Abstract
In the yeast Saccharomyces cerevisiae, a precursor form of aminopeptidase I (prApe1) and α-mannosidase (Ams1) are selectively transported to the vacuole through the cytoplasm-to-vacuole targeting pathway under vegetative conditions and through autophagy under starvation conditions. Atg19 plays a central role in these processes by linking Ams1 and prApe1 to Atg8 and Atg11. However, little is known about the molecular mechanisms of cargo recognition by Atg19. Here, we report structural and functional analyses of Atg19 and its paralog, Atg34. A protease-resistant domain was identified in the C-terminal region of Atg19, which was also conserved in Atg34. In vitro pulldown assays showed that the C-terminal domains of both Atg19 and Atg34 are responsible for Ams1 binding; these domains are hereafter referred to as Ams1-binding domains (ABDs). The transport of Ams1, but not prApe1, was blocked in atg19Δatg34Δ cells expressing Atg19ΔABD, indicating that ABD is specifically required for Ams1 transport. We then determined the solution structures of the ABDs of Atg19 and Atg34 using NMR spectroscopy. Both ABD structures have a canonical immunoglobulin fold consisting of eight β-strands with highly conserved loops clustered at one side of the fold. These facts, together with the results of a mutational analysis, suggest that ABD recognizes Ams1 using these conserved loops.
Keywords: Autophagy, NMR, Protein Degradation, Protein Structure, Trafficking, Yeast
Introduction
Autophagy is an intracellular degradation system conserved among eukaryotes from yeasts to mammals. During autophagy, double membrane structures called autophagosomes sequester a portion of the cytoplasm and fuse with the vacuole (lysosome in the case of mammalian autophagy) to deliver their inner contents into the organelle lumen (1). As the contents of autophagosomes are indistinguishable from their surrounding cytoplasm (2), autophagy has long been considered to be a nonselective catabolic pathway. Recent studies, however, have provided evidence for the selective degradation of various targets by autophagy. In autophagy-deficient neuronal cells, intracellular protein aggregates accumulate and eventually lead to neurodegeneration, suggesting that autophagy selectively degrades harmful protein aggregates (3, 4). Damaged or superfluous organelles, such as mitochondria and peroxisomes, and even intracellular infectious pathogens are also selectively degraded by autophagy (5–9). Thus, autophagy selectively eliminates various harmful targets, thereby protecting cells and organisms from disease (10). These autophagic functions appear to rely on precise cargo-sorting mechanisms to load autophagosomes with appropriate cytosolic materials for elimination. Deciphering these mechanisms in molecular detail is critical to the development of remedies for autophagy-related human diseases; however, current knowledge regarding these issues remains limited. Thus, it is important to study the process of selective autophagy in an experimentally more amenable model system, such as yeasts.
In the budding yeast Saccharomyces cerevisiae, α-mannosidase (Ams1)3 and a precursor form of aminopeptidase I (prApe1) are selectively transported to the vacuole through the cytoplasm-to-vacuole targeting (Cvt) pathway under vegetative conditions and through autophagy under starvation conditions (11, 12). The Cvt pathway is topologically and mechanistically similar to autophagy (11–13); therefore, studies on the molecular mechanisms of cargo recognition in the Cvt pathway will provide insight into the basic mechanism of selective autophagy.
The cargoes of the Cvt pathway, prApe1 and Ams1, directly interact with a receptor protein, Atg19, to form the Cvt complex (14, 15). Atg11 interacts with Atg19 to recruit the Cvt complex to the preautophagosomal structure, the functional entity involved in Cvt vesicle formation (15–17). Atg19 further interacts with Atg8, which is localized at the preautophagosomal structure, using the Atg8 family-interacting motif (18, 19) to induce formation of the Cvt vesicle (15, 18). Thus, Atg19 plays a critical role in the selective transport of prApe1 and Ams1 to the vacuole. Recently, Suzuki et al. (37) identified Atg34, an Atg19 paralog, as an additional receptor protein for Ams1 that functions only under starvation conditions. However, the molecular mechanism of specific cargo recognition by Atg19 and Atg34 has not been clarified. Previous studies have shown that the prApe1-binding domain of Atg19 is located in the region from amino acid residues 153 to 191 that contains the predicted coiled coil motif, and the Ams1-binding domain of Atg19 is located in a region within amino acid residues 192–387 (15).
Here, we report structural and functional analyses of Atg19 and Atg34. Ams1 binding domains (ABDs) were identified in Atg19 and Atg34 by limited proteolysis and in vitro pulldown assay. In vivo studies revealed that the Atg19 ABD is specifically required for Ams1 transport to the vacuole via the Cvt pathway. Furthermore, we determined the solution structures of the ABDs of both Atg19 and Atg34 using NMR spectroscopy. Both ABD structures consist of eight β-strands that fold into a canonical immunoglobulin fold. In vivo studies showed that a histidine residue conserved in the Atg19 and Atg34 ABDs is critical for Ams1 delivery to the vacuole. These structures provide a basis for elucidating the molecular mechanism of cargo recognition during selective autophagy.
EXPERIMENTAL PROCEDURES
Sample Preparation
Full-length Atg19 and the three domains of Atg19, the N-terminal domain (residues 1–123), the coiled coil domain (residues 124–253), and the C-terminal domain (residues 254–367), were amplified by polymerase chain reaction (PCR) and inserted into a pGEX-6p-1 vector to produce glutathione S-transferase (GST) fusion proteins (GE Healthcare). The expression vectors of the C-terminal domain of Atg34 (residues 246–358), the N-terminal propeptide of prApe1 (residues 1–20), and prApe1 were prepared in a manner similar to those of Atg19. Expression and purification of Ams1 as well as the construction of the Ams1 expression vector were described elsewhere (20). Mutations leading to the indicated amino acid substitutions were introduced by PCR-mediated site-directed mutagenesis. All of the constructs were sequenced to confirm their identities and expressed in Escherichia coli strain BL21 (DE3) cells and cultured in 2× YT medium (yeast extract, 10 g/liter; trypton, 16 g/liter; sodium chloride, 5 g/liter). After cell lysis by sonication, GST-fused proteins were purified by affinity chromatography using a glutathione-Sepharose 4B column (GE Healthcare) followed by excision of GST from the proteins with PreScission protease (GE Healthcare). For full-length Atg19 and the coiled coil domain, further purification was performed using a Superdex 200 gel filtration column (GE Healthcare) eluted with 20 mm Tris-HCl, pH 8.0 and 150 mm NaCl. For other proteins, further purification was performed using a Superdex 75 gel filtration column (GE Healthcare) eluted with 20 mm Tris-HCl, pH 8.0 and 150 mm NaCl.
Limited Proteolysis
185 μl of 1.0 mg/ml Atg19 in 20 mm Tris-HCl, pH 8.0 and 150 mm NaCl was preincubated at 16 °C for 15 min and further incubated at 16 °C for 15 min after V8 protease (1 μg) was added for digestion. The Atg19, before and after digestion, was subjected to SDS-PAGE, and protein bands were detected by Coomassie Brilliant Blue staining. Then the N-terminal sequences of the ∼15-kDa products were analyzed by a PPSQ-21 protein sequencer (Shimadzu). The molecular masses of digestion products were analyzed by a Voyager-DETM PRO MALDI-TOF mass spectrometer (Applied Biosystems).
GST Pulldown Binding Assays
All the pulldown assays were performed as follows. The purified GST-fused proteins and purified GST-free proteins as well as glutathione-Sepharose 4B beads were simultaneously incubated for 10 min at 4 °C. After washing the beads three times with phosphate-buffered saline, bound proteins were eluted with 10 mm glutathione in 50 mm Tris-HCl, pH 8.0. The eluates were subjected to SDS-PAGE, and then the protein bands were detected with Coomassie Brilliant Blue staining.
Strains and Media
Standard methods were used for yeast manipulation (21). Cells were grown in SD + casamino acid medium (0.17% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate, 0.5% casamino acid, and 2% glucose) with appropriate supplements. Autophagy was induced in growth medium containing 400 ng/ml rapamycin (Sigma). The expression plasmids of the Atg19ΔABD and Atg19 mutants with amino acid substitutions were prepared by PCR using the pRS316-based plasmid containing the ATG19 gene as a template. Successful introduction of the deletion or the mutation was confirmed by sequencing. These plasmids were introduced into atg19Δ::KAN cells on a SEY6210 background (MATα leu2 ura3 his3 trp1 lys2 suc2).
Vacuole Preparations
Yeast vacuoles were isolated on a Ficoll step gradient as described previously (22) with minor modifications. Cells were grown to early log phase, then 550–800 A600 units were harvested, and spheroplasts were prepared. Spheroplasts suspended in 2.5 ml of 15% Ficoll solution were disrupted with a polycarbonate filter (3.0-μm pore, 47 mm), and the lysed cell solution was loaded in the bottom of an ultraclear SW41 tube (Beckman Coulter, Fullerton, CA) and overlaid with 8, 4, and 0% Ficoll solutions. Vacuoles were collected from the 0%/4% float interface with a Pasteur pipette after spinning for 1.5 h at 110,000 × g at 4 °C. Vacuoles were checked under a microscope.
Activity Assay of Ams1
Cell lysates and harvested vacuoles were assayed for Ams1. Ams1 activity was determined based on the established protocol (23) with minor modifications. Samples were treated with Triton X-100 (2.5% final concentration), and the volume was then brought up to 400 μl with distilled H2O. 100 μl of 5× substrate mixture (200 mm sodium acetate, pH 6.5, and 2 mm 4-metylumbelliferyl-α-d-mannopyranoside) was added to start the reaction, and the samples were then incubated for 1 h at 37 °C. The reaction was stopped with 200 μl of 10% trichloroacetic acid, and any particulates were spun down in a microcentrifuge for 5 min. An equal volume of 1 m glycine, pH 10.4 was added to neutralize the reaction prior to fluorescence measurement at 450 nm. Results from assays for each strain were tabulated from three independent vacuole preparations.
Immunoblot Analysis
To detect Ape1 in Fig. 2B, cell lysates were prepared by breaking cells with glass beads in 250 mm Tris-HCl, pH 9.0, 10 mm MgSO4, 10 μm ZnSO4, and 1 mm PMSF. Anti-Ape1 antiserum (24) was used as a primary antibody. Signals were detected using Immobilon Western (Millipore) with an ECL Mini-Camera (GE Healthcare). In the case of Fig. 6B, cell lysates were prepared by NaOH/2-mercaptoethanol extraction with a slight modification (25). An anti-Atg19 antiserum was raised against bacterially expressed Atg19 and affinity-purified using the antigen. To detect GFP and Ape1, anti-GFP antiserum (Invitrogen) and anti-Ape1 antiserum were used, respectively, as a primary antibody. Signals were detected using an ECL system with a LAS4000 bioimaging analyzer (Fujifilm).
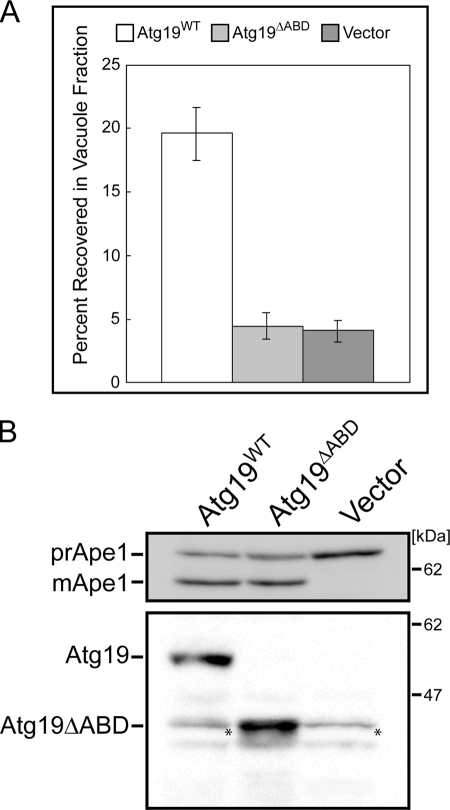
FIGURE 2.
Atg19ΔABD is defective in transporting Ams1 to vacuole. A, activity assay of Ams1. Vacuoles were isolated on Ficoll step gradients as described under “Experimental Procedures.” The level of Ams1 activity in the vacuole fraction of atg19Δ cells expressing Atg19ΔABD can be seen to be much lower than that of atg19Δ cells expressing wild-type Atg19 and is as low as that of atg19Δ cells containing control vectors. Vacuoles were purified at least three times for each strain, and the total enzyme activity recovered in the vacuole fraction was divided by the total activity loaded on the gradient to obtain the percentage of recovery. The values and the error bars are the means and the standard deviations of three independent experiments, respectively. B, top, monitoring Ape1 maturation under growth conditions using a CEN plasmid pRS316 as an expression vector for wild-type Atg19 and Atg19ΔABD. The lysates were separated by SDS-PAGE and subjected to immunoblotting with anti-Ape1 antibody. Bottom, expression levels of wild-type Atg19 and Atg19ΔABD were detected by immunoblotting using anti-Atg19 antibody. Asterisks indicate nonspecific bands.
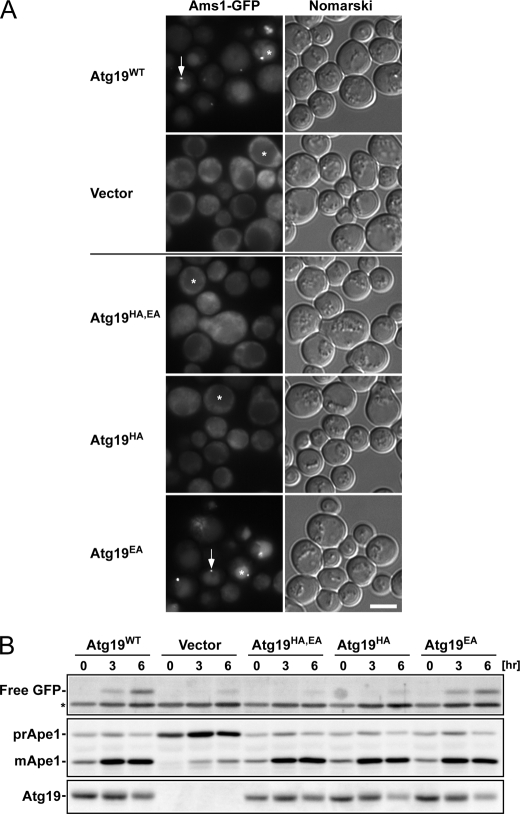
FIGURE 6.
Activity of Atg19 ABD mutants in vivo. A, localization of Ams1-GFP in atg19Δatg34Δ cells expressing Atg19 ABD mutants. Cells were grown in SD + casamino acid medium and then treated with rapamycin for 6 h. Arrows indicate dot structures in the cytoplasm. Asterisks indicate the vacuoles. Scale bar, 5 μm. B, Western blot analyses. Cells used in A were collected at the times indicated after rapamycin addition, and lysates were prepared by the alkaline lysis method. Top, free GFP, produced by the hydrolysis of Ams1-GFP in the vacuole, was detected by immunoblotting using anti-GFP antibody. The asterisk indicates a nonspecific band. Middle, Ape1 maturation was monitored by immunoblotting using anti-Ape1 antibody. Bottom, the accumulation level of Atg19 mutants was detected by immunoblotting using anti-Atg19 antibody.
Microscopy
Fluorescence microscopy was performed using a total internal reflection fluorescence microscopy system (Olympus) equipped with a UPlanSApo 100× oil objective (numerical aperture, 1.40) and a CoolSNAP HQ charge-coupled device camera (Nippon Roper) as described previously (26). A blue laser (Sapphire 488-20, Coherent) was used for excitation of GFP. A U-MNIBA2 mirror unit, from which the excitation filter was removed, was used for GFP visualization. Images were acquired using MetaMorph software (Molecular Devices).
Preparation of Uniformly Labeled Proteins
Uniformly labeled ABDs of Atg19 and Atg34 were expressed and purified as described above except that M9 medium containing [15N]ammonium chloride and d-[13C]glucose was used. The NMR samples for the structure determination were prepared for 0.75 mm Atg19 ABD in 25 mm sodium phosphate, pH 7.0 and 100 mm NaCl and for 0.77 mm Atg34 ABD in 25 mm sodium phosphate, pH 7.0, 100 mm NaCl, and 2 mm DTT.
NMR Measurements
The NMR spectra were obtained at 25 °C on Varian UNITY INOVA 600 and 800 spectrometers. The data were processed using the NMRPipe program (27) and analyzed using the Sparky program.4 Two- and three-dimensional spectra were obtained for the assignment of the protein backbone and side chain 1H, 15N, and 13C resonances. Backbone resonances were assigned using the two-dimensional 1H-15N HSQC and three-dimensional HNCA, HN(CO)CA, HNCACB, CBCA(CO)NH, HNCO, HNCAHA, and HBHA(CO)NH spectra. The aliphatic side chain atoms were assigned using two-dimensional 1H-13C HSQC and three-dimensional HCCH TOCSY spectra. Aromatic side chain atoms were assigned using the two-dimensional 1H-13C HSQC, (Hb)Cb(CgCd)Hd, and (Hb)Cb(CgCdCe)He and three-dimensional HCCH TOCSY spectra.
Structural Determination
The three-dimensional 15N-edited NOESY and 13C-edited NOESY spectra (tmix = 100 ms) were measured to obtain NOE distance constraints. The Atg19 ABD and Atg34 ABD structures were determined using the CANDID/CYANA 2.1 program (28). Structural statistics for the best 20 Atg19 ABD and Atg34 ABD structures are shown in Table 1.
TABLE 1.
Structural statistics for Atg19 and Atg34 ABDs
r.m.s., root mean square; r.m.s.d., root mean square difference.
| Atg19 ABD | Atg34 ABD | |
|---|---|---|
| NOE distance constraints | ||
| Total | 3,275 | 2,374 |
| Short range, |i − j| ≤ 1 | 1,660 | 1,414 |
| Medium range, 1 < |i − j| < 5 | 260 | 168 |
| Long range, |i − j| ≥ 5 | 1,355 | 792 |
| Structural coordinate r.m.s.d. | ||
| Residue range | 260–360 | 251–282, 289–343 |
| Backbone (Å) | 0.28 | 0.50 |
| Heavy atom (Å) | 0.73 | 0.97 |
| Residual NOE violations | ||
| Number >0.3 Å | 4 | 3 |
| Maximum (Å) | 0.57 | 0.37 |
| r.m.s. (Å) | 0.0093 ± 0.0008 | 0.0061 ± 0.0009 |
RESULTS
Identification of Ams1-binding Domains of Atg19 and Atg34
Stable domains in Atg19 were identified by limited proteolysis of the full-length Atg19 (supplemental Fig. S1), which showed that Atg19 comprises at least two structurally stable domains, the N-terminal domain (residues 1–123) and the C-terminal domain (residues 254–367). The region between the N-terminal and C-terminal stable domains (residues 124–253) of Atg19 contains a predicted coiled coil between amino acids 160 and 187; therefore, it is named the coiled coil domain. These three domains were cloned and overexpressed in E. coli BL21 (DE3) as GST-fusion proteins.
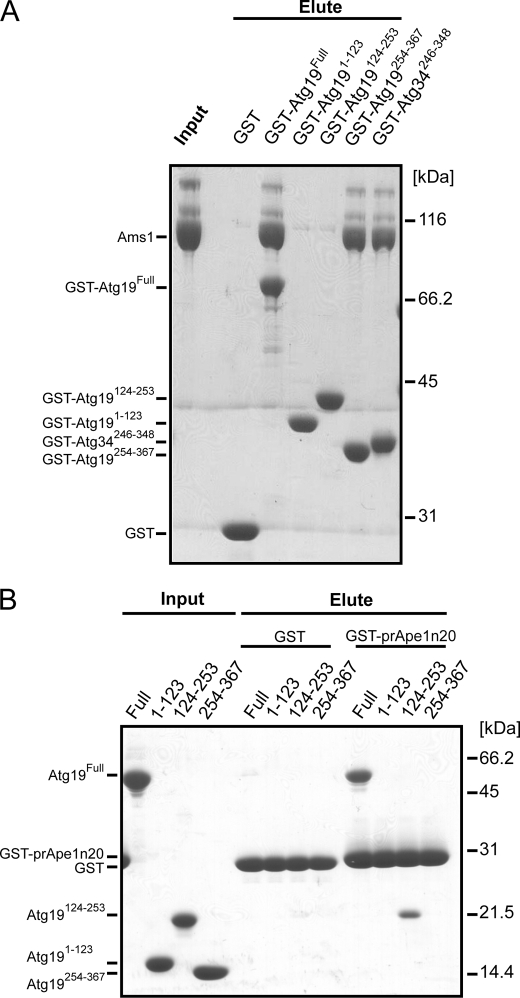
To elucidate the Ams1 binding properties of these domains, we performed an in vitro pulldown assay using the GST fusion proteins and Ams1, which showed that only the C-terminal domain is responsible for Ams1 binding (Fig. 1A). This result is consistent with those of a previous study that showed residues 192–387 of Atg19 to be responsible for Ams1 binding (15). Therefore, the C-terminal domain was named the ABD. Sequence alignment of Atg19 with Atg34, which has been recently identified as an additional receptor protein for Ams1 (see the accompanying paper by Suzuki et al. (37)), suggested that the Atg19 ABD is also conserved in Atg34 with high sequence similarity (see Fig. 5A). We then performed an in vitro pulldown assay using the GST fusion protein of the putative Atg34 ABD with Ams1, which showed that the putative Atg34 ABD is actually responsible for Ams1 binding (Fig. 1A). These results suggest that the ABD is sequentially and functionally conserved between Atg19 and Atg34.
FIGURE 1.
Identification of prApe1- and Ams1-binding domains of Atg19 and Atg34. A, in vitro pulldown assay between three GST-fused domains of Atg19, the GST-fused C-terminal domain of Atg34, and Ams1. The input and eluted proteins were subjected to SDS-PAGE and detected by Coomassie Brilliant Blue staining. B, in vitro pulldown assay between GST-fused prApe1 propeptide (residues 1–20; Ape1n20) and the three domains of Atg19. The input and eluted proteins were subjected to SDS-PAGE and detected by Coomassie Brilliant Blue staining.
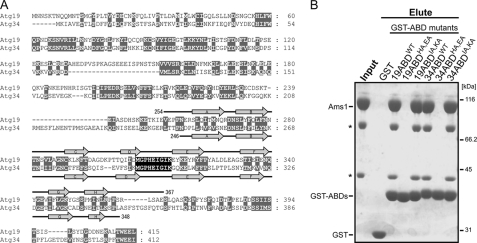
FIGURE 5.
A, sequence alignment between Atg19 and Atg34. Gaps are introduced to maximize the similarity. Conserved or type-conserved residues are shaded gray. Secondary structure elements of the Atg19 and Atg34 ABDs are shown above and below the sequence, respectively. Residues of the DE loop of the ABDs are perfectly conserved (shaded black). B, in vitro pulldown assay between GST-fused Atg19 ABD mutants, GST-fused Atg34 ABD mutants, and Ams1. The input and eluted proteins were subjected to SDS-PAGE and detected by Coomassie Brilliant Blue staining. Asterisks indicate degradation products of Ams1.
An additional in vitro pulldown assay showed that the coiled coil domain, but not the other two domains, of Atg19 is responsible for binding with the propeptide of prApe1 (Fig. 1B). This is consistent with the results of a previous report that showed the prApe1-binding domain of Atg19 is located in the region from amino acid residues 153 to 191 (15).
Atg19 ABD Is Responsible for Transporting Ams1 into Vacuole through Cvt Pathway
The direct interaction of ABD with Ams1 led us to study its possible involvement in the transport of Ams1 to the vacuole. The localization of Ams1 in yeast was studied using the subcellular fractionation method reported previously (29). Vacuoles were isolated from atg19Δ cells expressing wild-type Atg19 or Atg19ΔABD or containing empty vectors as a control under vegetative conditions, and the Ams1 transport to the vacuole was monitored by its enzymatic activity. Expression of wild-type Atg19 and Atg19ΔABD was confirmed by immunoblotting using anti-Atg19 antibody (Fig. 2B). In atg19Δ cells expressing wild-type Atg19, Ams1 activity in the vacuole fraction was ∼4 times higher than that in atg19Δ cells containing control vectors (Fig. 2A), which is consistent with previously reported results (14). Interestingly, in atg19Δ cells expressing Atg19ΔABD, Ams1 activity in the purified vacuole fraction was comparable with that in atg19Δ cells (Fig. 2A). These results suggest that the Atg19 ABD is required for Ams1 transport to the vacuole through the Cvt pathway. Another cargo protein, prApe1, is translated in the cytoplasm and processed into a mature form in the vacuole. Therefore, prApe1 transport to the vacuole can be evaluated by monitoring its maturation. As shown in Fig. 2B, the defect of Ape1 maturation in atg19Δ cells was restored by the expression of Atg19ΔABD, suggesting that Atg19 ABD is dispensable for the transport of prApe1 to the vacuole through the Cvt pathway.
Solution Structures of Atg19 and Atg34 ABDs
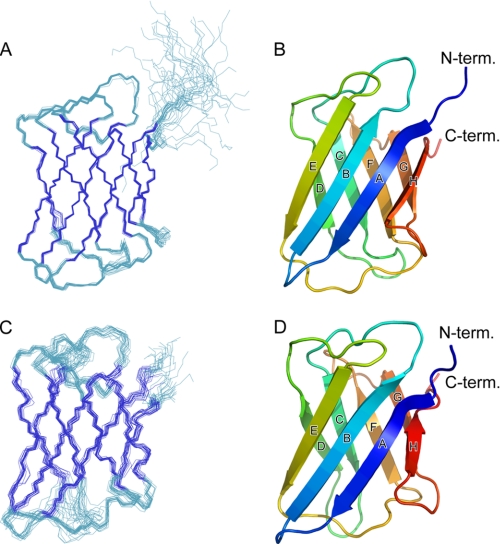
The Atg19 and Atg34 ABD structures were determined in solution using NMR spectroscopy (Fig. 3). Both ABDs comprise eight β-strands (A–H) among which A, B, E, and H form an antiparallel β-sheet, and the surface of this sheet faces a second antiparallel β-sheet comprising C, D, F, and G, thus forming a typical immunoglobulin-like β-sandwich fold. All the residues have a well converged conformation except for the N and C termini and the CD loop (the loop connecting strands C and D) of the Atg34 ABD. To investigate structural similarity with other proteins, the ABD structures were applied to the Dali server (30). A number of proteins, most of which have poor sequence similarity with the Atg19 and Atg34 ABDs (below 10% in sequence identity), were found to have a fold similar to those seen in the ABDs but with minor differences in topology. Among them, the C2 domain of Kibra (Protein Data Bank code 2Z0U), a WW domain-containing protein, has an eight β-stranded immunoglobulin fold with a topology identical to that of the ABDs.
FIGURE 3.
Solution structures of Atg19 and Atg34 ABDs. A, best fit superposition of backbone atoms of 20 NMR-derived structures of the Atg19 ABD. Structures are fitted in a well ordered region (residues 261–360). Strands are shown in blue. B, ribbon diagram of the Atg19 ABD structure. Secondary structural elements are labeled. C, best fit superposition of backbone atoms of 20 NMR-derived structures of the Atg34 ABD. Structures are fitted in a well ordered region (residues 252–282 and 289–343). Strands are shown in blue. D, ribbon diagram of the Atg34 ABD structure. Secondary structural elements are labeled. All of the structural figures were prepared using PyMOL.
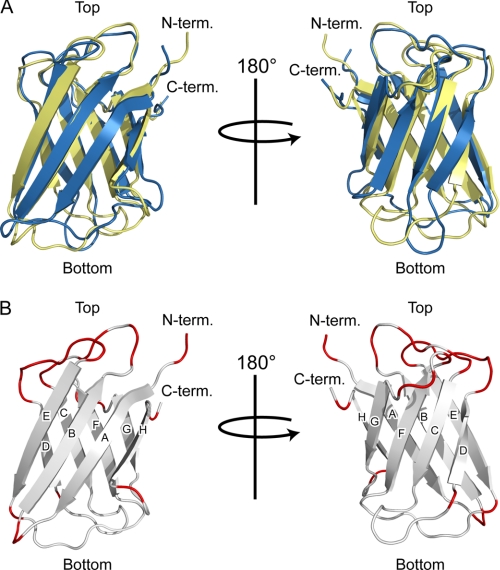
The Atg19 and Atg34 ABD structures are very similar to each other with a root mean square difference of 2.1 Å for 102 residues (the Z-score calculated by the Dalilite program is 12.8). Relatively large structural differences are observed in the loops located at the bottom of the immunoglobulin fold (Fig. 4A). In contrast, the loops located at the top of the immunoglobulin fold have a similar conformation between the Atg19 and Atg34 ABDs (Fig. 4A). Furthermore, the residues comprising the top loops are well conserved between the Atg19 and Atg34 ABDs in contrast to the less conserved sequence in the bottom loops (Fig. 4B). Considering that the Atg19 and Atg34 ABDs recognize the same target (Ams1), these sequentially and structurally conserved loops (those connecting strands B and C, D and E, and F and G) are potent candidates for Ams1 recognition sites. Among these, the DE loop is completely conserved (Figs. 4B and 5A); therefore, we introduced mutations at the exposed residues in the DE loop and performed an in vitro pulldown assay. The in vitro pulldown assay using four alanine-substituted ABD mutants, Atg19 ABDHA,EA (substitution of His-310 and Glu-311 with alanines), Atg19 ABDIA,KA (substitution of Ile-314 and Lys-315 with alanines), Atg34 ABDHA,EA (substitution of His-296 and Glu-297 with alanines), and Atg34 ABDIA,KA (substitution of Ile-300 and Lys-301 with alanines), showed that His-310 and/or Glu-311 of the Atg19 ABD (His-296 and/or Glu-297 of the Atg34 ABD) are essential for Ams1 recognition, whereas Ile-314 and Lys-315 of the Atg19 ABD (Ile-300 and Lys-301 of the Atg34 ABD) are not essential (Fig. 5B).
FIGURE 4.
Structural comparison between Atg19 and Atg34 ABDs. A, ribbon diagrams of the superposition of the Atg19 and Atg34 ABDs. The Atg19 and Atg34 ABDs are colored yellow and blue, respectively. Left and right are related by a 180° rotation along the vertical axis. B, ribbon diagrams of the Atg19 ABD in which the loop residues conserved between Atg19 and Atg34 are colored red. Left and right are related by a 180° rotation along the vertical axis.
Conserved Residues between Atg19 and Atg34 ABDs Required for Ams1 Recognition
Because the HA+EA mutation in the Atg19 and Atg34 ABDs abrogated binding of Ams1 in vitro, we studied the effect of His-310 and/or Glu-311 mutations on the transport of Ams1 into the vacuole. The localization of GFP-fused Ams1 in atg19Δatg34Δ cells expressing Atg19 mutants under autophagy-inducing conditions was observed (6 h after treatment with rapamycin) (Fig. 6). Three sets of alanine-substituted mutants, Atg19HA,EA, Atg19HA, and Atg19EA, were prepared and expressed in atg19Δatg34Δ cells using a centromere plasmid pRS316. The expression levels of the Atg19 mutants were comparable with that of wild-type Atg19 (Fig. 6B). As shown in Fig. 6A, in atg19Δatg34Δ cells expressing wild-type Atg19 or Atg19EA, GFP was detected as bright fluorescent dot structures at the perivacuolar region as indicated by the arrows. GFP also stained the vacuolar lumen (Fig. 6A, asterisks), suggesting that Ams1-GFP was localized in the Cvt complex and was properly transported into the vacuoles. In contrast, in atg19Δatg34Δ cells expressing Atg19HA,EA or Atg19HA, GFP localized to the cytoplasm (Fig. 6A), showing that Ams1-GFP could neither be localized in the Cvt complex nor transported into the vacuole. Transport of Ams1-GFP into the vacuole can be monitored by detecting the amount of free GFP as GFP is released from Ams1-GFP in the vacuole (Fig. 6B). The amount of free GFP in atg19Δatg34Δ cells expressing Atg19HA,EA or Atg19HA was negligible as was the case for cells containing the control vectors (Fig. 6B), which is consistent with the results obtained by fluorescence microscopy. In contrast, the defect of Ape1 maturation in atg19Δatg34Δ cells was restored by the expression of wild-type Atg19, Atg19HA,EA, Atg19HA, or Atg19EA under growth and autophagy-inducing conditions (Fig. 6B), showing that the present mutations in Atg19 did not affect the transport of prApe1 into the vacuole via either the Cvt pathway or autophagy. These results indicate that the conserved His residue in the Atg19 ABD (His-310) plays a critical role in Ams1 recognition and that the Ams1 binding of the Atg19 ABD is essential for Ams1 transportation to the vacuole. Similar experiments using Atg34 mutants showed that the conserved His residue in the Atg34 ABD (His-296), which corresponds to His-310 in the Atg19 ABD, plays a similar critical role in Ams1 recognition (see the accompanying paper by Suzuki et al. (37)).
DISCUSSION
Atg19 and Atg34 Share a Domain Specific to Ams1 Recognition
In this study, we have identified the Ams1-binding domain in both Atg19 and Atg34 (Fig. 1A) and shown that both ABDs are responsible for recognizing Ams1 and that this recognition is crucial for transporting Ams1 into the vacuole via both the Cvt pathway and autophagy (Figs. 2 and 6; see also the accompanying paper by Suzuki et al. (37)). Intriguingly, neither the deletion of the ABD from Atg19 nor mutations in the Atg19 ABD affected prApe1 transport to the vacuole. These data suggest that the formation of the prApe1-Atg19 complex and its localization to the preautophagosomal structure, both of which are required for the transport of prApe1 to the vacuole, are independent of the recognition of Ams1 by Atg19. We concluded that the function of the ABD is specialized in anchoring Ams1 to the prApe1-Atg19 complex to achieve efficient transport of Ams1 to the vacuole.
Limited proteolysis of Atg19 showed that Atg19 comprises two stable domains, the N-terminal domain and the ABD. In addition to these two domains, Atg19 possesses the coiled coil domain responsible for prApe1 binding at the region between the N-terminal domain and ABD and the Atg8 family-interacting motif at its extreme C terminus (18, 19). An in vitro pulldown assay between GST-fused prApe1, Atg19, and Ams1 showed that Ams1 binds to prApe1 through Atg19, showing that Atg19 can simultaneously bind both Ams1 and prApe1 on one molecule (supplemental Fig. S2). Thus, the architecture of Atg19 is formed by linking structurally and functionally independent modules together; this characteristic may also apply to Atg34. In mammals, p62, an autophagic receptor for ubiquitinated protein aggregates, also comprises multiple modules including PB1, ubiquitin-associated domain, and Atg8 family-interacting motif, and each module has its specific binding partner (31–33). Thus, the architecture formed by linking multiple modules together appears to be a conserved feature among autophagic receptors.
ABDs Have an Immunoglobulin Fold and May Interact with Ams1 in Manner Similar to That of Camelid Antibodies and Monobodies
We determined the solution structures of the Atg19 and Atg34 ABDs using NMR (Fig. 3, A–D). The ABDs in Atg19 and Atg34 have a β-sandwich fold that is observed in a variety of immunoglobulins and immunoglobulin-like domains responsible for recognizing various proteins. Because antibodies generally recognize antigens using the hypervariable loops from both the VH and VL regions, their mode of binding with their antigens should be different from that of monomeric ABDs with Ams1. Interestingly, the ABD-Ams1 interaction could be similar to that observed between camelid antibody fragments and their antigens as camelid antibodies lack a light chain and function as a monomer where hypervariable loops of the VH are responsible for antigen binding (34). It could also be similar to the interaction of monobodies, artificially designed proteins that use a fibronectin type III domain as a scaffold, with their targets because monobodies interact with their targets using similar loops in their monomeric immunoglobulin fold (35). Camelid antibody fragments and monobodies interact with their target proteins using the loops clustered at one side of their immunoglobulin fold; these loops correspond to the BC, DE, and FG loops of the ABDs (supplemental Fig. S3). Therefore, it is suggested that the ABDs also interact with Ams1 using these three loops. The conserved His residue of the DE loops of Atg19 and Atg34 was actually shown to be crucial for Ams1 recognition (Fig. 6; see also the accompanying paper by Suzuki et al. (37)).
To date, it has been thought that proteins are non-selectively degraded by autophagy. However, recent proteomics analyses have identified proteins that are selectively degraded by autophagy (36). Although the recognition mechanism of these target proteins by autophagy has not been established, autophagy-specific receptor proteins possessing an ABD-like fold might be responsible for such recognition. Identification and structural analysis of other autophagy-specific receptor proteins are required for further clarification of the molecular mechanism of specific cargo recognition during autophagy.
Supplementary Material
Acknowledgment
We are grateful to Mayumi Morimoto for technical assistance.
This work was supported in part by grants-in-aid for scientific research on priority areas and the Targeted Proteins Research Program from the Ministry of Education, Culture, Sports, Science and Technology, Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
The atomic coordinates and structure factors (codes 2KZB and 2KZK) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco.
- Ams1
- α-mannosidase
- ABD
- α-mannosidase-binding domain
- Ape1
- aminopeptidase I
- Cvt
- cytoplasm-to-vacuole targeting
- HSQC
- heteronuclear single quantum correlation
- prApe1
- precursor form of aminopeptidase I
- SD
- synthetic defined
- TOCSY
- total correlation spectroscopy.
REFERENCES
- 1.Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. (2009) Nat. Rev. Mol. Cell Biol. 10, 458–467 [DOI] [PubMed] [Google Scholar]
- 2.Baba M., Takeshige K., Baba N., Ohsumi Y. (1994) J. Cell Biol. 124, 903–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Mizushima N. (2006) Nature 441, 885–889 [DOI] [PubMed] [Google Scholar]
- 4.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. (2006) Nature 441, 880–884 [DOI] [PubMed] [Google Scholar]
- 5.Goldman S. J., Taylor R., Zhang Y., Jin S. (2010) Mitochondrion 10, 309–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manjithaya R., Nazarko T. Y., Farré J. C., Subramani S. (2010) FEBS Lett. 584, 1367–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakagawa I., Amano A., Mizushima N., Yamamoto A., Yamaguchi H., Kamimoto T., Nara A., Funao J., Nakata M., Tsuda K., Hamada S., Yoshimori T. (2004) Science 306, 1037–1040 [DOI] [PubMed] [Google Scholar]
- 8.Gutierrez M. G., Master S. S., Singh S. B., Taylor G. A., Colombo M. I., Deretic V. (2004) Cell 119, 753–766 [DOI] [PubMed] [Google Scholar]
- 9.Noda T., Yoshimori T. (2009) Int. Immunol. 21, 1199–1204 [DOI] [PubMed] [Google Scholar]
- 10.Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008) Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hutchins M. U., Klionsky D. J. (2001) J. Biol. Chem. 276, 20491–20498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baba M., Osumi M., Scott S. V., Klionsky D. J., Ohsumi Y. (1997) J. Cell Biol. 139, 1687–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynch-Day M. A., Klionsky D. J. (2010) FEBS Lett. 584, 1359–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott S. V., Guan J., Hutchins M. U., Kim J., Klionsky D. J. (2001) Mol. Cell 7, 1131–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shintani T., Huang W. P., Stromhaug P. E., Klionsky D. J. (2002) Dev. Cell 3, 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. (2001) EMBO J. 20, 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J., Huang W. P., Stromhaug P. E., Klionsky D. J. (2002) J. Biol. Chem. 277, 763–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noda N. N., Kumeta H., Nakatogawa H., Satoo K., Adachi W., Ishii J., Fujioka Y., Ohsumi Y., Inagaki F. (2008) Genes Cells 13, 1211–1218 [DOI] [PubMed] [Google Scholar]
- 19.Noda N. N., Ohsumi Y., Inagaki F. (2010) FEBS Lett. 584, 1379–1385 [DOI] [PubMed] [Google Scholar]
- 20.Watanabe Y., Noda N. N., Honbou K., Suzuki K., Sakai Y., Ohsumi Y., Inagaki F. (2009) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 571–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams A., Gottschling D. E., Kaiser C. A., Stearns T. (eds) (1998) Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 22.Haas A., Scheglmann D., Lazar T., Gallwitz D., Wickner W. (1995) EMBO J. 14, 5258–5270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Opheim D. J. (1978) Biochim. Biophys. Acta 524, 121–130 [DOI] [PubMed] [Google Scholar]
- 24.Suzuki K., Kamada Y., Ohsumi Y. (2002) Dev. Cell 3, 815–824 [DOI] [PubMed] [Google Scholar]
- 25.Horvath A., Riezman H. (1994) Yeast 10, 1305–1310 [DOI] [PubMed] [Google Scholar]
- 26.Suzuki K., Kubota Y., Sekito T., Ohsumi Y. (2007) Genes Cells 12, 209–218 [DOI] [PubMed] [Google Scholar]
- 27.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 28.Herrmann T., Güntert P., Wüthrich K. (2002) J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 29.Yoshihisa T., Ohsumi Y., Anraku Y. (1988) J. Biol. Chem. 263, 5158–5163 [PubMed] [Google Scholar]
- 30.Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamark T., Perander M., Outzen H., Kristiansen K., Øvervatn A., Michaelsen E., Bjørkøy G., Johansen T. (2003) J. Biol. Chem. 278, 34568–34581 [DOI] [PubMed] [Google Scholar]
- 32.Vadlamudi R. K., Joung I., Strominger J. L., Shin J. (1996) J. Biol. Chem. 271, 20235–20237 [DOI] [PubMed] [Google Scholar]
- 33.Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007) J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 34.Muyldermans S. (2001) J. Biotechnol. 74, 277–302 [DOI] [PubMed] [Google Scholar]
- 35.Koide A., Bailey C. W., Huang X., Koide S. (1998) J. Mol. Biol. 284, 1141–1151 [DOI] [PubMed] [Google Scholar]
- 36.Onodera J., Ohsumi Y. (2004) J. Biol. Chem. 279, 16071–16076 [DOI] [PubMed] [Google Scholar]
- 37.Suzuki K., Kondo C., Morimoto M., Ohsumi Y. (2010) J. Biol. Chem. 285, [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.