Abstract
The specificity in phosphorylation by kinases is determined by the molecular recognition of the peptide target sequence. In Saccharomyces cerevisiae, the protein kinase A (PKA) specificity determinants are less studied than in mammalian PKA. The catalytic turnover numbers of the catalytic subunits isoforms Tpk1 and Tpk2 were determined, and both enzymes are shown to have the same value of 3 s−1. We analyze the substrate behavior and sequence determinants around the phosphorylation site of three protein substrates, Pyk1, Pyk2, and Nth1. Nth1 protein is a better substrate than Pyk1 protein, and both are phosphorylated by either Tpk1 or Tpk2. Both enzymes also have the same selectivity toward the protein substrates and the peptides derived from them. The three substrates contain one or more Arg-Arg-X-Ser consensus motif, but not all of them are phosphorylated. The determinants for specificity were studied using the peptide arrays. Acidic residues in the position P+1 or in the N-terminal flank are deleterious, and positive residues present beyond P-2 and P-3 favor the catalytic reaction. A bulky hydrophobic residue in position P+1 is not critical. The best substrate has in position P+4 an acidic residue, equivalent to the one in the inhibitory sequence of Bcy1, the yeast regulatory subunit of PKA. The substrate effect in the holoenzyme activation was analyzed, and we demonstrate that peptides and protein substrates sensitized the holoenzyme to activation by cAMP in different degrees, depending on their sequences. The results also suggest that protein substrates are better co-activators than peptide substrates.
Keywords: Cyclic AMP (cAMP), Peptide Arrays, Protein Kinases, Protein Kinase A (PKA), Signal Transduction, Sacharomyces cerevisiae, Phosphorylation, Substrate
Introduction
Protein phosphorylation is one of the most fundamental mechanisms for signal transduction. The specificity of signal transduction depends upon the ability of each kinase to precisely phosphorylate particular sites on specific substrate proteins. Protein kinase A (PKA)3 is one of the best characterized members of this protein family (1). Phosphorylation of its substrate targets is known to be critical for regulating a multitude of cellular processes, including metabolism, gene transcription, ion flux, growth, and cell death (2).
The specificity in phosphorylation is determined by at least two major factors: peptide specificity and substrate and kinase localization. The effectiveness of protein phosphorylation by protein kinases in general, including PKA, is believed to depend on the primary structure of the protein around the phosphorylation site; synthetic peptides have thus been used to define the consensus phosphorylation sequences for PKA (3–5). The basic consensus phosphorylation sites for this enzyme are Arg-Arg-X-(Ser/Thr), (Arg/Lys)-X-X-(Ser/Thr), and (Arg/Lys)-X-(Ser/Thr) (3, 4, 6, 7).
The molecular recognition of the peptide sequence surrounding the phosphorylated residue of a substrate usually is referred to as the “peptide specificity” of a protein kinase. The phosphorylated residue is conventionally named P0, the substrate residues immediately N- and C-terminal to the P0 position as P−1 and P+1, respectively, and so on. These substrate residues bind to their corresponding subsites on the catalytic sites of the protein kinases.
Statistical analysis of proteins known to be phosphorylated has shown that many of the phosphorylation sites in the substrate proteins do not correspond exactly with the complete consensus sequence determinant (8). A screening of 150 physiological substrates of mammalian PKA, unique gene products for which one or more phosphorylation sites are known, permitted the evaluation of consensus sequences for PKA in a physiological context (9). The results indicated strong preferences for arginines at P-2 and/or P-3. The canonical Arg-Arg-X-Ser was the most abundant consensus sequence, representing around one-half of all sites. Though arginines are the preferred basic residues at P-2 and P-3, one every six sites contains at least one Lys at these positions, indicating a lower efficiency in the in vivo use of this determinant. Serine is by far preferred over threonine as the phosphate acceptor; other more subtle preferences at positions outside of the P-2 and P-3 positions also emerge upon closer inspection. There is a slight preference for Arg at the P-4 to P-7 positions and an increased occurrence of the hydrophobic residues Phe, Ile, Leu, or Val at P+1 (9). There also is an increased frequency of the small residues Ser, Gly, and Pro at P-1. These positional biases are consistent with numerous in vitro studies utilizing peptides based on well characterized substrates such as the porcine pyruvate kinase LRRASLG heptapeptide kemptide, with biochemical and crystallographic analysis of PKA inhibitor peptide interactions (10, 11), as well as through the characterization of PKA substrates in highly complex peptide libraries (12, 13). Loog et al. (14) compared in a mammalian model, the specificity of parent proteins with their short peptide derivatives using the hepatic L-pyruvate kinase as substrate and demonstrated that mutations made in the protein had lesser effect than the corresponding mutation made in the peptides, whereas the amino acid preference and the overall specificity pattern remained similar in both cases.
Whereas the substrate specificity determinants for mammalian PKA have been analyzed extensively, the specificity determinants for other organisms are much less understood. Denis et al. (15) have analyzed Saccharomyces cerevisiae PKA substrate determinants using synthetic peptides derived from the local phosphorylation site sequence around Ser230 in the yeast transcriptional activator ADR1 and shown common specificity determinants with mammalian PKA substrates on the proximal N-terminal side and up to the +4 position of the C-terminal side of the phosphoacceptor.
Previous results from our group have demonstrated that in yeast, a PKA substrate such as pyruvate kinase 1 (Pyk1) is less efficient as substrate than a peptide containing the phosphorylatable Ser22 residue despite having a higher affinity (16). Therefore, the behavior of a protein kinase toward its substrate could be different depending on whether the substrate is an entire protein or the peptide derivative indicating that the ability of a kinase to modify a protein at a specific site is influenced by its structural context.
In S. cerevisiae, PKA is a key regulator of cell growth and proliferation (17, 18). The regulatory subunit is encoded by the BCY1 gene, and the C subunit is encoded by the TPK1, TPK2, and TPK3 genes. Yeast is a good model with which to search for and identify PKA substrates as its genome is completely known. Several PKA substrates have been described, and, very recently, proteomic approaches that simplify the search for new potential PKA substrates have been reported (19–21). However, further work is needed to demonstrate that the candidate proteins identified are indeed PKA substrates and to identify their target sequences. The three Tpk1, Tpk2, and Tpk3 catalytic subunits appear to have both overlapping and distinct cellular functions (22–25). The recent global analysis of protein phosphorylation in vitro in yeast (21) indicated that Tpk1, Tpk2, and Tpk3 recognized 256, 29, and 79 substrates, respectively; however, only eight were recognized by all three kinases, and 39 were recognized by two of the three suggesting that each kinase has unique substrate specificity.
The classic model of PKA activation proposes that the interaction of cAMP with R subunit decreases the affinity between R and C, and, as a consequence, the holoenzyme dissociates, thus freeing the catalytic subunit to phosphorylate its targets. However, several reports suggest that cAMP does not fully dissociate the holoenzyme, and a new role for the substrates has been proposed in PKA activation (26–29).
A complete understanding of the biological role of the S. cerevisiae PKA requires the complete characterization of the enzyme and of its targets. In this paper, we assess the sequence features surrounding the phosphorylation sites of three protein substrates. We demonstrate that Tpk1 and Tpk2 have the same selectivity and catalytic activity toward a number of peptidic and protein substrates. We clearly demonstrate that both peptide and protein substrates sensitize the holoenzyme to activation by cAMP.
EXPERIMENTAL PROCEDURES
Yeast Strains and Growth Media
Yeast media were prepared as described (30). Strains were grown on rich medium containing 2% bactopeptone, 1% yeast extract, and 2% galactose (YPGal), or 2% glucose (YPG), or 2% raffinose (YPR). Synthetic medium containing 0.67% yeast nitrogen base without amino acids, 2% glucose, plus the necessary additions to fulfill auxotrophic requirements (SD) were used to maintain the selectable plasmids. Solid medium contained 2% agar. Yeast strains used were as follows: KT 1115, Mata leu2 ura3 his3 pep4Δ; NTH1-HA-His, KT1115 + pBG1805-NTH1-HA-His; PYK1-HA-His, KT1115 + pBG1805-PYK1-HA-His; PYK2-HA-His, KT1115 + pBG1805-PYK2-HA-His; JT20454(W), Mata his3 leu2 ura3 trp1 tpk1::ade8 tpk2::HIS3 tpk3::TRP1 msn2::LEU2 msn4::TRP1 + pPYK101; 033c TAP, S288C, MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0; S330:W303, Mata tpk2::tpk1::KmR; S331:W303, Mata tpk2::tpk3::KmR; S332:W303, Mata tpk3::tpk1::KmR; S330 BCY1-TAP:W303, Mata tpk2::tpk1::KmR BCY1TAP; S331 BCY1-TAP:W303, Mata tpk2::tpk3::KmR BCY1TAP; and S332 BCY1-TAP:W303, Mata tpk2::tpk3::KmR BCY1TAP.
Pyk1, Pyk2, and Nth1 Expression and Purification
Nth1 (neutral trehalase)-His, Pyk1-His, and Pyk2-His fusion proteins were purified from a KT1115 strain transformed with BG1805 plasmid containing cDNA from the following ORFs YDR001C, YAL038W, or YOR347C, under a Gal1 promoter (Open Biosystems). For the expression 25-ml SD−Ura, 2% raffinose media culture was grown overnight and then diluted in 200 ml SD−Ura, 2% raffinose media. The culture was grown up to A600 = 1.2 and added to 100 ml of 3× YP, 6% galactose, to a final concentration of 1× YP, 2% galactose and harvested after overnight incubation. The pellets were stored at −70 °C until used. For the purification, the cells were lysed by vortexing with glass beads. The lysates were clarified by centrifugation at 15,000 × g to remove the cellular debris. Resin nickel-nitrilotriacetic acid (Qiagen) was added, and the mix was incubated overnight at 4 °C. The resin was washed with buffer (50 mm NaH2PO4, 300 mm NaCl, 20 mm imidazole, 0.05% Tween 20, pH 8). The proteins were eluted with elution buffer (50 mm NaH2PO4, 300 mm NaCl, 250 mm imidazole, 0.05% Tween 20, pH 8). Pyk1 protein without tag was purified from the S. cerevisiae strain JT20454 containing pPYK101 vector (plasmid containing Pyk1 cDNA under its own promoter), using a previously described method (31). The yeast strain overexpresses Pyk1 when grown in 2% glucose media. Briefly, the semipurification of the enzyme involved two chromatography steps, DEAE-cellulose, and phosphocellulose and ammonium sulfate precipitation. The precipitated protein was stored at −20 °C.
PKA Holoenzyme Preparation
The yeast PKA holoenzyme (TPKs-Bcy1) was purified from a yeast tandem affinity purification (TAP)-fusion strain that contains the ORF of TAP-tagged Bcy1 (YJL033W; Open Biosystems). The standard TAP procedure (32) was applied but using only one specific affinity purification step: binding to IgG-Sepharose 4B through the protein A IgG binding domains of the ORF. This source of immobilized holoenzyme was used for PKA activation experiments.
Catalytic Subunit Purification
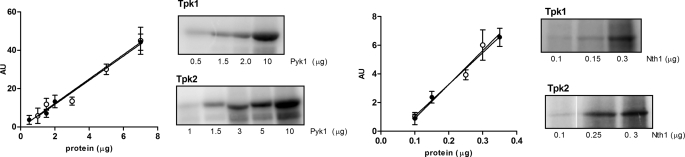
The yeast Tpk1 or Tpk2 enzymes were purified from yeast TAP-fusion strains, which express only one isoform of Tpk and the TAP-tagged Bcy1, constructed essentially as described (33). Briefly, the fragment containing the C-terminal coding region of Bcy1 fused to the TAP epitope including the HIS3 selectable marker was amplified by PCR using genomic DNA from the Bcy1-TAP strain using the primers F 5′-AGACCATGATTATTTCGGTG-3′ and R 5′-GTAGTAACAGCAGTAGTAGA-3′. The strains S331 and S332 were transformed with the PCR products by the lithium acetate method. Integration at the corresponding locus was tested by PCR. Expression of the protein fusions was tested by Western blot using anti-TAP or anti-Bcy1 antibodies. Tpk1 or Tpk2 enzymes were purified using the standard TAP procedure using the first affinity purification with IgG-Sepharose 4B. After exhaustive washing, Tpk1 or Tpk2 were eluted with 50 mm cAMP. These sources of pure enzyme were used immediately for phosphorylation experiments without storage. For the experiments in which the source of enzyme was the mixture of the three Tpk enzymes, the enzyme purification was made using the same protocol, but the strain used in this case was a Bcy1 TAP-tagged wild-type strain containing the three functional Tpk isoforms. Tpk1 and Tpk2 units from purified enzymes were defined as the pmol of enzyme that transfer 1 pmol of phosphate to kemptide/min, at 30°C, under the standard assay conditions. The amount of each enzyme was estimated by SDS-PAGE silver staining against a calibration curve of BSA. The identity of each enzyme after purification was verified by mass spectrometric data from tryptic digestion of SDS-PAGE gel bands obtained using a MALDI-TOF-TOF spectrometer, Ultraflex II (Bruker) from the mass spectrometry facility Centro de Estudios Químicos y Biológicos por Espectrometría de Masa, Facultad de Ciencias Exactas y Naturales, Argentina. The catalytic turnover number of Tpk1 and Tpk2 was calculated using the protein concentration estimated by SDS-PAGE and the catalytic activity using kemptide at saturating concentration as substrate of the same purification sample.
PKA Assays
PKA activity was determined by assay of phosphotransferase activity with kemptide, Ser22, Nth1-1, Nth1-2, or Pyk1 peptides as substrates. The assay was initiated by mixing different amounts of PKA sources (either soluble Tpk1, Tpk2, or purified immobilized Tpk-BCY1 holoenzyme) with 15 mm Tris-HCl, (pH 7.5), 0.1 mm EGTA, 0.1 mm EDTA, 10 mm 2-mercaptoethanol, phosphatase inhibitor mixture (10 mm NaF, 10 mm Na4P2O710H2O, 0.1 mm Na3VO4, 0.1 mm (NH4)6MO7O24) and EDTA-free protease inhibitor (buffer A), 15 mm MgCl2, 0.1 mm [γ-32P]ATP (1000 dpm/pmol) plus the protein or peptide substrates at the concentrations indicated in each experiment, plus variable concentrations of cAMP, when added. A standard assay, for unit definition, contains a saturating concentration of 300 μm kemptide. Incubations were for 10 min at 30 °C. PKA assays were linear with time and enzyme protein concentration. PKA activity is expressed in pmol of phosphate incorporated into substrate/min at 30 °C. When using peptides as substrates, aliquots of the incubation mixture were processed according to the phosphocellulose paper method (34). For protein substrates, aliquots of the reaction mixture were analyzed through SDS-PAGE. The incorporation of phosphate into the substrate was determined by digital image analysis and further densitometric quantitation and is expressed in arbitrary units. Alternatively, the amount of incorporation of phosphate into the substrate was determined by scintillation counting of the phosphorylated protein band excised from SDS-PAGE gels. For the holoenzyme activation assays, immobilized purified PKA holoenzymes were used for measurement of kinase activity at various cAMP concentrations as described above using different substrates and calculating the A0.5 value for cAMP under each condition. Two statistics analysis tests were applied to validate the curves. The D'Agostino and Pearson normality test, which indicated that the curves, showed deviation not significant from the model. We also applied the ALLFIT program, which is used for the statistical analysis of families of sigmoidal dose-response curves using the four-parameter logistic equations. All curves had a good statistical significance with p > 0.05, indicating the quality of the curve fit. The F ratio test was not significant within each of the two groups of proteins (Nth1 and Ser22 on one side and Pyk1, Pyk2, and kemptide on the other) indicating that the data shared the two parameters: IC50 and slope. However, the F ratio test corresponding to the comparison of the IC50 and slope from one group of peptides (Nth1 and Ser 22) with the other (Pyk1, Pyk2 and kemptide) is significant.
Peptide Array
Cellulose-bound peptide arrays were prepared automatically according to standard spot synthesis protocols by using a spot synthesizer (Abimed Analysen-Technik, Langenfeld, Germany) by Susan Taylor's laboratory, Department of Chemistry and Biochemistry, University of California at San Diego). The membranes were first wet in ethanol and further placed in phosphorylation buffer (50 mm Tris-HCl pH 7.4, 10 mm 2-mercaptoethanol, 0.1 mm EDTA, 0.1 mm EGTA, 100 mm NaCl, 15 mm MgCl2) containing 0.2 mg/ml BSA overnight at room temperature. Membranes were then incubated at 30 °C for 45 min in phosphorylation buffer containing 1 mg/ml BSA, 100 mm NaCl, and 100 μm ATP. The phosphorylation reaction was performed in phosphorylation buffer plus 0.2 mg/ml BSA, 100 μm ATP, 200 μCi [γ-32P]ATP, and 150 pmol of Tpk subunits during 1 h at 30 °C. The membrane was rinsed 10 × 15 min with 1 m NaCl, 3 × 5 min with H2O, 3 × 15 min with H3PO4 5%, and 3 × 5 min with H2O and 3 × 2 min with ethanol. The membrane was dried and subjected to digital image analysis (Bio-Imaging Analyzer Bas-1800II and Image Gauge version 3.12, FujiFilm).
RESULTS
Peptides Derived from Nth1, Pyk1, and Pyk2 Are Phosphorylated by Tpk1 and Tpk2
A global analysis of in vitro protein phosphorylation in yeast has shown that Tpks display differential protein selectivity (21). To define which are the differences in the target sequences that determine the unique substrate specificity for each Tpk, we assessed the kinetic behavior of peptide substrates derived from three proteins, Pyk1 (pyruvate kinase 1), Pyk2 (pyruvate kinase 2), and Nth1 (neutral trehalase) that have been identified as physiological substrates of PKA in yeast (16, 35–37). Both Pyk1 and Nth1 proteins have been described in the global analysis of protein phosphorylation in yeast as being selective substrates for Tpk1; in fact, the selectivity of Tpk2 was limited to a few numbers of proteins suggesting a strict recognition of substrates (21). As a first step, we corroborated the in vitro phosphorylation of the three proteins by a purified mixture of Tpks as an enzyme source, using purified His-Nth1, His-Pyk1, and His-Pyk2 as substrates (supplemental Fig. 1).
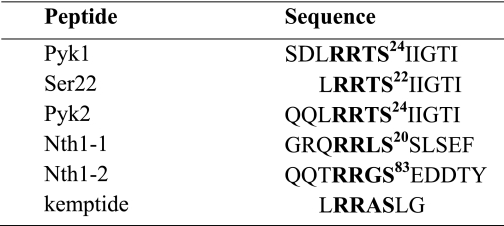
Pyk1 and Pyk2 have only one RRXS motif in their primary sequence. We have demonstrated previously that Pyk1 is a Tpk1 substrate and that the major PKA phosphorylation site is Ser22 (16). The sequence surrounding the Ser22 position in Pyk1 is conserved in the yeast isoform Pyk2 at Ser24. We have demonstrated that Pyk2 protein also is phosphorylated in vitro by Tpk1 with a better specificity constant than Pyk1 (35). To design the peptides for this study, we have selected the amino acid sequences from P-6 to P+4 surrounding the phosphorylation site P0, as these residues are described to be influenced by direct interaction with the kinase in the catalytic site (9, 10). We also include a shorter peptide, Ser22, common to both Pyk1 and Pyk2. Nth1 also has been defined as a PKA substrate (36, 37). The Nth1 amino acid sequence shows three potential phosphorylation sites, Ser20, Ser21, and Ser83 included in a consensus PKA phosphorylation motif RRXS. Genetic results have correlated the phosphorylation of all the three serines with trehalase activation by PKA (37). A recent global study of in vivo yeast phosphoproteome detects peptides containing Ser phosphorylated at these three positions for Nth1 (20). Table 1 summarizes the sequences of the peptides designed for the kinetic study.
TABLE 1.
Peptides derived from protein substrates
Sequence of peptides derived from each protein were as follows: Pyk1 and Ser22 from pyruvate kinase 1 and Pyk2 from pyruvate kinase 2 and Nth1-1/Nth1-2 from neutral trehalase. The consensus RRXS is in boldface as well as the phosphorylatable serine residue (P0); the subscript number corresponds to the position of that residue in the complete protein primary sequence.
Purified preparations of Tpk1 and Tpk2 were used as an enzyme source to measure their kinetic parameters with the different peptide substrates. The purification of Tpk3 from a strain expressing Bcy1-TAP and only the Tpk3 isoform was unsuccessful, even though the purification was made from very large amounts of cultures, and the activity was assayed with different concentrations of substrate and/or ATP.
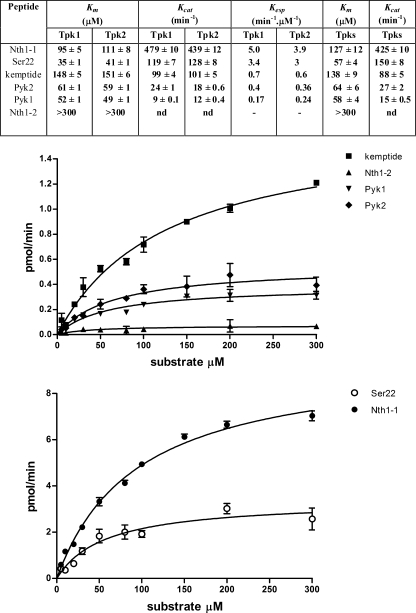
The results of Km, kcat, and the derived Kesp values are summarized in a Table inserted within Fig. 1. Several important conclusions are derived from these results. The first is that the Km and kcat for Tpk1 and Tpk2 with all the assayed peptides are the same, indicating no selectivity between the two isoforms. A catalytic turnover number of 3 s−1 has been estimated using kemptide as substrate for Tpk1 and Tpk2. This value is almost 10-fold lower than the turnover of mammalian C subunits, which is in the range of 20 s−1 (38).
FIGURE 1.
Steady-state kinetic parameters. Purified preparations of Tpk1, Tpk2, or Tpks mixture were used to determine Km, kcat, and Kesp values for the substrate peptides of Table 1. Top, table summarizing the kinetic values determined for each enzyme source; bottom, two representative experiments obtained using the mixture of Tpks.
The second and important conclusion is that although all the substrates include a RRXS consensus motif, the Kesp were very different. The analysis includes the Nth1-2 peptide with which no phosphorylation could be detected, at least with concentrations of up to 300 μm. The best substrate was Nth1-1, and the worst was Pyk1. The reason for the difference lies mainly in their kcat. It is interesting to note that Pyk1 and Pyk2 showed a different behavior, even though they only differ in two amino acids in positions P-5 and P-6. The peptide Ser22, being shorter than Pyk1 in only two residues in the N terminus, was a much better substrate than Pyk1. Kemptide, the prototype of PKA peptide substrate, was a substrate of intermediate performance, but in this case due to its high Km. This result was unexpected, because both proteins, Nth1 and Pyk1, had been described as being phosphorylated only by the Tpk1 isoform (21). Although the phosphorylation of a peptide derived from a protein sequence does not necessarily imply whole protein phosphorylation, we considered these results as a first indication that these proteins could be phosphorylated by both Tpks.
We then assessed the phosphorylation by Tpk1 and Tpk2 of two of the three proteins in study, Pyk1 and Nth1, to analyze whether the proteins also were substrates for the two isoforms as occurred with the peptides derived from them. Fig. 2 shows the phosphorylation of increasing concentrations of Pyk1 and Nth1 by the same amount of Tpk1 or Tpk2. The slopes of these curves, an approximation to the Kesp for each protein, are very similar for both isoforms of Tpk, indicating that both protein substrates are equally recognized by each. The slopes obtained for Nth1 are higher than the ones corresponding to Pyk1, indicating that Nth1 is a better substrate. These results agree with those obtained with the corresponding peptides derived from these two proteins.
FIGURE 2.
Phosphorylation of Pyk1 and Nth1 with Tpk1 and Tpk2. Different amounts of each protein were submitted to phosphorylation by purified Tpk1 or Tpk2. The phosphorylated samples were subjected to SDS-PAGE. The incorporation of phosphate into the substrate was determined by digital image analysis and followed by densitometric quantitation and is expressed in arbitrary units (AU). The graphical data correspond to the mean of two replicas, and the picture corresponds to a representative experiment.
Results from Mazon et al. (39) indicated that a strain containing only the TPK3 gene had undetectable phosphorylating PKA activity and that almost no TPK3 mRNA could be detected. These results indicate that Tpk3 is the isoform that is expressed at the lowest level. In agreement with these results, we could not detect by mass spectrometry peptides derived from Tpk3 in a purified preparation of a mixture of Tpks from a WT strain. We therefore predicted and confirmed that this mixture of Tpks would exhibit the same kinetic parameters than the ones of the isolated Tpk1 and Tpk2. Typical kinetic curves are shown in Fig. 1, and the data are summarized in the inset table. These results can be interpreted as an indication that either Tpk3 levels are very low and do not contribute significantly to the total PKA activity and/or that if it does, it has the same kinetic parameters as the other two isoforms. The results obtained with the mixture of Tpks validated the use of this mixture as enzyme source for further experiments.
Residues Determinant for Tpk Recognition
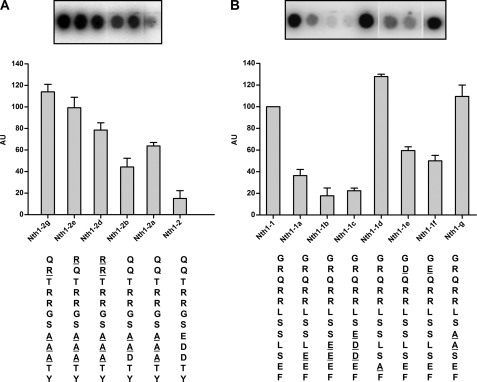
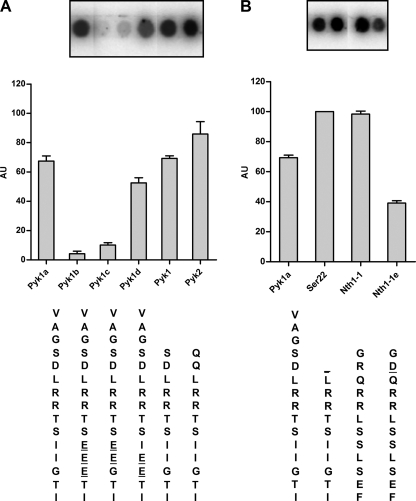
To evaluate which are the determinants for the recognition of a peptide by yeast PKA, we used the peptide array methodology and a mixture of Tpks as an enzyme source. We first investigated the reason why the peptide Nth1-2 was not phosphorylated even though it includes an RRXS consensus motif. The first observation is that the sequence of this peptide does not have a bulky hydrophobic residue in position P+1 as expected; instead, it has three acidic residues at P+1, P+2, and P+3. The P+1 residue lies in the vicinity of a hydrophobic pocket that is built up by the side chains of Leu198, Pro202, and Leu205 in mammalian C subunit. As a consequence, a large hydrophobic residue was found to be favored at this substrate position (5, 13, 40). These residues and the hydrophobic pocket also are conserved in Tpks. Fig. 3A shows that a peptide that has a replacement of the P+1, P+2, and P+3 acidic residues by A (Nth1-2a) was phosphorylated efficiently. Nth1-2b with only one Asp in P+3 also was phosphorylated but less efficiently than Nth1-2a. These results suggest that an acidic residue in the place that should interact with a hydrophobic pocket is a strong impediment. To test this hypothesis, we changed the residues in P+1, P+2, and P+3 of Pyk1a, which has its natural context, into acidic residues (Pyk1b, Pyk1c, and Pyk1d). The results of Fig. 4A show that even though an acidic context in these regions always impairs phosphorylation, the inclusion of an acidic residue at P+1 is crucial. The peptide Pyk1b was a poor substrate, like Nth1-2. The same results were observed when the sequence of the peptide Nth1-1, the best of the assayed peptide substrates (Fig. 1), was mutated introducing acidic residues in P+1, P+2, and P+3 (Nth1-1a, Nth1-1b, and Nth1-1c) The results, shown in Fig. 3B, show that an acidic residue at P+1 or P+2 has stronger inhibitory consequences than in P+3. We can thus conclude that the presence of acidic residues in the C-terminal flank of P0 is deleterious for its phosphorylation. The mutations by two Ala at positions P+1 and P+2 from Nth1-1 (Nth1-1g) had no effect on the phosphorylation of this peptide compared with the natural one. From this observation, we can conclude that the phosphorylatable Ser is the one at P0, as we had supposed, and not the one at P+1, and that the presence of a bulky hydrophobic residue for the P+1 position is not so critical for yeast PKA.
FIGURE 3.
Residues determinant for Tpk recognition in Nth1-1 and Nth1-2. Peptides derived from Nth1-1 and Nth1-2 synthesized on membrane arrays were incubated with Tpks mixture as a kinase source and submitted to phosphorylation as described under “Experimental Procedures.” The incorporation of phosphate into the peptides, expressed in arbitrary units (AU), was determined by digital image analysis followed by densitometric quantitation. The array images correspond to a representative experiment, and the bar graph data correspond to the mean of three replicas expressed relative to the spot intensity of Nth1-1. The spots shown in the insets in A and B correspond all of them to the same membrane array-phosphorylation assay and were assembled by cutting and pasting the spots of interest to facilitate the comparisons.
FIGURE 4.
Residues determinant for Tpk recognition in Pyk1 and Pyk2. Peptides derived from Pyk1 and Pyk2 synthesized on membrane arrays were incubated with Tpks mixture as kinase source and submitted to phosphorylation as described under “Experimental Procedures.” The incorporation of phosphate into the substrate, expressed in arbitrary units (AU), was determined by digital image analysis followed by densitometric quantitation. The array images correspond to a representative experiment, and the bar graph data correspond to two or three replicas, expressed relative to the spot intensity of Ser22. The spots shown in the insets in A and B correspond to the same membrane array-phosphorylation assay and were assembled by cutting and pasting the spots of interest to facilitate the comparisons.
The requirements for positive charged residues in the N-terminal flank of the phosphorylation site, at positions P-2 and P-3, with a preference for Arg, have been established as main determinants for the specificity in PKA for a long time (5). More recent reports on prediction of protein kinase A phosphorylation sites (13, 42) confirmed and generalized these early predictions and established that the requirements for positively charged residues are highest for residues in positions P-2 and P-3 but can be detected as far as six to eight residues prior to the phosphorylated serine. Yeast PKA has been shown to have the same requirements of basic residues at positions P-2 and P-3 (Arg is a better residue at position P-2 than Lys) (15). Another key residue is the Ser in position P0; the replacement of Ser by Thr is very deleterious in a yeast PKA susbtrate (15). We have confirmed in this work that this requirements are also valid for the peptide Nth1; very low phosphorylation levels were observed when the Ser at P0 was replaced by Thr, and when the Arg-Arg sequence at P-2 and P-3 were replaced by Lys-Lys, Arg-Lys, or Lys-Arg (data not shown).
We also have investigated the importance of residues N-terminal to the P0 and to the conserved Arg at P-2 and P-3. As the peptide Nth1-2 has neutral residues in positions P-5 and P-6 (Gln), we have assayed the effect of changing these amino acids to basic ones. In agreement with what has been observed for mammalian PKA substrates, the addition of extra Arg at these positions (Nth1-2d, Nth1-2e, and Nth1-2f) makes peptide Nth1-2a a better substrate (Fig. 3A). In accordance with this observation, when the natural Arg at P-5 from Nth1-1 was replaced by an acidic residue (Nth1-1e or Nth1-1f), the peptides showed lower phosphorylation (Fig. 3B). From the results shown in Fig. 4A, we can infer that the reason why peptide Pyk2 is a better substrate than Pyk1 and Pyk1-1a (a longer version of Pyk1) is the lack of the acidic residue at P-5. The same reasoning can be applied for the peptide Ser22, a shorter and better version of Pyk1 that does not contain the P-5 residue in its sequence; the improvement seems to be the lack of this acidic residue from the natural Pyk sequence (Fig. 4B). We therefore conclude that the presence of acidic residues C-terminal to P0 makes a nonpermissive sequence environment and that the inclusion of these residues N-terminal to P0 somehow impairs the behavior of the peptide as substrate, whereas on the contrary, the presence of positive residues in the N-terminal flank beyond P+2 and P+3 favors the phosphorylation.
No clear consensus sequence has been established for the C-terminal flank of the phosphorylatable Ser/Thr, with exception of position +1 with its large hydrophobic amino acid and a slight preference for small, flexible residues at positions +2 and +3 (42). Few data were available from the literature regarding P+4 until recently, as this position was missing in the resolved three-dimensional structures (43). However, the structures of PKA holoenzymes crystallized from a complex between mammalian Cα with N-terminal deletion mutants of both RI (44) and RII (45) are now available. In these structures, the interaction of the inhibitory site (IS) of the R subunits with the catalytic subunit surface is defined clearly. A close inspection of a thorough alignment of mammalian and fungal R subunits4 shows that the conservation of the IS sequence is extremely high. Mammalian R subunits have an RRA(S/A)V(C/S)AE, with a variation in the P0 position according to whether it is an RI that has a pseudosubstrate IS sequence or an RII that has a Ser that is autophosphorylated. The position at P+2 also changes according to the isoform. In fungi, the highly predominant IS sequence is RRTSVS(G/A)E; particularly in S. cerevisiae, the Bcy1 IS sequence is RRTSVSGE. As can be seen, the acidic residue at position P+4 is absolutely conserved; the corresponding amino acid in the C subunit of the crystal structure with RI is a Lys (Lys83), which is in a region with high sequence conservation in all the C subunits. Particularly, this Lys is conserved in Tpk1, Ttpk2, and Tpk3 (127, 110, and 128, respectively). The IS in the R subunits is one of the regions of interactions of R subunits with C that contribute to the high affinity between RC in the holoenzymes. It is reasonable to assume that protein substrates share with R the determinants of interaction in the short surrounding of the phosphorylatable amino acid. Therefore, an acidic residue in P+4 a can be considered as a favorable position. It is noticeable that in the case of the substrates we have analyzed in this work, Nth1-1, the best of the substrates, has a Glu at P+4.
Peptide Substrate Participation in Holoenzyme Activation
It has been widely accepted that cAMP activates the PKA holoenzyme by dissociating the regulatory and catalytic subunits, thus freeing the catalytic subunit to phosphorylate its targets. It has been demonstrated recently that although the addition of a molar excess of cAMP to the type Iα PKA holoenzyme causes partial dissociation, it is only upon addition of a PKA peptide substrate, together with cAMP that full dissociation occurs. The authors also demonstrate that substrate plays a differential role in the activation of type I versus type II holoenzymes, which could explain some important functional differences between PKA isoforms (27, 28). We have shown, some years ago, that peptide substrates were involved in the activation of a fungal PKA by cAMP (29).
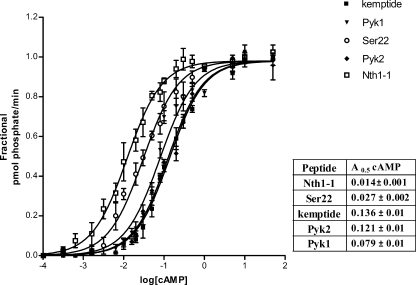
We assayed whether the activation of S. cerevisiae holoenzyme by cAMP could also be influenced by different peptide substrates, namely Pyk1, Pyk2, Nth1-1, Ser22, and kemptide. To evaluate whether the different peptidic substrates could sensitize PKA toward cAMP in a different way depending on their sequence, we assessed the cAMP dependence of the holoenzyme activity in the presence of saturating concentrations of each peptide. Fig. 5 shows the activation curves for the different peptide substrates, as well as a Table summarizing the A0.5 values for cAMP. The curves can be compared because all of the peptides were used at 3-fold their Km; for the sake of clarity, the curves were normalized relative to their respective maximum value. The first observation is that the values of A0.5 for cAMP range from 0.014–0.136 μm, almost a difference of one order. The peptides can be roughly divided in two groups according to the cAMP concentration needed for their half maximal phosphorylation. One group of peptides, comprised by Ser22 and Nth1-1 with an A0.5 for cAMP in the range of 0.01–0.03 μm and a second group including Pyk1, Pyk2, and kemptide with A0.5 values ranging from 0.08 to 0.14 μm. From these results, we can conclude that peptide substrates sensitize the yeast PKA holoenzyme to cAMP and that the better substrates (those with higher Kesp, see Fig. 1) are those that promote lower concentrations of cAMP for activation and therefore have a higher sensitizing effect on the enzyme.
FIGURE 5.
Peptide substrates enhance the activation of yeast PKA by cAMP. Purified holoenzyme (Tpks-Bcy1) was incubated with increasing concentrations of cAMP for 10 min at 30 °C with 100 μm [γ-32P]ATP and the peptide substrates in saturating concentrations (3-fold the Km values). PKA activation curve for each substrate is represented relative to the activity with that substrate at 100 μm cAMP. Shown graphical data and the A0.5 values for cAMP for each peptide in the inset table correspond to the mean ± S.D. from n = 4. Statistical analysis of sigmoidal dose response curves were made using the ALLFIT program and D'Agostino and Pearson normality test. All curves had significance with p > 0.05 (see also “Experimental Procedures”).
Whole Protein Substrate Sensitizes PKA toward Activation by cAMP
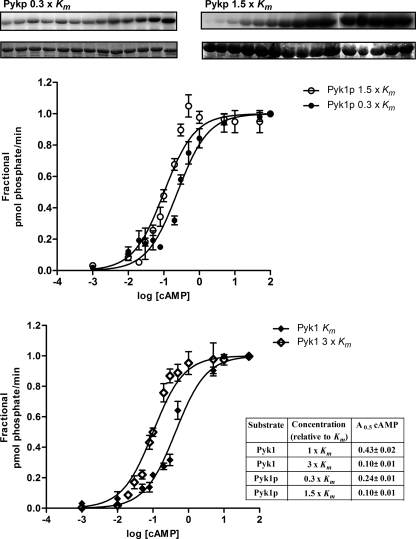
To analyze whether a physiological substrate whose activity is regulated by phosphorylation by PKA sensitizes the yeast holoenzyme to cAMP, we assayed the cAMP dependence of PKA in the presence of two concentrations of the Pyk1 protein and several concentrations of cAMP. The assays were conducted in parallel with the same enzyme preparation using Pyk1 peptide substrate for comparison and validation of the results. Fig. 6 shows that the A0.5 for cAMP when using Pyk1 protein as substrate changed according to the concentration of substrate in the assay: 0.10 μm at a concentration of 1.5 × Km and 0.24 μm at 0.3 × Km. For comparison, in a phosphorylation reaction in which Pyk1 peptide substrate was used at 1× Km and 3× Km, the A0.5 values for cAMP were 0.43 and 0.1 μm, respectively. These results show that both protein and peptide substrates sensitize the holoenzyme to cAMP and suggest that the protein is a better activator, as 0.1 μm cAMP was needed for half-maximal activation with 1.5 × Km of the protein substrate, but 2-fold higher concentration of peptide was needed to attain the same A0.5 for cAMP.
FIGURE 6.
Comparison of protein and peptide enhancement of yeast PKA activation by cAMP. The phosphorylation of Pyk1 protein without tag (Pyk1p) by purified yeast holoenzyme (Tpks-Bcy1) was determined after a 10-min incubation at 30 °C with 100 μm [γ-32P]ATP. The protein concentration was 1.5 × Km (13.5 μm) or 0.3 × Km (2.7 μm). The phosphorylation of Pyk1 protein was visualized by SDS-PAGE and digital image analysis. The amount of incorporation of phosphate into the substrate was determined by scintillation counting of the phosphorylated protein band excised from SDS-PAGE gels. The same enzyme preparation was assayed for holoenzyme activation in the presence of two concentrations of the peptide substrate Pyk1; 1 × Km (50 μm) and 3 × Km (150 μm). Pyk1 protein Km = 9 μm and Pyk1 peptide Km = 50 μm. The data of A0.5 value showed in the inset (table) corresponds to the mean ± S.D. from n = 3. Statistical analysis of sigmoidal dose response curves were made using the ALLFIT program and DÁgostino and Pearson normality test. All curves had significance with p > 0.05 (see also “Experimental Procedures”).
DISCUSSION
Several experimental studies and various computational approaches have been employed extensively in search of substrates and prediction of specific phosphorylation sites both for mammalian (13, 14) and yeast (21, 15, 19, 42) systems, both for kinases in general (4, 8, 13, 21) or for PKA in particular (14, 15, 19, 42). Our results contribute to the phosphorylation dynamics in S. cerevisiae. For yeast PKA isoforms Tpk1, Tpk2, and Tpk3, differential physiological roles have been proposed and demonstrated (22, 25). Strains lacking all three are nonviable; however, those containing any one of the three Tpks are able to propagate, indicating that each, although specific for some functions, is genetically redundant for cell growth (22). In the global in vitro analysis of protein phosphorylation in yeast, a differential substrate phosphorylation between Tpk1, Tpk2, and Tpk3 was observed (21). In this work, we indicated that the vast majority of proteins (87.7%) were recognized by only one of the Tpks, concluding that each kinase had unique substrate specificity depending on the primary structure of the protein around the phosphorylation site. These results are being taken, since published, as a hallmark of the intrinsic differential selectivity of the three Tpks toward substrates. However, our results are in contradiction with this conclusion, at least regarding Nth1 and Pyk1 proteins, two proteins that had been shown to be only Tpk1 substrates in that work. Here, we show the kinetic behavior of Tpk1 and Tpk2 toward three physiological yeast substrates, Pyk1, Pyk2, and Nth1, at the peptide and protein level. To our surprise, the isoforms did not show any difference in the kinetic parameters measured, not only with the peptides but with the protein substrates, and therefore the Kesp for each substrate was the same for Tpk1 and Tpk2. It is interesting that the kcat estimated for both Tpk1 and Tpk2, using kemptide as substrate, was of 3 s−1 10-fold lower than the value for mammalian C, reported as 20 s−1 (38).
The analysis of some aminoacidic determinants in the peptides derived from Pyk1, Pyk2, and Nth1 confirm results that had been previously suggested in the literature, namely that an Arg at positions P-2 and P-3 is very important and that extra positive charge on the N-terminal flank of P0 is beneficial; on the contrary, a negative charge in this position is detrimental for activity. The presence of an acidic residue in the P+1 position, as in the natural context of Ser83 in Nth1 (Nth1-2), severely impairs the phosphorylation of the peptide. This characteristic should now be included in the predictions of probable phosphorylation sites for yeast PKA kinases. In contradiction with our prediction that Ser83 is not a good phosphorylation site for yeast PKA, a phosphopeptide containing Ser83 in Nth1 has been identified in a global proteomic in vivo study of the yeast phosphoproteome (20); however, this kind of study does not give information regarding the kinase responsible for the phosphorylation of the observed phosphopeptides. We also include a new feature for the prediction of yeast PKA phosphorylation sites; an acidic residue in P+4, as occurs in the IS of the R subunits, is beneficial for substrate recognition. We have antecedents from our own work and others, that in the case of Pyk1 and choline kinase, in vitro phosphorylation of peptides derived from the proteins correlate with the in vivo site phosphorylated by PKA (16, 46).
While we were writing this article, a new report was published (47) in which, using an in vitro peptide screening approach, the authors could determine consensus phosphorylation site motifs targeted by 61 of the 122 kinases in S. cerevisiae. Their results indicate that PKA, as shown for Tpk1, Tpk2, and Tpk3 have only three clear determinants in the phosphorylation sequence: Ser in P0, and Arg in P-2 and P-3. A close inspection of the peptide arrays for each Tpk shows that the three arrays are identical and that no differential amino acid selectivity is suggested for each isoform. Our results presented in this work go a little further in the conclusions that can be obtained. Not only is the sequence selectivity for Tpk1 and Tpk2 the same, but the catalytic activity is identical in both isoforms.
The cellular targeting of PKA and the existence of microdomains of cAMP provide spatial and temporal specificity in the mediation of biological effects controlled by the cAMP-PKA pathway. We have demonstrated that the localization of the three Tpks under different physiological conditions is different (48) Thus, although both Tpk1 and Tpk2 isoforms have the same substrate specificity in vitro, they will not necessarily phosphorylate the same substrate in vivo. There are antecedents of this type of behavior for Tpk1 and Tpk2 (23). Both isoforms of PKA have been shown to have different roles in pseudohyphal differentiation. However, they were both capable of the in vitro phosphorylation of the two transcriptional regulators, Flo8 and Sfl1, involved in this phenotype. We concluded that the differences in substrate and kinase intracellular localization likely contributed to the unique activating function of Tpk2 in pseudohyphal differentiation compared with the inhibitory role of Tpk1.
It has been demonstrated that both the mammalian type Iα and the type IIβ holoenzymes are more stable in the presence of cAMP than thought previously and that the substrate can enhance the dissociation of PKA type I at low, physiologically relevant concentrations of cAMP but failed to affect the interaction of the subunits of type II kinase (26, 27). In our laboratory, we have demonstrated the participation of the peptide substrate in the activation of a fungal (Mucor rouxii) holoenzyme by cAMP (29). It was interesting to analyze whether the substrate could participate in the PKA holoenzyme activation in S. cerevisiae. Using an immobilized yeast holoenzyme formed by Bcy1 and a mixture of Tpks, we demonstrate in this work that both the peptide and protein substrates sensitize the holoenzyme to activation by cAMP. The protein substrate Pyk1 at a concentration of 2.7 or 3.5 μm (in the same order as physiological concentrations) can enhance the dissociation of yeast PKA at physiological cAMP concentrations (0.03–0.13 μm). The results show that there is a correlation between the behavior of the peptides as sensitizers to cAMP activation and their Kesp for the isolated C subunit; suggesting that the sensitization occurs at the level of competition between the substrate and the IS region of the regulatory subunit. However, interactions through different domains either in C or R, as has been suggested by us and others (49, 50), might be participating in the activation by a protein substrate, because the whole Pyk1 protein was a better activator than the peptide derived from it. Previous results from our group give a clear indication that activation of PKA by cAMP within the cell might be a regulatable process. We have measured PKA activity in situ in permeabilized cells, preserving its intracellular localization, and compared this value with total PKA activity measured in crude extracts in vitro and found that in situ, one can measure around 1–2% of the total PKA activity (41). These data indicate that there is a very high inhibitory effect of the R subunit within the cell and that the PKA is not freely accessible to the peptide substrate used in the in situ phosphorylation assay. The local cAMP concentration of the cAMP generated endogenously as well as the endogenous substrate concentrations might be key factors in the activation mechanism of the PKA holoenzyme. Substrate availability as well as substrate sensitization of the holoenzyme to cAMP can therefore be considered as a fine regulation mechanism in the activation of PKA.
In conclusion, the specificity of the substrate recognition by a kinase depends on a dynamic interrelationship of several factors: the amino acid sequence around the phosphorylation amino acid, the substrate and kinase localization and expression levels, and the presence or absence of kinase anchor proteins that limit the kinase-substrate interactions. In particular the substrate specificity for the isoforms Tpk1, Tpk2 and Tpk3 in yeast does not seem to rely on the sequence determinants around the phosphorylation sequence nor on a difference in kcat for each isoform. A new point of regulation to be considered and that might contribute to isoform selectivity is the participation of the substrate in the activation of PKA by cAMP.
Supplementary Material
Acknowledgments
We thank Susan Taylor (University of California, San Diego) for the generous gift of the peptide arrays and C. J. Allison for the skillful preparation of the arrays.
This work was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica (PICT 38212), from the University of Buenos Aires (UBA X-197), and from the Consejo Nacional de Investigaciones Científicas y Técnicas (PIP5239).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
S. Moreno, personal communication.
- PKA
- protein kinase A
- TAP
- tandem affinity purification
- IS
- inhibitory site
- SD
- synthetic defined medium.
REFERENCES
- 1.Taylor S. S., Kim C., Vigil D., Haste N. M., Yang J., Wu J., Anand G. S. (2005) Biochim. Biophys. Acta 1754, 25–37 [DOI] [PubMed] [Google Scholar]
- 2.Walsh D. A., Van Patten S. M. (1994) FASEB J. 8, 1227–1236 [DOI] [PubMed] [Google Scholar]
- 3.Kennelly P. J., Krebs E. G. (1991) J. Biol. Chem. 266, 15555–15558 [PubMed] [Google Scholar]
- 4.Zetterqvist O., Ragnarsson U., Engstrom L. (1990) Peptides and Protein Phosphorylation (Kemp B. E. ed) pp. 171–210, CRC Press, Boca Raton, FL [Google Scholar]
- 5.Kemp B. E., Graves D. J., Benjamini E., Krebs E. G. (1977) J. Biol. Chem. 252, 4888–4894 [PubMed] [Google Scholar]
- 6.Pearson R. B., Kemp B. E. (1991) Methods Enzymol. 200, 62–81 [DOI] [PubMed] [Google Scholar]
- 7.Kreegipuu A., Blom N., Brunak S. (1999) Nucleic Acids Res. 27, 237–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kreegipuu A., Blom N., Brunak S., Järv J. (1998) FEBS Lett. 430, 45–50 [DOI] [PubMed] [Google Scholar]
- 9.Shabb J. B. (2001) Chem. Rev. 101, 2381–2411 [DOI] [PubMed] [Google Scholar]
- 10.Walsh D. A., Glass D. B., Mitchell R. D. (1992) Curr. Opin Cell Biol. 4, 241–251 [DOI] [PubMed] [Google Scholar]
- 11.Taylor S. S., Knighton D. R., Zheng J., Ten Eyck L. F., Sowadski J. M. (1992) Annu. Rev. Cell Biol. 8, 429–462 [DOI] [PubMed] [Google Scholar]
- 12.Tegge W., Frank R., Hofmann F., Dostmann W. R. (1995) Biochemistry 34, 10569–10577 [DOI] [PubMed] [Google Scholar]
- 13.Songyang Z., Blechner S., Hoagland N., Hoekstra M. F., Piwnica-Worms H., Cantley L. C. (1994) Curr. Biol. 4, 973–982 [DOI] [PubMed] [Google Scholar]
- 14.Loog M., Oskolkov N., O'Farrell F., Ek P., Järv J. (2005) Biochim. Biophys. Acta 1747, 261–266 [DOI] [PubMed] [Google Scholar]
- 15.Denis C. L., Kemp B. E., Zoller M. J. (1991) J. Biol. Chem. 266, 17932–17935 [PubMed] [Google Scholar]
- 16.Portela P., Moreno S., Rossi S. (2006) Biochem. J. 396, 117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herman P. K. (2002) Curr. Opin Microbiol. 5, 602–607 [DOI] [PubMed] [Google Scholar]
- 18.Thevelein J. M., de Winde J. H. (1999) Mol. Microbiol. 33, 904–918 [DOI] [PubMed] [Google Scholar]
- 19.Budovskaya Y. V., Stephan J. S., Deminoff S. J., Herman P. K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13933–13938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gnad F., de Godoy L. M., Cox J., Neuhauser N., Ren S., Olsen J. V., Mann M. (2009) Proteomics 9, 4642–4652 [DOI] [PubMed] [Google Scholar]
- 21.Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., Fasolo J., Guo H., Jona G., Breitkreutz A., Sopko R., McCartney R. R., Schmidt M. C., Rachidi N., Lee S. J., Mah A. S., Meng L., Stark M. J., Stern D. F., De Virgilio C., Tyers M., Andrews B., Gerstein M., Schweitzer B., Predki P. F., Snyder M. (2005) Nature 438, 679–684 [DOI] [PubMed] [Google Scholar]
- 22.Toda T., Cameron S., Sass P., Zoller M., Wigler M. (1987) Cell 50, 277–287 [DOI] [PubMed] [Google Scholar]
- 23.Pan X., Heitman J. (2002) Mol. Cell. Biol. 22, 3981–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chevtzoff C., Vallortigara J., Avéret N., Rigoulet M., Devin A. (2005) Biochim. Biophys. Acta 1706, 117–125 [DOI] [PubMed] [Google Scholar]
- 25.Palomino A., Herrero P., Moreno F. (2006) Nucleic Acids Res. 34, 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang S., Fletcher W. H., Johnson D. A. (1995) Biochemistry 34, 6267–6271 [DOI] [PubMed] [Google Scholar]
- 27.Viste K., Kopperud R. K., Christensen A. E., Døskeland S. O. (2005) J. Biol. Chem. 280, 13279–13284 [DOI] [PubMed] [Google Scholar]
- 28.Vigil D., Blumenthal D. K., Brown S., Taylor S. S., Trewhella J. (2004) Biochemistry 43, 5629–5636 [DOI] [PubMed] [Google Scholar]
- 29.Zaremberg V., Donella-Deana A., Moreno S. (2000) Arch. Biochem. Biophys. 381, 74–82 [DOI] [PubMed] [Google Scholar]
- 30.Sherman F., Fink G., Hicks J. B. (1981) Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31.Mesecar A. D., Nowak T. (1997) Biochemistry 36, 6803–6813 [DOI] [PubMed] [Google Scholar]
- 32.Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. (2001) Methods 24, 218–229 [DOI] [PubMed] [Google Scholar]
- 33.Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K., (2003) Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 34.Roskoski R., Jr. (1983) Methods Enzymol. 99, 3–6 [DOI] [PubMed] [Google Scholar]
- 35.Portela P., Howell S., Moreno S., Rossi S. (2002) J. Biol. Chem. 277, 30477–30487 [DOI] [PubMed] [Google Scholar]
- 36.Uno I., Matsumoto K., Adachi K., Ishikawa T., (1983) J. Biol. Chem. 258, 10867–10872 [PubMed] [Google Scholar]
- 37.Wera S., De Schrijver E., Geyskens I., Nwaka S., Thevelein J. M., (1999) Biochem. J. 343, 621–626 [PMC free article] [PubMed] [Google Scholar]
- 38.Lieser S. A., Aubol B. E., Wong L., Jennings P. A., Adams J. A. (2005) Biochim. Biophys. Acta 1754, 191–199 [DOI] [PubMed] [Google Scholar]
- 39.Mazón M. J., Behrens M. M., Morgado E., Portillo F. (1993) Eur. J. Biochem. 213, 501–506 [DOI] [PubMed] [Google Scholar]
- 40.Moore M. J., Adams J. A., Taylor S. S. (2003) J. Biol. Chem. 278, 10613–10618 [DOI] [PubMed] [Google Scholar]
- 41.Portela P., Moreno S. (2006) Cell. Signal. 18, 1072–1086 [DOI] [PubMed] [Google Scholar]
- 42.Neuberger G., Schneider G., Eisenhaber F. (2007) Biology Direct 2, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knighton D. R., Zheng J. H., Ten Eyck L. F., Xuong N. H., Taylor S. S., Sowadski J. M. (1991) Science 253, 414–420 [DOI] [PubMed] [Google Scholar]
- 44.Kim C., Cheng C. Y., Saldanha S. A., Taylor S. S. (2007) Cell 130, 1032–1043 [DOI] [PubMed] [Google Scholar]
- 45.Wu J., Brown S. H., von Daake S., Taylor S. S. (2007) Science 318, 274–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi M. G., Kurnov V., Kersting M. C., Sreenivas A., Carman G. M. (2005) J. Biol. Chem. 280, 26105–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mok J., Kim P. M., Lam H. Y., Piccirillo S., Zhou X., Jeschke G. R., Sheridan D. L., Parker S. A., Desai V., Jwa M., Cameroni E., Niu H., Good M., Remenyi A., Ma J. L., Sheu Y. J., Sassi H. E., Sopko R., Chan C. S., De Virgilio C., Hollingsworth N. M., Lim W. A., Stern D. F., Stillman B., Andrews B. J., Gerstein M. B., Snyder M., Turk B. E. (2010) Sci. Signal 3, ra12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tudisca V., Recouvreux V., Moreno S., Boy-Marcotte E., Jacquet M., Portela P. (2010) Eur. J. Cell Biol. 89, 339–348 [DOI] [PubMed] [Google Scholar]
- 49.Deminoff S. J., Ramachandran V., Herman P. K. (2009) Genetics 182, 529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rinaldi J., Ocampo J., Rossi S., Moreno S. (2008) Arch. Biochem. Biophys. 480, 95–103 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.