Abstract
The Sonic hedgehog (Shh) signaling pathway controls a variety of developmental processes and is implicated in tissue homeostasis maintenance and neurogenesis in adults. Recently, we identified Ulk3 as an active kinase able to positively regulate Gli proteins, mediators of the Shh signaling in mammals. Here, we provide several lines of evidence that Ulk3 participates in the transduction of the Shh signal also independently of its kinase activity. We demonstrate that Ulk3 through its kinase domain interacts with Suppressor of Fused (Sufu), a protein required for negative regulation of Gli proteins. Sufu blocks Ulk3 autophosphorylation and abolishes its ability to phosphorylate and positively regulate Gli proteins. We show that Shh signaling destabilizes the Sufu-Ulk3 complex and induces the release of Ulk3. We demonstrate that the Sufu-Ulk3 complex, when co-expressed with Gli2, promotes generation of the Gli2 repressor form, and that reduction of the Ulk3 mRNA level in Shh-responsive cells results in higher potency of the cells to transmit the Shh signal. Our data suggests a dual function of Ulk3 in the Shh signal transduction pathway and propose an additional way of regulating Gli proteins by Sufu, through binding to and suppression of Ulk3.
Keywords: Protein Phosphorylation, Protein-Protein Interactions, Serine-threonine Protein Kinase, Signal Transduction, Transcription Factors, Fused, Sonic Hedgehog, Ulk3
Introduction
The evolutionarily conserved Hedgehog (Hh)3 signaling pathway controls a variety of developmental processes through regulation of cell proliferation and differentiation (1). In adults, the pathway is implicated in tissue homeostasis maintenance and stem cell proliferation (2, 3). Because inappropriate activation of the HH pathway contributes to various congenital abnormalities and tumorigenesis in humans, investigations of the molecular signaling mechanisms should shed light not only on developmental but also on important pathological issues. The hh signaling pathway was initially discovered in Drosophila melanogaster (4). Molecular mechanisms of the Hh signal transduction have been intensively investigated using fly, fish, chick, and rodent models. Despite well described mechanisms of hh signaling in Drosophila, intracellular events induced by Sonic Hedgehog (Shh, one of three mammalian homologues of hh) have remained unclear in many aspects.
The Hh signaling is initiated by binding of the morphogen Hh to its receptor Patched1 (Ptch1), a 12-pass transmembrane protein. This results in the attenuation of the inhibitory effect of Ptch1 on another transmembrane protein, Smoothened (Smo), allowing the latter to transfer the signal into the cell through induction of the signaling machinery responsible for activation of the Hh target genes (3). Initiation of the signaling occurring on the cell membrane is rather similar in invertebrates and vertebrates (5, 6).
In vertebrates the Shh signal aims at controlling the activities of transcription factors Gli1, Gli2, and Gli3 (6, 7). Gli1 is the strongest activator and the Gli1 gene is a transcriptional target of Shh activity. As Gli1 is generally not expressed in non-stimulated cells, it serves as a marker of Shh activity and is thought to contribute to the maintenance of signaling (8, 9). Both Gli2 and Gli3 contain an N-terminal repressor domain and a C-terminal activator domain, whereas Gli3 is the strongest repressor and Gli2 is a primary activator of the Shh target genes (10). In the absence of Shh, full-length Gli2 and Gli3 are subjected to proteosomal degradation or undergo partial proteolysis resulting in generation of C terminal-truncated repressor forms, Gli2/3Rep (11, 12). In Shh-stimulated cells the proteolysis is repressed and full-length Gli proteins are converted to transcriptional activators, GliAct, followed by their translocation to the nucleus where they take part in transcriptional activation of target genes. In fact, it has been suggested that the balance between activator and repressor forms of Gli proteins determines the transcriptional outcome (13, 14).
Genetic studies suggest that the PEST domain containing protein Sufu is a major negative regulator of Gli proteins in mammals (15, 16). Notably, sufu−/− mutant flies are viable (17), whereas Sufu-deficient mice die at 9.5 days post coitus with multiple defects resulting from abnormal up-regulation of Hh signaling (15, 16). Mammalian Sufu controls Gli proteins by direct binding and sequestering them in the cytoplasm (18–20). This interaction is believed to contribute to the generation of Gli2/3Rep that is preceded by the phosphorylation of full-length Gli2/3 by PKA, glycogen synthase kinase 3β, and casein kinase 1 (12, 21–24). However, recent findings have demonstrated that Sufu binding to full-length Gli2 and Gli3 protects them from complete proteosomal degradation, which in turn contributes to the accumulation of a pool of Gli2 and Gli3 proteins ready to be converted to transcriptional activators (25). The dual function of Sufu suggests the existence of several pools of Sufu, regulating Gli proteins context dependently.
In Drosophila, the divergent functions of Gli proteins are performed by one hh signal-dependent transcription factor, Cubitus interruptus (26). The activity of C. interruptus is controlled by a multimolecular complex associated with microtubules (so-called Hedgehog signaling complex or HSC). HSC contains a scaffolding protein costal2 (cos2), and putative serine/threonine kinase fused (fu), and sufu (27–30). HSC, through cos2 and sufu, binds C. interruptus and controls its stability, subcellular localization, and activity in an hh signal-dependent way (31, 32). In the absence of hh ligand, HSC is responsible for retaining the full-length C. interruptus in an inactive state and also participates in the generation of a C-terminal-truncated repressor form of C. interruptus. In the hh-stimulated cells, HSC dissociates, and full-length C. interruptus is released to perform transcriptional activation.
Fu is known as one of the central regulators of C. interruptus activity. Genetic studies suggest a positive role of fu, as loss of fu leads to Hh pathway activation (33, 34). Indeed, the predominant role of fu is to antagonize the negative effect of sufu (35). Fu and sufu are able to interact, and the fu domain responsible for this interaction has been mapped to amino acids residues 306–436 (27). Fu comprises an N-terminal kinase domain and a C-terminal regulatory domain and has been shown to play both kinase activity-dependent and -independent (regulatory) roles (36–38). In the absence of hh ligand, fu is inactive and subjected to autoinhibition through its regulatory domain (39). This domain is also required for processing of full-length C. interruptus into the transcriptional repressor form (37). However, when the pathway is activated, fu becomes phosphorylated (40). Moreover, it has been shown that phosphorylation of cos2, sufu, and smo in response to the pathway activation depends on fu kinase activity and full activation of the hh pathway requires fu kinase (28, 31, 41–43). Thus, fu plays an Hh ligand-dependent dual role in regulation of C. interruptus activity.
At present, a similar Gli-containing signaling complex has not been described in vertebrates. In contrast to the comprehensive role of fu, its mammalian homologue Stk36 has no or a limited role in the Shh pathway, as it is dispensable for proper embryonic development in mouse (44, 45). Although Stk36 was shown to some extent rescue the negative effect of Sufu on Gli-dependent transcription (46), no evidence of physical interaction between Sufu and Stk36 has been provided.
Recently, we have demonstrated that serine/threonine kinase ULK3 is able to regulate GLI proteins positively in a kinase activity-dependent manner (47). ULK3 shares sequence similarity with serine/threonine kinases fu and STK36. We have shown that ULK3 directly phosphorylates GLI proteins, enhances their transcriptional activity in cell culture, and promotes the nuclear translocation of GLI1.
In this study, we demonstrate that ULK3 kinase has a intramolecular self-regulation property. In addition to the previously demonstrated phosphorylation/activation of GLI, ULK3 has a kinase-independent regulatory role in the SHH pathway. By RNAi, it is demonstrated that reduction of the Ulk3 mRNA level results in a higher response of cells to the Shh signal. We show that ULK3 through its kinase domain (KD) physically interacts with SUFU, and that the ULK3-Sufu complex is sensitive to SHH signaling. Interaction with Sufu inhibits the catalytic activity of ULK3, preventing its autophosphorylation and subsequent phosphorylation of GLI proteins. Moreover, the SUFU-ULK3 complex, when co-expressed with GLI2, induces generation of the GLI2 repressor form, whereas none of those proteins alone is able to do that. Finally, we propose a model unraveling the role of Ulk3 in the regulation of Gli proteins.
EXPERIMENTAL PROCEDURES
Expression Constructs
ULK3FLAG, ULK3(K139R), and GLI2FLAG were described in Ref. 47, GLI2GFP was described in Ref. 48, and SUFUmyc described in Ref. 20. The pSV40-β-gal construct used for luciferase assay data normalization was described in Ref. 49. pCI-GFP (Promega, Madison, WI) and pBABEpuro (Addgene, Cambridge, MA) were kindly provided by Dr. Rune Toftgård. The sequence for small interfering RNA1 (siRNA1, 5′-TATCTACCTCATCATGGAG-3′) was chosen to be specific for human, mouse, and rat Ulk3 nt 258–276 and the sequence of siRNA2 (5′-ACGAAACATCTCTCACTTGGA-3′) was specific for mouse and rat Ulk3 nt 390–410 (numbering is given relative to translation initiation codon ATG). Expression constructs siRNA1pSUPER and siRNA2pSUPER were generated according to the pSUPER RNAi System Protocol (OligoEngine, Seattle, WA). The part of mouse Ulk3 cDNA containing the sequences specific for siRNAs was amplified by PCR from mouse hippocampus cDNA and cloned into the ULK3FLAG construct between StuI and Eco72I sites (nt 91 and 513, respectively). Constructs ULK3(Δ301–365), ULK3(Δ373–446), ULK3-KD (amino acids 1–270), and ULK3-CT (amino acids 271–472) were generated from ULK3FLAG by PCR using the Pfu Turbo DNA polymerase system (Stratagene, La Jolla, CA) according to the manufacturers' recommendations. Construct pULK3-Ubi was generated by subcloning the ULK3 coding sequence from ULK3FLAG into modified bacterial expression vector pET24d (Novagen, Darmstadt, Germany) containing the sequence encoding decahistidine-tagged Saccharomyces cerevisiae ubiquitin protein.
Antibodies
The following antibodies were used for WB: M2 anti-FLAG-HRP (Sigma), anti-GFP-HRP (Rockland), anti-GFP (Clontech, Saint-Germain-en-Laye, France), and H-300 anti-Sufu (Santa Cruz, CA). Antibodies used for IP were C-15 anti-Sufu (Santa Cruz), anti-FLAG-M2 (Sigma), and c-myc 9E10 (Santa Cruz) conjugated with agarose. The secondary antibodies used were HRP-conjugated goat anti-mouse and goat anti-rabbit Ig (Jackson Laboratories, West Grove, PA).
Bacterial Expression and Purification of Proteins
SHH is described in Ref. 50. ULK3-Ubi fusion protein was expressed in Escherichia coli strain BL21-CodonPlusTM-RP at 30 °C in LB broth containing 10% glycerol and induced overnight with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside. Fusion protein was purified using Ni-CAM HC resin (Sigma) according to the manufacturer's recommendations except that all buffers contained 10% glycerol.
Cells Culture
HEK293, NIH3T3, and Shh-Light2 (Shh-L2) cells were propagated as previously described (47). Stable cell lines were generated as described in Ref. 50 and propagated in Shh-L2 cell medium supplemented with 2 μg/ml of puromycin (Sigma). Approximately 24 h prior to transfections or induction with SHH, the cells were plated onto appropriate growth dishes. Rat cerebellar granular cells (RCGCs) were isolated from P6 rat pups, as described (51), and plated on dishes pre-coated with 0.1 mg/cm2 of poly-l-lysin (Sigma). Cells were propagated in Neurobasal medium supplemented with B-27 (Invitrogen), 78 ng/ml of d-glucose, 20 mm KCl (Sigma), 20 μm glutamine, and 100 μg/ml of penicillin/streptomycin (Invitrogen). All cells were grown at 37 °C and 5% CO2.
Overexpression Studies
Shh-L2 cells and stable cell lines derived from Shh-L2, HEK293, and NIH3T3 cells were transfected using polyethylenimine transfection agent (PEI) (Inbio, Tallinn, Estonia) as described (50). After transfection, HEK293 and NIH3T3 cells were propagated in the normal growth medium for 48 h. Shh-L2 cells were handled as previously described and subjected to luciferase assay (50). The obtained firefly luciferase data were normalized with β-galactosidase values. For assessment of efficiency of siRNAs, HEK293 cells were co-transfected with the Ulk3FLAG construct, pCI-GFP plasmid, and either siRNA1pSUPER or siRNA2pSUPER constructs (40 + 40 ng + 1500 ng/3-cm plate, respectively). Stable cell clones were co-transfected with 100 ng of Ulk3FLAG and 50 ng of pCI-GFP on a 3-cm plate. RCGCs were transfected using Amaxa Rat Neuron Nucleofector kit (Lonza, Basel, Switzerland) according to the manufacturer's instructions. We used 2 μg of plasmid per ∼2 ml of RCGCs. For detection of the GLI2 repressor form, HEK293 cells were transfected with 1 μg of GLI2GFP, 0.6 μg of ULK3FLAG or ULK3(K139R), and 0.4 μg of SUFUmyc plasmids or the respective empty vectors. Alternatively, the FLAG-tagged GLI2 construct was used in combination with His-tagged ULK3 and SUFUmyc encoding plasmids. Cells were incubated 48 h, lysed using RIPA lysis buffer supplemented with Protease inhibitor mixture (Roche Applied Science), and subjected to WB using anti-GFP antibody or M2 anti-FLAG antibody.
Quantitative Real Time PCR
Total RNA from Shh-L2 and stable cell lines was isolated using RNAqueous kit (Ambion, Austin, TX) and total RNA from RCGC was isolated using the RNeasy micro kit (Qiagen) according to the manufacturer's instructions. cDNA was synthesized from 1 μg of total RNA using SuperScriptIII reverse transcriptase (Invitrogen) according to the manufacturer's recommendations. The levels of Ulk3 and Gli1 mRNAs and mRNA of the housekeeping gene Hprt used for normalization were detected in triplicates by quantitative real time PCR (qRT-PCR) using qPCR Core kit for SYBR Green (Eurogentec, Seraing, Belgium) with Lightcycler 2.0 (Roche Applied Science) according to the manufacturer's instructions. Data were analyzed with Lightcycler 4.05 software (Roche). The data are expressed as the average mean ± S.E. of three independent measurements. The following primers were used: Ulk3 sense, 5′-ACGAAACATCTCTCACTTG-3′; Ulk3 antisense, 5′-TGCTGGGCAAAGCCAAAGTC-3′; Gli1 sense, 5′-ACGTTTGAAGGCTGTCGGAA-3′; Gli1 antisense, 5′-CACACGTATGGCTTCTCATT-3′; Hprt sense, 5′-CAGTCCCAGCGTCGTGATTA-3′; and Hprt antisense, 5′-AGCAAGTCTTTCAGTCCTGTC-3′.
Statistical Analysis
Statistical analysis was carried out and p values were calculated using t test (Two Sample Assuming Equal Variances).
Immunoprecipitation
Cells were lysed and immunoprecipitation (IP) was performed as previously described (47). HEK293 cells were transfected with FLAG-tagged constructs expressing ULK3, ULK3(K139R), ULK3(Δ301–365), ULK3(Δ373–446), or ULK3-KD and the myc-tagged construct expressing SUFU (each 2.5 μg) on 6-cm plates. For negative controls, expression constructs were substituted with the respective empty vectors. FLAG-tagged constructs were immunoprecipitated using anti-FLAG-M2 affinity gel and myc-tagged SUFU was precipitated using anti-myc affinity gel according to the manufacturer's instructions. Cell lysates were incubated with the antibodies for 2 h at room temperature with gentle agitation. Immunocomplexes were subjected to WB using M2 anti-FLAG-HRP antibody and H-300 anti-myc antibody. Endogenous Sufu was immunoprecipitated from NIH3T3 cells using C-15 anti-Sufu antibody conjugated with agarose beads. Cells were transiently transfected with 10 μg of ULK3FLAG or empty vector and, if indicated, induced with 12 nm SHH for 48 h on 10-cm plates. Cell lysates were incubated with the antibody overnight at 4 °C with gentle agitation. Immunocomplexes were subjected to WB using H-300 anti-Sufu antibody or M2 anti-FLAG-HRP antibody.
In Vitro Kinase Assay
In vitro kinase assay was performed as described (47). Briefly, FLAG-tagged ULK3, ULK3(K139R), ULK3-KD, ULK3-CT, and GLI2FLAG proteins were overexpressed in HEK293 cells, immunoprecipitated using anti-FLAG-M2 affinity gel, washed 3 times with TBS, 2 times with kinase buffer and resuspended in 30 μl of kinase buffer. Aliquots of 2 μl were used for in vitro kinase assay. One-half of the myc immunocomplexes was washed twice with kinase buffer and subjected to in vitro kinase assay in the presence of ULK3-Ubi and separately immunopurified FLAG-tagged ULK3, if indicated.
Mass Spectrometry Analysis
For detection of ULK3 autophosphorylation sites, ULK3FLAG and ULK3(K139R) were expressed in HEK293 cells. The proteins were immunopurified using anti-FLAG-M2 affinity gel. ULK3FLAG and ULK3-Ubi proteins were subjected to in vitro kinase assay. The reaction was stopped by adding Laemmli buffer containing 100 mm DTT. ULK3FLAG protein, not subjected to the in vitro kinase assay, was used as a control. Proteins were resolved by SDS-PAGE and visualized by Coomassie Blue staining. The bands were excised from the gel and in-gel digested with modified sequencing grade trypsin (Promega), as described previously (52). Peptides from in-gel-digested samples were purified with StageTips1 and analyzed by LC-MS/MS using an Agilent 1200 series nanoflow system (Agilent Technologies, Santa Clara, CA) connected to a LTQ Orbitrap mass spectrometer (Thermo Electron, Bremen, Germany) equipped with a nanoelectrospray ion source (Proxeon, Odense, Denmark). Up to five data-dependent MS/MS spectra were acquired in centroid in the linear ion trap for each FTMS full-scan spectrum. Fragment MS/MS spectra from raw files were extracted as MSM files and then merged to peak lists using Raw2MSM version 1.72 selecting the top six peaks for 100 Da. MSM files were searched with the Mascot 2.2 search engine (Matrix Science, London, UK) against the protein sequence data base composed of ULK3 sequences and common contaminant proteins such as trypsin, keratins etc.
RESULTS
Silencing of Ulk3 Gene Expression by RNAi Suggests a Negative Role of Ulk3 in the Transduction of Shh Signal
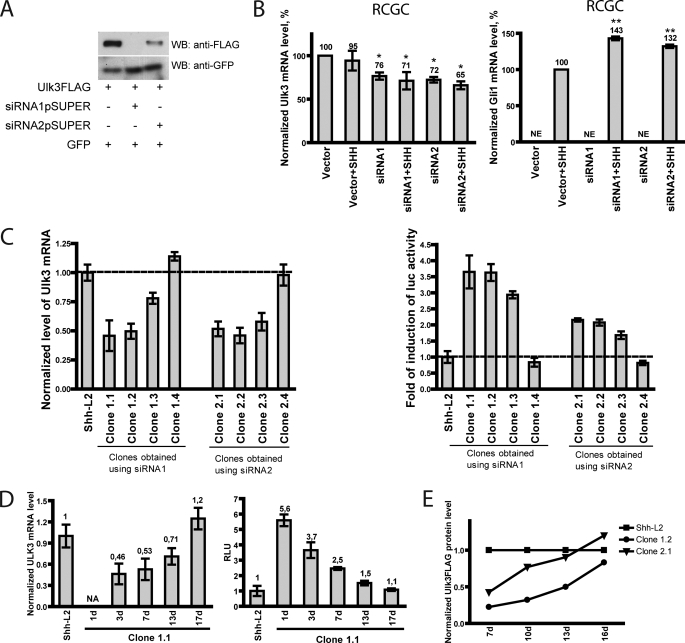
To investigate the role of Ulk3 in Shh signal transduction, we suppressed Ulk3 expression by RNAi. We designed two siRNA-expressing constructs, siRNA1pSUPER and siRNA2pSUPER. The effectiveness of the siRNAs was estimated by overexpressing Ulk3FLAG with siRNA1pSUPER and siRNA2pSUPER constructs in HEK293 cells. pCI-GFP plasmid expressing GFP was co-transfected to estimate the efficiency of transfection. Expression of FLAG-tagged Ulk3 and GFP was detected by WB analysis using anti-FLAG and anti-GFP antibodies, respectively. The experiment was repeated three times and the data of a representative experiment are shown in Fig. 1A. Both siRNAs were able to suppress Ulk3 expression, whereas siRNA1 demonstrated higher efficiency than siRNA2.
FIGURE 1.
Suppression of Ulk3 gene expression suggests a negative role of Ulk3 in Shh signal transduction. A, constructs expressing Ulk3-specific siRNA1 and siRNA2 were co-expressed with FLAG-tagged Ulk3 and GFP in HEK293 cells and cell lysates were subjected to WB analysis. Both siRNAs suppress expression of Ulk3. B, RCGCs were transiently transfected with the construct expressing Ulk3-specific siRNA1 or siRNA2 and stimulated with SHH protein. The Ulk3 mRNA level was measured by qRT-PCR and normalized with the Hprt mRNA level (left panel) along with measurements of Gli-induced luciferase activity (right panel). The cells transfected with the siRNA-expressing constructs were compared with the cells transfected with empty vector *, p < 0.05; **, p < 0.01 (NE, not expressed). The data are presented as average mean ± S.E. of three measurements obtained from three independent experiments. C, suppression of Ulk3 gene expression is achieved in cell lines stably expressing siRNA1 (clones 1.1, 1.2, and 1.3) and siRNA2 (clones 2.1, 2.2, and 2.3) (left panel). The Ulk3 mRNA level is reduced most effectively (∼50%) in clones 1.1, 1.2, 2.1, and 2.2. The expression level of Ulk3 mRNA, normalized with Hprt mRNA, in the parental cell line Shh-L2 is set as 1 and the values in other cell lines are normalized accordingly. Clones expressing the lower level of Ulk3 mRNA demonstrate a higher induction of Gli-dependent luciferase gene expression under influence of SHH compared with control cell lines Shh-L2, clones 1.4 and 2.4 (right panel). The data are presented as average mean ± S.E. of three independent measurements. D, prolonged propagation of stable cell clone 1.1 expressing the Ulk3-specific siRNA1. Left panel shows Ulk3 mRNA levels and the right panel shows the luciferase activities in cells induced with SHH protein. NA, not analyzed. The data are presented as the average mean ± S.E. of three independent measurements. E, cell lines stably expressing Ulk3-specific siRNA1 and siRNA2 (clones 1.2 and 2.1, respectively) and Shh-L2 cells were transfected at different time points during propagation with Ulk3FLAG and GFP encoding constructs. Cell lysates were analyzed with WB using anti-FLAG and anti-GFP antibodies. The levels of overexpressed proteins were quantified and the Ulk3FLAG protein level was normalized with the level of GFP expression. Ulk3FLAG protein level in Shh-L2 cells at each time point was calculated as 1.
To study the effect of Ulk3 silencing on Shh signal transduction, we used RCGCs that are known to be Shh responding cells (53). RCGCs were isolated from P6 rat pups and immediately transfected with an empty vector, siRNA1- or siRNA2-encoding constructs in two replicates. One replicate of each transfection was induced by the SHH protein. The level of Ulk3 and Gli1 mRNA was measured in triplicates using qRT-PCR and normalized by Hprt mRNA expression level. The average mean of three independent experiments ± S.E. is shown in Fig. 1B. The normalized level of Ulk3 mRNA is shown in the left panel. The level of Ulk3 mRNA in non-induced cells transfected with empty vector was set as 100%. Addition of SHH did not affect the level of Ulk3. Transient expression of both siRNAs could suppress Ulk3 mRNA expression by ∼30%. Addition of SHH did not significantly affect the extent of Ulk3 silencing.
Normalized level of Gli1 mRNA in the same samples is shown in the right panel of Fig. 1B. As the level of Gli1 mRNA was below the detection limit in non-induced cells, we concluded that Gli1 is not expressed in RCGCs. The level of Gli1 mRNA in cells transfected with empty vector and induced with SHH was considered 100%. Transfection of siRNA1pSUPER and siRNA2pSUPER constructs followed by SHH induction, elevated the Gli1 mRNA expression levels by 43 and 32%, respectively. It should be noted that siRNA1, as the more potent silencer of Ulk3 expression, triggered higher induction of Gli1 mRNA expression.
To corroborate our findings in RCGCs and achieve higher levels of suppression of Ulk3 mRNA expression, we generated stable cell lines expressing the Ulk3-specific siRNA1 and siRNA2 in the Shh-responsive cell line Shh-L2. We co-transfected either of the two siRNA constructs together with the pBABEpuro construct into Shh-L2 cells. In total 38 puromycin-resistant clones (23 clones obtained using siRNA1 and 15 clones obtained using siRNA2) from two independent experiments were picked, propagated, and divided in three parts: the first part was plated and induced by SHH to be analyzed using luciferase assay, the second part was frozen for total RNA isolation and the third part was propagated further.
The levels of Ulk3 mRNA and mRNA of the housekeeping gene Hprt used for normalization were measured using qRT-PCR. The level of Ulk3 mRNA, normalized by Hprt mRNA expression, in Shh-L2 cells was taken as 1. Suppression of Ulk3 mRNA was achieved in 6 clones: clones 1.1, 1.2, and 1.3 were obtained using siRNA1 and clones 2.1, 2.2, and 2.3 were obtained using siRNA2 (Fig. 1C, left panel). Ulk3 mRNA was suppressed most effectively (by ∼50%) in clones 1.1, 1.2, 2.1, and 2.2. The level of Ulk3 mRNA in clones 1.4 and 2.4 is shown as an additional control, as the level of Ulk3 mRNA was similar to that in the parental Shh-L2 cells. We also analyzed the level of Gli1 mRNA in stable and control cell lines. However, it did not correlate with changes in Ulk3 mRNA levels (data not shown).
Luciferase activity of three independent replicates was obtained and normalized with alkaline phosphatase values. SHH-dependent induction of luciferase activity in Shh-L2 cells was considered as 1. All clones expressing the lower level of Ulk3 mRNA had higher potency in the induction of Gli-dependent luciferase gene expression compared with the control cell line Shh-L2, clones 1.4 and 2.4 (Fig. 1C, right panel). However, although Ulk3 mRNA levels were similar in clones expressing siRNA1 and siRNA2, clones obtained using the more potent siRNA1 (Fig. 1A) demonstrated higher induction of Gli-dependent luciferase activity compared with clones obtained using siRNA2 (3.5 versus 2.2 times above the controls, respectively). This suggests that siRNA1 might suppress the expression of Ulk3 not only at the transcriptional but also at the translational level.
Continuing the analysis of clones stably expressing Ulk3-specific siRNAs, we found that during propagation of the cell lines, the Ulk3 mRNA levels constantly increased with time, reaching the level of the control cell line within 2.5 weeks. A total of 4 clones obtained from two independent experiments were analyzed for Ulk3 mRNA expression and induction of luciferase activity during 2.5 weeks of culturing. All clones showed the same tendency and the results of representative clone 1.1 are shown in Fig. 1D, as compared with parental Shh-L2 cells. The Ulk3 mRNA level increased from the initial 46 to 120% during 14 days (Fig. 1D, left panel), in concert with reduction of luciferase activity under the influence of SHH (Fig. 1D, right panel).
Due to the lack of working antibody against Ulk3, we were unable to show the endogenous Ulk3 protein levels in the stable cell lines. Therefore we transfected Ulk3FLAG and GFP encoding constructs into clones 1.2 and 2.1 and Shh-L2 cells. The levels of exogenously added Ulk3 and GFP proteins were analyzed by WB using antibodies against FLAG tag and GFP, respectively. The experiment was repeated 4 times. The obtained bands were quantified using ImageQuant TL software. Ulk3 protein levels, normalized with GFP levels, are shown in Fig. 1E and supplemental Fig. S1. The lowest amount of Ulk3 protein was detected in clone 1.2, whereas clone 2.1 showed a moderately reduced level of Ulk3 and the control cell line demonstrated the highest level of the protein. These results are perfectly in line with data of the transcription assay and higher effectiveness of the siRNA1 construct in suppressing the expression of Ulk3. Similarly to the increase of the Ulk3 mRNA level in stable cell lines in time, we found that the level of exogenous Ulk3 protein was increased in time, reaching the level in Shh-L2 after 2 weeks.
Taken together, our results show that reduction of the Ulk3 mRNA level in cells triggers a stronger response to SHH signal. Thus, our data from RNAi experiments suggest that Ulk3 is functioning as a negative regulator of the Shh pathway.
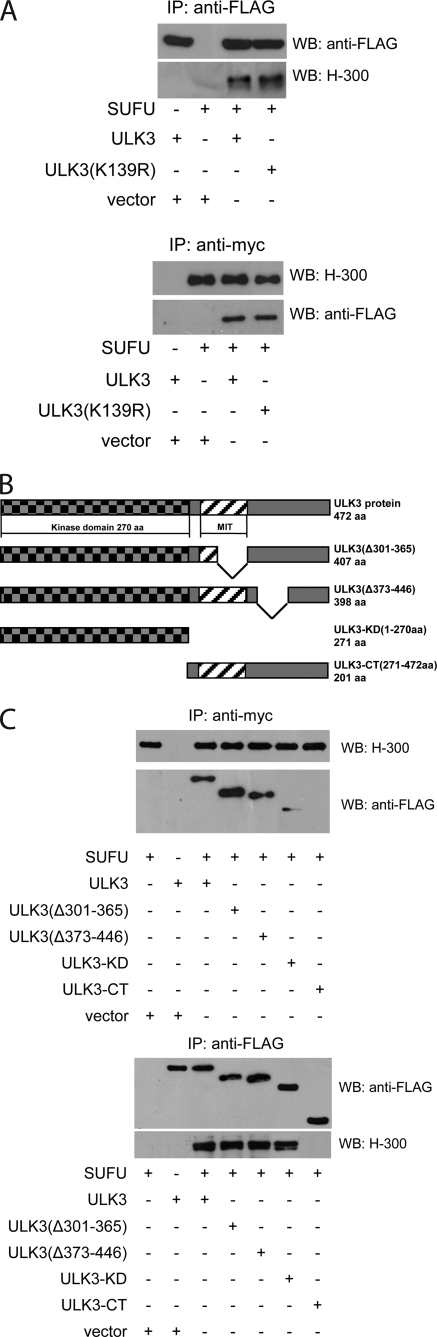
ULK3 Binds SUFU through Its Kinase Domain
ULK3 shares similarity with fu, and Drosophila proteins fu and sufu form a complex. Because SUFU is the major negative regulator of SHH signaling in mammals, we investigated if ULK3 is able to interact physically with SUFU protein. We overexpressed the FLAG-tagged WT and kinase-deficient ULK3 proteins and myc-tagged SUFU or respective empty vectors in HEK293 cells and immunoprecipitated them. The immunocomplexes were subjected to WB analysis using antibodies against FLAG tag and SUFU protein. The experiment was repeated 3 times, and the results of a representative experiment are shown in Fig. 2A. SUFU protein was detected in ULK3 as well as in ULK3(K139R) immunocomplexes (upper panel). Also, ULK3 and ULK3(K139R) proteins were detected in SUFU immunocomplexes (lower panel). Similar amounts of ULK3 and ULK3(K139R) were coimmunoprecipitated with SUFU suggesting that WT and kinase-deficient ULK3 bind SUFU with equal efficiency.
FIGURE 2.
ULK3 physically interacts with SUFU through its KD. A, FLAG-tagged WT and kinase-deficient ULK3 proteins were co-expressed with myc-tagged SUFU in HEK293 cells and immunoprecipitated using M2-a-FLAG or c-myc 9E10 affinity gel, respectively. Immunocomplexes were subjected to WB using a-FLAG and H-300 a-Sufu antibodies. B, schematic presentation of ULK3 proteins tested in the current study. MIT, domain contained within microtubule interacting and trafficking molecules. C, FLAG-tagged WT ULK3 and its deletion mutants were co-expressed with myc-tagged SUFU in HEK293 cells and immunoprecipitated using M2-a-FLAG or c-myc 9E10 affinity gel, respectively. Immunocomplexes were subjected to WB using a-FLAG and H-300 a-Sufu antibodies.
To investigate further the interaction of SUFU and ULK3, we attempted to determine the domain of ULK3 responsible for this interaction. First, we performed homology analysis of ULK3 and fu protein sequences using the DNAMAN sequence alignment algorithm and ClustalW program (EBI, EMBL). Regardless of the overall dissimilarity of the carboxyl termini of fu and ULK3, the analysis revealed two regions of homology between ULK3 and fu. Homologous regions were mapped to the fu domain, responsible for interaction with sufu (amino acids residues 306–436) (27). The first region corresponded to residues 310–365 and the second to residues 398–433 of ULK3. Bioinformatic analysis of the secondary structure of ULK3 (PredictProtein, EXPASY) predicted several α-helixes in those regions, which might participate in the interaction with Sufu. Based on the bioinformatic analysis, we generated two expression constructs encoding FLAG-tagged ULK3 deletion mutants, ULK3(Δ301–365) and ULK3(Δ373–446). We also generated the expression constructs encoding the kinase domain (KD) and C-terminal domain (CTD) of ULK3 (ULK3-KD and ULK3-CT, respectively). The structure of the resulting proteins is depicted in Fig. 2B.
To reveal the domain of ULK3 responsible for interaction with SUFU, we co-expressed the FLAG-tagged ULK3 or its deletion mutants with SUFUmyc in HEK293 cells. The proteins were immunoprecipitated using anti-FLAG and anti-myc antibodies and subjected to WB analysis (Fig. 2C). We detected ULK3wt, ULK3(Δ301–365), ULK3(Δ373–446), and ULK3-KD in SUFU immunoprecipitates (upper panel) and SUFU protein in ULK3wt, ULK3(Δ301–365), ULK3(Δ373–446), and ULK3-KD immunocomplexes (lower panel). No significant difference was found in the efficiency of interaction of WT ULK3 or deletion mutants ULK3(Δ301–365) and ULK3(Δ373–446) with SUFU. However, we found that ULK3-KD was able to interact with SUFU, but the efficiency of their interaction was lower compared with that of WT ULK3 and SUFU, as indicated on the upper panel. ULK3-CT did not interact with SUFU.
Our data indicate that the ULK3 domain responsible for interaction with SUFU is not homologous to the fu interaction domain with sufu. Instead, ULK3 interacts with SUFU at least partly through its KD.
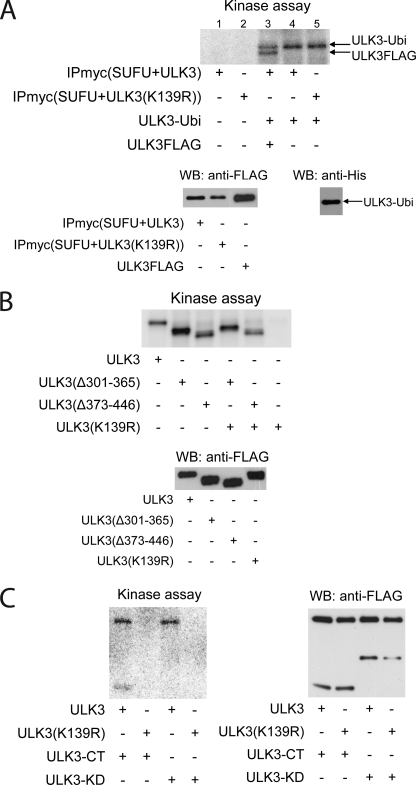
ULK3 C-terminal Autophosphorylation Is Blocked by SUFU Binding
Keeping in mind that fu, being in HSC, is catalytically inactive, we tested if ULK3, being in complex with SUFU, exhibits autophosphorylation activity. Myc-tagged SUFU and FLAG-tagged ULK3 or its catalytically inactive mutant ULK3(K139R) were co-expressed in HEK293 cells, subjected to immunoprecipitation using anti-myc antibody, followed by in vitro kinase assay (Fig. 3A, upper panel). The presence of the respective proteins in the immunoprecipitates was detected by WB and is shown in the lower panel of Fig. 3A. No ULK3 autophosphorylation activity was detected in the immunocomplexes (Fig. 3A, lane 1), suggesting that the kinase activity of ULK3 was abolished by interaction with SUFU. In contrast, bacterially expressed ULK3-Ubi or separately immunoprecipitated ULK3FLAG were able to self-phosphorylate themselves, indicating that autophosphorylation per se was not affected under these conditions (Fig. 3A, lanes 3–5). However, ULK3-Ubi neither phosphorylated SUFU-bound ULK3 nor ULK3(K139R) (Fig. 3A, lanes 4 and 5, respectively). These findings show that the kinase activity of ULK3 may be completely blocked by interaction with SUFU. In addition, we have found that trans-phosphorylation of ULK3 bound by SUFU does not occur. These data suggests that ULK3 phosphorylation takes place only within one molecule.
FIGURE 3.
Physical interaction of ULK3 with SUFU abolishes C-terminal autophosphorylation of ULK3. A, upper panel, immunocomplexes obtained using anti-myc antibody and containing myc-tagged SUFU and FLAG-tagged ULK3 or ULK3(K139R) were subjected to in vitro kinase assay in the presence of [γ-32P]ATP (lanes 1 and 2). Bacterially expressed and purified His-tagged ULK3-Ubi and separately immunopurified FLAG-tagged ULK3 proteins were added to the immunocomplexes prior to the kinase assay (lanes 3–5). Lower panel, the presence of the proteins in the immunoprecipitates was detected by WB using the respective antibody. B, upper panel, FLAG-tagged ULK3 proteins were overexpressed in HEK293 cells, immunopurified using anti-FLAG antibody, and subjected to in vitro kinase assay in the presence of [γ-32P]ATP. Lower panel shows the presence of the proteins detected by WB using anti-FLAG antibody. C, left panel, FLAG-tagged WT ULK3, kinase-deficient ULK3(K139R), ULK3-CT, and ULK3-KD were overexpressed in HEK293 cells, immunopurified using anti-FLAG antibody, and subjected to in vitro kinase assay in the presence of [γ-32P]ATP (CT, carboxyl terminus). Right panel, the presence of the proteins in the in vitro kinase reactions was detected by WB using anti-FLAG antibody.
To further confirm the lack of trans-phosphorylation activity of ULK3 kinase, we expressed FLAG-tagged WT ULK3, its deletion mutants ULK3(Δ301–365) and ULK3(Δ373–446), and kinase-deficient ULK3(K139R) in HEK293 cells and immunoprecipitated the proteins. We mixed the kinase-competent immunoprecipitates with lower molecular mass (either ULK3(Δ301–365) or ULK3(Δ373–446), 46 and 45 kDa, respectively) with the ULK3(K139R) (53 kDa) immunoprecipitate and subjected the complexes to in vitro kinase assay. Using this kind of mixing, we were able to distinguish between autophosphorylation and trans-phosphorylation activity. As shown in the upper panel of Fig. 3B, none of the kinase-competent ULK3 proteins were able to phosphorylate the ULK3(K139R) protein, but retained the autophosphorylation activity. The presence of all proteins tested in the immunocomplexes was confirmed by WB (Fig. 3B, lower panel).
Because ULK3 loses its autocatalytic activity, when bound to SUFU, we were interested in determining which residues of ULK3 are autophosphorylated. We subjected bacterially expressed and purified protein (ULK3-Ubi), FLAG-tagged ULK3, and ULK3(K139R) proteins immunopurified from mammalian cells to in vitro kinase assay. Immunopurified ULK3FLAG protein that did not undergo the in vitro kinase reaction was used as a negative control. The proteins were resolved by SDS-PAGE, excised from the gel, and in-gel digested with trypsin. Phosphorylated residues were mapped using LC-ESI-MS/MS in two technical replicates. Sequence coverage, representing the percentage of the entire protein sequence within the identified peptides, was 71% in ULK3FLAG and 75% in ULK3-Ubi. In contrast to the negative controls, autophosphorylated ULK3 proteins contained four phosphorylated residues: Ser-300, Ser-350, Ser-384, and Ser-464 (Table 1). Our analysis shows that ULK3 phosphorylates itself in the CTD.
TABLE 1.
Autophosphorylation sites in ULK3 protein
Bacterially expressed and purified ULK3-Ubi overexpressed in HEK293 cells and immunopurified ULK3FLAG and ULK3(K139R) proteins were subjected to in vitro kinase assay. ULK3FLAG protein unexposed to the kinase assay was used as a negative control. The proteins were trypsinized followed by LC-ESI-MS/MS analysis of phosphopeptides. Four phosphorylated serine residues situated in C-terminal part of WT ULK3 proteins are identified. The numbers are given relative to the first methionine residue of ULK3. The Mascot score is given as S = −10 × log(P), where P is the probability that the observed match is a random event.
| Peptide | Phosphorylated residue | Mascot score |
|---|---|---|
| KDQEGDSAAALSLYCK | Ser-300 | 69 |
| AIVSSSNQALLR | Ser-350 | 58 |
| LLAALEVASAAMAK | Ser-384 | 56 |
| EGLSESVR | Ser-464 | 93 |
To confirm the MS data, we subjected the immunopurified ULK3-KD and ULK3-CT to in vitro kinase assay in the presence of immunopurified WT ULK3 or kinase-deficient ULK3(K139R). No phosphorylation was detected in the ULK3-KD protein when mixed with WT ULK3 or ULK3(K139R) (Fig. 3C, left panel, lanes 3 and 4). However, when we mixed immunopurified WT ULK3 protein with ULK3-CT, we observed phosphorylation of both proteins (Fig. 3C, left panel, first lane). No phosphorylation of ULK3-CT was observed when it was mixed with ULK3(K139R) (Fig. 3C, left panel, second lane). The presence of all proteins tested in the in vitro kinase reaction was confirmed using WB (Fig. 3C, right panel).
These data confirm the MS findings and show that ULK3 autophosphorylation sites lie within the CTD of the protein. Furthermore, these data suggest that the C-terminal-phosphorylated residues of ULK3 are masked, preventing trans-phosphorylation by another ULK3 molecule. When the KD of ULK3 is removed, the phosphorylation sites become exposed and are readily accessible to another ULK3 molecule.
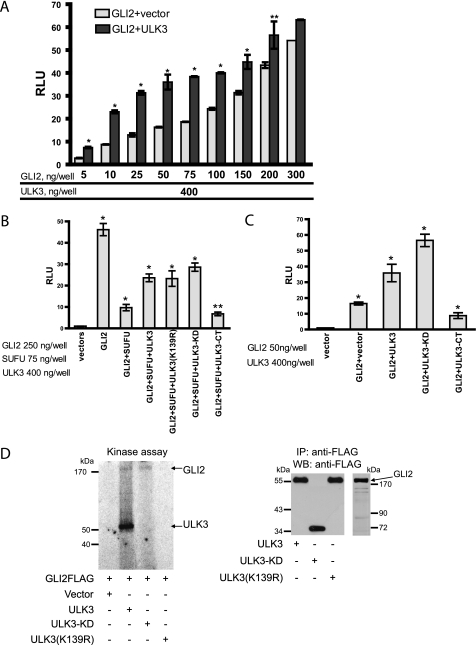
Luciferase Assay Suggests the Dual Role for ULK3
To get further insight into the biological meaning of the SUFU-ULK3 interaction, we performed an GLI-dependent transcriptional activation assay in Shh-L2 cells. In overexpression studies, SUFU is shown to elicit a strong negative effect on GLI proteins (20). In contrast, ULK3 is able to enhance significantly the transcriptional activity of GLI1 and GLI2 proteins in a kinase activity-dependent way (47). Thus, we assessed the potency of WT ULK3, ULK3(K139R), ULK3-KD, and ULK3-CT to rescue the negative effect of SUFU on GLI2 transcriptional activity. Because deletion mutants ULK3(Δ301–365) and ULK3(Δ373–446) were functionally highly similar to WT ULK3 (Figs. 2C and 3B, and data not shown), we did not test them in the luciferase assay.
First, we established the concentration curve for GLI2GFP co-expressed with the constant amount of WT ULK3 to find the amount of GLI2GFP plasmid sufficient to achieve the saturated level of Gli-dependent luciferase activity. The total DNA amount was kept constant by co-transfection with a compensatory amount of empty vector. Average induction of GLI2-dependent luciferase activity ± S.E. in the presence or absence of ULK3FLAG is shown in Fig. 4A. Our data showed that ULK3 was able to enhance the transcriptional activity of overexpressed GLI2 ∼3–2.5 times when co-expressed with low concentrations of GLI2GFP (5–75 ng of GLI2GFP plasmid per well). An increase of GLI2GFP plasmid amount (100–300 ng/well) led to a decrease of additional activation of Gli-dependent luciferase expression by ULK3 (1.6–1.2 times) because the level of induction approached the maximum.
FIGURE 4.
ULK3 relieves the inhibitory effect of SUFU on GLI2 transcriptional activity due to its KD but independently of its kinase activity. A, different amounts of the GLI2GFP-encoding construct and constant amount of the ULK3-encoding construct or respective empty vector were co-overexpressed in Shh-L2 cells and Gli-dependent luciferase activity was measured. Transfected DNA amount was kept constant by compensation of GLI2GFP plasmid with pCI-GFP vector. *, p value < 0.001 and **, p value < 0.05 GLI2 versus GLI2 + ULK3. The data are presented as average mean ± S.E. of three replicates obtained from three independent experiments. B, SUFU and ULK3 (WT, kinase-deficient, or deletion mutants) and GLI2 (250 ng/well) were co-expressed in Shh-L2 cells. Induction of Gli-dependent luciferase activity by empty vector was set as 1. GLI2 induces luciferase activity 46 times above the control (*, p value < 0.001 GLI2 versus empty vector). SUFU represses transcriptional activity of GLI2 (*, p value < 0.001 GLI2 versus GLI2 + SUFU). This repression is partly relieved by WT, kinase-deficient ULK3, and ULK3-KD (*, p value < 0.001, GLI2 + SUFU versus GLI2+SUFU+ULK3/ULK3(K139R)/ULK3-KD) and is not relieved, but more inhibited, by ULK3-CT (**, p value < 0.05, GLI2 + SUFU versus GLI2 + SUFU + ULK3-CT). The data are presented as average mean ± S.E. of three replicates obtained from three independent experiments. C, GLI2 (50 ng/well), ULK3, ULK3-KD, and ULK3-CT or the respective empty vector were co-expressed in Shh-L2 cells. Induction of luciferase activity by the empty vector was set as 1. GLI2 induces luciferase activity 16 times above the vector (*, p value < 0.001). ULK3 and ULK3-KD enhance GLI2 transcriptional activity (*, p value < 0.001). ULK3-KD potentiates the transcriptional activator function of GLI2 stronger than ULK3 (♦, p value < 0.001). Overexpression of ULK3-CT leads to inhibition of GLI2 transcriptional activity (*, p value < 0.001). The data are presented as average mean ± S.E. of three replicates obtained from three independent experiments. D, FLAG-tagged ULK3, ULK3(K139R), ULK3-KD, and GLI2 were overexpressed in HEK293 cells, immunopurified, and subjected to in vitro kinase assay in the presence of [γ-32P]ATP (left panel). Right panel shows the presence of the proteins detected by WB using anti-FLAG antibody.
To investigate the role of ULK3 and its KD and CTD in GLI2 regulation in the presence of SUFU, we overexpressed a high amount of GLI2GFP (250 ng of GLI2GFP construct per well) with SUFUmyc and FLAG-tagged ULK3, ULK3(K139R), ULK3-KD, and ULK3-CT proteins (or respective empty vectors) in Shh-L2 cells and analyzed the protein for Gli-dependent transcriptional activation (Fig. 4B). GLI2 was able to induce luciferase activity ∼46 times above control (induction of luciferase expression by empty vector taken as 1). Co-transfection with SUFU inhibited GLI2-induced luciferase activity 4.7 times. Addition of ULK3, ULK3(K139R), or ULK3-KD could partially rescue the negative effect of SUFU on GLI2 (2.4, 2.4, and 2.9 times, respectively). WT and kinase-deficient ULK3 demonstrated an equal effect in the assay suggesting the existence of a kinase activity-independent function of ULK3 in the regulation of GLI2. Notably, the effectiveness of ULK3-KD in restoring of GLI2 transcriptional activator properties was higher regardless on its lower affinity to SUFU compared with WT ULK3 (Fig. 2C, upper panel) suggesting the possibility of an inhibitory function of the CTD. Indeed, ULK3-CT could not rescue the inhibitory effect of SUFU on GLI2. Moreover, addition of ULK-CT to GLI2 and SUFU resulted in stronger inhibition of the GLI2 transcriptional activity (6.8 versus 4.7 times). ULK3-CT also inhibited GLI2 transcriptional activity 1.6 times (supplemental Fig. 2A). Expression of GLI2 with ULK3, ULK3(K139R), or ULK3-KD plasmids could additionally activate Gli-dependent luciferase expression ∼1.2 times (supplemental Fig. 2A).
To test the effect of ULK3, ULK3-KD, and ULK3-CT on GLI2 transcriptional activity in non-saturated conditions, we co-transfected 50 ng of GLI2GFP and ULK3 constructs (or respective empty vectors) into Shh-L2 cells and analyzed the induction of luciferase activity (Fig. 4C). Induction of luciferase expression by empty vector was set as 1. GLI2 induced luciferase activity 16 times. ULK3 was able to enhance GLI2 transcriptional activity ∼2.2 times and ULK3-KD enhanced GLI2 transcriptional activity 3.5 times. ULK3-CT, in contrast, inhibited GLI2-induced luciferase activity 2 times. Induction of the Gli-dependent luciferase by ULK3, ULK3-KD, and ULK3-CT in Shh-L2 cells is demonstrated in supplemental Fig. 2B. ULK3 was able to stimulate the luciferase activity 2.3 times above the induction by the empty vector, ULK3-KD, 3.4 times, and ULK3-CT had no effect on Gli-dependent luciferase activity.
Previously we have shown that ULK3 directly phosphorylates GLI proteins. Here we tested if ULK3-KD is able to phosphorylate the GLI2 protein. We overexpressed FLAG-tagged ULK3, ULK3(K139R), ULK3-KD, and GLI2 in HEK293 cells, immunopurified them, and subjected them to in vitro kinase assay in the presence of [γ-32P]ATP. ULK3-KD, despite its failure to autophosphorylate, was able to phosphorylate the GLI2 protein (Fig. 2D). These data suggest that ULK3-KD positively regulates GLI2 transcriptional activity through direct phosphorylation.
Taken together, our overexpression studies in Shh-L2 cells suggests that the ULK3 interaction with SUFU partly restores GLI2 transcriptional activity inhibited by SUFU. Furthermore, we demonstrate that ULK3 may have a dual role in regulation of GLI2 transcriptional activity. KD of ULK3 binds to SUFU and is able to phosphorylate and positively regulate GLI2. The CTD of ULK3 inhibits GLI2 transcriptional activity.
ULK3-SUFU Complex Promotes Generation of GLI2 Repressor Form
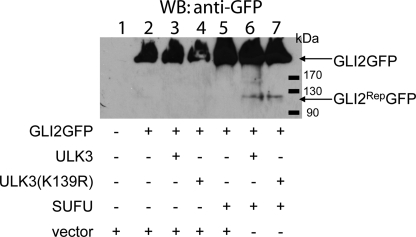
Our data showing the inability of ULK3 to entirely restore GLI2 transcriptional activity inhibited by SUFU, lead us to the hypothesis that the function of the ULK3-SUFU complex may be linked to generation of the GLI2Rep form. To test that, we co-expressed GLI2GFP protein with WT or kinase-deficient FLAG-tagged ULK3 and myc-tagged SUFU proteins in HEK293 cells. Alternatively, the constructs encoding FLAG-tagged GLI2 and His-tagged ULK3 were used. Cell lysates were subjected to WB analysis using anti-GFP or anti-FLAG antibodies. The experiment using both GLI2 constructs was repeated three times. The data of representative experiments obtained using anti-GFP and anti-FLAG antibodies are shown in Fig. 5 and supplemental Fig. 3, respectively. GLI2GFP has a molecular mass of 210 kDa (Fig. 5). We failed to detect the GLI2RepGFP form when full-length GLI2GFP was expressed alone or together with either of the ULK3 proteins or SUFU (Fig. 5, lanes 2–5). SUFU significantly stabilizes full-length GLI2, which is consistent with previously published results (25) (Fig. 5, lane 5 versus lane 2, and supplemental Fig. 3, right panel). Proteins with a molecular mass of about 120 kDa were detected in the presence of ULK3 (either WT or kinase-deficient mutant) combined with SUFU (Fig. 5, lanes 6 and 7). The molecular weight of the protein corresponded to the expected molecular weight of the GLI2Rep-GFP fusion protein. In the case of FLAG-tagged GLI2 (170 kDa), a repressor form with a molecular mass of about 90 kDa was detected (supplemental Fig. 3, left panel). However, using anti-FLAG antibody, we could detect low amounts of GLI2RepFLAG in the samples obtained from cells transfected with GLI2FLAG alone or together with SUFU. In the case of co-overexpressing of GLI2FLAG with WT or kinase-deficient ULK3, GLI2RepFLAG was undetectable. Because GLI2Rep was detected in cases when SUFU was co-expressed both with the WT and kinase-deficient mutant of ULK3, we conclude that the kinase activity of ULK3 is not required for the generation of GLI2Rep.
FIGURE 5.
ULK3-SUFU complex induces the generation of GLI2Rep. GLI2GFP fusion protein was expressed alone (lane 2), in combination with WT and kinase-deficient FLAG-tagged ULK3 (lanes 3 and 4), or myc-tagged SUFU (lane 5) in HEK293 cells. GLI2RepGFP protein is detected in the case of co-expression of GLI2GFP with SUFU combined with ULK3 or ULK3(K139R) by WB using GFP antibody (lanes 6 and 7, respectively).
ULK3-Sufu Complex Dissociates under SHH Signal
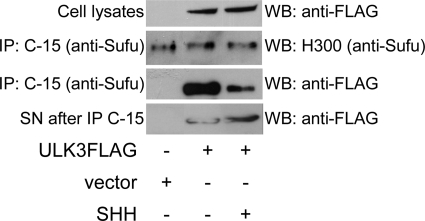
Next, we asked whether overexpressed ULK3 was able to act in a dominant-negative manner and form a SHH signal-dependent complex with endogenous Sufu. We overexpressed FLAG-tagged ULK3 (or respective empty vector) in Shh-responsive NIH3T3 cells and induced the cells with SHH protein. Endogenous Sufu was immunoprecipitated using an anti-Sufu C-15 antibody. The obtained cell lysates, immunocomplexes, and IP supernatants were subjected to WB using anti-FLAG antibody to detect the presence of ULK3 in the samples. The experiment was repeated 3 times and data of a representative experiment are shown in Fig. 6. The efficiency of IP reactions was estimated using H-300 anti-Sufu antibody. The amount of Sufu protein detected in the immunocomplexes was similar in all samples. Lysates of the transfected cells contained equal amounts of ULK3 protein. ULK3 protein was co-immunoprecipitated with endogenous Sufu from both non-induced and SHH-induced cells, indicating the ability of ULK3 to effectively form a complex with endogenous Sufu. However, immunoprecipitates from non-induced cells contained significantly more ULK3 protein compared with cells induced with SHH (Fig. 6). In contrast, the supernatants obtained after IP with Sufu antibody contained more ULK3 in SHH-induced cells. These data indicate that ULK3 is able to form a complex with endogenous Sufu. This complex is responsive to SHH signaling, because SHH triggers its dissociation.
FIGURE 6.
Interaction of ULK3 with endogenous Sufu is regulated by SHH. FLAG-tagged ULK3 was overexpressed in NIH3T3 cells in the absence or presence of SHH. Endogenous Sufu was immunoprecipitated using C-15 affinity gel. The amount of the precipitated Sufu protein was detected using H-300 a-Sufu antibody. ULK3 protein was detected using a-FLAG antibody. SN, supernatant.
DISCUSSION
Recently we have reported the identification of serine/threonine kinase ULK3 as a positive regulator of GLI proteins. ULK3 was identified based on its sequence similarity with serine/threonine kinases fu and STK36, a putative homologue of fu. We have previously shown that ULK3 is able to enhance the activity of GLI1 and GLI2 proteins in a kinase activity-dependent manner.
In this article we demonstrate that ULK3 has intramolecular self-regulatory properties. We show that ULK3 autophosphorylation occurs at four serine residues (Ser-300, Ser-350, Ser-384, and Ser-464) situated outside of the KD. Those sites are hidden in catalytically inactive ULK3 and unexposed to other ULK3 molecules. However, deletion of the KD results in conformational changes making CTD available for intermolecular phosphorylation by ULK3. Deletion of CTD results in generation of the catalytically active KD that is able to phosphorylate its substrate, GLI2. Thus, autophosphorylation of ULK3 may involve conformational changes resulted in exposure of CTD to KD and consequently in generation of the catalytically active kinase. A similar mechanism of self-regulation control has been proposed for ULK1 and ULK2 kinases (53) and seems to be conserved between members of the ULK family.
To enhance our understanding of the function of ULK3 in mammalian cells, we applied RNAi to test the overall requirement of Ulk3 for Shh signal transduction. We used two independent experimental models. First, we transiently transfected 2 different expression constructs encoding Ulk3-specific siRNAs, the more effective siRNA1 and the less effective siRNA2, into RCGCs. Second, we suppressed Ulk3 gene expression in the Shh-L2 cell line by stable transfection of the siRNA1 and siRNA2 constructs. The maximum suppression of the endogenous Ulk3 mRNA expression level was ∼30% in RCGC and 50% in stable cell lines. We failed to get cells with a lower level of Ulk3 mRNA. One possible reason may be the fact that reduction of the Ulk3 mRNA level leads to S-phase arrest (54). Therefore, it is possible that the cells with a lower level of Ulk3 mRNA were not able to grow sufficiently to be picked and analyzed.
In both experimental models the cells were induced by SHH and the induction of the Gli1 mRNA expression level was assessed, in Shh-L2-derived stable cell lines indirectly by measuring the Gli-dependent luciferase activation, and in RCGCs, directly, using qRT-PCR. Unexpectedly, reduction of the Ulk3 mRNA level resulted in an elevated response of cells to Shh reflected by an increased level of Gli1 mRNA. In fact, the level of Gli1 mRNA induction in response to SHH is in correlation with the Ulk3 mRNA level in the cells. One possible explanation of these results is that Ulk3 may be involved in the negative regulation of some component(s) of the Shh pathway, for instance, in generation of Gli2/3Rep in non-induced cells.
Because Sufu is known as a negative regulator of Gli proteins and is able to form a complex with fu (19), we tested the possibility that ULK3 acts in concert with SUFU in regulating GLI proteins. Indeed, our coimmunoprecipitation analysis demonstrates a physical interaction of ULK3 with Sufu proteins of human and mouse origin. Moreover, we demonstrate that KD of ULK3 is responsible for the interaction with SUFU. Transcriptional regulation experiments in the SHH-responsive cell line give functional meaning to this interaction, because ULK3 partially rescues the inhibitory effect of SUFU on GLI2-dependent transcription. However, ULK3 could not recover GLI2-induced luciferase activity to the initial level regardless of its molar superiority above SUFU, suggesting, analogically to the RNAi results, the existence of an ULK3-dependent mechanism negatively regulating the GLI2 protein. In fact, we found that CTD of ULK3 elicits a negative effect on GLI2 transcriptional activity. Deletion of this domain results in generation of a more potent transcriptional co-activator form of ULK3, ULK3-KD, that positively regulates GLI2 via direct phosphorylation.
Here, we further demonstrate the involvement of the novel ULK3-SUFU complex in the generation of GLI2Rep, and both proteins are required for this. As expected, the ULK3 kinase activity is not required for GLI2Rep generation.
Although Sufu was previously thought to be merely a negative regulator of Gli proteins by sequestering them in the cytoplasm, its function in protecting the full-length Gli2/3 proteins from proteolytic degradation has been described recently (25). We also observed the stabilizing effect of SUFU on full-length GLI2. However, here we show that SUFU is also needed for GLI2Rep generation. Neither SUFU nor ULK3 alone can induce the generation of GLI2Rep, but acting in concert, they promote C-terminal processing of full-length GLI2 probably through recruiting PKA, glycogen synthase kinase 3β, and casein kinase 1 protein kinases responsible for the initiation of proteolytic cleavage of GLI2/3. However, the precise mechanism of GLI2Rep generation with the help of the SUFU-ULK3 complex remains to be investigated further.
Hitherto, apart from Gli proteins, three mammalian proteins, SAP18, Galectin3, and Cdc2l1, have been identified as proteins able to physically interact with Sufu (55–57). Besides, Sufu has been shown to move to primary cilia in response to Shh signaling (58). Thus, Sufu is thought to shuttle within the cell, interacting with different proteins essential for enhancement or suppression of its regulatory functions.
We show that ULK3 is able to interact not only with overexpressed, but also with endogenous SUFU protein. Moreover, ULK3 autophosphorylation activity is completely abolished in the complex with SUFU. The rational explanation of this phenomenon is that ULK3 interacts with Sufu through its KD.
In this study we show that the ULK3-Sufu complex is responsive to the SHH signal, which induces its dissociation. An analogous mechanism is described in the case of HSC in Drosophila where HSC dissociates under influence of the Hh ligand (31). Taking into account the data showing that catalytically active ULK3 is able to phosphorylate and positively regulate its substrates, GLI proteins (47), we suggest a model for the actions of Ulk3 in the Shh pathway (Fig. 7).
FIGURE 7.
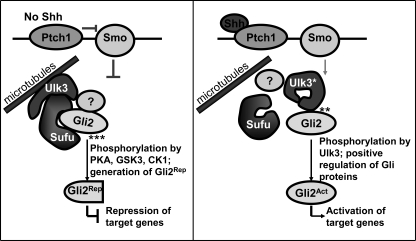
A model of Ulk3 function in Shh signaling pathway. In the absence of Shh, Sufu forms a complex with Ulk3 that possibly interacts with cytoskeleton through its MIT domain. The complex binds full-length Gli2 through Sufu, protecting it from proteosomal degradation. Ulk3 is not catalytically active but plays a regulatory role. The Ulk3-Sufu complex contributes to C-terminal processing of Gli2 probably through recruiting PKA, glycogen synthase kinase 3β (GSK3β), and casein kinase 1 (CK1) kinases to full-length Gli2. This results in generation of Gli2Rep that may enter the nucleus and inhibit its target gene expression. In the presence of Shh, Ulk3-Sufu-Gli2 complex dissociates, Ulk3 activates itself by autophosphorylation and phosphorylates full-length Gli2. This contributes to generation of Gli2Act, which translocates to the nucleus and activates its target genes. Other molecule(s) indicated as a question mark (?), for instance, Stk36, may participate in the regulation of Gli2 activity by converting it to transcriptional activator or repressor forms depending on the cellular context and strength of Shh signaling.
We propose that Ulk3 is part of a Shh signal-dependent cytoplasmic complex regulating the activity and processing of at least the Gli2 protein. In the absence of Shh ligand, the kinase domain of Ulk3 is bound by Sufu, therefore Ulk3 is catalytically inactive and unable to phosphorylate and positively regulate itself and Gli proteins. The complex binds full-length Gli2 through Sufu and induces the generation of the C-terminal-truncated transcriptional repressor Gli2Rep. Through its putative MIT domain (a domain contained within microtubule interacting and trafficking molecules), Ulk3 may be a link between Sufu/Gli2 and the cytoskeleton. In the presence of Shh, the Ulk3-Sufu-Gli2 complex dissociates. Ulk3 is released from the complex and may be able to phosphorylate itself. It may also phosphorylate the full-length Gli2, activating it and promoting its nuclear translocation. Gli2Act activates the transcription of its target genes, for instance Gli1. In the case of the activated pathway, Ulk3 plays a kinase activity-dependent positive role.
Our experiments show that the role of Ulk3 in the regulation of the Shh pathway involves eliciting both kinase activity-dependent and -independent effects on Gli transcription factors. The positive regulation is implemented by catalytically active Ulk3, whereas the negative regulation does not depend on Ulk3 kinase activity. That resembles in many aspects the model describing implementation of the fu-sufu-cos2-Ci complex in the Drosophila Hh pathway. Taking into account the sequence similarity of fu and Ulk3 proteins and the similarity of their function in Drosophila and mammalian Hh pathways, respectively, we suggest that Ulk3 is a structural and functional homologue of fu.
Supplementary Material
Acknowledgments
We are very grateful to Dr. Rune Toftgård and Dr. Csaba Finta (Center for Nutrition and Toxicology, Karolinska Institute, Sweden) for providing pCI-GFP and pBABEpuro vectors and C-15 anti-Sufu antibody. We thank Priit Pruunsild (Tallinn University of Technology) for providing mouse hippocampus cDNA.
This work was supported in part by Estonian Science Foundation Grants 7279 (to T. Ø.) and 8116 (to P. K.). Mass spectrometric analyses were supported in part by the European Regional Development Fund through the Center of Excellence in Chemical Biology (Institute of Technology, University of Tartu).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- Hh
- Hedgehog
- CTD
- carboxyl-terminal domain
- fu
- Fused
- GliAct
- activator form of Gli proteins
- Gli2/3Rep
- repressor form of Gli2/3 protein
- IP
- immunoprecipitation
- KD
- kinase domain
- RCGC
- rat cerebellar granular cells
- Shh
- Sonic Hedgehog
- Shh-L2
- Shh-LIGHT2
- Stk36
- serine/threonine kinase 36
- Sufu
- Suppressor of Fused
- Ulk3
- unc-51-like kinase 3
- WB
- Western blot
- WT
- wild-type
- HSC
- Hedgehog signaling complex
- nt
- nucleotide.
REFERENCES
- 1.Hooper J. E., Scott M. P. (2005) Nat. Rev. Mol. Cell. Biol. 6, 306–317 [DOI] [PubMed] [Google Scholar]
- 2.Beachy P. A., Karhadkar S. S., Berman D. M. (2004) Nature 432, 324–331 [DOI] [PubMed] [Google Scholar]
- 3.Ingham P. W., McMahon A. P. (2001) Genes Dev. 15, 3059–3087 [DOI] [PubMed] [Google Scholar]
- 4.Nüsslein-Volhard C., Wieschaus E. (1980) Nature 287, 795–801 [DOI] [PubMed] [Google Scholar]
- 5.Varjosalo M., Li S. P., Taipale J. (2006) Dev. Cell. 10, 177–186 [DOI] [PubMed] [Google Scholar]
- 6.Huangfu D., Anderson K. V. (2006) Development 133, 3–14 [DOI] [PubMed] [Google Scholar]
- 7.Jacob J., Briscoe J. (2003) EMBO Rep. 4, 761–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai C. B., Auerbach W., Lee J. S., Stephen D., Joyner A. L. (2002) Development 129, 4753–4761 [DOI] [PubMed] [Google Scholar]
- 9.Bai C. B., Joyner A. L. (2001) Development 128, 5161–5172 [DOI] [PubMed] [Google Scholar]
- 10.Sasaki H., Nishizaki Y., Hui C., Nakafuku M., Kondoh H. (1999) Development 126, 3915–3924 [DOI] [PubMed] [Google Scholar]
- 11.Wang B., Fallon J. F., Beachy P. A. (2000) Cell 100, 423–434 [DOI] [PubMed] [Google Scholar]
- 12.Pan Y., Bai C. B., Joyner A. L., Wang B. (2006) Mol. Cell. Biol. 26, 3365–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stecca B., Ruiz I. A. A. (2010) J. Mol. Cell. Biol. 2, 84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruiz i Altaba A., Mas C., Stecca B. (2007) Trends Cell. Biol. 17, 438–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper A. F., Yu K. P., Brueckner M., Brailey L. L., Johnson L., McGrath J. M., Bale A. E. (2005) Development 132, 4407–4417 [DOI] [PubMed] [Google Scholar]
- 16.Svärd J., Heby-Henricson K., Henricson K. H., Persson-Lek M., Rozell B., Lauth M., Bergström A., Ericson J., Toftgård R., Teglund S. (2006) Dev. Cell. 10, 187–197 [DOI] [PubMed] [Google Scholar]
- 17.Préat T. (1992) Genetics 132, 725–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding Q., Fukami S., Meng X., Nishizaki Y., Zhang X., Sasaki H., Dlugosz A., Nakafuku M., Hui C. (1999) Curr. Biol. 9, 1119–1122 [DOI] [PubMed] [Google Scholar]
- 19.Pearse R. V., 2nd, Collier L. S., Scott M. P., Tabin C. J. (1999) Dev. Biol. 212, 323–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kogerman P., Grimm T., Kogerman L., Krause D., Undén A. B., Sandstedt B., Toftgård R., Zaphiropoulos P. G. (1999) Nat. Cell. Biol. 1, 312–319 [DOI] [PubMed] [Google Scholar]
- 21.Pan Y., Wang C., Wang B. (2009) Dev. Biol. 326, 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang B., Li Y. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 33–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y., Gallaher N., Goodman R. H., Smolik S. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia J., Amanai K., Wang G., Tang J., Wang B., Jiang J. (2002) Nature 416, 548–552 [DOI] [PubMed] [Google Scholar]
- 25.Chen M. H., Wilson C. W., Li Y. J., Law K. K., Lu C. S., Gacayan R., Zhang X., Hui C. C., Chuang P. T. (2009) Genes Dev. 23, 1910–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aza-Blanc P., Ramírez-Weber F. A., Laget M. P., Schwartz C., Kornberg T. B. (1997) Cell 89, 1043–1053 [DOI] [PubMed] [Google Scholar]
- 27.Monnier V., Dussillol F., Alves G., Lamour-Isnard C., Plessis A. (1998) Curr. Biol. 8, 583–586 [DOI] [PubMed] [Google Scholar]
- 28.Lefers M. A., Wang Q. T., Holmgren R. A. (2001) Dev. Biol. 236, 411–420 [DOI] [PubMed] [Google Scholar]
- 29.Robbins D. J., Nybakken K. E., Kobayashi R., Sisson J. C., Bishop J. M., Thérond P. P. (1997) Cell 90, 225–234 [DOI] [PubMed] [Google Scholar]
- 30.Stegman M. A., Vallance J. E., Elangovan G., Sosinski J., Cheng Y., Robbins D. J. (2000) J. Biol. Chem. 275, 21809–21812 [DOI] [PubMed] [Google Scholar]
- 31.Ruel L., Rodriguez R., Gallet A., Lavenant-Staccini L., Thérond P. P. (2003) Nat. Cell. Biol. 5, 907–913 [DOI] [PubMed] [Google Scholar]
- 32.Lum L., Zhang C., Oh S., Mann R. K., von Kessler D. P., Taipale J., Weis-Garcia F., Gong R., Wang B., Beachy P. A. (2003) Mol. Cell. 12, 1261–1274 [DOI] [PubMed] [Google Scholar]
- 33.Préat T., Thérond P., Lamour-Isnard C., Limbourg-Bouchon B., Tricoire H., Erk I., Mariol M. C., Busson D. (1990) Nature 347, 87–89 [DOI] [PubMed] [Google Scholar]
- 34.Préat T., Thérond P., Limbourg-Bouchon B., Pham A., Tricoire H., Busson D., Lamour-Isnard C. (1993) Genetics 135, 1047–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pham A., Therond P., Alves G., Tournier F. B., Busson D., Lamour-Isnard C., Bouchon B. L., Préat T., Tricoire H. (1995) Genetics 140, 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thérond P., Alves G., Limbourg-Bouchon B., Tricoire H., Guillemet E., Brissard-Zahraoui J., Lamour-Isnard C., Busson D. (1996) Genetics 142, 1181–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ascano M., Jr., Nybakken K. E., Sosinski J., Stegman M. A., Robbins D. J. (2002) Mol. Cell. Biol. 22, 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alves G., Limbourg-Bouchon B., Tricoire H., Brissard-Zahraoui J., Lamour-Isnard C., Busson D. (1998) Mech. Dev. 78, 17–31 [DOI] [PubMed] [Google Scholar]
- 39.Ascano M., Jr., Robbins D. J. (2004) Mol. Cell. Biol. 24, 10397–10405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thérond P. P., Knight J. D., Kornberg T. B., Bishop J. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 4224–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nybakken K. E., Turck C. W., Robbins D. J., Bishop J. M. (2002) J. Biol. Chem. 277, 24638–24647 [DOI] [PubMed] [Google Scholar]
- 42.Claret S., Sanial M., Plessis A. (2007) Curr. Biol. 17, 1326–1333 [DOI] [PubMed] [Google Scholar]
- 43.Dussillol-Godar F., Brissard-Zahraoui J., Limbourg-Bouchon B., Boucher D., Fouix S., Lamour-Isnard C., Plessis A., Busson D. (2006) Dev. Biol. 291, 53–66 [DOI] [PubMed] [Google Scholar]
- 44.Merchant M., Evangelista M., Luoh S. M., Frantz G. D., Chalasani S., Carano R. A., van Hoy M., Ramirez J., Ogasawara A. K., McFarland L. M., Filvaroff E. H., French D. M., de Sauvage F. J. (2005) Mol. Cell. Biol. 25, 7054–7068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen M. H., Gao N., Kawakami T., Chuang P. T. (2005) Mol. Cell. Biol. 25, 7042–7053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murone M., Luoh S. M., Stone D., Li W., Gurney A., Armanini M., Grey C., Rosenthal A., de Sauvage F. J. (2000) Nat. Cell. Biol. 2, 310–312 [DOI] [PubMed] [Google Scholar]
- 47.Maloverjan A., Piirsoo M., Michelson P., Kogerman P., Osterlund T. (2010) Exp. Cell. Res. 316, 627–637 [DOI] [PubMed] [Google Scholar]
- 48.Agren M., Kogerman P., Kleman M. I., Wessling M., Toftgård R. (2004) Gene 330, 101–114 [DOI] [PubMed] [Google Scholar]
- 49.Tsanev R., Tiigimägi P., Michelson P., Metsis M., Østerlund T., Kogerman P. (2009) FEBS Lett. 583, 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maloveryan A., Finta C., Osterlund T., Kogerman P. (2007) J. Cell. Commun. Signal. 1, 165–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rieff H. I., Raetzman L. T., Sapp D. W., Yeh H. H., Siegel R. E., Corfas G. (1999) J. Neurosci. 19, 10757–10766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hellman U. (2000) Exs 88, 43–54 [DOI] [PubMed] [Google Scholar]
- 53.Chan E. Y., Longatti A., McKnight N. C., Tooze S. A. (2009) Mol. Cell. Biol. 29, 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Daub H., Olsen J. V., Bairlein M., Gnad F., Oppermann F. S., Körner R., Greff Z., Kéri G., Stemmann O., Mann M. (2008) Mol. Cell. 31, 438–448 [DOI] [PubMed] [Google Scholar]
- 55.Cheng S. Y., Bishop J. M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5442–5447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paces-Fessy M., Boucher D., Petit E., Paute-Briand S., Blanchet-Tournier M. F. (2004) Biochem. J. 378, 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Evangelista M., Lim T. Y., Lee J., Parker L., Ashique A., Peterson A. S., Ye W., Davis D. P., de Sauvage F. J. (2008) Sci. Signal. 1, ra7. [DOI] [PubMed] [Google Scholar]
- 58.Haycraft C. J., Banizs B., Aydin-Son Y., Zhang Q., Michaud E. J., Yoder B. K. (2005) PLoS Genet. 1, e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.