Abstract
p400/mDomino is an ATP-dependent chromatin-remodeling protein that catalyzes the deposition of histone variant H2A.Z into nucleosomes to regulate gene expression. We previously showed that p400/mDomino is essential for embryonic development and primitive hematopoiesis. Here we generated a conditional knock-out mouse for the p400/mDomino gene and investigated the role of p400/mDomino in adult bone marrow hematopoiesis and in the cell-cycle progression of embryonic fibroblasts. The Mx1-Cre- mediated deletion of p400/mDomino resulted in an acute loss of nucleated cells in the bone marrow, including committed myeloid and erythroid cells as well as hematopoietic progenitor and stem cells. A hematopoietic colony assay revealed a drastic reduction in colony-forming activity after the deletion of p400/mDomino. Moreover, the loss of p400/mDomino in mouse embryonic fibroblasts (MEFs) resulted in strong growth inhibition. Cell-cycle analysis revealed that the mDomino-deficient MEFs exhibited a pleiotropic cell-cycle defect at the S and G2/M phases, and polyploid and multi-nucleated cells with micronuclei emerged. DNA microarray analysis revealed that the p400/mDomino deletion from MEFs caused the impaired expression of many cell-cycle-regulatory genes, including G2/M-specific genes targeted by the transcription factors FoxM1 and c-Myc. These results indicate that p400/mDomino plays a key role in cellular proliferation by controlling the expression of cell-cycle-regulatory genes.
Keywords: Cell Cycle, Chromatin Remodeling, Gene Knockout, Hematopoiesis, Myc, Ep400, FoxM1, H2AZ, mDomino, p400
Introduction
The alteration of chromatin structure and function is a critical process in the transcriptional activation or repression of various cell-type-specific or developmentally regulated genes and is mainly executed by covalent histone modification and ATP-hydrolysis-dependent chromatin remodeling. All of the ATP-dependent chromatin remodelers are multisubunit protein complexes containing a SWI2/SNF2 family ATPase subunit, which plays a central role in chromatin-remodeling activities (1, 2). p400/mammalian Domino (p400/mDomino,3 Gene symbol: Ep400), which was identified as an interaction partner for adenovirus E1A (3) and a myeloid-specific transcription factor, MZF-2A (4), is an SWR1-class chromatin-remodeling ATPase that is homologous to the yeast Swr1p and Drosophila Domino proteins (5, 6). The p400/mDomino-containing protein complex consists of more than 10 subunits, including the Tip60 histone acetyltransferase and a PI3K family protein kinase TRRAP (3, 7, 8). The SWR1-class remodelers are responsible for the regulated exchange of selective histone H2A variants, such as H2A.Z, with the canonical H2A in nucleosomes (5, 9–11). This histone-exchanging activity plays a key role in the epigenetic regulation of gene expression as well as in DNA repair (12–16).
p400/mDomino is known to interact physically and/or functionally with growth-regulating transcription factors, such as Myc, p53, E2F, and adenovirus E1A (3, 17–20). In primary human fibroblasts and osteosarcoma-derived U2OS cells, the knockdown of p400/mDomino results in cell-cycle arrest at the G1 phase with the induction of p21 expression (21, 22). At the p21 promoter, p400/mDomino colocalizes with p53 at a p53-binding site and with c-myc at a TATA region to regulate p21 expression (16, 19). Thus, p400/mDomino appears to regulate both the p53-dependent and c-Myc-modulated expression of p21, which blocks cell proliferation and leads to cellular senescence. However, a recent study showed that the siRNA-mediated knockdown of p400/mDomino in mouse cells reduces the proliferation rate of embryonic stem cells without p21 up-regulation, and has only a modest effect on the cell cycle of embryonic fibroblasts (23). This discrepancy in the effect of p400 knockdown on cellular proliferation could be attributable to cell-type-specific roles of p400/mDomino, or different levels of residual p400/mDomino protein might have resulted in the distinct phenotypes in these knockdown experiments.
We previously reported that mice with an N-terminally deleted p400/mDomino mutation die in utero with a defect in primitive erythropoiesis, indicating that the mDomino complex is essential for early development (24). In this study, we generated conditional knock-out mice to examine the role of p400/mDomino in adult mice. The induced deletion of mDomino in mice led to lethality within 2 weeks, accompanied by the rapid loss of bone marrow cells, including hematopoietic progenitor/stem cells. Analysis of the cell-cycle profile of the mDomino-deleted fibroblasts showed that mDomino was essential for cell-cycle progression. Gene expression analysis revealed that the mDomino deletion caused the reduced expression of cell-cycle-regulatory genes that are targets of the transcription factors FoxM1 and c-Myc. These results indicated that mDomino plays a key role in cellular proliferation by regulating the expression of genes involved in cell-cycle progression.
EXPERIMENTAL PROCEDURES
Construction of Targeting Vectors and Generation of Conditional Knock-out Mice
FLPe transgenic mice (25) were kindly provided by Dr. S. Itohara (Brain Science Institute, RIKEN, Japan) through the RIKEN BioResource Center (Tsukuba, Japan). E2a-Cre (26) and Mx1-Cre transgenic mice (27) were from the Jackson Laboratory. A targeting vector was constructed in which exon 15 of the mDomino gene was flanked with two loxP sites (Fig. 1A). In brief, a loxP site was inserted into intron 14, and an FRT- and loxP-flanked neomycin-resistance gene (NeoFRT cassette) was inserted into intron 15 of the mDomino gene. The diphtheria toxin A fragment gene (DTA cassette) for negative selection was ligated to the 3′-end of the 3′-homologous arm.
FIGURE 1.
p400/mDomino is essential for embryonic development. A, schematic illustration of the exon organization of the p400/mDomino gene and targeting strategy. A targeting vector was designed in which exon 15 was flanked by two loxP sites, and an FRT/loxP-flanked Neo cassette (NeoFRT) was inserted into intron 15. Removal of the NeoFRT cassette by FLPe recombinase generated the mDomfl allele. Cre-mediated recombination between the loxP sites generated the exon-15-deleted allele (mDomΔ). The positions of the primers used for genotyping are indicated by short arrows. B, Southern blot analysis of a correctly targeted ES clone. Genomic DNA from parental (R1) or targeted (Dom) ES cells was digested with ApaI and AflII, and analyzed by Southern blotting using the probe indicated by the hatched box in A. The WT and targeted mDom alleles were predicted to result in 8.6- and 10.6-kb bands, respectively, as shown in A. C, PCR genotyping of mDomfl/fl and mDomΔ/+ mice. D, genotype analysis of the progeny from mDomΔ/+ heterozygous matings.
mDom+/NeoFRT mice were produced as described previously (28). In brief, mouse R1 embryonic stem (ES) cells were transfected with the targeting vector by electroporation, and G418-resistant clones were screened for homologous recombination by PCR. ES clones carrying the single mDomino-targeted allele were injected into BDF1 blastocysts, which were then implanted into recipient ICR female mice. Chimeric mice with a high ES contribution were crossed to C57BL/6 females to yield mDom+/NeoFRT mice. Germ line transmission was identified by coat color and then confirmed by PCR. To remove the NeoFRT cassette, mDom+/NeoFRT heterozygotes were crossed with CAG-FLPe transgenic mice (25), generating mDom+/fl mice. To delete the mDomfl allele in vivo, poly(I):poly(C) (pI:pC, BD Biosciences) was given by one intraperitoneal injection (5 mg/kg body weight) every other day, for a total of three injections. All the mice were housed in a specific pathogen-free facility at Kyoto University, and all animal experiments were carried out in accordance with protocols approved by Kyoto University (Kyoto, Japan).
Genomic DNA for PCR was prepared from tail snips. The genotype of the mDomino gene was determined by PCR using a mixture of three specific primers: an exon-14 sense primer (5′-ATTGGAAAATCCAACACCAAGGA-3′) and two antisense primers for the wild-type (WT) exon 15 (5′-GTCTCGGAGAGCACCATACAACAAAGATGG-3′) and its floxed allele (5′-CCCTGGGATGCCTGCAAGCTTATAACTTCG-3′).
For Southern hybridization, genomic DNA extracted from parental and mDom+/NeoFRT ES cells was digested with restriction enzymes, separated by agarose gel electrophoresis, and transferred to a BA85 nitrocellulose filter (Schleicher & Schuell). Hybridization was carried out using a 32P-labeled probe (Fig. 1A).
Flow Cytometry and Western Blotting
For flow cytometric analyses, antibodies against the following proteins were purchased from BD Pharmingen: CD3 (145–2C11), CD4 (L3T4), CD8 (53–6.7), c-Kit (2B8), Sca-1 (D7), Mac-1 (M1/70), B220 (RA3–6B2), Gr-1 (RB6–8C5), FcRII/III (2.4G2), Ter119, and CD71 (C2). The flow cytometric characterization of bone marrow hematopoietic cells was performed as described before (29).
To detect endogenous mDomino protein, mouse embryonic fibroblasts (MEFs) were lysed directly in Laemmli sample loading buffer and heated for 30 min at 85 °C and for 5 min at 95 °C. The samples were separated by electrophoresis on a 5% SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane filter (Millipore). Immunoblotting analysis was carried out using the enhanced chemiluminescence system (Millipore) with a rabbit anti-Dom-C polyclonal antibody (4) and peroxidase-conjugated goat anti-rabbit immunoglobulin (Dako).
Histology
To prepare bone marrow sections, femurs were fixed with 4% paraformaldehyde in 0.1 m sodium phosphate buffer (pH 7.2, SPB) containing 4% sucrose, and then incubated at room temperature for 24 h in Morse's solution (0.4 m sodium citrate and 22.5% formic acid) for decalcification, and embedded in paraffin. The blocks were sectioned at 4 μm, deparaffinized, and subjected to staining with hematoxylin and eosin.
To detect apoptotic cells, tissues were fixed in 0.1 m SPB containing 4% sucrose and 4% paraformaldehyde at 4 °C for 2 h, then transferred to 0.1 m SPB containing 10% sucrose at 4 °C for 4 h, and then to 0.1 m SPB containing 20% sucrose at 4 °C overnight. The fixed tissues were embedded in OCT compound (Sakura), quickly frozen, sectioned at 8 μm, and mounted onto APS-coated glass slides (Matsunami). For TUNEL staining, the fixed sections were stained with an ApopTag fluorescein in situ apoptosis detection kit from Chemicon International, and the nuclei were counterstained with DAPI. The sections were mounted with Fluoromount (Calbiochem) and were visualized by fluorescence microscopy (Keyence BIOREVO).
Cell Culture and Establishment of CreER MEFs
Primary mDomfl/fl MEFs were prepared from E13.5 embryos and immortalized according to the 3T3 protocol. The MEFs were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin (hereafter “culture medium”) in a humidified atmosphere. The pMXs-CreERT2-IRES-EGFP-puroR plasmid (CreER refers to a fusion of protein Cre with the estrogen receptor) was transfected into the packaging cell line Plat-E (30) using FuGENE transfection reagent (Roche Diagnostics), and the transfected cells were cultured for 2 days to produce a helper-free retrovirus encoding CreERT2. The culture supernatant was recovered and used directly as a retrovirus stock. Immortalized MEFs (1 × 105 cells) in 10-cm dishes were infected by adding 10 ml of the culture medium containing 10 μg/ml Polybrene and 1% of the retrovirus stock. To delete the mDomfl allele from mDomfl/fl:CreER MEFs, the cells were treated with 7.5 nm 4-hydroxytamoxifen (OHT) for 8 h, washed with the culture medium, and further incubated in culture medium under a humidified atmosphere. To establish MEFs expressing exogenous mDomino, the mDomfl/fl;CreER MEFs were transfected with the pNEF-DomHA plasmid encoding a neomycin-resistance gene and the C-terminally HA-tagged mDomino cDNA (1/2-disrupted Myc-mDomino-HA (4)) by electroporation using the Amaxa system (Amaxa), and stable transfectants were selected by G418 resistance.
Cell-cycle Analysis
Cells were trypsinized and fixed in 70% ethanol overnight at −20 °C, treated with 100 μg/ml RNase A (Sigma) in phosphate-buffered saline (PBS) for 30 min at room temperature, and then incubated with 40 μg/ml propidium iodide in PBS for 30 min on ice. Data were acquired using a FACSCalibur (BD Biosciences) and analyzed using Flow-Jo software (TreeStar, Inc.).
Microarray Analysis
For DNA microarray analysis, RNA was extracted from Domfl/fl;CreER MEFs that were left untreated or treated with 7.5 nm OHT for 8 h and then cultured for 2 days. Biotin labeling of complementary RNA was performed using the GeneChip 3′ IVT Express kit (Affymetrix). The biotinylated RNA was fragmented and hybridized to Mouse Gene 430 2.0 chips (Affymetrix) as per the manufacturer's protocol. Both raw image (.dat) and intensity (.cel) files were generated using Gene Chip Operating Software (Affymetrix). All the data, including the signal intensity of each gene, were determined with Microarray Analysis Suite Version 5.0 (Affymetrix). The overall signal intensity of each array was normalized so that the average would be 500. The -fold change analysis was done using the average of OHT-untreated MEFs (n = 3) relative to the average of OHT-treated MEFs (n = 3). p values were calculated using Student's t test. The data were filtered for a change in expression exceeding 2.0-fold and a p value of <0.015. In addition, in the assessment of down-regulated genes, genes presenting with a negative value in at least one of the three profiles or with an average intensity of <100 were deleted in the profiles of OHT-untreated MEFs.
Real-time PCR for the Quantitative Analysis of mRNA
For the reverse-transcribed (RT)-PCR reaction, cDNA was synthesized from DNase I-treated total RNA (0.5 μg) using an oligo(dT) primer and Superscript III (Invitrogen) in a 10-μl reaction mixture. Quantitative real-time RT-PCR was carried out using the LightCycler 480 SYBR Green system (Roche Diagnostics), as described before (31), under the following conditions: 10 s at 95 °C, 10 s at 60 °C, and 20 s at 72 °C for 40 cycles. The primer sets used were: 5′-AGCGTTAAGCAGGAACTGGA-3′ and 5′-TCTGCTGTGATTCCAAGTGC-3′ for FoxM1, 5′-TCACTTCTGGCTACATCCCC-3′ and 5′-ATAGGACTCCGTGCCATCAC-3′ for PLK1, 5′-TGAGGAGAAGCAGTGAGGAA-3′ and 5′-CTGAGAGGTATTCTTAGCCT-3′ for CENP-F, 5′-GGGAGAACTTTCCAGGTGTC-3′ and 5′-AGAGAGACTCATCGAGCGAG-3′ for Skp2, 5′-GTGGGTCAGCGCCACACCTC-3′ and 5′-GGGAGAGGCGCTTGTGCAGG-3′ for p53, 5′-TGGCTGGCGGTAAGGCTGGA-3′ and 5′-ACGTCCGTGGCTGGTTGTCC-3′ for H2A.Z, 5′-GGACTGTATGTGGAGCGGTT-3′ and 5′-GAATCGGACGAGGTACAGGA-3′ for c-Myc, and 5′-CCCAGCGCCTGGCCTATGTG-3′ and 5′-TGCAGTCCGGTCCTCCCCAG-3′ for E2F1. All the PCR data were normalized to standards and expressed as copy numbers of target mRNA per nanogram of total RNA.
RESULTS
Generation of p400/mDomino Conditional Knock-out Mice
The targeting construct that was designed to delete exon 15 of the mDomino gene by Cre-mediated recombination is shown in Fig. 1A. The deletion of exon 15 from the mDomino locus is expected to remove the catalytic center of the ATPase domain and to generate a mutant mDomino that lacks a major portion (encoded by exons 15–52) of the protein (4). We used an FRT- and loxP-flanked Neo cassette (NeoFRT) and a DTA cassette as positive and negative selection markers, respectively, for homologous recombination. Mouse ES cell clones containing the mDomino-targeted allele (mDomNeoFRT) were identified by PCR (not shown) and Southern blot analyses (Fig. 1B) and were used to generate mice carrying the mDomNeoFRT allele in the germ line. Then, the mDomNeoFRT mice were crossed with CAG-FLPe transgenic mice (25) to remove the NeoFRT cassette. The resulting mDomino allele containing two loxP sites in introns 14 and 15 (the “floxed” allele) was designated as mDomfl (Fig. 1, A and C). mDomfl/fl mice were obtained from heterozygous matings at a Mendelian ratio and were phenotypically indistinguishable from their wild-type (WT) or heterozygous littermates, indicating that the mDomfl allele was functional (data not shown). We also crossed mDomNeoFRT mice with E2a-Cre transgenic mice (26), to obtain an mDomino allele lacking exon 15 in the germ line, which was designated as mDomΔ (Fig. 1, A and C). Heterozygous mDomΔ/+ mice were born and developed normally, but no homozygous mutant offspring were born from the intercross of mDomΔ/+ mice, indicating that the homozygous mDomΔ/Δ mutation is lethal during embryonic development (Fig. 1D). We have not determined whether mDomΔ/Δ embryos die in a very early stage of embryogenesis, or can develop at least to embryonic day 8.5 as observed in the homozygous embryos expressing the N-terminally deleted mDomino mutant (24).
Acute Loss of Bone Marrow Hematopoietic Cells Caused by the Induced Deletion of mDomino
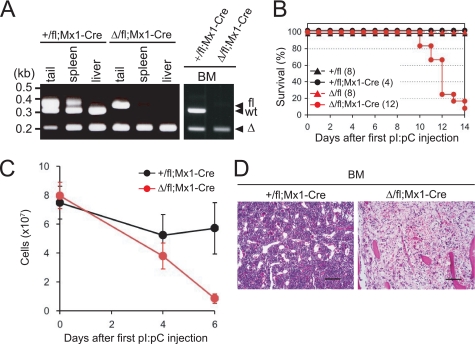
To generate mice in which the mDomino gene was inducibly inactivated, mDomfl mice were crossed with Mx1-Cre transgenic mice (Mx1-Cre), which carry the Cre gene under the control of the Mx1 promoter. The Mx1-Cre gene is induced in adult mice by the administration of poly(I):poly(C) (pI:pC) via interferon (IFN) induction (27). To estimate the efficiency of the mDom deletion in adult tissues, pI:pC was administered intraperitoneally to mDom-floxed mice three times, once each on days 0, 2, and 4. Two days after the last pI:pC injection, genomic DNA from the tail, spleen, liver, and bone marrow (BM) was analyzed by PCR. Although the deletion of the floxed exon 15 of the mDomfl allele was inefficient in the tail, an efficient deletion of the floxed allele (50–70%) was observed in the spleen, and almost complete deletion was achieved in the liver and BM in the mDomΔ/fl;Mx1-Cre and the mDom+/fl;Mx1-Cre mice (Fig. 2A).
FIGURE 2.
Mx1-Cre-mediated conditional deletion of mDomino leads to rapid mortality and a loss of bone marrow cells. Mice were given three intraperitoneal injections of pI:pC, one each on days 0, 2, and 4, and then analyzed. A, PCR genotyping of DNA from the tail, spleen, liver, and BM was performed on day 6 for the pI:pC-treated mDom+/fl;Mx1-Cre and mDomΔ/fl;Mx1-Cre mice. B, survival of the pI:pC-treated mDomΔ/fl;Mx1-Cre mice (n = 12) and control mice (n = 20 in total) was observed daily. C, mDom+/fl;Mx1-Cre mice (black, n = 3) and mDomΔ/fl;Mx1-Cre mice (red, n = 3) were treated with pI:pC, and the total number of nucleated BM cells in all the femurs and tibiae from a single mouse was determined on the indicated days. D, hematoxylin and eosin staining of a paraffin-fixed section of the femur 8 days after the first pI:pC injection. Scale bars represent 100 μm.
To explore the role of mDomino in adult mice, mDomΔ/fl;Mx1-Cre mice were injected with pI:pC, as described above. This treatment resulted in the death of almost all of the mDomΔ/fl;Mx1-Cre mice within 14 days after the first pI:pC injection, whereas no mortality was observed in any of the pI:pC-injected control mice (mDom+/fl;Mx1-Cre, mDomΔ/fl, and mDom+/fl) (Fig. 2B). Our previous study showed that mice with the N-terminally deleted mDom mutation die during embryonic development with defects in primitive hematopoiesis (24). Therefore, to investigate the roles of mDomino in adult hematopoiesis, we examined the BM phenotypes of the pI:pC-injected mDomΔ/fl;Mx1-Cre mice on days 4 and 6 (i.e. 0 and 2 days after the last pI:pC injection, respectively), and found a drastic reduction of nucleated cells (Fig. 2C). Hematoxylin-eosin staining revealed the obvious disappearance of BM hematopoietic cells, except for the anucleate mature erythrocytes (Fig. 2D). These results indicated that the proliferation and maintenance of hematopoietic cells in the BM are severely impaired by the inactivation of mDomino.
To determine which of the BM hematopoietic lineages was affected by the mDomino deficiency, we analyzed the BM cells of pI:pC-treated mDomΔ/fl mice by flow cytometry. The treatment of control mice, such as mDom+/fl;Mx1-Cre mice (Fig. 3, A and B) and mDomΔ/fl mice (free of Mx1-Cre, data not shown), with pI:pC resulted in a significant reduction of bone marrow B220+ B cells and Ter119+ erythroid cells but not of Mac-1+ or Gr-1+ myeloid cells, indicating that IFN (and/or pI:pC itself) has some deleterious effect on the lymphoid and erythroid lineages but not on the myeloid lineage. In contrast, the deletion of mDomino in BM cells resulted in a significant reduction of the Mac-1+Gr-1mid-lo monocyte/macrophage lineage and Mac-1+Gr-1hi granulocyte lineage (Fig. 3, A and B). The CD71+Ter119mid-hi erythroid progenitors were also diminished when compared with the pI:pC-injected control mice. The reduction in nucleated hematopoietic cells was accompanied by a dramatic increase in CD71−Ter119high mature erythrocytes, which comprised nearly 80% of the non-adherent cells in the BM cavity (Fig. 3, A and B). Furthermore, lineage-marker-negative (Lin−)c-kit+Sca-1+ hematopoietic stem cells and Lin−c-kit+Sca-1− progenitor cells were lost in the pI:pC-treated mDomΔ/fl;Mx1-Cre mice (Fig. 3C), indicating that mDomino is necessary for the proliferation and/or maintenance of hematopoietic progenitor/stem cells as well as of committed myeloid cells. To address whether the mDomino-deleted hematopoietic cells underwent apoptosis, we analyzed BM cells using the TUNEL assay. One day after the second pI:pC injection, the BM cells from mDomΔ/fl;Mx1-Cre mice displayed more TUNEL-positive cells than the control (Fig. 3D). Similar results were obtained by immunohistochemical staining for active caspase-3 (data not shown), indicating that the rapid extinction of nucleated hematopoietic cells from the mDom-deleted BM is, at least in part, due to an increase in their apoptotic cell death.
FIGURE 3.
Loss of BM hematopoietic cells by mDomino deletion. A, total number of BM hematopoietic subsets (n = 3, mean ± S.E.) in pI:pC-treated (+) or untreated (−) mice on day 4. The number of cells in each subset was calculated from the total number of nucleated BM cells and the flow cytometric analysis of hematopoietic subsets shown in B. The hematopoietic subsets were defined by lineage markers as follows: B cells (B220+), erythroblasts (CD71hiTer119mid and CD71lo-hiTer119hi), erythrocytes (CD71−Ter119hi), monocytes (Mac-1+Gr-1mid), and granulocytes (Mac-1+Gr-1hi). B, flow cytometric profiles of BM cells from pI:pC-treated (+) or untreated (−) mice on day 4. C, flow cytometric analysis of lineage-marker-negative (Lin−) hematopoietic stem and progenitor cells (left), and the total number of Lin−Sca-1+c-Kit+(LSK) stem cells (right) among the BM cells from pI:pC-injected mice. D, apoptotic cell death in the mDomino-deleted BM cells. Cryosections of BM were prepared from pI:pC-injected mice on day 3. The BM samples were stained with TUNEL and counterstained with DAPI (left). The TUNEL-positive cells were counted, and the percentage of DAPI-positive cells that were TUNEL-positive was calculated (right). E and F, in vitro hematopoietic colony assay of mDomΔ/fl;Mx1-Cre BM cells under the induced deletion of mDomino by IFN-β. BM cells (2 × 104 nucleated cells) from mDom+/fl;Mx1-Cre or mDomΔ/fl;Mx1-Cre mice were cultured using the MethoCult M3434 system (Stem Cell Technologies) in 35-mm culture dishes in the presence or absence of 100 units/ml mouse IFN-β (Merck), in duplicate. E, three colonies from each of the IFN-treated or untreated (−) cultures of mDom+/fl;Mx1-Cre BM cells were analyzed for mDomino deletion by PCR. F, BM cells were cultured in the presence (filled bars) or absence (open bars) of IFN-β, and colonies of the colony-forming unit (CFU)-GEMM (granulocyte/erythrocyte/macrophage/megakaryocyte) and CFU-GM (granulocyte/macrophage), and the burst-forming unit of erythroid (BFU-E) containing more than 30 cells were counted after 12 days of culture.
Next, we examined the effect of mDomino deletion on the in vitro growth and differentiation of erythroid and myeloid progenitors using a multilineage colony assay. The BM cells from mDomΔ/fl;Mx1-Cre mice or control mDom+/fl;Mx1-Cre mice were cultured in methylcellulose medium containing multiple cytokines in the presence or absence of IFN-β. The presence of 100 units/ml IFN-β, which effectively deleted the mDomfl allele (Fig. 3E), did not have any deleterious effect on the hematopoietic colony formation (CFU-GM, BFU-E, and CFU-GEMM) of control BM cells (Fig. 3F, left), but resulted in a drastic reduction in the colony formation of the mDomΔ/fl;Mx1-Cre cells (Fig. 3F, right). PCR genotyping of the colonies that survived in the IFN-treated mDomΔ/fl;Mx1-Cre BM culture revealed that the small number of surviving colonies arose from cells that escaped the induced deletion of the floxed mDom gene (data not shown). These results indicated that mDomino-deleted hematopoietic progenitors rapidly, and cell autonomously, lose the ability to proliferate.
mDomino Is Necessary for the Cell-cycle Progression of Embryonic Fibroblasts
The rapid proliferation arrest and/or apoptosis in mDom-deleted BM hematopoietic cells suggested that mDomino could be involved generally in the cell growth regulation. To examine this possibility, we prepared MEFs from mDomfl/fl mice and control mDomfl/+ mice, and immortalized them by the 3T3 passage method. To inducibly inactivate the mDomfl allele, the immortalized MEFs were infected with a recombinant retrovirus expressing Cre-ERT2 (hereafter simply CreER), which can be activated by treatment with OHT (32). In the CreER-expressing MEFs, the mDomfl allele was deleted within a day after OHT treatment (Fig. 4A), and the mDomino protein disappeared concomitantly from the cell lysates (Fig. 4B). The OHT-induced deletion of mDomino resulted in the rapid and strong growth inhibition of mDomfl/fl;CreER MEFs but not of the vector control mDomfl/fl MEFs (Fig. 4C). Another control cell line, mDom+/fl;CreER MEF, which was established by the CreER-retrovirus infection of heterozygous mDomfl/+ MEFs, grew normally even after the induced activation of CreER (Fig. 4D). These results indicated that the growth of MEFs is not affected by the addition of OHT or by the activation of the CreER protein and that mDomino is essential for their proliferation. In addition, microscopic examination revealed the emergence of a bi- or multi-nucleated population among the mDomino-deleted MEFs that had an enlarged cell body, abnormal nuclear morphology, and micronuclei (Fig. 4, E and F).
FIGURE 4.
Strong growth inhibition of conditional mDomino-deficient embryonic fibroblasts. A, PCR analysis of the mDomfl deletion by OHT. mDomfl/fl;CreER MEFs were left untreated (−) or treated with 7.5 nm OHT for 8 h, washed and further cultured, and then analyzed for the status of the mDomfl allele on the indicated days. B, Western blot analysis of the mDomino protein in the whole cell lysate of mDomfl/fl;CreER MEFs treated with OHT. C and D, growth defect of mDomino-deficient MEFs. Four types of retrovirally transduced MEF lines, mDomfl/fl;CreER (left), mDomfl/+;CreER (right), and their respective vector controls, were left untreated (−) or treated with OHT (+), and then cultured in the normal medium. At the indicated times, the cells were trypsinized and counted. Data are the means ± S.D. of three separate experiments. E and F, nuclear abnormalities observed in mDomino-deficient MEFs. E, Wright-Giemsa staining of the OHT-treated or untreated (−) mDomfl/fl;CreER MEFs after 6 days of culture. Scale bars represent 100 μm. F, DAPI staining of OHT-treated mDomfl/fl;CreER MEFs (upper). Arrowhead indicates cells with micronuclei. The DAPI image is merged with its bright-field image (lower). Scale bars represent 25 μm.
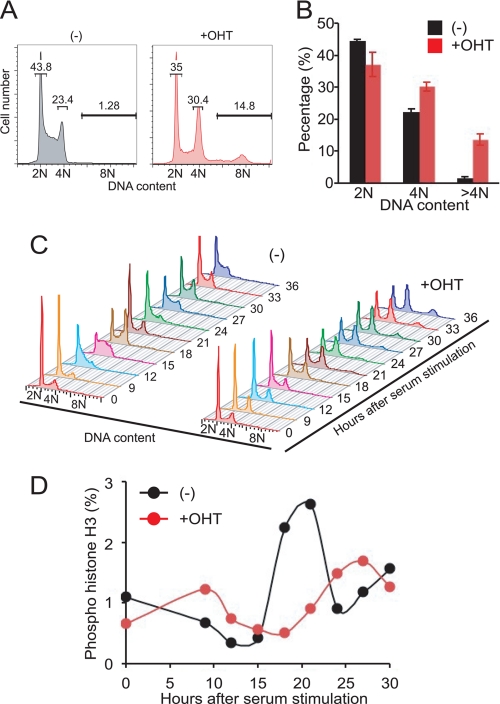
The above results suggested that mDomino plays an important role in cell-cycle regulation. We therefore analyzed the cell-cycle profiles of mDomino-deleted MEFs by flow cytometry. As shown in Fig. 5 (A and B), the mDomfl/fl;CreER MEFs treated with OHT showed a significant increase in the population with 4N DNA content and the emergence of polyploid (>4N) cells. To examine the effect of the mDomino deficiency on cell-cycle progression, mDomfl/fl;CreER MEFs were treated with OHT, driven into quiescence by serum starvation, and then released back into the cell-cycle by the addition of serum. As shown in Fig. 5C, the mock-treated cells synchronously entered the S phase 12–15 h after serum stimulation, reached the G2/M phase at about 18 h, and returned to the G1 phase at 21 h. In contrast, MEFs treated with OHT displayed a small delay in entering the S phase and a slow but continuous increase in a population with 4N DNA content. This 4N population seemed to include not only the G2/M population but also an aberrant cell population that had initiated a new round of DNA synthesis without cytokinesis (endoreduplication), which resulted in the accumulation of tetraploid cells with ∼8N DNA content. To examine whether mDomino-deficient MEFs entered mitosis, MEFs re-stimulated with serum were stained with a phospho-Ser10-specific anti-histone H3 antibody, and quantified by flow cytometry (Fig. 5D). Mock-treated MEFs showed a sharp M-phase peak 18–20 h after stimulation, whereas the mDomino-deficient MEFs showed a delayed entry into mitosis. These results indicated that the depletion of mDomino causes the malfunction of the cell-cycle regulation at multiple steps, especially in the G2/M phase, and eventually leads to the aberrant mitosis known as “mitotic catastrophe.”
FIGURE 5.
mDomino-deficient MEFs exhibit polyploidy and defective cell-cycle progression. A, the DNA content of OHT-treated or untreated (−) mDomfl/fl;CreER MEFs was determined by flow cytometry after propidium iodide staining. B, the percentage of cells with 2N, 4N, and >4N DNA content in A was determined in three independent experiments. C and D, cell-cycle profile of mDomfl/fl;CreER MEFs. Cells were treated with (+) or without (−) OHT for 8 h, serum-starved for 24 h, and then released from the starvation by stimulation with 10% fetal bovine serum. The cells were then collected at the indicated times and subjected to the flow cytometric analysis of their DNA content by propidium iodide staining (C), or of the mitotic index by staining with a phospho-Ser-10-specific, anti-histone H3 antibody (D).
p400/mDomino was shown to participate in the adenovirus-E1A-induced transformation process through interaction with the N-terminal region of E1A (3). To examine whether expression of E1A affects the cell cycle defect caused by mDomino deletion, mDomfl/fl;CreER MEFs were infected with retroviral constructs expressing wild-type 12 S E1A or dl1102(Δ26–35) mutant that was defective in interaction with mDomino. Resulting MEFs that overexpressed E1A or dl1102 grew faster than the parental MEFs or vector control cells, but were growth-arrested soon after the OHT treatment (supplemental Fig. 1), suggesting that overexpression of E1A protein cannot compensate for the mDomino deletion.
To confirm that the observed phenotypes of mDomino-deleted MEFs were caused by the absence of mDomino protein and not by other unexpected events, such as genomic recombination caused by CreER, mDomfl/fl;CreER MEFs were transfected with an expression plasmid for HA-tagged mDomino (pNEF-DomHA), and stable cell lines were analyzed. As shown in Fig. 6, mDomfl/fl;CreER cells expressing mDomino-HA displayed normal proliferation, cell-cycle distribution, and nuclear morphology irrespective of the OHT-induced deletion of the endogenous mDomino. This showed that the cell-cycle deficiency could be rescued by the exogenous expression of the wild-type mDomino protein and was, therefore, due to the absence of mDomino.
FIGURE 6.
Rescue of the cell-cycle phenotype by the exogenous expression of mDomino cDNA. mDomfl/fl;CreER MEFs were transfected with an mDomino expression plasmid, and stable transformants were cultured with or without OHT treatment. A, growth curve of the transformant MEFs treated with (+) or without (−) OHT. Data are the means ± S.D. of three separate experiments. B, cell-cycle profiles of the transformants treated without or with OHT and then cultured for 6 days. C, Wright-Giemsa staining of the transformants grown as in B. Scale bars represent 100 μm.
Impairment of Cell-cycle Regulatory Gene Expression in mDomino-deficient Cells
The p400/mDomino chromatin-remodeling complex is known to interact physically or functionally with cell-cycle-regulatory transcription factors, including c-Myc, E1A, p53, and E2F (3, 17–20). To gain insight into the molecular function of mDomino in cell-cycle regulation, we characterized the gene expression profiles of mDomfl/fl;CreER MEFs treated or untreated with OHT by DNA microarray analysis (n = 3 for both untreated and OHT-treated cells), which revealed that 770 genes were differentially (≥2.0-fold) expressed by mDomino depletion, including 191 down-regulated genes (supplemental Table 1) and 579 up-regulated genes (data not shown). A gene annotation analysis revealed that, among the 93 genes whose expression was reduced to <45% by OHT treatment, many (27 genes) are characterized as cell-cycle-related genes, such as E2F2, E2F7, E2F8, CENP-F (cenpf), Skp2, Nek2, and Cyclin A2 (ccna2) (Table 1). Moreover, 17 of the 93 genes are known Myc targets (supplemental Table 1) (Myc Cancer Gene, available on-line).
TABLE 1.
Cell-cycle-related genes affected by mDomino deletion
DNA microarray analysis of OHT-treated (+OHT) or untreated (−) mDomfl/fl;CreER MEFs were performed as described under “Experimental Procedures.” Among the 93 genes whose expression was reduced to <45% by OHT treatment (supplemental Table 1), genes that are characterized as “cell cycle” are listed.
| Gene symbol | -fold change | +OHT | (−) | p value | Accession number |
|---|---|---|---|---|---|
| E2f2 | 3.5 | 30 | 107 | 7.33E-05 | BB543028 |
| Mybl2 | 3.2 | 208 | 666 | 2.42E-04 | NM_008652 |
| Cenpf | 3.2 | 97 | 310 | 1.02E-02 | BB667318 |
| Psrc1 | 3.0 | 288 | 878 | 4.66E-04 | NM_019976 |
| Fbxo5 | 3.0 | 618 | 1851 | 7.31E-05 | AK011820 |
| E2f7 | 2.7 | 557 | 1517 | 3.98E-03 | BG069355 |
| Cdc6 | 2.7 | 564 | 1531 | 4.25E-05 | NM_011799 |
| Ccna2 | 2.6 | 2615 | 6868 | 2.04E-03 | X75483 |
| Ccnf | 2.6 | 144 | 378 | 3.59E-03 | BB089717 |
| Rbl1 | 2.6 | 178 | 465 | 2.64E-04 | U27178 |
| E2f8 | 2.6 | 336 | 876 | 2.80E-05 | BM247465 |
| Rcc1 | 2.6 | 470 | 1213 | 5.99E-06 | NM_133878 |
| 2810433K01Rik | 2.5 | 581 | 1461 | 2.32E-04 | NM_025581 |
| Mapk12 | 2.5 | 134 | 336 | 6.35E-04 | BC021640 |
| Cdca3 | 2.5 | 1732 | 4277 | 1.52E-03 | BI081061 |
| Uhrf1 | 2.4 | 192 | 470 | 6.75E-03 | BB702754 |
| Suv39h2 | 2.4 | 353 | 854 | 7.45E-04 | NM_022724 |
| Plk1 | 2.4 | 742 | 1787 | 3.42E-05 | NM_011121 |
| Rassf1 | 2.4 | 83 | 198 | 2.07E-03 | BB385028 |
| Cdc25a | 2.4 | 590 | 1405 | 1.46E-02 | C76119 |
| Nek2 | 2.4 | 339 | 802 | 3.80E-04 | NM_010892 |
| Skp2 | 2.3 | 221 | 518 | 1.43E-02 | AV259620 |
| Sgol1 | 2.3 | 314 | 732 | 3.92E-03 | BB410537 |
| Incenp | 2.3 | 87 | 201 | 2.06E-04 | AV301185 |
| Mcm3 | 2.3 | 234 | 531 | 1.02E-03 | BI658327 |
| Cdc20 | 2.2 | 2367 | 5311 | 3.80E-03 | NM_023223 |
| Ncapg2 | 2.2 | 1610 | 3593 | 1.26E-03 | NM_133762 |
To confirm the microarray data and to characterize the time course of mRNA expression during cell-cycle progression, we carried out quantitative RT-PCR analysis of the genes for Skp2, PLK1, CENP-F, and FoxM1. As reported previously, the expressions of these genes were dependent on cell-cycle progression in the control mDomfl/fl;CreER MEFs (Fig. 7A). In cells treated with OHT, the expression levels of these genes were low, and they were not clearly induced after the serum stimulation. We have also compared expression of c-Myc, E2F1, p53, and H2A.Z genes in the OHT-treated or -untreated cells. Expression levels of p53 and H2A.Z in the mDomino-deleted cells were consistently lower than those in the untreated cells. Unexpectedly, the expression of c-Myc and E2F1 genes was rather up-regulated in the mDomino-deficient MEFs (Fig. 7B), suggesting that mDomino may be involved in repression of these genes by a direct mechanism or by an indirect, feedback pathway. RT-PCR analysis of the mDomino mRNA showed that the expression of mDomino in normal cells was independent of the cell cycle (supplemental Fig. 2). Finally, we examined whether the impaired expression of the cell-cycle-related genes could be rescued by the exogenous expression of mDomino protein. The mDomfl/fl;CreER MEFs expressing mDomino-HA exhibited a normal expression of the cell-cycle-regulated genes irrespective of OHT treatment (Fig. 7C). In contrast, the expression of c-Myc, which was up-regulated in the mDom-deficient cells, was reduced to normal by the mDomino expression (Fig. 7D). Together, these results indicated that mDomino plays an essential role in the expression of various genes that are involved in cell-cycle regulation and suggested that the impaired expression of those genes causes the aberrant cell-cycle progression, growth arrest, and mitotic catastrophe of the mDomino-deficient cells.
FIGURE 7.
Expression of cell-cycle-related genes in mDomino-deleted MEFs. A and B, mDomfl/fl;CreER MEFs were untreated (−) or treated (+) with OHT, serum-starved for 24 h, and then re-stimulated with 10% fetal bovine serum. At the indicated times, the RNA was extracted and analyzed for expression of the indicated genes by quantitative RT-PCR. C and D, parental mDomfl/fl;CreER MEFs and mDomino-expressing transformants (fl/fl;CreER + Dom) were untreated (−) or treated (+) with OHT, cultured for 3 days, and then subjected to expression analysis by quantitative RT-PCR.
DISCUSSION
In this report, we generated conditional knock-out mice of the p400/mDomino gene. These mice died within 2 weeks of pI:pC-induced, Mx1-Cre-mediated mDomino depletion. We showed that the mDomino deletion in BM cells resulted in the rapid loss of committed myeloid and erythroid cells as well as hematopoietic progenitor/stem cells. The extinction of hematopoietic cells from the BM was probably due to both a decrease in the proliferation and an increase in the apoptotic cell death of the mDomino-deleted cells, followed by their clearance and replacement by mature erythrocytes. A hematopoietic colony formation assay demonstrated that the IFN-induced deletion of mDomino strongly inhibited the growth of all the colony-forming progenitors, including the myeloid, erythroid, and megakaryocytic lineages, suggesting that the impaired hematopoiesis was cell autonomous and not caused by a defect in the hematopoiesis-supporting environment in the BM.
The strong growth inhibition of the mDomino-deficient hematopoietic cells prompted us to investigate the cell proliferation using embryonic fibroblasts from mDomfl/fl mice. The induced deletion of the mDomino gene led to an acute and strong inhibition of the cellular proliferation of MEFs. Cell-cycle analysis revealed that the mDomino-deficient MEFs exhibited pleiotropic cell-cycle defects, including delayed and insufficient entry into the S phase, increase in the G2/M phase, and the accumulation of polyploid and/or multinucleated cells with micronuclei. These phenotypes are very similar to those of Trrap-deleted MEFs, in which a spindle checkpoint failure leads to mitotic catastrophe (33), suggesting that the polyploid and/or multinucleated population of mDom-deficient cells might arise from a failure in mitotic checkpoint followed by endoreduplication of the DNA without cytokinesis. In Saccharomyces cerevisiae, the Swr1 complex is involved in chromosome segregation and plays an important role in chromosome stability (34). In addition, the TRRAP-Tip60 complex, which also contains p400/mDomino, is known to be required for the recruitment and accumulation of DNA repair proteins at sites of DNA double-strand break (35). Thus, p400/mDomino may also be involved in maintaining chromosome integrity.
In the human osteosarcoma U2OS cell line and primary fibroblasts, the shRNA-mediated knockdown of p400/mDomino results in the induction of cell-cycle inhibitor p21, cell-cycle arrest at G1, and premature senescence (21, 22). However, the mDomino-deleted MEFs, which also showed strong growth inhibition, did not show up-regulated p21 expression (data not shown), suggesting that the loss of mDomino can block the cell cycle by a mechanism other than the p53-p21 pathway. Our DNA microarray analysis showed that the expression levels of many cell-cycle-related genes were significantly reduced in the mDomino-deleted MEFs. Interestingly, among the strongly affected genes, CENP-F, Nek2, and Plk1 are representative G2/M-specific genes that are regulated by the transcription factor FoxM1 (36–38), which was also among the strongly reduced genes. This finding indicates that the cell-cycle phenotypes of the mDomino-deficient MEFs were at least partly due to the impaired expression of FoxM1 and its target genes. In this regard, it is noteworthy that FoxM1-deficient MEFs exhibit mitotic malfunction similar to our results (36). However, the exogenous overexpression of FoxM1 cDNA in mDomfl/fl;CreER MEFs failed to rescue the cell-cycle phenotype (data not shown), suggesting that p400/mDomino is responsible for the regulated expression not only of FoxM1 targets but also of other cell-cycle-related genes. A candidate transcription factor of note is the oncoprotein c-Myc, which interacts with the Tip60-TRRAP-p400/mDomino complex and regulates the expression of various genes positively or negatively, possibly through the histone-acetyl-transferring and/or the H2A.Z-exchanging activities of the complex (3, 17, 19). This hypothesis is consistent with the results of our microarray analysis (supplemental Table 1), in which the strongly affected genes included several Myc targets, such as E2F2, Mybl2, Cdc6, Cyclin A, and FoxM1 (39–43). To elucidate whether p400/mDomino is directly involved in the regulatory expression of these genes, we need to investigate physical and functional interactions of the p400/mDomino chromatin-remodeling complex with the c-Myc and/or FoxM1 on the promoter region of their target genes by chromatin immunoprecipitation analyses using cell-cycle-synchronized fibroblasts, in our future experiments.
Taking our present results together with our previous studies, we have shown that p400/mDomino is essential for hematopoiesis of both the yolk sac and the adult BM. The acute extinction of hematopoietic cells is probably due to the strong growth inhibition and apoptotic cell death of the mDomino-deleted cells. We also provide evidence that p400/mDomino plays an essential role in the cell-cycle progression of fibroblasts, probably through its transcriptional activation of cell-cycle-regulatory genes, especially the targets of FoxM1 and c-Myc. Because p400/mDomino exerts its chromatin-remodeling function via its H2A.Z-exchange activity, the interaction of p400/mDomino with c-Myc (and possibly with FoxM1) might regulate the deposition and/or eviction of the H2A.Z variant at the promoter nucleosome of cell-cycle-regulatory genes (14, 16, 44). Future studies on the interaction between transcription factors and the p400/mDomino complex and their role in the H2A.Z-positioning mechanism will provide insight into the biological significance of the H2A.Z-exchanging machinery in cell growth and differentiation.
Supplementary Material
Acknowledgments
We are grateful to Dr. Shigeyoshi Itohara and the RIKEN BioResource Center for providing the FLPe transgenic mice. We also thank Dr. Toru Nakano for the Cre-ERT2 construct, and Dr. Joe S. Mymryk for the E1A cDNAs.
This work was supported in part by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and Table 1.
- mDomino
- mammalian Domino
- MEF
- mouse embryonic fibroblast
- pI:pC
- poly(I):poly(C)
- SPB
- sodium phosphate buffer
- ER
- estrogen receptor
- OHT
- 4-hydroxytamoxifen
- CreER
- a fusion of protein Cre with the estrogen receptor
- BM
- bone marrow
- TRRAP
- transformation/transactivation domain-associated protein.
REFERENCES
- 1.Saha A., Wittmeyer J., Cairns B. R. (2006) Nat. Rev. Mol. Cell Biol. 7, 437–447 [DOI] [PubMed] [Google Scholar]
- 2.Cairns B. R. (2007) Nat. Struct. Mol. Biol. 14, 989–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuchs M., Gerber J., Drapkin R., Sif S., Ikura T., Ogryzko V., Lane W. S., Nakatani Y., Livingston D. M. (2001) Cell 106, 297–307 [DOI] [PubMed] [Google Scholar]
- 4.Ogawa H., Ueda T., Aoyama T., Aronheim A., Nagata S., Fukunaga R. (2003) Genes Cells 8, 325–339 [DOI] [PubMed] [Google Scholar]
- 5.Kobor M. S., Venkatasubrahmanyam S., Meneghini M. D., Gin J. W., Jennings J. L., Link A. J., Madhani H. D., Rine J. (2004) PLoS Biol. 2, E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruhf M. L., Braun A., Papoulas O., Tamkun J. W., Randsholt N., Meister M. (2001) Development 128, 1429–1441 [DOI] [PubMed] [Google Scholar]
- 7.Cai Y., Jin J., Florens L., Swanson S. K., Kusch T., Li B., Workman J. L., Washburn M. P., Conaway R. C., Conaway J. W. (2005) J. Biol. Chem. 280, 13665–13670 [DOI] [PubMed] [Google Scholar]
- 8.Jin J., Cai Y., Yao T., Gottschalk A. J., Florens L., Swanson S. K., Gutiérrez J. L., Coleman M. K., Workman J. L., Mushegian A., Washburn M. P., Conaway R. C., Conaway J. W. (2005) J. Biol. Chem. 280, 41207–41212 [DOI] [PubMed] [Google Scholar]
- 9.Krogan N. J., Keogh M. C., Datta N., Sawa C., Ryan O. W., Ding H., Haw R. A., Pootoolal J., Tong A., Canadien V., Richards D. P., Wu X., Emili A., Hughes T. R., Buratowski S., Greenblatt J. F. (2003) Mol. Cell 12, 1565–1576 [DOI] [PubMed] [Google Scholar]
- 10.Mizuguchi G., Shen X., Landry J., Wu W. H., Sen S., Wu C. (2004) Science 303, 343–348 [DOI] [PubMed] [Google Scholar]
- 11.Kusch T., Florens L., Macdonald W. H., Swanson S. K., Glaser R. L., Yates J. R., 3rd, Abmayr S. M., Washburn M. P., Workman J. L. (2004) Science 306, 2084–2087 [DOI] [PubMed] [Google Scholar]
- 12.Jin J., Cai Y., Li B., Conaway R. C., Workman J. L., Conaway J. W., Kusch T. (2005) Trends Biochem. Sci. 30, 680–687 [DOI] [PubMed] [Google Scholar]
- 13.van Attikum H., Gasser S. M. (2005) Nat. Rev. Mol. Cell Biol. 6, 757–765 [DOI] [PubMed] [Google Scholar]
- 14.Squatrito M., Gorrini C., Amati B. (2006) Trends Cell Biol. 16, 433–442 [DOI] [PubMed] [Google Scholar]
- 15.Morrison A. J., Shen X. (2009) Nat. Rev. Mol. Cell Biol. 10, 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svotelis A., Gévry N., Gaudreau L. (2009) Biochem. Cell Biol. 87, 179–188 [DOI] [PubMed] [Google Scholar]
- 17.Frank S. R., Parisi T., Taubert S., Fernandez P., Fuchs M., Chan H. M., Livingston D. M., Amati B. (2003) EMBO Rep. 4, 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taubert S., Gorrini C., Frank S. R., Parisi T., Fuchs M., Chan H. M., Livingston D. M., Amati B. (2004) Mol. Cell. Biol. 24, 4546–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gévry N., Chan H. M., Laflamme L., Livingston D. M., Gaudreau L. (2007) Genes Dev. 21, 1869–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu J., Ruhf M. L., Perrimon N., Leder P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9381–9386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan H. M., Narita M., Lowe S. W., Livingston D. M. (2005) Genes Dev. 19, 196–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tyteca S., Vandromme M., Legube G., Chevillard-Briet M., Trouche D. (2006) EMBO J. 25, 1680–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fazzio T. G., Huff J. T., Panning B. (2008) Cell 134, 162–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueda T., Watanabe-Fukunaga R., Ogawa H., Fukuyama H., Higashi Y., Nagata S., Fukunaga R. (2007) Genes Cells 12, 581–592 [DOI] [PubMed] [Google Scholar]
- 25.Kanki H., Suzuki H., Itohara S. (2006) Exp. Anim. 55, 137–141 [DOI] [PubMed] [Google Scholar]
- 26.Williams-Simons L., Westphal H. (1999) Transgenic Res. 8, 53–54 [DOI] [PubMed] [Google Scholar]
- 27.Kühn R., Schwenk F., Aguet M., Rajewsky K. (1995) Science 269, 1427–1429 [DOI] [PubMed] [Google Scholar]
- 28.Ueda T., Watanabe-Fukunaga R., Fukuyama H., Nagata S., Fukunaga R. (2004) Mol. Cell. Biol. 24, 6539–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weischenfeldt J., Damgaard I., Bryder D., Theilgaard-Mönch K., Thoren L. A., Nielsen F. C., Jacobsen S. E., Nerlov C., Porse B. T. (2008) Genes Dev. 22, 1381–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morita S., Kojima T., Kitamura T. (2000) Gene Ther. 7, 1063–1066 [DOI] [PubMed] [Google Scholar]
- 31.Iida S., Kohro T., Kodama T., Nagata S., Fukunaga R. (2005) J. Leukoc. Biol. 78, 481–490 [DOI] [PubMed] [Google Scholar]
- 32.Feil R., Wagner J., Metzger D., Chambon P. (1997) Biochem. Biophys. Res. Commun. 237, 752–757 [DOI] [PubMed] [Google Scholar]
- 33.Herceg Z., Hulla W., Gell D., Cuenin C., Lleonart M., Jackson S., Wang Z. Q. (2001) Nat. Genet. 29, 206–211 [DOI] [PubMed] [Google Scholar]
- 34.Krogan N. J., Baetz K., Keogh M. C., Datta N., Sawa C., Kwok T. C., Thompson N. J., Davey M. G., Pootoolal J., Hughes T. R., Emili A., Buratowski S., Hieter P., Greenblatt J. F. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13513–13518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murr R., Loizou J. I., Yang Y. G., Cuenin C., Li H., Wang Z. Q., Herceg Z. (2006) Nat. Cell Biol. 8, 91–99 [DOI] [PubMed] [Google Scholar]
- 36.Laoukili J., Kooistra M. R., Brás A., Kauw J., Kerkhoven R. M., Morrison A., Clevers H., Medema R. H. (2005) Nat. Cell Biol. 7, 126–136 [DOI] [PubMed] [Google Scholar]
- 37.Laoukili J., Stahl M., Medema R. H. (2007) Biochim. Biophys. Acta 1775, 92–102 [DOI] [PubMed] [Google Scholar]
- 38.Fu Z., Malureanu L., Huang J., Wang W., Li H., van Deursen J. M., Tindal D. J., Chen J. (2008) Nat. Cell Biol. 10, 1076–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menssen A., Hermeking H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6274–6279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fernandez P. C., Frank S. R., Wang L., Schroeder M., Liu S., Greene J., Cocito A., Amati B. (2003) Genes Dev. 17, 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Z., Van Calcar S., Qu C., Cavenee W. K., Zhang M. Q., Ren B. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8164–8169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menssen A., Epanchintsev A., Lodygin D., Rezaei N., Jung P., Verdoodt B., Diebold J., Hermeking H. (2007) Cell Cycle 6, 339–352 [DOI] [PubMed] [Google Scholar]
- 43.Herold S., Herkert B., Eilers M. (2009) Nat. Rev. Cancer 9, 441–444 [DOI] [PubMed] [Google Scholar]
- 44.Martinato F., Cesaroni M., Amati B., Guccione E. (2008) PLoS One 3, e3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.