:5000 (IF); mouse monoclonal dystrophin antibody Dy4/6D3 (Novocastra NCL-DYS1, Leica Biosystems Newcastle Ltd., Newcastle Upon Tyne, UK) at 1:20 (IF); rabbit polyclonal CAPN3 ab RP2 (Triple Point Biologics, Inc., Forest Grove, OR) at 1:5000 (WB); rabbit polyclonal GFP ab (Abcam plc, Cambridge, UK) at 1:2500 (WB); and rabbit polyclonal actin ab (Sigma) at 1:400 (WB).
For IF staining of muscle sections, secondary antibodies conjugated with Alexa Fluor dyes (Molecular Probes, Invitrogen) were used at 1:500. For staining of cultured cardiomyocytes, preabsorbed secondary antibodies conjugated with cyanine dyes (Cy2, Cy3, and Cy5) were used at 1:100 and unabsorbed Cy3-anti-mouse antibody at 1:500 (all from Jackson ImmunoResearch, West Grove, PA).
In Western blotting, HRP-conjugated secondary antibodies (Dako Denmark A/S, Glostrup, Denmark) were used at 1:5000 for ECL detection, and IRDye-labeled secondary antibodies (LI-COR Biosciences, Lincoln, NE) were used at 1:10,000 for fluorescent detection. Agarose-conjugated goat polyclonal anti-V5 and anti-Myc antibodies (Novus Biologicals, Inc., Littleton, CO) and EZview Red Anti-c-Myc Affinity Gel (Sigma) were used for coimmunoprecipitation.
Coimmunoprecipitation
Coimmunoprecipitation was performed according to a protocol modified from Lykke-Andersen (35). COS-1 cells were seeded on 6-well plates at 200,000 cells per well and cultured in DMEM containing 10% FCS, 2 mm l-glutamine, and penicillin/streptomycin. The following day, the cells were cotransfected with appropriate plasmid constructs using 1 μg of DNA and FuGENE 6 transfection reagent (Roche Applied Science). Two days after transfection, the cells were washed with PBS, scraped into PBS, and pelleted at 200 × g, 10 min. The cells were lysed in 200 μl of ice-cold freshly prepared hypotonic gentle lysis buffer (10 mm Tris-HCl, pH 7.5, 10 mm NaCl, 2 mm EDTA, 0.5% Triton X-100, 1× Complete Protease Inhibitor Mixture (Roche Applied Science)) and incubated on ice for 10 min. NaCl was added to the final concentration of 150 mm, and the lysates were incubated on ice for a further 5 min. After pelleting the insoluble material (15 min at 16,000 × g, 8 °C), a total lysate sample was removed from each supernatant.
Antibody-conjugated agarose beads (original slurry volume 10 μl) were prewashed twice with NET-2 (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.05% Triton X-100), then combined to 170 μl of cleared lysate, and rotated overnight at 8 °C. The beads were collected by centrifugation (2 min at 82 × g, 8 °C) and washed three to eight times with 500 μl of ice-cold NET-2. The immunoprecipitated proteins were eluted in 25 μl of 2× SDS sample buffer by heating 5 min at 95 °C. The eluates and total lysate samples were analyzed by Western blotting. Chemiluminescent detection was performed with either Immun-Star HRP substrate (Bio-Rad) or Pierce SuperSignal West Femto maximum sensitivity substrate (Thermo Fisher Scientific Inc., Rockford, IL).
CAPN3-Myospryn Coexpression Studies
For coexpression studies of CAPN3 and myospryn, 911 cells (human retinoblasts) were seeded on 6-well plates at 200,000 cells per well and cultured in DMEM containing 10% FCS. After 24 h, the cells were cotransfected with a total of 3 μg of plasmid constructs using FuGENE 6 transfection reagent and cultured for 24 h.
Cells were lysed for 30 min in 40 μl of lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 2 mm EGTA, 0.1% Triton X-100, 1× Complete Protease Inhibitor Mixture, 2 μm E64 (Sigma)). The lysate was centrifuged for 10 min at 14,000 × g in 4 °C. The supernatant was mixed with LDS NuPAGE buffer (Invitrogen) supplemented with 100 mm DTT and denatured 10 min at 70 °C.
Fifty micrograms of proteins were separated by SDS-PAGE and transferred onto PVDF membranes. The membranes were blocked with Odyssey blocking buffer (LI-COR Biosciences) diluted 1:1 in PBS, probed with appropriate primary and IRDye-labeled secondary antibodies, and visualized using an Odyssey Infrared Laser Imaging System (LI-COR Biosciences).
Neonatal Rat Cardiomyocyte Cultures
Primary cardiomyocyte cultures from neonatal rat hearts were isolated using the neonatal cardiomyocyte isolation system (Worthington). Cell culture and transient transfections using the Escort III reagent (Sigma) were performed as described previously (36). The cells were transfected with four different myospryn constructs (Myc-CMYA53811–CT, GFP-CMYA53811–CT, Myc-MD9, and GFP-MD9) and the titin construct HA-is6-M10, either alone or in different myospryn/titin combinations.
After transfection, the cells were cultured for 1–5 days either in maintenance medium (74.7% DMEM, 18.6% medium M199, 3.8% horse serum, penicillin/streptomycin, 4 mm l-glutamine, 100 μm phenylephrine, 10 μm AraC for 1- or 2-day cultures) or in transfection medium with antibiotics (73% DBSS-K medium, 21% medium M199, 4% horse serum, 4 mm l-glutamine, penicillin/streptomycin for 5-day cultures). The cells were fixed on culture dishes with 4% PFA/PBS for 10 min at room temperature.
Muscle Material and Immunofluorescence Stainings
Hind limb muscles (tibialis cranialis, soleus, and gastrocnemius) were dissected from wild-type C57BL/6 mice sacrificed for other experimental purposes. Muscles were prepared for IF staining as described by Locke et al. (37), with slight modifications. Briefly, the muscles were stretched to 1.5× length to increase sarcomeric resolution, pinned on cork, fixed by immersion in 4% PFA/PBS for 30 min at room temperature, and cryoprotected at 8 °C in increasing concentrations of glucose in PBS (10% for 1 h, 20% for 1 h, and 30% overnight). The muscles were embedded in Tissue-Tek O.C.T. medium (Sakura Finetek Europe B.V., Zoeterwoude, The Netherlands), snap-frozen in liquid-nitrogen-chilled isopentane, and cut into 8-μm longitudinal sections. The sections were air-dried for 20 min room temperature before immunofluorescence staining.
Human control muscle (tibialis anterior) was obtained from an amputated leg, with informed consent from the donor. The muscle pieces, held in stretched position with Alm retractors, were immersion-fixed and cryoprotected as described above for the mouse muscles and then frozen and sectioned. Optionally, sections were postfixed in 4% PFA/PBS for 10 min at room temperature prior to immunofluorescence staining.
For nonstretched samples, skeletal muscle biopsies were obtained from control and LGMD2J patients. The samples were freshly embedded in Tissue-Tek, frozen in liquid nitrogen-chilled isopentane, and cut into 8-μm longitudinal sections. Prior to immunofluorescence staining, the sections were fixed in 4% PFA/PBS for 10 min at room temperature.
All muscle sections were permeabilized with 0.2% Triton X-100/PBS for 10 min, blocked with 5% BSA/PBS for 30 min, and subjected to indirect immunofluorescence staining with appropriate antibodies diluted in 1% BSA/PBS. Cultured neonatal rat cardiomyocytes, fixed onto culture dishes, were permeabilized with 0.2% Triton X-100/PBS for 5 min, washed with PBS, and stained with antibodies diluted in 1% BSA/Gold buffer (20 mm Tris-HCl, pH 7.5, 155 mm NaCl, 2 mm EGTA, 2 mm MgCl2).
All antibody incubations were performed in a humid chamber for 1–2 h at room temperature or overnight at 4 °C, followed by washes with PBS. In addition to antibodies, phalloidin conjugated to Alexa Fluor 488 (Molecular Probes, Invitrogen) was used for visualizing the actin cytoskeleton and DAPI for nuclear staining. After staining, the samples were mounted in Gel Mount (Sigma; for tissue sections) or Lisbeth's mounting medium (30 mm Tris-HCl, pH 9.5, 70% glycerol, 5% n-propyl gallate; for cardiomyocytes).
Proximity Ligation Assays
For in situ PLA, mouse muscle sections were prepared and permeabilized as for immunofluorescence stainings. PLA experiments were then performed with the Duolink kit (Olink Bioscience, Uppsala, Sweden) according to the manufacturer's instructions. Briefly, the sections were blocked with Duolink blocking reagent, incubated with appropriate primary antibodies (Des122 alone or together with T51 or T41) for 1–2 h room temperature, probed with anti-rabbit plus and anti-mouse minus PLA probes (at 1:10) for 1 h at 37 °C, and stained with the Duolink fluorescent detection kit 563. Primary antibodies and PLA probes were diluted in Duolink antibody dilution buffer. Washes were done in low buffered TBST (10 mm Tris-HCl, 150 mm NaCl, 0.05% Tween 20, pH 7.4). In negative control experiments performed in parallel, either one or both primary antibodies were omitted. Alexa Fluor 488-conjugated phalloidin, added 1:100 to the primary antibody solution, served as a counterstain.
Confocal Microscopy
Confocal microscopy was performed using LSM 510 Meta confocal microscopes equipped with diode, argon, and HeNe lasers, and 63×/NA 1.4 and 40×/NA 1.3 objectives (Carl Zeiss MicroImaging GmbH, Göttingen, Germany). Image processing and analysis were done using the LSM 510 Meta 3.2 software (Carl Zeiss MicroImaging), Adobe Photoshop CS2 9.0.2 (Adobe Systems Inc., San Jose, CA), and ImageJ 1.41o (W. S. Rasband, ImageJ, rsb.info.nih.gov/ij).
Protein Modeling
A structural model of the C-terminal FN3 domain of myospryn was generated by homology modeling at the Swiss-Model workspace (38), using the Protein Data Bank structure 2dmk as template.
RESULTS
Myospryn Interacts with C-terminal Titin in the Yeast Two-hybrid System
To identify proteins interacting with M-band titin, we performed a yeast two-hybrid interaction screen on a skeletal muscle cDNA library using the ultimate C terminus of titin (construct pGBKT7-M10 WT) as bait. Screening of 10.56 million colonies yielded 84 interacting prey clones, coding for 21 different proteins. Six known proteins were represented by more than one in-frame prey clone as follows: myospryn (CMYA5), phosphoglucomutase 1 (PGM1), Ran-binding protein M (RanBPM), RING finger protein 1 (RING1), formin homology 2 domain containing 1 (FHOD1), and kinectin 1 (KTN1). These were regarded as the most plausible titin ligands and chosen for further yeast two-hybrid and coimmunoprecipitation studies. Conclusive supporting evidence was obtained for myospryn (see below), and PGM1 is currently under investigation. The four others were judged as probable false-positive hits.
Myospryn was encoded by two prey clones interacting with the titin M10 bait. The longer one (CMYA53811–CT) included the 259 C-terminal amino acid residues, covering the latter of the two fibronectin type 3-like (FN3) domains and the SPRY domain. The shorter clone (CMYA53860–CT) of 210 amino acids covered the C-terminal half of the FN3 domain followed by SPRY (Fig. 1C).
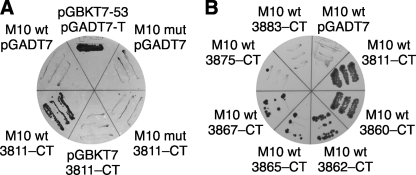
In pairwise Y2H studies, prey constructs pGADT7-CMYA53811–CT and pGADT7-CMYA53860–CT, corresponding to the originally identified myospryn clones, were tested against wild-type and FINmaj mutant versions of the pGBKT7-M10 bait construct. Both myospryn preys produced reporter signals of similar intensities with the wild-type bait, but not with the mutant (Fig. 2A), suggesting that the signal resulted from a specific interaction between myospryn and the M10 domain. In negative control experiments, testing the bait constructs against the empty prey vectors or vice versa produced no reporter signals, demonstrating that the observed signals resulted from an interaction between the titin and myospryn moieties of the constructs rather than autoactivation.
FIGURE 2.
Yeast two-hybrid studies of titin and myospryn. A, myospryn prey construct pGADT7-CMYA53811–CT (3811–CT) was tested against wild-type (wt) and FINmaj mutant (mut) titin M10 baits on an SD-LWHA plate cultured for 4 days. Yeast growth, indicating activity of HIS3 and ADE2 reporter genes, demonstrated interaction with wild type but not with FINmaj bait. In negative controls, where the same constructs were tested against empty bait (pGBKT7) and prey (pGADT7) vectors, no yeast growth was visible. Yeast carrying pGBKT7–53 and pGADT7-T served as a positive control. B, various myospryn prey constructs were tested in Y2H against the wild-type (wt) titin M10 bait. On an SD-LWHA plate cultured for 11 days, CMYA53811–CT and CMYA53860–CT displayed comparable reporter signals, whereas the shorter constructs (3862–CT, 3865–CT, and 3867–CT) showed signals of lower intensities, correlating with construct length (compare with Fig. 1C). CMYA53875–CT showed a marginally detectable signal, and CMYA53883–CT did not allow any yeast growth. None of the prey constructs allowed yeast growth when tested against the FINmaj mutant M10 bait or the empty bait vector pGBKT7 (data not shown).
Additional pairwise Y2H studies with a series of shorter myospryn prey constructs served to define the myospryn region involved in the interaction with titin M10 (Figs. 1C and 2B). The three constructs N-terminally up to seven amino acids shorter than the original clone 3860–CT all showed reporter signals similar to the original prey clones but weaker. Further N-terminal shortening (construct 3875–CT) resulted in an even weaker signal, barely distinguishable from the background. None of the other constructs produced a detectable signal. These findings indicate that both the C-terminal part (Gln3867–CT) of the latter FN3 domain and the SPRY domain of myospryn are needed for titin M10 binding. In addition, the weaker reporter signals obtained with some of the constructs suggest that some amino acid residues directly involved in the interaction are situated between Leu3862 and Tyr3875, a region that according to protein modeling forms a surface loop flanked by stretches of the β-strand (Fig. 1D).
Myospryn Interacts with Calpain 3 in the Yeast Two-hybrid System
To identify proteins interacting with CAPN3, another yeast two-hybrid screen was performed with a bait encompassing the domain III and is2 of human CAPN3. Screening of 106 million prey clones produced 76 interacting clones, 6 of which encoded myospryn fragments. These clones, differing only slightly in length, covered the BBox′, coiled coil, and the first FN3 domains of myospryn (Fig. 1C).
Coimmunoprecipitation Confirms the Interactions of Myospryn with Titin and CAPN3
To verify the interactions of titin and calpain 3 with myospryn in vitro, we performed coimmunoprecipitation (CoIP) of proteins coexpressed in COS-1 cells. Epitope-tagged bait proteins were immunoprecipitated with agarose-coupled antibodies, and related protein constructs not recognized by the antibodies served as negative control baits.
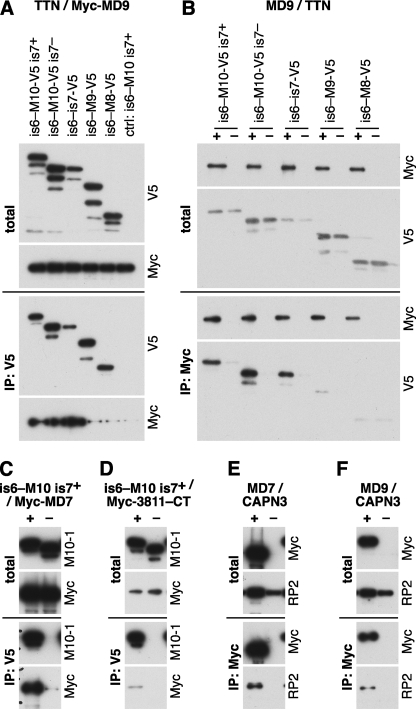
The C-terminally V5-tagged titin construct is6-M10-V5 is7+ WT efficiently coimmunoprecipitated the myospryn construct Myc-MD9 (Fig. 3A). In the reverse experiment, Myc-MD9 coimmunoprecipitated is6-M10-V5 is7+ WT and the corresponding untagged construct at a similar efficiency (data not shown), demonstrating that the interaction in CoIP depended on the titin moiety of the constructs and not on the epitope tag. Also, the other myospryn constructs Myc-MD7 and Myc-CMYA53811–CT showed binding to is6-M10-V5 is7+ WT (Fig. 3, C and D). Myc-MD7 was more efficiently precipitated than Myc-MD9, although their amounts in the lysates were comparable. The short Myc-CMYA53811–CT construct performed variably, probably due to its lower solubility.
FIGURE 3.
Coimmunoprecipitation studies of myospryn with titin and CAPN3. Titin, myospryn, and CAPN3 constructs were expressed in COS-1 cells and immunoprecipitated (IP) with V5 or Myc epitope tag antibodies. Samples of total cell lysates and immunoprecipitates were analyzed by Western blotting using antibodies against V5 and Myc tags, titin M10 (M10-1), and CAPN3 (RP2). A and B, titin constructs covering the regions is6-M10 (is7+ and is7− isoforms), is6-is7, is6-M9, and is6-M8 tested against myospryn MD9. A, V5-tagged titin constructs and the untagged is6-M10 is7+ construct (negative control) coexpressed with Myc-MD9 and immunoprecipitated with V5 antibodies. B, MD9 constructs with a Myc (+) or GFP (−) tag coexpressed with V5-tagged titin constructs and immunoprecipitated with Myc antibodies. Myc-MD9 showed binding to the three longest titin constructs in both setups. C and D, titin constructs is6-M10-V5 is7+ WT (+) or is6-M10 is7+ WT (−) coexpressed with Myc-MD7 (C) or Myc-CMYA53811–CT (D) and immunoprecipitated with V5 antibodies. Both myospryn constructs were coimmunoprecipitated with the tagged titin construct. E and F, Myospryn constructs MD7 (E) or MD9 (F) with a Myc (+) or GFP (−) tag, coexpressed with CAPN3C129S and immunoprecipitated with Myc antibodies. CAPN3C129S was coimmunoprecipitated with both myospryn constructs.
The GFP-M10 construct, including only the M10 domain of titin, did not conclusively show binding to myospryn constructs in the same CoIP conditions that allowed the interaction for is6-M10 is7+ (data not shown), suggesting that the myospryn interaction was not restricted to the M10 domain. Therefore, we tested binding of Myc-MD9 to a series of V5-tagged titin constructs of different lengths. The three longest constructs, is6-M10-V5 is7+, is6-M10-V5 is7−, and is6-is7-V5, all bound Myc-MD9 at comparable efficiencies, whereas the shorter constructs is6-M9-V5 and is6-M8-V5 only showed marginal or no binding (Fig. 3, A and B). This indicates that both M9 and is7, in addition to the M10 domain, participate in the interaction with myospryn.
To determine the effect of the FINmaj titin mutation on the interaction with myospryn in vitro, we compared the wild-type and FINmaj versions of the is6-M10-V5 (is7+ and is7−) constructs in CoIP. In contrast to our Y2H results, the mutation was not able to prevent coimmunoprecipitation of the titin and myospryn constructs (data not shown). Rather, the mutant constructs precipitated the different myospryn fragments more efficiently than the wild-type constructs and were also more efficiently coimmunoprecipitated by Myc-MD9 in the reverse experiments.
In a similar CoIP setup, myospryn was tested against the proteolytically inactive calpain 3 mutant C129S. Both the Myc-MD7 and Myc-MD9 constructs were able to coimmunoprecipitate CAPN3C129S from cell lysates, supporting the interaction of myospryn with calpain 3 (Fig. 3, E and F).
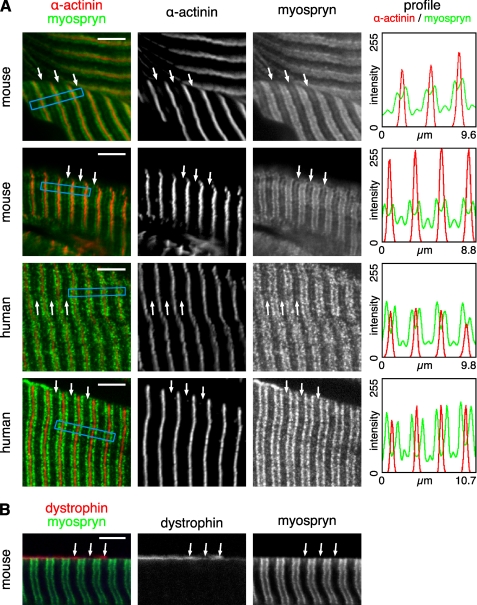
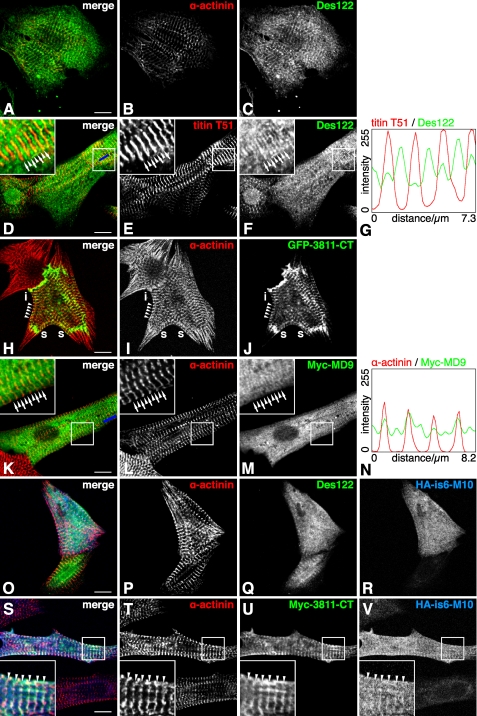
Myospryn Is Partly Localized at the M-band Level in Skeletal Muscle Sections
Confocal microscopy of longitudinal sections from stretched and immersion-fixed mouse and human muscles showed myospryn predominantly localized as broad doublet bands flanking the Z-discs. In addition, a fainter striation was occasionally present at the M-band level (Fig. 4A). The observed staining pattern was similar to that previously seen with the same antibody and protocol.4 Costaining for dystrophin showed that the M-band myospryn stripes were most prominent at, but not restricted to, the subsarcolemmal level (Fig. 4B). Such a trend was not apparent with the major, Z-disc-flanking, myospryn striations.
FIGURE 4.
Immunofluorescence staining of muscle sections. Cryosections from stretched and immersion-fixed mouse and human muscles were stained for myospryn, α-actinin, and dystrophin and analyzed by confocal microscopy. A, in both mouse and human muscle, myospryn was predominantly localized as doublet bands flanking the Z-disc (marked by α-actinin). A fainter signal was variably present at the M-band level (arrows). Staining profiles of α-actinin (red) and myospryn (green), measured from the regions boxed with blue in the merged images, demonstrate periodic myospryn staining flanking the Z-discs and at the M-bands. The images show single optical sections of 0.78 μm. B, as demonstrated by the double staining with dystrophin, the M-band myospryn stripes (arrows) were most prominent at the subsarcolemmal level. The images show a single optical section of 1.05 μm. Scale bars, 5 μm.
To determine whether myospryn localization was affected by the homozygous titin mutation in LGMD2J, we compared muscle biopsies from healthy humans and LGMD2J patients (data not shown). The overall myospryn staining pattern resembled that seen in the immersion-fixed muscles and showed no obvious difference between control and LGMD2J muscles. However, in all these biopsy samples, the major doublet bands appeared fuzzier, and we were unable to discern the weak M-band myospryn stripes, which may reflect a lower resolution of the nonstretched samples or better preservation of the fine structure in the immersion-fixed muscle. Hence, subtle alterations of myospryn localization in LGMD2J muscles, especially in the M-band region, could not be reliably assayed from the material available.
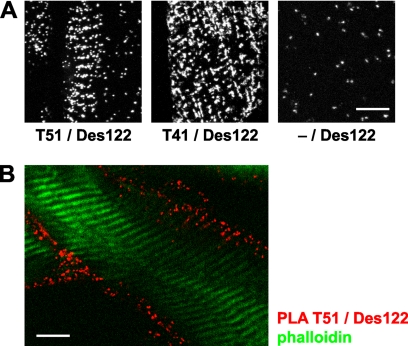
To obtain further evidence for the localization of myospryn at M-bands and for its interaction with titin, we studied mouse muscle sections with in situ PLA, a technique that allows the detection of the physical proximity of antibody-binding sites. The combination of the myospryn ab Des122 with either of the titin antibodies (T51 or T41) produced more PLA signals than seen in negative controls, demonstrating close proximity (<40 nm) of myospryn and M-band titin. The PLA signals appeared predominantly at the fiber periphery, and occasionally they showed a clear striated pattern with a spacing matching the sarcomeric length (Fig. 5). The fact that these striations arose in PLA assays with M-band-specific titin antibodies strongly suggests that they were localized at the M-band level.
FIGURE 5.
In situ proximity ligation assays. Mouse muscle sections were subjected to PLA stainings with the antibodies T51 or T41 against M-band titin and Des122 against myospryn and analyzed by confocal microscopy. Punctate fluorescent signals indicate physical proximity of the two different primary antibodies. A, antibody combinations T51/Des122 and T41/Des122 produced striated patterns of PLA signals, indicating periodic localization of myospryn in close proximity to M-band titin. Fewer PLA signals were produced in the negative control staining without the titin antibody (−/Des122). The images are maximum intensity projections through 13 optical sections of 0.78 μm. B, PLA signals between M-band titin and myospryn were most abundant at the fiber periphery, as shown here for the antibody pair T51/Des122 (red). Phalloidin counterstain (green) visualizes the thin filaments. The image shows a single optical section of 0.78 μm. Scale bars, 10 μm.
Transfected Myospryn Can Localize to M-bands in Cultured Cardiomyocytes
As a complementary approach, we used cultured neonatal rat cardiomyocytes for studying the localization of endogenous and transfected myospryn and their possible colocalization with M-band titin. Neonatal rat cardiomyocytes represent a fully differentiated and transfectable muscle cell in vitro, thus providing a good cell model for localization studies of myofibrillar proteins.
Endogenous myospryn, detected with the Des122 antibody in untransfected cells, typically showed reticular localization, possibly associated with SR, together with sarcomeric striations of variable intensity (Fig. 6, A–C). The sarcomeric localization was most prominent at the Z-discs, but in a small subset of cells a weaker striation, colocalizing with C-terminal titin, was present at the M-bands (Fig. 6, D–G).
FIGURE 6.
Confocal microscopy of neonatal rat cardiomyocytes. Neonatal rat cardiomyocytes, either transfected with different myospryn and titin constructs or untransfected, were stained with antibodies against sarcomeric α-actinin, myospryn, C-terminal titin, or epitope tags. A–C, in untransfected cells cultured for 3 days, endogenous myospryn localized typically in a reticular pattern with a variable degree of Z-disc striations (A, merge; B, α-actinin; C, myospryn Des122). D–G, in some cells, endogenous myospryn was seen at the M-bands (arrows) as well as Z-discs (arrowheads) (D, merge; E, titin T51; F, myospryn Des122; G, staining profile measured along the blue bar in D). H–J, GFP-CMYA53811–CT localized typically at the Z-discs (arrowheads), stress-fiber-like structures (s), and intercalated discs (i) (H, merge; I, α-actinin; J, GFP). K–N, Myc-MD9 showed reticular localization with comparably strong striations at the Z-discs and M-bands (K, merge; L, α-actinin; M, Myc; N, staining profile measured along the blue bar in K). O–R, in cells transfected with HA-is6-M10 alone, the titin construct typically showed a diffuse localization (O, merge; P, α-actinin; Q, myospryn Des122; R, HA). S–V, in cotransfections, HA-is6-M10 sometimes showed partial colocalization with the CMYA53811–CT constructs at the Z-discs (arrowheads), as shown here by Myc-CMYA53811–CT and HA-is6-M10 (S, merge; T, α-actinin; U, Myc; V, HA). (D–G, H–J, O–R, and S–V, 1 day post-transfection; K–N, 2 days post-transfection). Scale bars, 10 μm.
The myospryn constructs of two different lengths (3811–CT and MD9) differed clearly in their localization. Although there was cell-to-cell variation within the transfection experiments, typical localization patterns for each construct were discernible. The shorter CMYA53811–CT constructs were predominantly targeted to the sarcomeric Z-discs, stress fiber-like structures (regions of developing sarcomeres) and intercalated discs (Fig. 6, H–J). The degree of Z-disc localization varied considerably, with almost exclusive Z-disc staining in some cells and a less defined localization in others. The Z-disc pattern was clearest in short term cultures (fixed one or 2 days post-transfection). After 5 days in culture, some cells showed a similar pattern, whereas others exhibited abnormal morphology possibly due to toxic effects of the overexpressed protein.
The longer MD9 constructs typically adopted a reticular pattern with somewhat denser staining in the perinuclear region. In some cells, striations were evident as well, and where present, these were comparably strong in the Z-discs and M-bands (Fig. 6, K–N). The striated patterns seemed to become clearer and more frequent during the culture, with the striations most conspicuous 2–5 days after transfection.
The Myc- and GFP-tagged versions of both myospryn constructs behaved essentially in a similar fashion, demonstrating that the tags did not have a major effect on protein localization. Cells expressing GFP-tagged constructs did beat normally, indicating that the overexpression of C-terminal myospryn did not have a dramatic harmful effect on sarcomeric function. In the long term cultures, however, some transfected cells showed signs of cytotoxic effects.
To find out whether overexpression of C-terminal titin could affect the localization of myospryn or vice versa, we transfected cardiomyocytes with the HA-tagged titin construct HA-is6-M10 alone or together with the different myospryn constructs. As judged on a relatively high number of dead transfected cells, HA-is6-M10 overexpression seemed to have toxic effects, perhaps on the cells expressing the construct at the highest levels. Healthy-looking transfected cells could be found as well; in those, expression of the titin construct did not have any obvious effect on the localization of transfected or endogenous myospryn nor on the general appearance of sarcomeric markers.
In singly transfected cells cultured 1–2 days, HA-is6-M10 showed a diffuse localization throughout the cell and occasional association with actin filaments, but typically without striated sarcomeric staining (Fig. 6, O–R). By contrast, in cells cotransfected with the Z-disc-targeting CMYA53811–CT construct, HA-is6-M10 did show increased association with the Z-discs (Fig. 6, S–V). Although not directly reflecting a natural situation, this ability of myospryn to modulate the localization of the titin construct further supports a direct interaction of the proteins. Also in general, the titin and myospryn constructs in double-transfected cells seemed to adopt similar localization patterns, suggesting a tendency for the proteins to associate.
Myospryn Is a CAPN3 Substrate and May Stabilize CAPN3 in Vitro
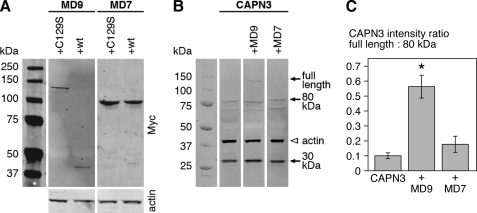
To study the functional consequences of the CAPN3-myospryn interaction, we coexpressed Myc-tagged myospryn constructs MD7 and MD9 with wild-type or inactive C129S versions of YFP/CFP-tagged CAPN3 in 911 cells.
When coexpressed with the inactive CAPN3C129S, Myc-MD9 was detected in cell lysates as expected (Fig. 7A). Coexpression with the proteolytically active construct, however, resulted in the disappearance of full-length Myc-MD9 from lysates, accompanied by the appearance of a smaller (∼40 kDa) N-terminal protein fragment. In contrast, the shorter Myc-MD7 construct was unaffected by CAPN3 coexpression. This demonstrates proteolytic cleavage of myospryn MD9 but not MD7 by CAPN3.
FIGURE 7.
Myospryn-CAPN3 coexpression studies. Myc-tagged myospryn constructs MD9 and MD7 were coexpressed with active (WT) or inactive (C129S) YFP/CFP-tagged CAPN3 in 911 cells. Samples of cell lysates were analyzed by Western blotting using antibodies against Myc, GFP, and actin. A, coexpression of active CAPN3 with Myc-MD9 resulted in the disappearance of full-length Myc-MD9 and appearance of a 40-kDa N-terminal MD9 fragment, demonstrating cleavage of MD9 by CAPN3. The MD7 construct was unaffected by CAPN3. B, compared with cells expressing CAPN3 alone or together with Myc-MD7, cells coexpressing Myc-MD9 showed a higher amount of the full-length calpain construct, suggesting attenuated autolysis of CAPN3. In the blot, several N- and C-terminal autolytic fragments of the calpain construct are visible, as the GFP antibody recognizes the fluorescent protein tags at both ends of the construct. Arrows indicate the full-length calpain construct at ∼140 kDa (corresponding to CAPN3 of 94 kDa plus the two tags) and the most prominent autolytic fragments at 80 and 30 kDa. The 80-kDa band results from cleavage at the IS1 region and corresponds to the 55-kDa autolytic fragment of CAPN3 (31) with the C-terminal CFP tag. The 30-kDa fragment, resulting from cleavage at the NS region, includes a short N-terminal calpain peptide preceded by the YFP tag. Actin is shown as a loading control. C, intensity ratios of the full-length CAPN3 construct to the 80-kDa autolytic fragment were quantified from cells expressing CAPN3 construct alone or together with the myospryn constructs. Cells coexpressing Myc-MD9 showed a significantly higher ratio (p = 0.0495, Mann-Whitney U test), suggesting stabilization of CAPN3. Mean ratios ± S.E. are shown; n = 3 for each group.
Proteolytically active CAPN3 normally undergoes rapid autolytic activation, which was demonstrated by a low level of the full-length CAPN3 construct and presence of autolytic calpain fragments in the cell lysates. However, in cells cotransfected with Myc-MD9, the total amount of full-length CAPN3 construct and its ratio to the 80-kDa autolytic fragment were higher, suggesting increased stability of CAPN3 (Fig. 7, B and C). Again, the effect was only observed with Myc-MD9 and not Myc-MD7.
DISCUSSION
Our results demonstrate direct molecular interactions of myospryn with the M-band part of the sarcomeric protein titin and with the skeletal muscle-specific protease calpain 3. We also show localization of endogenous and transfected myospryn at the M-band level, compatible with the suggested interaction with M-band titin. Fig. 8 places the novel interactions in the context of the known interaction networks of titin and myospryn.
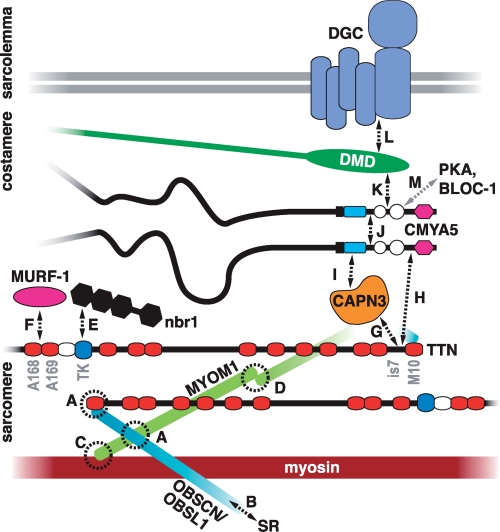
FIGURE 8.
Interaction network of M-band titin. The figure models the layout of titin and its associated proteins in the M-band and summarizes the molecular interaction network centered at M-band titin in skeletal muscle. The C termini of titin molecules from the adjacent half-sarcomeres overlap in the M-band. Interactions between the titin M10 domain, obscurin/obscurin-like 1 (OBSCN/OBSL1) and myomesin 1 (MYOM1), connect the three proteins as a ternary complex (A) (30). C-terminal obscurin links the myofibrils to the SR via ankyrin (B) (43). Myomesin 1 cross-links the thick filaments through its interaction with myosin (C) (44) and homodimerization (D) (45). The other members of the myomesin family (myomesin 2 or M-protein, and myomesin 3) also localize to the M-band, but their possible interactions with titin are not clarified. The known signaling functions of M-band titin reside at the M-band periphery. Nbr1 binds to the titin kinase (TK) domain and transmits information about the mechanical activity of titin to pathways controlling protein turnover (E) (3). The muscle ring finger protein MURF-1 binds the titin domains A168–A169 (F); this interaction has been proposed to have both regulatory and structural functions (46, 47). CAPN3 binds titin through the is7 region (G) (12). As shown in this study, both C-terminal titin (H) and CAPN3 (I) can bind myospryn (CMYA5), but it remains to be determined whether these interactions take place simultaneously at the M-band. Myospryn self-associates (J) (21), suggesting that it may act as a dimer. At the costamere level, myospryn interacts with dystrophin (K) (27), which in turn connects to the dystrophin-associated glycoprotein complex (DGC) at the sarcolemma (L) (48). Also protein kinase A (PKA) and the dysbindin subunit of the biogenesis of lysosome-related organelles-complex 1 (BLOC-1) are reported myospryn ligands that may interact with myospryn at the M-band level (M) (21, 26). The model for the organization of titin, myomesin, obscurin was adapted from Fukuzawa et al. (30).
Both of the novel interactions take place at the evolutionarily conserved C-terminal tripartite motif-like part of myospryn, a region previously reported to interact with dysbindin (21), α-actinin (23), desmin (24), and dystrophin (27), and to be involved in myospryn self-association (21). We did not look into whether these interactions can take place simultaneously, but given the large number of reported binding partners, some of the interactions may be mutually exclusive.
The reported self-association (21) and the presence of a coiled-coil region suggest that myospryn could act as a dimer. Although our interaction results should be equally valid for myospryn monomers and dimers, a different dimerization propensity could account for some of the differences seen between the long and short myospryn constructs in CoIP and transfection studies.
Titin-Myospryn Interaction
The minimal protein regions needed for the titin-myospryn interaction in Y2H are the M10 domain of titin and a C-terminal part of myospryn comprising a partial FN3 and the SPRY domain. According to our CoIP studies, however, the entire region M9-is7-M10 can participate in myospryn binding, and the interacting region might extend even more N-terminally. The location of the minimal binding regions in the C-terminal ends of titin and myospryn could thus place the interacting molecules side-by-side in a parallel fashion. It should be noted that although the titin is7 region does seem to have myospryn-binding potential, efficient binding of both is7+ and is7− constructs to myospryn in CoIP shows that the interaction is not isoform-specific.
Immunofluorescence microscopy and PLA experiments suggest that the interaction between M-band titin and myospryn takes place primarily at the myofiber periphery. This is in accordance with the predominantly subsarcolemmal localization of myospryn (21). As myospryn has been reported to associate with costameres (23, 24), the interaction of titin with myospryn could provide a link between the peripheral myofibrils and the costamere-like structures present at the M-band level (39). The nature of this interaction could be either structural or regulatory. Partial localization of the PKA RII subunits to the M-band level (26) suggests that the role of myospryn there could be related to its protein kinase A-anchoring protein function.
CAPN3-Myospryn Interaction
The interaction with myospryn involves the domain III and is2 of CAPN3, regions thought to be important for substrate recognition. Indeed, our coexpression studies indicated proteolysis of the myospryn construct MD9 by CAPN3 in vitro. The size of the N-terminal fragment places the cleavage site at 70–80 kDa from the myospryn C terminus, ∼40–140 amino acid residues N-terminally from the BBox′ domain. The MD7 construct might just be short enough to escape the cleavage, which would explain the different behavior of the two myospryn constructs. Cleavage by CAPN3 could serve to regulate the turnover, interactions, localization, or activity of myospryn. However, for most of the CAPN3 substrates identified in vitro, there is no evidence for cleavage in physiological conditions. Whether CAPN3 actually cleaves myospryn in vivo remains to be elucidated.
Stabilization of CAPN3 by MD9, suggested by our coexpression studies, provides additional support for the interaction of the proteins, and it also implies possible regulation of CAPN3 by myospryn. A plausible mechanism would be that anchoring to myospryn directly protects CAPN3 from autolysis, as has previously been shown for binding of CAPN3 to titin (12, 40). Alternatively, the stabilization could be indirectly mediated, for example by PKA.
The predominant localization pattern of myospryn (doublet bands flanking the Z-disc in skeletal muscle and a single Z-disc-overlapping band in cardiomyocytes) correlates with the position of the T-tubules, and localization of myospryn in this region has been suggested (41). CAPN3 was recently reported to have a structural, nonproteolytic role in the calcium release complex at the triads, the contact sites of T-tubules and SR (42). Localization of both myospryn and CAPN3 in this compartment hints that their interaction could take place there. Another attractive possibility is that the proximity of the CAPN3- and myospryn-binding sites in titin reflects a ternary interaction between the proteins in the M-band region.
Implications for Muscular Dystrophies
The involvement of myospryn and the PKA pathway in Duchenne muscular dystrophy has already indicated that myospryn may play a role in the pathomechanism of one type of muscular dystrophy. The novel interactions of myospryn with M-band titin and CAPN3, proteins associated with TMD/LGMD2J and LGMD2A, may suggest that myospryn could be of wider importance in the pathogenesis of muscle disease.
Y2H suggested that the FINmaj titin mutation could directly affect the interaction with myospryn. In light of our CoIP results, direct disruption of the binding is not likely, probably because the interaction is not limited to the mutated M10 domain. The loss of C-terminal titin epitopes and the secondary CAPN3 deficiency in LGMD2J muscle suggests, however, that the TMD/LGMD2J-causing mutations ultimately lead to the absence of the entire titin C terminus. This will obviously compromise any protein interactions taking place in the region, including that with myospryn, whether or not they are directly affected by the mutation. Downstream consequences of the altered titin-myospryn interaction could be relevant for the pathogenesis of titinopathies.
Besides CAPN3 and myospryn, the titinopathy mutations are known to affect the giant structural and signaling protein obscurin (OBSCN) and its smaller homologue obscurin- like 1 (OBSL1). Ternary interactions with the titin M10 domain and the M-band cross-linking protein myomesin are required for the proper localization of OBSCN/OBSL1 at the M-band. TMD/LGMD2J mutations directly disrupt these interactions, and obscurin is mislocalized in LGMD2J muscle (30). The role of these interactions in the pathomechanisms of TMD and LGMD2J needs to be addressed by further studies.
For LGMD2A, impaired cleavage of CAPN3 substrates is thought to be the most important pathogenetic factor (16), but involvement of the nonproteolytic functions of CAPN3 has been also suggested (42). The substrate(s) important for the pathogenesis are unknown. If myospryn is a proteolytic target for CAPN3 in vivo, loss of its cleavage may be one contributing factor in the molecular pathogenesis of calpainopathy. On the other hand, if myospryn acts upstream of calpain 3, its perturbation may have even more profound effects on the calpain-mediated regulatory proteolysis in muscle.
Acknowledgments
We thank Prof. Derek Blake (Cardiff University) and Dr. Matthew Benson (University of Oxford) for providing the Des122 antibody and the original MD7 and MD9 constructs and for helpful discussions; Prof. Mathias Gautel (King's College London) for critical reading of the manuscript; Dr. Atsushi Fukuzawa (King's College London) for the pEGFP-M10 construct; Kirsi Mänttäri and Outi Kokkonen (Biocentrum Helsinki Two-hybrid Facility, University of Helsinki) for performing the M10 interaction screen; Prof. Tomi Mäkelä (University of Helsinki) for the pAMC and pAHC vectors; Sanna Huovinen, Hanna-Liisa Kojo, and Sini Penttilä for help with the muscle samples; and Marjut Ritala, Katri Leinonen, Anni Riihiaho, pH Jonson, and Helena Luque for technical assistance. Confocal microscopy was partly carried out at the Molecular Imaging Unit, University of Helsinki.
This work was supported by Association Française Contre les Myopathies (to B. U. and I. R.), the Folkhälsan Research Foundation, the Academy of Finland, The Sigrid Jusélius Foundation (to B. U.), and University of Helsinki Research Foundation and EMBO Short Term Fellowship ASTF 13-08 (to J. S.).
A. Vihola and B. Udd, unpublished data.
M. Benson and D. Blake, personal communication.
- TMD
- tibial muscular dystrophy
- CoIP
- coimmunoprecipitation
- CT
- C terminus
- FN3
- fibronectin type 3-like domain
- CFP
- cyan fluorescent protein
- IF
- immunofluorescence
- LGMD
- limb-girdle muscular dystrophy
- MIM
- Mendelian Inheritance in Man
- PLA
- proximity ligation assay
- SPRY
- domain in SPlA and RYanodine receptor
- SR
- sarcoplasmic reticulum
- T-tubule
- transverse tubule
- WB
- western blotting
- Y2H
- yeast two-hybrid
- PFA
- paraformaldehyde
- ab
- antibody.
REFERENCES
- 1.Fürst D. O., Osborn M., Nave R., Weber K. (1988) J. Cell Biol. 106, 1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Granzier H., Labeit S. (2007) Muscle Nerve 36, 740–755 [DOI] [PubMed] [Google Scholar]
- 3.Lange S., Xiang F., Yakovenko A., Vihola A., Hackman P., Rostkova E., Kristensen J., Brandmeier B., Franzen G., Hedberg B., Gunnarsson L. G., Hughes S. M., Marchand S., Sejersen T., Richard I., Edström L., Ehler E., Udd B., Gautel M. (2005) Science 308, 1599–1603 [DOI] [PubMed] [Google Scholar]
- 4.Knöll R., Hoshijima M., Hoffman H. M., Person V., Lorenzen-Schmidt I., Bang M. L., Hayashi T., Shiga N., Yasukawa H., Schaper W., McKenna W., Yokoyama M., Schork N. J., Omens J. H., McCulloch A. D., Kimura A., Gregorio C. C., Poller W., Schaper J., Schultheiss H. P., Chien K. R. (2002) Cell 111, 943–955 [DOI] [PubMed] [Google Scholar]
- 5.Miller M. K., Bang M. L., Witt C. C., Labeit D., Trombitas C., Watanabe K., Granzier H., McElhinny A. S., Gregorio C. C., Labeit S. (2003) J. Mol. Biol. 333, 951–964 [DOI] [PubMed] [Google Scholar]
- 6.Hackman P., Vihola A., Haravuori H., Marchand S., Sarparanta J., De Seze J., Labeit S., Witt C., Peltonen L., Richard I., Udd B. (2002) Am. J. Hum. Genet. 71, 492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Udd B., Kääriänen H., Somer H. (1991) Muscle Nerve 14, 1050–1058 [DOI] [PubMed] [Google Scholar]
- 8.Udd B., Vihola A., Sarparanta J., Richard I., Hackman P. (2005) Neurology 64, 636–642 [DOI] [PubMed] [Google Scholar]
- 9.Van den Bergh P. Y., Bouquiaux O., Verellen C., Marchand S., Richard I., Hackman P., Udd B. (2003) Ann. Neurol. 54, 248–251 [DOI] [PubMed] [Google Scholar]
- 10.Hackman P., Marchand S., Sarparanta J., Vihola A., Pénisson-Besnier I., Eymard B., Pardal-Fernández J. M., Hammouda el-H., Richard I., Illa I., Udd B. (2008) Neuromuscul. Disord. 18, 922–928 [DOI] [PubMed] [Google Scholar]
- 11.Udd B., Partanen J., Halonen P., Falck B., Hakamies L., Heikkilä H., Ingo S., Kalimo H., Kääriäinen H., Laulumaa V., Paljärvi L., Rapola J., Reunanen M., Sonninen V., Somer H. (1993) Arch. Neurol. 50, 604–608 [DOI] [PubMed] [Google Scholar]
- 12.Sorimachi H., Kinbara K., Kimura S., Takahashi M., Ishiura S., Sasagawa N., Sorimachi N., Shimada H., Tagawa K., Maruyama K. (1995) J. Biol. Chem. 270, 31158–31162 [DOI] [PubMed] [Google Scholar]
- 13.Kinbara K., Sorimachi H., Ishiura S., Suzuki K. (1997) Arch. Biochem. Biophys. 342, 99–107 [DOI] [PubMed] [Google Scholar]
- 14.Haravuori H., Vihola A., Straub V., Auranen M., Richard I., Marchand S., Voit T., Labeit S., Somer H., Peltonen L., Beckmann J. S., Udd B. (2001) Neurology 56, 869–877 [DOI] [PubMed] [Google Scholar]
- 15.Suzuki K., Hata S., Kawabata Y., Sorimachi H. (2004) Diabetes 53, S12–S18 [DOI] [PubMed] [Google Scholar]
- 16.Duguez S., Bartoli M., Richard I. (2006) FEBS J. 273, 3427–3436 [DOI] [PubMed] [Google Scholar]
- 17.Kramerova I., Kudryashova E., Venkatraman G., Spencer M. J. (2005) Hum. Mol. Genet. 14, 2125–2134 [DOI] [PubMed] [Google Scholar]
- 18.Huang Y., de Morrée A., van Remoortere A., Bushby K., Frants R. R., Dunnen J. T., van der Maarel S. M. (2008) Hum. Mol. Genet. 17, 1855–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ojima K., Ono Y., Hata S., Koyama S., Doi N., Sorimachi H. (2005) J. Muscle Res. Cell. Motil. 26, 409–417 [DOI] [PubMed] [Google Scholar]
- 20.Richard I., Broux O., Allamand V., Fougerousse F., Chiannilkulchai N., Bourg N., Brenguier L., Devaud C., Pasturaud P., Roudaut C., Hillaire D., Passos-Bueno M. R., Zatz M., Tischfield J. A., Fardeau M., Jackson C. E., Cohen D., Beckmann J. S. (1995) Cell 81, 27–40 [DOI] [PubMed] [Google Scholar]
- 21.Benson M. A., Tinsley C. L., Blake D. J. (2004) J. Biol. Chem. 279, 10450–10458 [DOI] [PubMed] [Google Scholar]
- 22.Tkatchenko A. V., Piétu G., Cros N., Gannoun-Zaki L., Auffray C., Léger J. J., Dechesne C. A. (2001) Neuromuscul. Disord. 11, 269–277 [DOI] [PubMed] [Google Scholar]
- 23.Durham J. T., Brand O. M., Arnold M., Reynolds J. G., Muthukumar L., Weiler H., Richardson J. A., Naya F. J. (2006) J. Biol. Chem. 281, 6841–6849 [DOI] [PubMed] [Google Scholar]
- 24.Kouloumenta A., Mavroidis M., Capetanaki Y. (2007) J. Biol. Chem. 282, 35211–35221 [DOI] [PubMed] [Google Scholar]
- 25.Sarparanta J. (2008) J. Muscle Res. Cell. Motil. 29, 177–180 [DOI] [PubMed] [Google Scholar]
- 26.Reynolds J. G., McCalmon S. A., Tomczyk T., Naya F. J. (2007) Biochim. Biophys. Acta 1773, 891–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reynolds J. G., McCalmon S. A., Donaghey J. A., Naya F. J. (2008) J. Biol. Chem. 283, 8070–8074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Formstecher E., Aresta S., Collura V., Hamburger A., Meil A., Trehin A., Reverdy C., Betin V., Maire S., Brun C., Jacq B., Arpin M., Bellaiche Y., Bellusci S., Benaroch P., Bornens M., Chanet R., Chavrier P., Delattre O., Doye V., Fehon R., Faye G., Galli T., Girault J. A., Goud B., de Gunzburg J., Johannes L., Junier M. P., Mirouse V., Mukherjee A., Papadopoulo D., Perez F., Plessis A., Rossé C., Saule S., Stoppa-Lyonnet D., Vincent A., White M., Legrain P., Wojcik J., Camonis J., Daviet L. (2005) Genome Res. 15, 376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tiainen M., Ylikorkala A., Mäkelä T. P. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9248–9251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukuzawa A., Lange S., Holt M., Vihola A., Carmignac V., Ferreiro A., Udd B., Gautel M. (2008) J. Cell Sci. 121, 1841–1851 [DOI] [PubMed] [Google Scholar]
- 31.Taveau M., Bourg N., Sillon G., Roudaut C., Bartoli M., Richard I. (2003) Mol. Cell. Biol. 23, 9127–9135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ono Y., Shimada H., Sorimachi H., Richard I., Saido T. C., Beckmann J. S., Ishiura S., Suzuki K. (1998) J. Biol. Chem. 273, 17073–17078 [DOI] [PubMed] [Google Scholar]
- 33.Obermann W. M., Gautel M., Steiner F., van der Ven P. F., Weber K., Fürst D. O. (1996) J. Cell Biol. 134, 1441–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Ven P. F., Obermann W. M., Lemke B., Gautel M., Weber K., Fürst D. O. (2000) Cell Motil. Cytoskeleton 45, 149–162 [DOI] [PubMed] [Google Scholar]
- 35.Lykke-Andersen J. (2002) Mol. Cell. Biol. 22, 8114–8121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lange S., Auerbach D., McLoughlin P., Perriard E., Schäfer B. W., Perriard J. C., Ehler E. (2002) J. Cell Sci. 115, 4925–4936 [DOI] [PubMed] [Google Scholar]
- 37.Locke M., Tinsley C. L., Benson M. A., Blake D. J. (2009) Hum. Mol. Genet. 18, 2344–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 39.Porter G. A., Dmytrenko G. M., Winkelmann J. C., Bloch R. J. (1992) J. Cell Biol. 117, 997–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ono Y., Torii F., Ojima K., Doi N., Yoshioka K., Kawabata Y., Labeit D., Labeit S., Suzuki K., Abe K., Maeda T., Sorimachi H. (2006) J. Biol. Chem. 281, 18519–18531 [DOI] [PubMed] [Google Scholar]
- 41.Tinsley C. L., Esapa C. T., Benson M. A., Waite A. J., Locke M., Blake D. J. (2005) Neuromuscul. Disord. 15, 717 [Google Scholar]
- 42.Kramerova I., Kudryashova E., Wu B., Ottenheijm C., Granzier H., Spencer M. J. (2008) Hum. Mol. Genet. 17, 3271–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bagnato P., Barone V., Giacomello E., Rossi D., Sorrentino V. (2003) J. Cell Biol. 160, 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Obermann W. M., Gautel M., Weber K., Fürst D. O. (1997) EMBO J. 16, 211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lange S., Himmel M., Auerbach D., Agarkova I., Hayess K., Fürst D. O., Perriard J. C., Ehler E. (2005) J. Mol. Biol. 345, 289–298 [DOI] [PubMed] [Google Scholar]
- 46.Centner T., Yano J., Kimura E., McElhinny A. S., Pelin K., Witt C. C., Bang M. L., Trombitas K., Granzier H., Gregorio C. C., Sorimachi H., Labeit S. (2001) J. Mol. Biol. 306, 717–726 [DOI] [PubMed] [Google Scholar]
- 47.McElhinny A. S., Kakinuma K., Sorimachi H., Labeit S., Gregorio C. C. (2002) J. Cell Biol. 157, 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki A., Yoshida M., Hayashi K., Mizuno Y., Hagiwara Y., Ozawa E. (1994) Eur. J. Biochem. 220, 283–292 [DOI] [PubMed] [Google Scholar]