Abstract
Filamin A (FLNA) is an integrator of cell mechanics and signaling. The spreading and migration observed in FLNA sufficient A7 melanoma cells but not in the parental FLNA deficient M2 cells have been attributed to FLNA. In A7 and M2 cells, the normal prion (PrP) exists as pro-PrP, retaining its glycosylphosphatidyl-inositol (GPI) anchor peptide signal sequence (GPI-PSS). The GPI-PSS of PrP has a FLNA binding motif and binds FLNA. Reducing PrP expression in A7 cells alters the spatial distribution of FLNA and organization of actin and diminishes cell spreading and migration. Integrin β1 also binds FLNA. In A7 cells, FLNA, PrP, and integrin β1 exist as two independent, yet functionally linked, complexes; they are FLNA with PrP or FLNA with integrin β1. Reducing PrP expression in A7 cells decreases the amount of integrin β1 bound to FLNA. A PrP GPI-PSS synthetic peptide that crosses the cell membrane inhibits A7 cell spreading and migration. Thus, in A7 cells FLNA does not act alone; the binding of pro-PrP enhances association between FLNA and integrin β1, which then promotes cell spreading and migration. Pro-PrP is detected in melanoma in situ but not in melanocyte. Invasive melanoma has more pro-PrP. The binding of pro-PrP to FLNA, therefore, contributes to melanomagenesis.
Keywords: Cytoskeleton, Integrin, Prions, Protein-Protein Interactions, Signal Transduction, Filamin A
Introduction
Filamin A (FLNA)3 is a cytolinker, which links cell surface receptors, such as integrins to F-actin filaments, creating an orthogonal actin network that is important in maintaining membrane integrity, cell-cell, and cell-matrix interactions (1, 2). Despite its importance in cellular biology, FLNA is dispensable for cell-autonomous survival. Human melanoma cell lines, such as M2 and M3 do not express FLNA. These cells lack actin fiber bundles and are less mobile in vitro (3). The deficiency is restored in the A7 cell, which is derived from the M2 cell by transfection of a plasmid encoding human FLNA (4). This pair of isogenic cell lines has been used extensively for studying FLNA function. Biological properties observed in A7 cells, but not in M2 cells, are attributed solely to FLNA function (4–11).
The normal prion protein, PrP, is a highly conserved, widely expressed, GPI-anchored cell surface glycoprotein. Although the expression of PrP is essential for the pathogenesis of prion diseases (12, 13), its normal function remains unclear (14). PrP is first synthesized as pre-pro-PrP. After removing the N-terminal signal sequence and the C-terminal GPI-PSS in the endoplasmic reticulum (ER), a GPI anchor and two N-linked glycans are added, thus, forming a mature GPI-anchored PrP.
Recently, we discovered that in human pancreatic ductal adenocarcinoma (PDAC) cell lines, PrP existed as pro-PrP, retaining its GPI-PSS. Unexpectedly, the GPI-PSS of PrP contained a FLNA binding motif. The binding of pro-PrP to FLNA altered the cytoskeleton and signaling events, thus, providing a growth advantage to the PDAC cell lines (15). This conclusion is based on our findings that down-regulation of PrP expression in the PDAC cell lines reduces their proliferation and invasiveness in vitro and their growth in vivo as xenografts in nude mice.
In the PDAC cell lines the failure to remove the GPI-PSS of PrP is not due to a global defect in the GPI anchor machinery; CD55, a normally GPI-anchored protein, remained GPI-anchored in the PDAC cell lines. The FLNA binding motif furthermore is present only on the GPI-PSS of PrP and was absent on the GPI-PSS of 14 other normally GPI-anchored proteins (15). We also rule out the possibility that the failure to remove the GPI-PSS is due to a mutation in the coding region of PRNP.
In a normal human pancreas, PrP was only detectable in islet cells; neither acinar cells nor ductal cells had detectable PrP. About 40% of patients with PDAC, however, expressed PrP in their cancer. Most importantly, these patients had significantly shorter survival times compared with patients whose PDAC lacked PrP (15). We hypothesized that the presence of pro-PrP and its binding to FLNA provided a growth advantage to human pancreatic cancer cells, thus, enabling the tumor cell to become more aggressive. However, the underlying mechanism by which the binding of pro-PrP to FLNA provided a growth advantage to a cancer cell was not known.
The M2 and A7 melanoma cell lines offer a unique model for studying FLNA function. Here, we report that in M2 and A7 cells, PrP also exists as pro-PrP and binds FLNA. One of the best-characterized FLNA binding partners is integrin β1 (16, 17). In A7 cells, the binding of pro-PrP to FLNA enhances the binding of FLNA to integrin β1, ultimately regulating cell spreading and migration. These findings provide a novel mechanism by which the presence of pro-PrP modulates the functionality of FLNA and integrin β1. Finally, we show that pro-PrP is detected in melanoma in situ but not in normal melanocytes. Invasive melanoma in the dermal component has more pro-PrP. We hypothesize that the presence of pro-PrP may be important for the neoplastic transformation of melanocytes and the progression of melanoma. Prevention of this interaction may provide a novel therapeutic target for the treatment of human melanoma.
MATERIALS AND METHODS
Cell Lines
The generation and culture of the melanoma cell lines M2 and A7 have been described (3, 4).
Immunological and Other Reagents
Recombinant PrP (rPrP), recombinant pro-PrP, and other PrP mutant proteins were prepared using conventional molecular biological techniques as described (18). All the anti-PrP and control mAbs have been described (18). Mouse anti-FLNA, anti-human integrin β1, anti-talin, and anti-actin mAbs were purchased from Chemicon. Anti-LIMK1, anti-LIMK2, anti-cofilin, anti-p-cofilin, anti-focal adhesion kinase (FAK), anti-phosphorylated-focal adhesion kinase (Tyr-576, -577), anti-p-Src (Tyr-529), and rabbit anti-calnexin antibodies were purchased from Cell Signaling Technology. Rabbit anti-Src and rabbit anti-FLNA mAbs were purchased from Epitomics. Fluorescein isothiocyanate-labeled goat anti-mouse IgG antibody was purchased from Southern Biotech. All culture medium and supplements, Dulbecco's phosphate-buffered saline (DPBS), trypsin/EDTA, Alexa fluor 488-nm-conjugated goat anti-mouse Ig-specific antibody, Alexa fluor 555-nm-conjugated donkey anti-rabbit Ig-specific antibody, Texas Red-conjugated phalloidin, 4′,6-diamidino-2-phenylidole, dialactate (DAPI), and Bodipy™ F-C5 ceramide BSA complex were purchased from Invitrogen. Protein G-agarose beads were purchased from Roche Applied Science. Profound Co-Ip™ kit, EDTA-free protease inhibitor mixture, dimethyl suberimidate·2HCl, and SuperSignal® West Femto kit were purchased from Pierce. The Bio-Rad protein assay kit was purchased from Bio-Rad. Phenylmethanesulfonyl fluoride (PMSF), Triton X-100, Tween 20, brefeldin A (BFA), and phosphatidylinositol phospholipase C (PI-PLC) were purchased from Sigma.
Immunofluorescent Staining of Tumor Cell Lines for Confocal Microscopic Studies
Tumor cell lines were cultured in poly-d-lysine-coated glass-bottom Petri dishes (MatTek) overnight. Cells were then rinsed 3× with ice-cold DPBS and fixed in 4% paraformaldehyde for 15 min at 20 °C. PrP or FLNA was detected with anti-PrP mAb 8H4 or anti-FLNA mAb PM6/317 (0.01 μg/μl). Bound antibody was detected with an Alexa fluor 488-nm-conjugated goat anti-mouse Ig-specific antibody. For pro-PrP staining, cells treated similarly were stained with a 1:100 dilution of the affinity-purified rabbit anti-PrP-GPI-PSS antiserum. Bound rabbit antibody was detected with an Alexa fluor 555-nm-conjugated donkey anti-rabbit Ig-specific antibody. Nuclei were stained with DAPI. F-actin was detected with Texas Red-conjugated phalloidin. Samples were analyzed on a LSM 510 META confocal microscope at The Case Comprehensive Cancer Center, Image Core Facility. For double staining of FLNA and integrin β1, overnight-cultured cells were fixed as above and washed three times. Rabbit anti-FLNA and mouse anti-integrin β1 antibodies were added to the cultured cells as suggested by the manufacturers. Bound mouse and rabbit antibodies were detected with Alexa fluor 488-nm-conjugated goat anti-mouse and Alexa fluor 555-nm-conjugated donkey anti-rabbit antibodies. For double staining of pro-PrP and FLNA, affinity-purified rabbit anti-pro-PrP and mouse anti-FLNA were added (0.01 μg/μl). Bound antibodies were detected as above. Double staining of FLNA and PrP was performed as described previously (15). To locate PrP in Golgi or ER, PrP was detected as above, Golgi was detected with Bodipy™ F-C5 ceramide BSA complex, and ER was detected with rabbit anti-calnexin. Bound antibodies were detected with Alexa fluor 555-nm-conjugated donkey and rabbit antibody. Experiments were repeated at least twice with comparable results.
Phosphatidylinositol Phospholipase C, BFA Treatment of Tumor Cells, and Flow Cytometry Analysis
Tumor cells were seeded overnight as described. The next day tumor cells were first washed three times with ice-cold DPBS and then treated with trypsin/EDTA to prepare a single cell suspension of the tumor cells. After washing twice with DPBS, cells were incubated with phosphatidylinositol phospholipase C (500× dilution of 1 unit) at 37 °C for 1 h. At the end of the incubation, cells were washed twice with DPBS and then stained with control antibody or 8H4 as described (15). All samples were then analyzed in a BD Biosciences FACS™ flow cytometer. For BFA treatment of cells, 0.35 μm BFA were added in cultured cells for specified periods of time in culture condition. Cell surface PrP staining was performed as described in above.
Immunoblotting and Enzymatic Treatment of PrP
Tumor cells were seeded overnight, and lysates were prepared in lysis buffer containing 20 mm Tris (pH 7.5), 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 2.5 mm sodium pyrophosphate, 1 mm β-glycerol phosphate, 1 mm Na3VO4, and 1 mm PMSF, and an EDTA-free protease inhibitor mixture was added just before cell lysis. PrP was subjected to peptide N-glycosidase F treatment according to the protocols provided by the provider. After treatment, samples were separated on SDS-PAGE and immunoblotted with an anti-PrP mAb. To detect GPI-PSS on PrP, mAb 8B4 affinity-purified PrP was separated and detected with affinity-purified rabbit anti PrP-GPI-PSS.
To study signal transduction molecules change in PrP down-regulated cells, control and PrP knocking down cells were grown overnight. The next morning cells were rinsed three times with DPBS and incubated with Opti-MEM (Invitrogen) for 30 min. After aspirating off the medium, fresh culture medium were added, and the cells were incubated for additional 10 min. Cell lysate was made as above. Signal transduction molecules were detected using various mAbs and the protocols suggested by manufacturers.
To identify proteins that are bound to PrP, cell lysates were prepared in co-immunoprecipitation buffer (Cell Signaling Technology). Immunoprecipitation was performed with anti-PrP mAb 8B4 or control, irrelevant mAb D7C7 that was conjugated to-Sepharose beads. Bound proteins were eluted using IgG elution buffer (Pierce). The eluted proteins were then separated by SDS-PAGE (12% gel, Bio-Rad) and then immunoblotted with anti-FLNA or anti-integrin β1 mAb. Bound antibody was detected with a goat anti-mouse-horseradish peroxidase antibody using the chemiluminescence blotting system (Pierce). Similar approaches were used to determine whether FLNA co-purifies with integrin β1 using anti-integrin β1 mAb. Experiments were repeated at least twice with comparable results.
Competition of Co-immunoprecipitation with Synthetic Peptide
400 μl of cell lysate from each cell type was loaded into the mAb 8B4 column. Because the PrP GPI-PSS is rather insoluble, a KKRPK motif was added to the N terminus of the PrP to increase is solubility. Control peptide also has the KKRPK motif followed by 21 irrelevant amino acids. Synthetic peptides in the indicated amounts were also added as well as 4 μl of PMSF and 1 μl of DMSO/column. The columns were placed at 4 °C overnight with gentle rocking, and bound proteins eluted as described by us (15). Eluted proteins were separated in a 4–20% Tris-glycine gel, transferred to nitrocellulose membrane, and then blotted with anti-FLNA mAb (15).
Binding of Individual FLNA Domains to Pro-PrP
All the recombinant FLNA proteins and the individual FLNA domains were prepared as described (19). In in vitro pulldown assays, 250 ng of GST-tagged FLNA was mixed with 1.2 μg of recombinant PrP23–253 or recombinant rPrP mutants in 400 μl of binding buffer (20 mm Tris·HCl (pH 7.4), 150 mm NaCl, 1 mm EGTA, and 0.1% Tween 20). The tubes were rocked slowly and incubated at 20 °C for 1 h. Then 3 μg of anti-PrP mAb 8H4 was added and incubated for another hour with gentle rocking. 10 μl of protein G-agarose beads (pre-equilibrated with binding buffer) was then added for 30 min. The beads were washed ×5 with binding buffer for 5 min. Subsequently, beads were resuspended in 15 μl of 2× SDS loading buffer and boiled at 95 °C for 10 min. The proteins were separated on a 4–20% Tris-glycine gel and then transferred to nitrocellulose membrane. FLNA was detected with anti-GST tag mAb (Sigma, 1:1000 dilution, 4 °C overnight). After second antibody incubation and washing, the membrane was developed as described above. On the same membrane, input recombinant rPrPs were detected with anti-PrP mAb 8B4. All experiments were repeated at least twice with comparable results.
In Vitro Proliferation of Tumor Cell Lines
Single cell suspension of the tumor cells were prepared and counted. Equal numbers of the tumor cells (1 × 104) were then platted in a 96-well tissue culture plates in triplicates. At different times after culture, the cells from each well were harvested and counted. The results presented are the mean ± S.E. of two experiments.
In Silico Modeling of FLNA Domain 23 Bound to GPI-PSS Peptide
The complex of FLNA-23 with bound PrP GPI-PSS peptide was modeled with HOMODGE in BODIL (20) by using FLNA-21 with bound integrin β7 peptide (21) (PDB code 2BRQ) as a template structure. Intramolecular and intermolecular interactions at the interaction areas between FLNA-23 and bound PrP GPI-PSS peptide were optimized by using side-chain rotamer library (22) incorporated within BODIL.
Down-regulation of PrP Expression by shRNA
For the inducible system, we used the BLOCK-iT®-inducible H1 lentiviral RNAi system (Invitrogen) to generate PrP down-regulated M2 or A7 cell lines by following the manufacturer's guideline. The sequence of shRNA-10 has been described (15). The sequence of shRNA-12 was 5′-gctcagtatactaatgccctatctt-3′, the sequence of inducible control shRNA, (shRNA-Cont) was 5′-acccgacaacatttctgccaggttt-3′. For non-inducible knockdown, shRNA-10, shRNA-12, or a scramble sequence were directly transfected into M2 and A7 cells using protocols suggested by the manufacturer. 48 h after transfection 50 μg/ml Geneticin was added to the medium. The PrP expression levels were quantified by immunoblot with anti-PrP mAb. Experiments were repeated at least three times with independently derived, pooled, transfectants. The constitutive system was carried out as described (15).
Spreading Assay
To study the effects of PrP down-regulation on cell spreading, a single suspension of PrP down-regulated or control cells was seeded in a 24-well tissue culture plate in triplicate and cultured for 2 h. The cells were then counted in a Zeiss Axiovert microscope. The results presented were the mean ± S.D. of two experiments. To study peptide effects on cell spreading, cells were cultured with the PrP GPI-PSS synthetic peptide, KKRPKPPVILLISFLIFL-IVG (Peptide 2), or a control peptide, KKRPKDMDYLPRVPNQG-IIIN-PMLSD (Peptide2), overnight at specified concentrations. After that, single cell suspensions were prepared and counted. Same numbers of cells were plated in 24-well tissue culture plates in triplicate. Thirty minutes later the numbers of cells with adherent morphology or in suspension in each well were counted in a microscope. The results presented were the mean of the two experiments ± S.E.
Wound Healing Assay
To study the effects of PrP down-regulation on cell migration, shRNA-10 and control cells were seeded and allowed to grow to confluence then either induced or not induced. A wound was inflicted by scraping across the cell layer with a 200-μl sterile peptide tip. The cells were incubated for various lengths of time followed by imaging on a Zeiss Axiovert 200 microscope equipped with an AxioCam digital camera system. Average wound area was quantified in the picture using ImageJ software (means ± S.E. of the two experiments). Inhibition of cell migration was determined by comparing the healed area of control cells with the healed areas of PrP down-regulated cells. To study the effects of peptide on wound healing, the same approach was adopted. A7 cells grown to confluence were inflicted with a 200-μl pipette tip. Inhibition of cell migration was determined by comparing with the healed area of non-treated cells with the healed areas of cells treated either with various concentrations of the PrP GPI-PSS synthetic peptide or a control, irrelevant synthetic peptide. All peptides were added right after the creation of the wound. Percent inhibition was calculated as 100 × (one-healed area of treated cells/healed area of non-treated cells). Experiments were repeated twice with comparable results.
Immunohistochemical Staining of Tissue Biopsies
Unstained 5-μm sections were cut from the paraffin blocks of selected cases and de-paraffinized using standard techniques. Slides were treated with 1× sodium citrate buffer (diluted from 10 × heat-induced epitope retrieval buffer; Ventana-Bio Tek Solutions, Tucson, AZ) before heating for 20 min in a microwave. Slides were then cooled at room temperature for 20 min and incubated with 3% w/v H2O2 for 10 min. The anti-PrP mAb 8H4 or affinity-purified polyclonal rabbit anti-PrP GPI-PSS antibody was then added and incubated at room temperature for 1 h. An irrelevant monoclonal antibody or normal rabbit Ig was included in all the experiments as a negative control. After serial washing, a biotinylated goat anti-mouse Ig antibody or a biotinylated goat anti-rabbit Ig antibody was added followed by the avidin-biotin complex and 3,3′-diaminobenzidine chromagens (Dako Inc.). Sections were counterstained with hematoxylin. Each slide was coded and evaluated independently. The cytoplasmic and membrane staining intensity and distribution of each sample were graded as highly positive (>50% neoplastic cells stained strongly positive) or negative (<5% neoplastic cells stained). The study has been approved by the Institutional Review Board for Human Investigation of the University Hospital Case Medical Center, Cleveland, OH.
Statistics
Student's t test or analysis of variance test were applied, p < 0.05 was considered statistically significant.
RESULTS
PrP Exists as Pro-PrP in M2 and A7 Cells
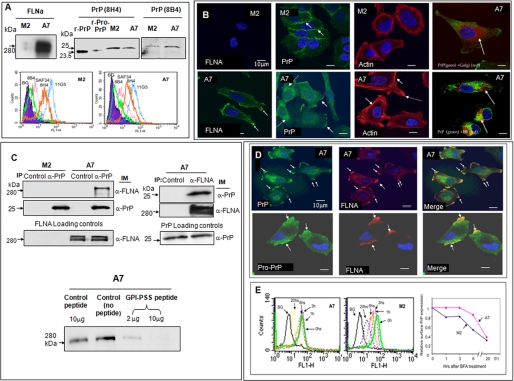
As expected, FLNA was detected only in A7 cells but not in M2 cells (Fig. 1A). However, both M2 and A7 cells expressed PrP. A glycosylated, GPI-anchored PrP has a molecular mass of about 34 kDa (15) (supplemental Fig. S1). In contrast, PrP from M2 and A7 cells migrated as a 26-kDa protein (Fig. 1A). PrP was present on the cell surface of M2 and A7 cells at a comparable level, as judged by immunofluorescent staining of M2 and A7 cells with multiple anti-PrP mAbs with distinct epitopes (Fig. 1A). Subsequently, we find that PrP is unglycosylated and exists as pro-PrP in M2 and A7 cells, as in PDAC. This conclusion is based on the following; 1) treatment of PrP with endoglycosidase-F did not alter its mobility in SDS-PAGE (supplemental Fig. S1B); 2) the cell surface PrP was resistant to phospholipase C (supplemental Fig. S1C); 3) the PrP reacted with a polyclonal antiserum that is specific for the GPI-PSS of PrP (supplemental Fig. S1D).
FIGURE 1.
Expression of FLNA and pro-PrP in M2 and A7 cells. A, immunoblots show that only A7 cells express FLNA. Both M2 and A7 cells express PrP. PrP from M2 and A7 cells has a molecular mass of about 26 kDa. A recombinant mature PrP23–231 (r-PrP) and a recombinant pro-PrP23–253 (r-pro-PrP) were included as molecular mass markers. Pro-PrP from A2 and M7 cells migrates slower than recombinant pro-PrP23–253. This likely reflects the conformational difference between recombinant pro-PrP23–253 and native pro-PrP. Recombinant pro-PrP23–253 has to be solubilized and refolded in urea, which might cause the protein to be more compacted. This experiment was repeated at least three times with comparable results. Histograms show that PrP on the cell surface of M2 and A7 cells reacts with multiple anti-PrP mAbs. The epitopes of the mAbs are shown in supplemental Fig. S1A. BG, background staining with irrelevant mAb D7C7. This experiment was repeated at least three times with comparable results. B, microscopic images show the expression of FLNA in A7 cells but not in M2 cells. The distributions of PrP and actin in M2 and A7 cells also differ. For organelle-specific markers, a rabbit anti-calnexin antibody was used to mark the ER, and a Bodipy™F-C5 ceramide-BSA was used to locate the Golgi. This experiment was repeated at least three times with comparable results. C, immunoblots show that in A7 cells PrP co-purifies with FLNA and vice versa. Loading controls show the levels of PrP or FLNA in cell lysates before co-immunoprecipitation (IP) and an immunoblot (IM). This experiment was repeated at least three times with comparable results. Immunoblots show that co-purification of FLNA with PrP can be competed with a PrP GPI-PSS synthetic peptide but not a control peptide. This result was confirmed at least twice. D, microscopic images show the co-localization of pro-PrP and FLNA in A7 cells. This result was confirmed at least three times. E, histograms show that cell surface PrP on A7 cells has a longer half-life. A7 and M2 cells were cultured with BFA for various lengths of time to prevent the transit of newly synthesized PrP to the cell surface. At different times after culture, cells were prepared and stained with an anti-PrP mAb, 8H4. The levels of cell surface PrP were then quantified by flow cytometry. Mean fluorescent intensities of control cells or cells treated with BFA at time 0 were arbitrarily defined as 1. This result was confirmed at least twice.
Results of immunofluorescent staining further confirms the presence of FLNA in A7 cells but not in M2 cells (Fig. 1B). In A7 cells, FLNA was associated with the inner membrane leaflet in the leading edges (Fig. 1B, bottom panel, arrows identify leading edges). The spatial distributions of PrP also differed noticeably in M2 and A7 cells. In M2 cells, most of the PrP was on the cell membrane, in areas of cell-cell contact (Fig. 1B, arrows show cell-cell contact areas). In contrast, PrP was concentrated in the membrane ruffle areas in A7 cells (Fig. 1B, solid arrows show leading edges) and in the cytosol (Fig. 1B, dashed arrows). The distribution of actin filaments also differed between A7 and M2 cells. In M2 cells, actin filaments were distributed around the periphery of the cells (Fig. 1B). In A7 cells, the actin filaments were better organized with readily identifiable stress fibers that were concentrated at the leading edges (Fig. 1B, solid arrows; the dashed arrow shows the leading edge) in the same area where PrP and FLNA were localized. In the cytosol, staining with organelle-specific markers revealed that some, but not all of the cytosolic PrP was associated with the Golgi and the ER (Fig. 1B, arrows show co-localization), suggesting the presence of PrP in other cellular compartments in addition to the Golgi and the ER.
Next we confirmed that pro-PrP indeed interacts with FLNA in A7 cells by co-immunoprecipitation. Immunoblotting of proteins co-purified with PrP using an anti-FLNA-specific mAb detects FLNA (Fig. 1C, left top panel). Conversely, immunoblotting of proteins co-purified with FLNA in A7 cells using an anti-PrP mAb identified PrP (Fig. 1C, right top panel). Finally, co-purification of PrP and FLNA was competitively inhibited by a synthetic peptide corresponding to the GPI-PSS of PrP but not by an irrelevant peptide (Fig. 1C, bottom panel). PrP and FLNA also co-localized in the leading edges of A7 cells (Fig. 1D, arrows show co-localization). Staining with the anti-PrP-GPI-PSS antibody showed the co-localization of pro-PrP and FLNA (Fig. 1D, arrows show co-localization).
Because cell surface PrP is physically associated with FLNA in A7 cells, we hypothesize that PrP on the cell surface of A7 cells may be more stable and, thus, have a longer half-life. We find that on the cell surface of M2 cells, PrP has a half-life of about 5–6 h. In contrast, the half-life of cell surface PrP on A7 cells is about 10–12 h (Fig. 1E). Thus, anchoring of PrP to FLNA indeed stabilizes PrP on the cell surface.
Identifying the Interacting Domains on Pro-PrP and FLNA
We find that a full-length FLNA1–24 dimer binds pro-PrP (15). Subsequently, we found that the pro-PrP binding domain on FLNA is located between domains 10 and 24 (supplemental Fig. S2). Next, we prepare individual FLNA domains with a GST tag, which allows the individual domains to dimerize, and then determined which domain binds pro-PrP. We found that pro-PrP binds domains 10, 16, 17, 18, 20, 21, or 23 but not domains 11, 19, 22, or 24 (Fig. 2A). Pro-PrP appears to bind domain 23 better than other FLNA domains.
FIGURE 2.
Identification of the pro-PrP binding domains on FLNA and the motif in the GPI-PSS that binds FLNA. A, immunoblots of in vitro pulldown assays show that recombinant pro-PrP23–253 (r-pro-PrP) binds to domains 10, 15, 16, 17, 18, 20, 21, and 23 but not domains 11, 19, 22, or 24 of FLNA. This result was confirmed at least three times. IP, immunoprecipitation; IM, immunoblot. B, immunoblots of in vitro pulldown assays show that pro-PrP lacking the last five residues is unable to bind FLNA. Furthermore, replacing two of the three polar residues in the last eight residues completely eliminated FLNA binding. This result was confirmed at least twice. C, sequence alignment shows that the FLNA binding site on the GPI-PSS of PrP from different mammalian species is highly conserved. D, shown is an in silico model of the binding interface between the GPI-PSS of PrP and domain 23 of FLNA. Amino acids at C and D strands of domain 23 of FLNA (gray carbon atoms, black letters and numbers) and PrP GPI-PSS (green carbon atoms, magenta letters and numbers) are shown as ball-and-stick. Multiple hydrogen bonds exist in the binding interface (small yellow dots).
The GPI-PSS of PrP is composed of 22 amino acids. We created recombinant pro-PrP proteins that were truncated at different positions in the C terminus to precisely pinpoint the binding motif (Fig. 2B). We found that pro-PrP lacking the last five residues was unable to bind FLNA.
The FLNA ligand binding interface has multiple hydrophobic, non-polar amino acid contact residues (19, 21, 23). Based on this information, we predict that Phe-246, Phe-249, or Leu-250 of the GPI-PSS may be critical for binding FLNA. This region is highly conserved among mammalian PrPs (Fig. 2C). To test this hypothesis, we replaced these non-polar residues either individually or in combination with polar residues, such as Trp or Tyr. Replacing residues 246, 249, or 250 (Fig. 2B, lanes 4–6) individually did not disrupt the binding of FLNA. However, replacing both 246 and 250 completely eliminated the FLNA binding activity (lane 7). Thus, non-polar residues at 246 and 250 are important for FLNA binding. An in silico model of a PrP GPI-PSS from residues 241–252 bound to domain 23 of FLNA (Fig. 2D) showed that in addition to the non-polar amino acids, hydrogen bonds involving other residues on the GPI-PSS and FLNA may also contribute to the interaction.
Interaction of PrP and FLNA Modulates the Cytoskeleton and Regulates Cell Spreading and Migration
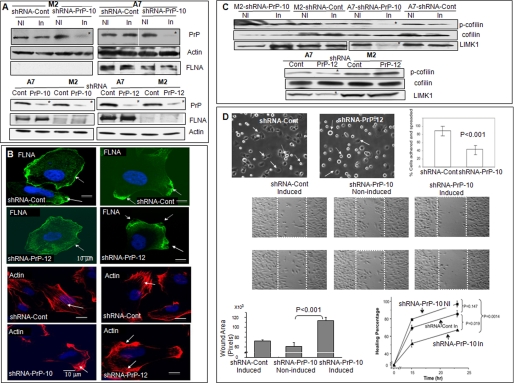
Next, we used PrP-specific shRNA to down-regulate PrP expression in M2 and A7 cells. One inducible PrP-specific sequence, shRNA-PrP-10, inhibited the expression of PrP in M2 and A7 cells by about 80–90% (Fig. 3A). A control-inducible shRNA (shRNA-Cont) inhibited PrP expression by less than 5% (Fig. 3A). Similarly, constitutively active PrP-specific shRNA-PrP-10 and shRNA-PrP-12 inhibited the expression of PrP by about 50–70% in M2 and A7 cells (Fig. 3A). In subsequent experiments, cells transfected with either inducible control shRNA or constitutively active “scrambled” shRNA were used as controls and are referred to as shRNA-Cont.
FIGURE 3.
Effects of reducing PrP expression in M2 and A7 cells. A, immunoblots show that down-regulation of PrP in M2 and A7 cells reduces the levels of PrP in both cell lines but does not reduce the expression level of FLNA in A7 cells. Top panels show shRNA under the control of an inducible promoter. (shRNA-Cont, control; shRNA-PrP-10, prp knocking down sequence 10; NI, non-induced; In, induced). Bottom panels show constitutively active shRNA. This result was independently confirmed at least twice. B, microscopic photos show that down-regulation of PrP alters the spatial distribution of FLNA and organization of actin filaments in A7 cells. This result was confirmed twice. C, down-regulation of PrP reduces the expression levels of p-cofilin and LIMK1 in A7 cells but not in M2 cells. Top panels show shRNA under the control of an inducible promoter (Cont, knocking down control; PrP-12, PrP knocking down sequence 12). Bottom panels show constitutively active shRNAs. This result was confirmed twice. D, top panels, microscopic photos show that PrP-down-regulated A7-shRNA-PrP-12 cells are much less adhesive compared with control A7-shRNA-Cont cells. Solid arrows identify adherent cells, and dashed arrows identify non-adherent, floating cells. Original magnification (OM), 10×10. The graph shows quantification of the spreading results of another experiment comparing PrP down-regulated A7-shRNA-PrP-10 with control A7-shRNA-Cont cells. Results presented were the average of the two experiments ± S.D. Middle panels, microscopic photos show that PrP down-regulated A7 cells have reduced cellular migration in a wound-healing assay. Photos were taken at 15 h post-wound initiation. Two representative wells from each condition were shown. Areas between two white lines mark the original wound area. OM, 10×10. This result was confirmed twice. Bottom panels, bar graph, quantification of the wound-healing assay was taken at 15 h post-wound. PrP down-regulated A7 cells have a reduced would-healing capacity. Results presented were the average ± S.E. of two different experiments. Line graph, quantification of the would-healing assay at different time points after wound healing is shown. Results presented were the average of two different experiments ± S.E. p < 0.05 was considered to be statistically significant. The migration of shRNA-PrP-10-induced A7 cells is significantly slower (p = 0.0014) compared with shRNA-PrP-10 non-induced A7 cells.
Reducing PrP expression in A7 cells did not change the expression level of FLNA (Fig. 3A) but altered the spatial distribution of FLNA. In control cells, FLNA was either associated with the membrane (Fig. 3B, first row, see arrows) or concentrated at the leading edges (Fig. 3B, first row, see arrows). In some PrP down-regulated A7 cells, the level of membrane-associated FLNA was decreased (Fig. 3B, second row, see the arrow); in others, FLNA appears to dissociate from the inner membrane leaflet (Fig. 3B, second row, see arrows). These staining patterns are similar to the patterns seen in PrP down-regulated PDAC cell lines (15).
Next, we determine whether down-regulation of PrP alters actin filament organization in A7 cells. In control A7 cells, the actin filaments were well organized (Fig. 3B, third row, arrows identify actin filaments). In PrP down-regulated A7 cells, the actin filaments were less organized and concentrated in certain areas of the cells (Fig. 3B, fourth row, see arrows).
Cofilin regulates actin filament organization by controlling its polymerization and de-polymerization (24). Two kinases, LIMK1 and -2 phosphorylate and inactivate cofilin (25). Accordingly, we find that the levels of p-cofilin and LIMK1 were reduced by more than 80% (n = 3) in PrP down-regulated A7 cells (Fig. 3C, marked with as asterisk) but not in similarly down-regulated M2 cells. LIMK 2 is undetectable in M2 and A7 cells (not shown).
Binding of pro-PrP to FLNA may be the underlying mechanism by which A7 cells spread and migrate more efficiently than M2 cells. To test this, we first confirm that A7 cells indeed migrate more readily than M2 cells in a wound-healing assay (supplemental Fig. S3). Second, we demonstrate that down-regulation of PrP does not change the proliferation of M2 and A7 cells at 2 and 4 days after culture (supplemental Fig. S4). Finally, we compare the spreading and migration of PrP down-regulated A7 cells with control A7 cells. Reducing PrP expression greatly diminished the spreading (Fig. 3D) and migration of A7 cells (Fig. 3D). Therefore, the enhanced cell spreading and migration observed in A7 cells are indeed due to binding of pro-PrP to FLNA.
Pro-PrP Enhances Binding of FLNA to Integrin β1 and Regulates Cell Spreading and Migration
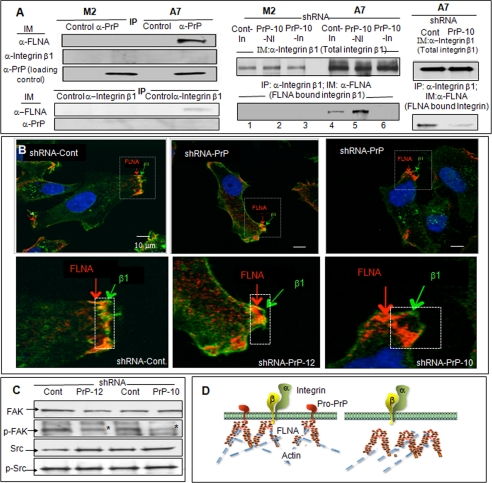
Integrins are bidirectional, signaling molecules that control cell spreading and migration (26, 27). FLNA binds to the cytoplasmic tail of the integrin β chain (16, 28, 29) and modulates cell adhesion and migration (17, 30, 31). We find that A7 cells have more integrin β1 than M2 cells. However, talin, another binding partner of integrin β1, was expressed at a comparable level (supplemental Fig. S5A). Because FLNA binds pro-PrP as well as integrin β1, we investigated whether FLNA, integrin β1 and PrP co-exist in a complex in A7 cells. Using immunofluorescent staining, it appears that some PrP and integrin β1 do co-localize (supplemental Fig. S5B). By co-immunoprecipitation, FLNA co-purifies with PrP, and FLNA also co-purifies with integrin β1. However, we are unable to co-purify integrin β1 with PrP (Fig. 4A). Thus, FLNA, integrin β1, and PrP appear not to co-exist in a stable, trimeric complex. They exist in two independent complexes, one containing FLNA and PrP, the other containing FLNA and integrin β1.
FIGURE 4.
Co-purification of FLNA and integrin β1 and the effects of down-regulation of PrP on integrin β1 and FLNA association. A, left panels, immunoblots (IM) show that PrP co-purifies (IP) with FLNA, and integrin β1 co-purifies with FLNA, but PrP does not co-purify with integrin β1 in A7 cells. This result was confirmed twice. Right panels, immunoblots show that down-regulation of PrP does not alter the expression levels of integrin β1 (top panel) but reduced the amounts of integrin β1 co-purified with FLNA (bottom panel) in the inducible shRNA model (lanes 1–6) and in the constitutively active model. This result was confirmed twice. B, microscopic images show co-localization of FLNA and integrin β1 in control A7 cells. But in PrP down-regulated A7 cells, integrin β1 is separated from FLNA. This result was independently confirmed twice. C, immunoblots show that down-regulation of PrP in A7 cells reduces the level of phosphorylated focal adhesion kinase (p-FAK) but not p-Src. This result was confirmed at least twice. D, a drawing shows that in A7 cells binding of FLNA to pro-PrP pulls FLNA closer to the inner membrane leaflet, which then promotes the binding to FLNA to integrin β1. When the level of PrP is reduced, FLNA retracts from the inner membrane leaflet, which disrupts the binding of FLNA to integrin β1.
Next, we investigates whether expression of PrP influences the level of FLNA bound to integrin β1. First, we showed that PrP down-regulated A7 cells and control A7 cells have comparable levels of cell surface integrin β1 (supplemental Fig. S6) as well as total integrin β1 (Fig. 4A, top panels). Next, we compared the amount of FLNA co-purified with integrin β1 in control A7 cells with the amount co-purified in PrP down-regulated A7 cells. We found that the amount of FLNA co-purified with integrin β1 is greatly reduced in A7 cells with inducible PrP-specific shRNA-PrP-10 as well as in A7 cells with constitutively active PrP-specific shRNA-PrP-10 (Fig. 4A, bottom panels). Results with shRNA-PrP-12 down-regulated A7 cells are comparable (not shown). Therefore, although down-regulation of PrP does not alter the expression of total integrin β1, it does reduce the amount of integrin β1 bound to FLNA. In PrP down-regulated A7 cells FLNA is dissociated from the inner membrane leaflets (Fig. 3B). We suggest that this spatial change may be the reason that less integrin β1 is bound to FLNA in PrP down-regulated A7 cells. This hypothesis is supported by results of immunofluorescent staining of integrin β1 and FLNA in PrP down-regulated A7 cells. In control cells, integrin β1 and FLNA co-localize at the leading edges (Fig. 4B, bottom photos were larger images of the dashed rectangle in the top photos). In PrP down-regulated A7 cells, FLNA is dissociated from the cell surface, and cell surface integrin β1 is no longer co-localized with FLNA.
The focal adhesion kinase is a critical component of the integrin-signaling cascade (32). Accordingly, the level of phosphorylated focal adhesion kinase was reduced by about 30–50% (n = 3) in PrP-down-regulated A7 cells (Fig. 4C, marked by an asterisk). However, the level of p-Src, which is also important in integrin-signaling, did not change. A hypothetical drawing that depicts the interactions between pro-PrP, FLNA and integrin β1 in control and in PrP down-regulated A7 cells is shown in Fig. 4D.
A PrP GPI-PSS Synthetic Peptide Blocks Cell Spreading and Migration
We reported earlier that a pentapeptide, KKRPK, had cell-penetrating capacity in a Ca2+-dependent manner (33). Recently we found that if the cells were incubated with the peptide for an extended period, the peptide entered the cells without Ca2+ (not shown). We hypothesized that if we added a KKRPK motif to the N terminus of the GPI-PSS, the peptide might be able to enter cells and competed for the binding of pro-PrP to FLNA.
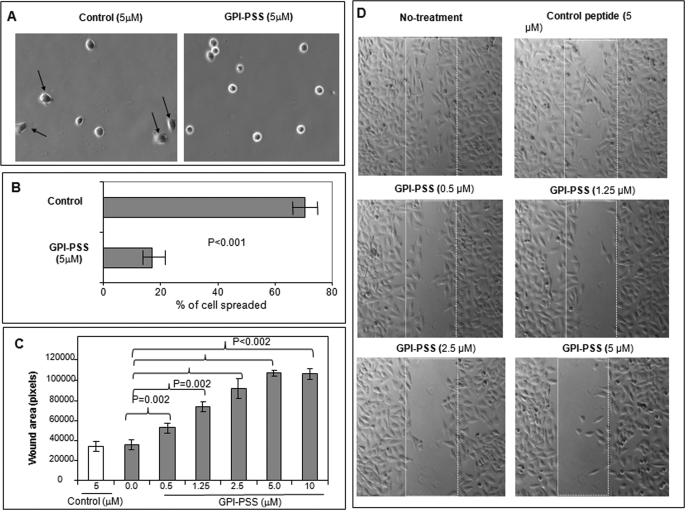
The KKRPK-GPI-PSS synthetic peptide was not toxic and did not alter cell surface expression of either PrP or integrin β1 (supplemental Fig. S7A) or the total level of PrP, FLNA, or integrin β1 in A7 cells (supplemental Fig. S7B). However, when A7 cells were incubated with the peptide, their spreading (Fig. 5, A and B) and migration (Fig. 5, C and D) were significantly reduced. Control peptide did not interfere with either cell spreading or migration. Thus, binding of pro-PrP to FLNA is important for A7 cell spreading and migration, and this interaction can be partially inhibited by the KKRPK-PrP-GPI-PSS synthetic peptide.
FIGURE 5.
A PrP-GPI-PSS synthetic peptide inhibits A7 cell spreading and migration. A, microscopic images show that incubation with a KKRPK-PrP-GPI-PSS synthetic peptide reduces A7 cell spreading. Arrows identify adherent cells. OM, 10×10. B, quantification of the cell spreading results. Results presented were the average ± S.E. of two experiments. OM, 10×10. C, quantification of a wound-healing assay at 15 h after the initiation of wound which shows that significant inhibition of cell migration could be achieved with as little as 0.5 μm concentrations of the peptide. Five images of every concentration were used to calculate the p values by analysis of variance one-way test. D, representative photographs of microscopic images taken at 15 h after initiation of wound, which show that incubation with a KKRPK-PrP-GPI-PSS synthetic peptide reduces A7 cell migration.
Pro-PrP Is Detected in Melanoma in Situ but Not in Melanocytes
Whether melanocyte or melanoma expresses PrP is not known. Therefore, we next investigate whether melanocytes or melanoma from patients with melanoma express PrP and, if they do, whether it is normal PrP or pro-PrP (Table 1). Melanocytes did not react with either the anti-PrP mAb (Fig. 6A, arrows (m) identify melanocytes, section-stained n = 5) or the polyclonal anti-GPI-PSS antibody (Fig. 6B, section-stained n = 5). Intra-dermal benign nevus also did not react with either the anti-PrP mAb (Fig. 6C, section-stained, n = 5) or the anti-GPI-PSS antibody (not shown, section-stained, n = 5). However, both melanoma in situ and invasive melanoma (Fig. 6, D–F, arrows (M) and dashed circles identify melanoma) showed moderate to strong anti-PrP mAb immunoreactivity. Invasive melanoma cells clearly showed the strongest membrane staining (Fig. 6F, magnified from the red dashed circle in D). The anti-GPI-PSS antibody also reacted strongly with melanoma in situ as well as invasive melanoma (Fig. 6, G–I). Invasive melanoma cells in the dermal component consistently had the strongest immunoreactivity (Fig. 6G). Staining with control antibodies only show background staining (not shown). Collectively, these results suggest that expression and accumulation of pro-PrP occurs during the neoplastic transformation of melanocyte; as the tumor cells invade the dermal component, they express higher levels of pro-PrP.
TABLE 1.
Human melanoma stained positive for PrP and pro-PrP
| Melanoma in situ (n = 11) |
Invasive melanoma (n = 9) |
||||
|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||
| PrP (n = 11)a | 11 | 0 | PrP (n = 9) | 9 | 0 |
| Pro-PrP (n = 7)b | 7c | 0 | Pro-PrP (n = 6) | 6d | 0 |
a Samples stained with anti-PrP mAb 8H4.
b Samples stained with anti-PrP GPI-PSS antibody.
c Of the 11 melanoma in situ samples, 7 were stained with the anti-GPI-PSS antibody; all 7 were positive.
d Of the nine invasive melanoma samples, six were stained with the anti-GPI-PSS antibody; all six were positive.
FIGURE 6.
Detection pro-PrP in melanoma in situ and invasive melanoma but not in melanocyte. A, immunostaining with anti-PrP mAb shows that PrP is undetectable in melanocytes (m). The brown spot identified by a dashed arrow is nonspecific staining. OM, ×400. B, melanocytes also do not react with the anti-GPI-PSS antibody. OM ×400; C, intra-dermal benign nevus does not react with anti-PrP mAb. OM × 400. D, Anti-PrP mAb reacts with melanoma in situ as well as invasive melanoma. OM ×200. E, anti-PrP mAb reacts with invasive melanoma cells (M) but not with melanocytes (m) in the same field ×400. F, invasive melanoma cells show strong cell surface staining. OM ×400. G, the anti-GPI-PSS antibody reacts with melanoma in situ and invasive melanoma. Invasive melanoma cells in the dermal component have the strongest staining. OM ×200. H, the anti-GPI-PSS antibody reacts with melanoma in situ. OM ×400. I, the anti-GPI-PSS antibody reacts strongly with invasive melanoma cells; OM ×400. The number of samples is listed in Table 1.
DISCUSSION
Biological responses observed in A7 cells, but not in M2 cells, have been attributed solely to the function of FLNA. Herein, we report that in A7 cells FLNA does not act alone. PrP modulates FLNA functionality by enhancing its binding to integrin β1, which then promotes melanomagenesis.
In M2 and A7 cells, PrP also exists as pro-PrP retaining its GPI-PSS. The reason that PrP retains its GPI-PSS in these cells is unknown. Sequencing of PRNP in these cells revealed no mutation (not shown). The synthesis, processing, and transit of GPI-anchored PrP are complex and not completely understood (34). Pro-PrP is undetectable in cells in which PrP is expressed as a GPI-anchored protein (15). Thus, the processing or turnover of pro-PrP must be rapid. Compared with the GPI-PSS of other GPI-anchored proteins, the GPI-PSS of PrP is intrinsically inefficient in accepting the lipid anchor (35). A slight defect in the GPI assembly pathway, therefore, will have a more dramatic effect on PrP than other GPI-anchored proteins. Deficiency in the ER quality control system or the proteasome may also contribute to the amassing of pro-PrP in these cells.
As expected in A7 cells, pro-PrP co-purified and co-localized with FLNA. Pro-PrP on the A7 cell surface had a longer half-life than the pro-PrP on M2 cells. This result is consistent with earlier reports showing that FLNA regulated the stability, internalization, and trafficking of its cell surface binding partners (6, 8–11). In addition, in A7 cells cytosolic PrP may also interact with FLNA. The significance of this interaction is less clear.
In vitro, recombinant Pro-PrP binds multiple individual, recombinant FLNA domains. Some of these binding sites are unique to pro-PrP, whereas others are shared between pro-PrP and other FLNA binding partners (19, 21, 23). However, it is not known whether in a living cell an intact dimeric FLNA can simultaneously bind multiple pro-PrP molecules. Based on our in silico model, it is also not clear why the mutant pro-PrP (L250Y/F246W), with two non-polar residues replaced, is unable to bind FLNA. In this model the Phe-246 points toward the solvent, away from the binding interface, and it should not disrupt the binding interface. One possible explanation may be the stoichiometry of the binding; one pro-PrP may bind more than one FLNA or vice versa. The non-polar residues may be important in these interactions.
Reducing PrP expression in A7 cells also altered the distribution of FLNA and organization of the actin filaments. In these cells, the levels of p-cofilin and LIMK1 were also reduced (Fig. 3C). Because the levels of p-cofilin and LIMK1 did not change in PrP down-regulated M2 cells, the effect of reduced p-cofilin in PrP down-regulated A7 cells was indeed dependent on the binding of pro-PrP to FLNA. If the effects were simply due to a reduction in PrP expression, a reduced p-cofilin level in PrP down-regulated M2 cells would have been observed. Down-regulation of PrP expression in A7 cells reduced their spreading and migration, suggesting that PrP was intimately involved in these cellular behaviors.
FLNA binds integrin and regulates cell spreading and migration (16, 17, 28, 30, 31). A7 cells have more β1 integrin than M2 cells. This increase is likely acquired during the in vitro selection processes during the establishment of the A7 cell line. Transient expression of FLNA does not up-regulate integrin β1 in M2 cells (not shown). We find that in A7 cells, FLNA/PrP and FLNA/integrin β1 exist in two distinct, but functionally linked, complexes. This conclusion is based on our findings that when PrP is down-regulated in A7 cells, the amount of integrin β1 co-purified with FLNA is greatly reduced. Down-regulation of PrP in A7 cells also moderately reduces the levels of p- focal adhesion kinase, which is critical in integrin signaling (32). It was reported earlier that the level of phosphorylated-focal adhesion kinase correlates with the degree of mobility in human melanoma cell lines (36). Src is important in integrin signaling (37, 38). However, the level of p-Src does not change in PrP down-regulated A7 cells. A more detailed biochemical analysis will be required to delineate the signaling pathways that are affected by pro-PrP.
We propose that pro-PrP binds and pulls FLNA closer to the inner membrane leaflet, enabling FLNA to bind more effectively to the short cytoplasmic tail of β1 integrin. Worthy of note is the fact that the motif on integrin β1 that binds FLNA is located at the N terminus, proximal to the inner membrane leaflet (16). We further posit that when PrP is down-regulated, FLNA is dislodged from the inner membrane leaflet, moving away from the integrin β chain. This disengagement is likely to affect the bidirectional functionality of integrin β1.
Competition between FLNA and talin for integrin β7 binding regulates cell migration, and an increase in FLNA binding inhibits cell migration (30). However, because integrin β7 is only expressed in leukocytes and FLNA binds to different integrin β chains with different affinities; thus, the relevance of these findings to our result is not clear. Another cellular complex that is important in regulating actin organization is the Arp2/3 complex (39, 40). However, M2 and A7 cells have comparable levels of Arp2/3 (41), and therefore, it is unlikely that the Arp2/3 complex plays a critical role in regulating the spreading and migration of M2 and A7 cells.
Incubation of A7 cells with a synthetic peptide, KKRPK-PrP-GPI-PSS, reduces cell spreading and migration. Hence, the binding of pro-PrP to FLNA does occur in intact cells. When PrP is down-regulated by shRNA, the expression level of PrP is also reduced. One may argue that a reduction in PrP may also contribute to a decline in cell spreading and migration. This scenario seems unlikely; the peptide blocks cell spreading and migration without affecting the level of PrP expression. This result also provides a “proof of principle” that the binding of pro-PrP to FLNA is ameliorable to intervention in a living cell. Potentially, we may be able to identify small molecules that can block this interaction. These molecules may have therapeutic value.
Little PrP is detected in normal human skin. PrP staining is confined mainly to epithelial cells and to sporadic mononuclear cells within the dermis (42). Normal melanocytes and melanocytes in benign nevus do not show PrP staining. On the other hand, PrP is readily detected in melanoma in situ and in invasive melanoma. In melanoma in situ and invasive melanoma the PrP exists as pro-PrP, as it reacts with the anti-GPI-PSS antibody. The intensity of staining furthermore increases from melanoma in situ to invasive melanoma, and invasive melanoma in the dermal component has the strongest immunoreactivity. Therefore, during the transition from melanocytes to melanoma, there is an up-regulation of PrP as well as accumulation of pro-PrP. The molecular mechanisms that regulate melanomagenesis are multifarious and not completely understood (43, 44). A much more detailed biochemical study will be required to elucidate the underlying mechanism that causes the accumulation of pro-PrP during the transformation of melanocytes.
Melanoma cells growing along the dermal-epidermal junctions as single cells are largely FLNA negative, whereas tumors cells in nests and dermal components show strong FLNA immunoreactivity (45). It is postulated that FLNA may promote melanoma cell motility during tissue invasion from the epidermis to the dermis. We further suggest that it is the interplay between pro-PrP, FLNA, and integrin that facilitates the invasion of melanoma cells. Tumor cells missing any one of the three components are less efficient in their invasion.
Whether our findings in the expression of PrP in human melanoma have diagnostic or prognostic value will require an examination of a much larger cohort of melanoma patients, including different subtypes of melanoma and melanoma from different anatomical sites as well as at different stages of progression. Finally, in addition to FLNA and pro-PrP, integrin expression is also important in the progression of human melanoma (46). Thus, further examination of pro-PrP, FLNA, and integrin expression will provide new insights into the role these molecules play in human melanomagenesis.
Supplementary Material
Acknowledgments
We thank Pearl Ling, Emily Sy, and Sarah Sy for editorial assistance. We also thank Dr. Stephen Somach of MetroHealth System for providing some of the melanoma tissue sections.
This work was supported, in whole or in part, by National Institutes of Health Grant R21-CA133559-01 and Pilot Grant AR039750 (Skin Disease Research Center, NIAMS; to M.-S. S. and K. Cooper).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
- FLNA
- filamin A
- GPI
- glycosylphosphatidylinositol
- PSS
- peptide signal sequence
- PrP
- the normal cellular prion protein
- pro-PrP
- prion protein with its GPI-PSS
- ER
- endoplasmic reticulum
- PDAC
- pancreatic ductal adenocarcinoma
- DPBS
- Dulbecco's phosphate-buffered saline
- BFA
- brefeldin A
- OM
- original magnification.
REFERENCES
- 1.Stossel T. P., Condeelis J., Cooley L., Hartwig J. H., Noegel A., Schleicher M., Shapiro S. S. (2001) Nat. Rev. Mol. Cell Biol. 2, 138–145 [DOI] [PubMed] [Google Scholar]
- 2.Feng Y., Walsh C. A. (2004) Nat. Cell Biol. 6, 1034–1038 [DOI] [PubMed] [Google Scholar]
- 3.Byers H. R., Etoh T., Doherty J. R., Sober A. J., Mihm M. C., Jr. (1991) Am. J. Pathol. 139, 423–435 [PMC free article] [PubMed] [Google Scholar]
- 4.Cunningham C. C., Gorlin J. B., Kwiatkowski D. J., Hartwig J. H., Janmey P. A., Byers H. R., Stossel T. P. (1992) Science 255, 325–327 [DOI] [PubMed] [Google Scholar]
- 5.Awata H., Huang C., Handlogten M. E., Miller R. T. (2001) J. Biol. Chem. 276, 34871–34879 [DOI] [PubMed] [Google Scholar]
- 6.Lin R., Karpa K., Kabbani N., Goldman-Rakic P., Levenson R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5258–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He H. J., Kole S., Kwon Y. K., Crow M. T., Bernier M. (2003) J. Biol. Chem. 278, 27096–27104 [DOI] [PubMed] [Google Scholar]
- 8.Feng S., Reséndiz J. C., Lu X., Kroll M. H. (2003) Blood 102, 2122–2129 [DOI] [PubMed] [Google Scholar]
- 9.Liu G., Thomas L., Warren R. A., Enns C. A., Cunningham C. C., Hartwig J. H., Thomas G. (1997) J. Cell Biol. 139, 1719–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thelin W. R., Chen Y., Gentzsch M., Kreda S. M., Sallee J. L., Scarlett C. O., Borchers C. H., Jacobson K., Stutts M. J., Milgram S. L. (2007) J. Clin. Invest. 117, 364–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu T. N., He H. J., Kole S., D'Souza T., Agarwal R., Morin P. J., Bernier M. (2007) J. Biol. Chem. 282, 14816–14826 [DOI] [PubMed] [Google Scholar]
- 12.Prusiner S. B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weissmann C. (2004) Nat. Rev. Microbiol. 2, 861–871 [DOI] [PubMed] [Google Scholar]
- 14.Chiesa R., Harris D. A. (2009) PLoS Biol. 7, e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li C., Yu S., Nakamura F., Yin S., Xu J., Petrolla A. A., Singh N., Tartakoff A., Abbott D. W., Xin W., Sy M. S.. (2009) J. Clin. Invest. 119, 2725–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loo D. T., Kanner S. B., Aruffo A. (1998) J. Biol. Chem. 273, 23304–23312 [DOI] [PubMed] [Google Scholar]
- 17.Glogauer M., Arora P., Chou D., Janmey P. A., Downey G. P., McCulloch C. A. (1998) J. Biol. Chem. 273, 1689–1698 [DOI] [PubMed] [Google Scholar]
- 18.Yin S., Pham N., Yu S., Li C., Wong P., Chang B., Kang S. C., Biasini E., Tien P., Harris D. A., Sy M. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7546–7551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura F., Osborn T. M., Hartemink C. A., Hartwig J. H., Stossel T. P. (2007) J. Cell Biol. 179, 1011–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehtonen J. V., Still D. J., Rantanen V. V., Ekholm J., Björklund D., Iftikhar Z., Huhtala M., Repo S., Jussila A., Jaakkola J., Pentikäinen O., Nyrönen T., Salminen T., Gyllenberg M., Johnson M. S. (2004) J. Comput Aided Mol. Des. 18, 401–419 [DOI] [PubMed] [Google Scholar]
- 21.Kiema T., Lad Y., Jiang P., Oxley C. L., Baldassarre M., Wegener K. L., Campbell I. D., Ylänne J., Calderwood D. A. (2006) Mol. Cell 21, 337–347 [DOI] [PubMed] [Google Scholar]
- 22.Lovell S. C., Word J. M., Richardson J. S., Richardson D. C. (2000) Proteins 40, 389–408 [PubMed] [Google Scholar]
- 23.Playford M. P., Nurminen E., Pentikäinen O. T., Milgram S. L., Hartwig J. H., Stossel T. P., Nakamura F. (2010) J. Biol. Chem. 285, 17156–17165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bamburg J. R., Wiggan O. P. (2002) Trends Cell Biol. 12, 598–605 [DOI] [PubMed] [Google Scholar]
- 25.Scott R. W., Olson M. F. (2007) J. Mol. Med. 85, 555–568 [DOI] [PubMed] [Google Scholar]
- 26.Hynes R. O. (2002) Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 27.Ginsberg M. H., Partridge A., Shattil S. J. (2005) Curr. Opin. Cell Biol. 17, 509–516 [DOI] [PubMed] [Google Scholar]
- 28.Sharma C. P., Ezzell R. M., Arnaout M. A. (1995) J. Immunol. 154, 3461–3470 [PubMed] [Google Scholar]
- 29.Luo B. H., Carman C. V., Springer T. A. (2007) Annu. Rev. Immunol. 25, 619–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calderwood D. A., Huttenlocher A., Kiosses W. B., Rose D. M., Woodside D. G., Schwartz M. A., Ginsberg M. H. (2001) Nat. Cell Biol. 3, 1060–1068 [DOI] [PubMed] [Google Scholar]
- 31.Kim H., Sengupta A., Glogauer M., McCulloch C. A. (2008) Exp. Cell Res. 314, 834–846 [DOI] [PubMed] [Google Scholar]
- 32.Parsons J. T. (2003) J. Cell Sci. 116, 1409–1416 [DOI] [PubMed] [Google Scholar]
- 33.Yin S., Fan X., Yu S., Li C., Sy M. S. (2008) J. Biol. Chem. 283, 25446–25454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hegde R. S., Rane N. S. (2003) Trends Neurosci. 26, 337–339 [DOI] [PubMed] [Google Scholar]
- 35.Chen R., Knez J. J., Merrick W. C., Medof M. E. (2001) J. Cell. Biochem. 84, 68–83 [DOI] [PubMed] [Google Scholar]
- 36.Akasaka T., van Leeuwen R. L., Yoshinaga I. G., Mihm M. C., Jr., Byers H. R. (1995) J. Invest. Dermatol. 105, 104–108 [DOI] [PubMed] [Google Scholar]
- 37.Martin G. S. (2001) Nat. Rev. Mol. Cell Biol. 2, 467–475 [DOI] [PubMed] [Google Scholar]
- 38.Mitra S. K., Schlaepfer D. D. (2006) Curr. Opin. Cell Biol. 18, 516–523 [DOI] [PubMed] [Google Scholar]
- 39.Higgs H. N., Pollard T. D. (2001) Annu. Rev. Biochem. 70, 649–676 [DOI] [PubMed] [Google Scholar]
- 40.Soderling S. H. (2009) Sci. Signal 2, pe5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flanagan L. A., Chou J., Falet H., Neujahr R., Hartwig J. H., Stossel T. P. (2001) J. Cell Biol. 155, 511–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pammer J., Weninger W., Tschachler E. (1998) Am. J. Pathol. 153, 1353–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.High W. A., Robinson W. A. (2007) Adv. Dermatol. 23, 61–79 [DOI] [PubMed] [Google Scholar]
- 44.Miller A. J., Mihm M. C., Jr. (2006) N. Engl. J. Med. 355, 51–65 [DOI] [PubMed] [Google Scholar]
- 45.Bouffard D., Duncan L. M., Howard C. A., Mihm M. C., Jr., Byers H. R. (1994) Hum. Pathol. 25, 709–714 [DOI] [PubMed] [Google Scholar]
- 46.Edward M. (1995) Curr. Opin Oncol. 7, 185–191 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.