Abstract
CD44 is a major cell surface receptor for the large polydisperse glycosaminoglycan hyaluronan (HA). Binding of the long and flexible HA chains is thought to be stabilized by the multivalent nature of the sugar molecule. In addition, high and low molecular weight forms of HA provoke distinct proinflammatory and anti-inflammatory effects upon binding to CD44 and can deliver either proliferative or antiproliferative signals in appropriate cell types. Despite the importance of such interactions, however, neither the stoichiometry of multivalent HA binding at the cell surface nor the molecular basis for functional distinction between different HA size categories is understood. Here we report on the design of a supported lipid bilayer system that permits quantitative analysis of multivalent binding through presentation of CD44 in a stable, natively oriented manner and at controlled density. Using this system in combination with biophysical techniques, we show that the amount of HA binding to bilayers that are densely coated with CD44 increases as a function of HA size, with half-maximal saturation at ∼30 kDa. Moreover, reversible binding was confined to the smaller HA species (molecular weight of ≤10 kDa), whereas the interaction was essentially irreversible with larger polymers. The amount of bound HA decreased with decreasing receptor surface density, but the stability of binding was not affected. From a physico-chemical perspective, the binding properties of HA share many similarities with the typical behavior of a flexible polymer as it adsorbs onto a homogeneously attractive surface. These findings provide new insight into the multivalent nature of CD44-HA interactions and suggest a molecular basis for the distinct biological properties of different size fractions of hyaluronan.
Keywords: Biophysics, Cell Surface Receptor, Extracellular Matrix, Glycosaminoglycan, Leukocyte, Receptor Structure-Function, CD44, Hyaluronan, Supported Lipid Bilayer
Introduction
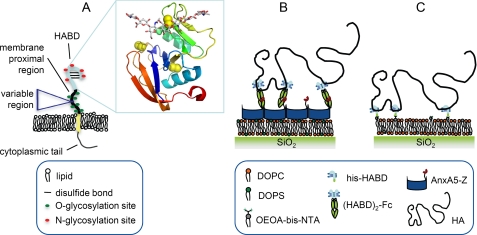
CD44 is a major cell surface receptor for the glycosaminoglycan hyaluronan (HA).2 It is a type I membrane glycoprotein that consists of a small cytoplasmic domain, a transmembrane domain, a highly glycosylated membrane-proximal region, and an N-terminal HA binding domain (HABD) (Fig. 1A). The HABD, which spans a sequence of about 150 amino acids, forms an α/β structure stabilized by three disulfide bonds that encompasses the Link module, a domain conserved among members of the hyaladherin superfamily (1, 2). The most common form of CD44, the so-called hematopoietic or standard form (CD44H/CD44s), has an apparent molecular weight of 80–95 kDa, which exceeds the total amino acid mass by 2–3-fold due to posttranslational N- and O-linked glycosylation. The membrane-proximal region is, additionally, subject to alternative splicing, which results in the expression of variant exons in various isoforms of CD44 (3, 4).
FIGURE 1.
A, schematic presentation of human CD44, embedded in the plasma membrane. The inset shows the crystal structure of the hyaluronan binding domain in complex with an HA octasaccharide (13) (Protein Data Bank code 2JCR), in which the protein is shown as a ribbon diagram, with disulfide bonds depicted as yellow spheres, and the HA is in a stick representation. B and C, schematic presentation of the design of the model systems. B, model system 1, based on (HABD)2-Fc. C, model system 2, based on His-HABD.
CD44 is found on many cell types, including leukocytes, chondrocytes, fibroblasts, and endothelial and epithelial cells and has been implicated in various processes, such as lymphocyte recruitment, T cell signaling, cell matrix adhesion, apoptosis, limb development, and tumor metastasis. The role of HA binding in these processes has been the subject of intensive studies (4–10). The propensity of CD44 to bind HA varies strongly across cell types and state of maturation, and such modulation of binding is thought to be functionally important.
Several mechanisms have been proposed for the regulation of receptor-mediated HA binding to CD44. One type of mechanism relates to changes in affinity between individual receptors and HA, exerted by modifications to CD44 glycosylation (1, 11–13). A second type of mechanism derives from the fact that hyaluronan molecules, by virtue of their repetitive structure (GlcNAcβ1–4GlcUA)n contain within them multiple (up to a few thousand) CD44 binding sites. Individual HA chains can thus interact simultaneously with many receptors on the cell surface. Moreover, changes in the density and arrangement of CD44 at the cell surface, including local enrichment or clustering, can affect the overall strength or avidity of such a multivalent interaction. A third mechanism proposes that changes in the conformational state of HA through the formation of protein macromolecular complexes (10, 14) or differences in HA chain length can influence both HA binding and its functional consequences. For example, low molecular weight HA produced during matrix turnover is known to have proinflammatory properties, whereas high molecular weight HA present in intact matrix induces immunosuppression, reflecting differences in the capacity of each size fraction either to activate Toll-like receptors or to bind CD44 and stimulate FoxP3+ CD25+ regulatory T cells (Treg), respectively (15–17). In addition, preparations of HA defined operationally as “low molecular weight” (20–500 kDa) have been reported to promote cell cycle progression via a CD44/Rac/ERK pathway in smooth muscle cells, whereas higher molecular weight fractions (>500 kDa) were shown to be inhibitory (18). The molecular basis for this HA size threshold phenomenon remains completely obscure. Moreover, the definition of “low molecular weight HA” lacks consistency in these studies, in some cases referring to oligomers that correspond to a single binding site for CD44 (16, 19) and in others to polymers with more than 100 binding sites (18, 20).
The affinity of CD44 for high molecular weight HA has been found to be rather weak, with a KD in the range of 10 to 100 μm (13, 21). It is therefore likely that much of the regulatory potential lies in the spatial arrangement of receptors on the cell surface. Apart from artificial manipulation of CD44 to assess the effects of dimerization (7, 22), there are currently no methods available to investigate the influence of receptor density in a controlled manner. Conventional in vitro binding assays are sensitive to interactions between individual ligand/receptor pairs but ignore the spatial confinement and arrangement of HA receptors in two dimensions that pertain within the cell membrane. Although cellular assays have provided valuable insights into overall patterns of regulation, the complexity of the living cell makes it difficult to disentangle the relative contributions of the different types of interactions outlined above. For example, CD44 isoforms with different affinities may co-exist on the same cell surface, and the average and local surface densities of these receptor populations are rarely known. As a consequence, it has been difficult to address the simple yet pertinent question of how many receptors are required to bind HA stably to the cell surface.
Here, we present a novel approach that overcomes many of the shortcomings of existing cell-based analyses and allows investigation of the multivalent interaction between CD44 and HA in a highly controlled manner. The method is based on the immobilization of CD44 on a planar solid support that is covered with a lipid bilayer. We employed several complementary and surface-sensitive biophysical techniques to quantify binding of HA and to characterize the stability and morphology of the HA films that are formed. Our results provide new insights into the multivalent nature of stable CD44-HA interactions and the role of HA as a flexible polymer in the regulation of binding. Furthermore, they have far reaching implications, allowing novel understanding of the molecular mechanisms underlying biological function.
EXPERIMENTAL PROCEDURES
Buffers
A Hepes buffer solution of 150 mm NaCl, and 10 mm HEPES, pH 7.4, in ultrapure water was used to prepare lipid vesicles and in all experiments reported. CaCl2 (2 mm) was added throughout all measurements related to model system 1 (see “Results”). NiCl2 (5 mm) was included in the incubation step leading to the formation of supported lipid bilayers (SLBs) containing bis-NTA-functionalized lipids.
Hyaluronan Preparations
Lyophilized HA with well defined molecular weights of 10 ± 2 kDa (PrimeHA-12), 30 ± 1.5 kDa (SelectHA-50), 262 ± 13 kDa (SelectHA-250), 450 ± 23 kDa (SelectHA-500), 1156 ± 58 kDa (SelectHA-1000), and 2400 ± 120 kDa (SelectHA-2500) was purchased from Hyalose (Oklahoma City, OK). HA8 and HA34 oligomers were prepared as reported before (23). For reconstitution, HA was dissolved in ultrapure water at a stock concentration of 1 mg/ml and gently shaken overnight. The stock solution was then aliquoted and stored at −20 °C. Before use, HA was diluted in the Hepes buffer to the desired concentration.
Expression and Purification of CD44 HABD Constructs
The (HABD)2-Fc construct was prepared in a modified version of the expression vector pCDM7Ig, which encodes the hinge, CH2, and CH3 regions of human IgG1. The modification consisted of a linker inserted into the BamHI site such that a 10-histidine tract followed by a TEV protease cleavage site (ENLYFQG) was placed in frame with the Fc coding sequence. The HABD region consisting of the first 170 residues of human CD44 (including the N-terminal signal peptide) was amplified using Pyrococcus furiosus (Pfu) DNA polymerase from full-length human CD44 cDNA using the forward and reverse primers huCD44-12 (5′-CGCGaagcttCGCTCCGGACACCATGGACAAG-3′) and huCD44-465 (5′-CGagatctGGGTAGATGTCTTCAGGATTCG-3′) that contain HindIII and BglII restriction sites (shown in lowercase type), respectively.
Appropriately digested PCR products were ligated into the modified pCDM7Ig vector that had been digested at the HindIII site and at a new BamHI site created at the 5′-end of the linker. Candidate recombinant plasmids were sequenced to confirm the integrity of the clones.
HEK 293T cells were transiently transfected and cultured in CHO-S-SFM II serum-free medium (Invitrogen) for 3 days before supernatant was collected and passed over protein A-Sepharose (Sigma), followed by elution of the Fc fusion with 0.1 m glycine, pH 3, and the addition of one-twentieth volume 2 m Tris-HCl, pH 8, to eluted fractions.
To produce His10-tagged monomers of HABD (His-HABD), DTT and EDTA were added (to final concentrations of 0.1 and 0.05 mm, respectively) to samples of (HABD)2-Fc prior to the addition of AcTEV protease (Invitrogen) at a ratio of 1 unit per 5 μg of protein and digested for 2 h at 20 °C before the removal of uncleaved fusion protein and digested Fc by passing the digest over protein A-Sepharose and removal of AcTEV protease by passing the sample over Ni2+-NTA-agarose (Invitrogen). Finally, the digested monomeric HABD was subjected to gel filtration using a HiLoad 26/60 Superdex 75 pg chromatography column (GE Healthcare).
Stock solutions of about 1 mg/ml HABD constructs were stored at −20 °C. Before use, (HABD)2-Fc and His-HABD were diluted in Hepes buffer with and without added calcium, respectively, to the desired concentrations.
Preparation of Other Proteins
A chimeric protein (AnxA5-Z) was constructed by fusing annexin A5 with a Z domain of protein A from Staphylococcus aureus, using standard molecular biology methods (24). The ZZ domain from the plasmid pEZZ-18 (GE Healthcare), which codes for a protein binding the Fc fragment of IgGs (25), was amplified by PCR, with primers to introduce NheI and BamHI sites at the 5′- and 3′-ends of the ZZ-coding sequence; the product was then cloned into the pGEF-A5 vector, which encodes full-length rat AnxA5 (26) to produce plasmid pGEF-A5ZZ. The insert encoding AnxA5-ZZ was then removed from pGEF-A5ZZ and cloned into the pET11b vector (GE Healthcare) at the XbaI and BamHI sites to produce plasmid pET11b-A5ZZ. A Z domain from this vector was amplified by PCR and cloned into the NheI/BamHI sites of pET11b-A5 to produce the desired plasmid, pET11b-A5Z.
Recombinant AnxA5 and AnxA5-Z were expressed in Escherichia coli BL21 (DE3) cells and purified on a Superdex-75 (GE Healthcare) size exclusion column, followed by MonoQ HR5/5 (GE Healthcare) anion exchange chromatography, as described previously for AnxA5 (26). Stock solutions of about 1 mg/ml AnxA5 constructs were stored at 4 °C and diluted in calcium-containing Hepes buffer prior to use.
Purified BRIC 235 monoclonal antibody (BRIC 235 mAb) was purchased from the International Blood Group Laboratory (Bristol, UK).
Substrate Preparation
Silica-coated QCM-D sensors (QSX303 and QSX335, Q-Sense AB, Västra Frölunda, Sweden) and silicon wafers with a native oxide layer of about 2-nm thickness (University Wafers, South Boston, MA) were immersed in an aqueous solution of 3% (w/v) SDS for 30 min, rinsed abundantly with ultrapure water, blow-dried in N2, and treated with UV/ozone (Bioforce Nanoscience, Ames, IA) for 25 min and stored in air. Glass coverslips (24 × 24 mm2) were immersed into Piranha solution (H2O2/H2SO4 = 1:3) for 1 h, extensively rinsed with ultrapure water, blow-dried with N2, and stored in sealed Petri dishes under nitrogen. Cleaned substrates were exposed to UV/ozone (25 min) prior to use.
Preparation of Lipids and Vesicles
Lyophilized 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS) were purchased from Avanti Polar Lipids (Alabaster, AL). Bis-NTA-functionalized octadec-9-enyl-octadecyl-amine (OEOA-bis-NTA) was kindly provided by Jacob Piehler (Osnabrück University, Germany) and prepared as described earlier (27). The divalent presentation of Ni2+-loaded NTA improves the binding stability of histidine-tagged proteins, with dissociation constants in the lower nanomolar range (28). Mixtures of DOPC/DOPS (3:1 molar ratio), DOPC/OEOA-bis-NTA (9:1), and pure DOPC were dissolved in chloroform, dried, resuspended in Hepes buffer, and homogenized as described earlier (29). Small unilamellar vesicles (SUVs) were obtained by sonication (30) or extrusion (50 nm final pore size) (31). SUV suspensions were stored at 4 °C under nitrogen and diluted to desired concentrations before use.
Quartz Crystal Microbalance with Dissipation Monitoring
QCM-D measures changes in resonance frequency, Δf, and dissipation, ΔD, of a sensor crystal upon interaction of (soft) matter with its surface. The QCM-D response is sensitive to the mass (including coupled water) and the mechanical properties of the surface-bound layer. Adsorption and interfacial processes were monitored in situ with subsecond time resolution. Measurements were performed with a Q-Sense E4 system (Q-Sense AB), in flow mode (30) and at a working temperature of 23 °C. Changes in dissipation and normalized frequency, Δf = Δfn/n, of the seventh overtone (n = 7; i.e. ∼35 MHz) are presented.
For sufficiently rigid biomolecular layers at high surface coverage, the film thickness can be estimated, to within an error of typically <20%, by the acoustic thickness, d = Δm/ρH2O = −C/ρH2O × Δf (32), where Δm, ρH2O = 1.0 g·cm−3, and C = 18.06 ng·cm−2·Hz−1 are, respectively, the adsorbed mass (including coupled water), the density of the bulk solution, and the mass sensitivity constant for a sensor crystal with a fundamental resonance frequency of 4.95 MHz. For very soft films, such as HA, this equation is not valid, and colloidal probe reflection interference contrast microscopy (RICM) was used for the thickness estimation instead.
Ellipsometry
Ellipsometry measures changes in the ellipsometric angles, Δ and Ψ, of polarized light upon reflection at a planar surface. We employed ellipsometry in situ, using silicon wafers as substrates that were installed in an open glass cuvette with continuously stirred sample solution, to quantify adsorbed biomolecular masses in a time-resolved manner.
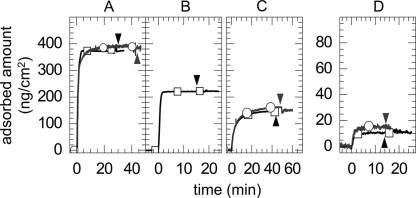
Measurements were conducted with a spectroscopic rotating compensator ellipsometer (M2000V, J. A. Woollam Co., Lincoln, NE), as described earlier (30). The custom built cuvette was cleaned in 3% SDS (30 min), rinsed with ultrapure water, and blow-dried with N2, passivated by immersion in an aqueous solution of 10 mg/ml bovine serum albumin (15 min), rinsed again with ultrapure water, and blow-dried with N2. Bound masses were determined by numerical fitting of the ellipsometric data to a multilayer model, as described elsewhere (30, 33). To validate our quantitative approach, we compared the experimentally determined adsorbed amounts for known films (see Fig. 5, A and B) with data from the literature. The amount of lipids in our SLBs (380 ± 10 ng/cm2; Fig. 5A) agreed within 10% error with previously reported data (34). The amount of membrane-bound AnxA5-Z (220 ng/cm2, or 5.0 pmol/cm2; see Fig. 5B) was also similar to expectations for a dense protein monolayer. A two-dimensional crystalline layer of AnxA5 with p6 symmetry, for example, has a surface density of 5.5 pmol/cm2 (35).
FIGURE 5.
Amounts of phospholipids, AnxA5-Z, CD44 HABDs, and HA bound during formation of model systems monitored by ellipsometry. Data are shown for the formation of both (HABD)2-Fc-coated (model system 1, black squares) and His-HABD-coated (model system 2, gray circles) bilayers, from which the stoichiometry of HA binding was estimated (Table 2). The individual steps were as follows: SLB-formation (A), the sequential adsorption of AnxA5-Z (B, model system 1 only), HABD capture (C), and HA250 binding (D). Each incubation step started at 0 min; concentrations as in Figs. 2 and 3 were employed; the start of rinsing in buffer is indicated (arrowheads in the respective panels).
Combination of QCM-D and Ellipsometry
Selected measurements (supplemental Fig. S1) were performed with a combined setup that simultaneously provides QCM-D and ellipsometric data on the same surface. To this end, a purpose-designed flow module (Q-Sense AB) was used. The technical implementation and the numerical fitting of the data are described elsewhere.3
Colloidal Probe Reflection Interference Contrast Microscopy
Colloidal probe RICM was used to estimate the thickness of surface-bound HA films. This microinterferometric technique measures the height at which a colloidal probe hovers above a transparent planar substrate (37). The model systems were assembled on glass coverslips, using a custom-built liquid cell. Polystyrene microspheres (Polysciences, Eppelheim, Germany) of ∼25-μm diameter were coated with a passivating polyethylene glycol layer of about 10 nm in thickness and used as colloidal probes. RICM images at three different wavelengths were acquired simultaneously to ensure unambiguous determination of distances up to 1 μm (31). Distances were determined with a simple model (parallel plate approximation with incident light parallel to the surface normal) (38). See the supplemental material for details of the measurements and data analysis.
RESULTS
Design of the Model Systems
Two different approaches were devised to immobilize the HABD of CD44 on SLBs (Fig. 1, B and C). In model system 1, two HA binding domains of CD44 were fused to an immunoglobulin Fc domain ((HABD)2-Fc). A bifunctional fusion protein, made of annexin A5 (AnxA5; a protein that binds to negatively charged lipids) and the Z-fragment of Protein A that binds the Fc domain, was used to immobilize the HABD dimer to SLBs containing the phospholipid DOPS. For model system 2, the (HABD)2-Fc construct was cleaved within the TEV protease site, located at the N-terminal side of the Fc hinge region, releasing His-HABD. HABD monomers were then directly immobilized on SLBs containing lipids with an Ni2+-loaded bis-NTA-functionalized headgroup. Thus, these two strategies provided both monomeric and dimeric CD44 HABD constructs for study. SLBs were chosen as a support because they are inert to nonspecific binding of most proteins and preserve the lateral mobility that characterizes membrane-bound proteins. All steps in the assembly of the model systems were monitored by QCM-D (see Figs. 2 and 3 and Table 1).
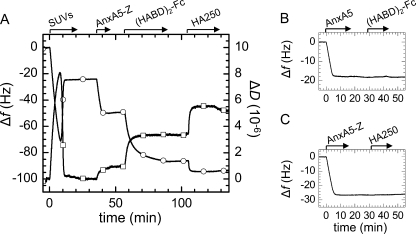
FIGURE 2.
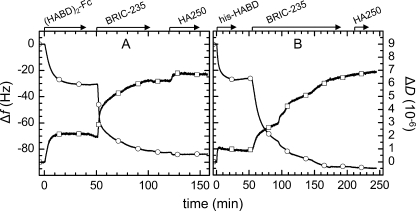
A, step-by-step assembly of model system 1, followed by QCM-D. Start and duration of all incubation steps is indicated (arrows). SLBs were formed by spreading of 50 μg/ml SUVs, made of DOPC and DOPS (3:1), to a silica surface. The two-phase behavior together with final changes in frequency (circles) of Δf = −25 Hz and in dissipation (squares) of ΔD < 0.3 × 10−6 characterize the formation of an SLB of good quality (36, 39). Responses for the sequential incubation of 10 μg/ml AnxA5-Z and 10 μg/ml (HABD)2-Fc indicate formation of stable monolayers. Binding of 10 μg/ml HA250 was readily detected. B, (HABD)2-Fc, incubated at 10 μg/ml, did not bind to native AnxA5, confirming that binding to AnxA5-Z was specific. C, HA250 (10 μg/ml) did not bind to AnxA5-Z, indicating that binding to (HABD)2-Fc was specific. For simplicity, SLB formation is not displayed for the assays in B and C, and only frequency shifts are shown.
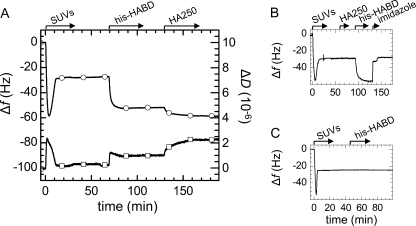
FIGURE 3.
A, construction of model system 2, followed by QCM-D. SLBs were formed by spreading of 50 μg/ml SUVs, made of DOPC and bis-NTA-functionalized lipids (9:1), to a silica surface. The final absolute frequency shift, |Δf| = 29 Hz, was slightly higher than in Fig. 2A, most likely due to the presence of the bulkier bis-NTA headgroups (33). His-HABD, incubated at 10 μg/ml, formed a stable monolayer, and binding of 10 μg/ml HA250 was readily detected. B, HA250 (10 μg/ml) did not adsorb to SLBs containing NTA functionality, and the bound monolayer of his-HABD could be fully eluted by 200 mm imidazole. Changes in Δf and ΔD at about 130 min do not reflect any changes on the surface but resulted from a change in the viscosity and/or density of the surrounding solution due to the presence of imidazole. C, His-HABD (10 μg/ml) did not bind to SLBs that were made of pure DOPC and thus lacked NTA functionality.
TABLE 1.
Final QCM-D responses for each individual step of the formation of the model systems
Values were determined after rinsing in buffer; S.E. values correspond to variations between two or more measurements, and experimental noise.
| Biomolecular film | Model system 1 |
Model system 2 |
||
|---|---|---|---|---|
| Δf | ΔD | Δf | ΔD | |
| Hz | ×10−6 | Hz | ×10−6 | |
| SLB | −25 ± 1 | 0.25 ± 0.1 | −29 ± 1 | 0.4 ± 0.2 |
| AnxA5-Z | −27 ± 1 | 1.0 ± 0.2 | ||
| CD44-HABD | −37 ± 2 | 2.5 ± 0.1 | −26 ± 2 | 1.0 ± 0.2 |
| HA250 | −7.5 ± 1 | 1.7 ± 0.2 | −7 ± 1 | 1.2 ± 0.1 |
Construction of Model System 1 (HABD Dimers)
Exposure of SUVs made of a mixture of DOPC and DOPS revealed the characteristic response for SLB formation (Fig. 2A) (39). Subsequent incubation with AnxA5-Z yielded an additional frequency shift of about −27 ± 1 Hz (Fig. 2A), or an acoustic thickness of 4.9 nm. It was previously shown that AnxA5-Z retains the capacity of native AnxA5 to bind to negatively charged SLBs in a stable and highly oriented manner, with the Z-fragment being exposed in the bulk solution (40). The acoustic thickness of 4.9 nm is in reasonable agreement with the expected thickness of the AnxA5 layer (2.8 nm) (35) plus the size of the Z-fragment (∼3 nm). Changes in dissipation of about 1.0 ± 0.2 × 10−6 were observed for AnxA5-Z, whereas only minor changes are typically found for native AnxA5 (26). We attribute the increased dissipation to the presence of a flexible linker that confers rotational freedom to the bulk exposed Z-fragment (41).
(HABD)2-Fc readily bound to AnxA5-Z-covered SLBs (Fig. 2A). Adsorption was initially fast but slowed down progressively until stabilization of frequency and dissipation after about 30 min. Binding was stable upon rinsing in buffer solution. (HABD)2-Fc did not bind to native AnxA5 (Fig. 2B), confirming that binding to AnxA5-Z was specific. The total acoustic thickness of the AnxA5-Z/(HABD)2-Fc layer was about 12 nm. For comparison, the lengths of the HABD (1, 13) and the Fc domain (42) are 3.5 and 7 nm, respectively, whereas the thickness of AnxA5 and the average size of the Z-fragment are each about 3 nm. Because the Z-fragment binds between Fc domains CH2 and CH3 (42), it is likely that about half of the Fc domain intercalates into the layer of Z-fragments. The resulting protein layer would then have a total thickness of about 13 nm, in reasonable agreement with the acoustic thickness. From the design of the protein fusion constructs and the QCM-D response, we conclude that the HA receptors are immobilized in a stable and suitably oriented manner on the surface.
HA with a selected molecular weight of 262 kDa (HA250) bound fast to the receptor-covered surface (Fig. 2A). Equilibrium was reached within less than 10 min, yielding Δf = −7.5 ± 1 Hz and ΔD = 1.7 ± 0.2 × 10−6. Although the interaction between HA and individual HABD domains is rather weak (KD ∼65 μm (21)), little or no desorption was observed upon rinsing (Fig. 2A). This provides a first indication that individual HA chains interact with several CD44 molecules to stabilize binding (i.e. the half-lives of the interaction are long). A small decrease in dissipation upon rinsing hints at a minor reorganization of the HA film upon removal of HA from the bulk solution.
Construction of Model System 2 (HABD Monomers)
SUVs that contained a fraction of bis-NTA-functionalized lipids formed stable SLBs (Fig. 3A). In a second step, his-HABD was added. In contrast to the dimeric HABD, the monomer adsorbed quickly (within 15 min). The acoustic thickness of the protein film, about 5 nm, corresponds well to the size of the HABD (1, 13). The magnitude of the dissipation shift, 1.0 ± 0.2 × 10−6, and the slight maximum at intermediate coverage are in accordance with the molecules being linked via a flexible linker to the SLB (41). The proteins remained stably bound upon rinsing in buffer but could be fully eluted in 200 mm imidazole (Fig. 3B). Also, His-HABD did not bind to SLBs that lacked NTA functionality (Fig. 3C). These controls confirmed that stable and specific anchorage is conferred by the protein's C-terminal His tag and the lipid's bis-NTA functionality.
The subsequent addition of HA250 resulted in shifts of Δf = −7 ± 1 Hz and ΔD = 1.2 ± 0.2 × 10−6. Again, binding was stable upon rinsing in buffer (Fig. 3A). We note that the ΔD/−Δf ratio of the HA films on monomeric and dimeric HABD were similar (0.17 and 0.23 × 10−6/Hz, respectively) but exceeded the values for the protein or lipid layers by at least 2.5-fold. This indicates that the receptor-bound HA chains assemble into a rather soft film and that the morphology of the HA film is only marginally, if at all, affected by the mode of presentation, monomeric or dimeric, of CD44 HABDs.
Authentic Nature of HA Binding to CD44-coated Bilayers
In order to validate the use of the synthetic bilayer system for analysis of CD44-HA interactions, we carried out a number of controls. HA250 did not show any binding when exposed to surfaces that lacked either (HABD)2-Fc (Fig. 2C) or His-HABD (Fig. 3B), indicating that binding to the receptors was specific. Furthermore, the addition of BRIC-235 mAb, an anti-HABD antibody that blocks HA binding to native CD44 on leukocytes (43), led to inhibition of most or all HA binding to both His-HABD and (HABD)2-Fc coated SLBs (Fig. 4). These findings indicate that the binding of HA to recombinant CD44 HABDs observed in coated SLBs resembles that of authentic full-length CD44 on the cell surface.
FIGURE 4.
Confirmation of authentic binding of HA to CD44-coated bilayers. BRIC-235 mAb blocks HA binding, as followed by QCM-D (circles, Δf; squares, ΔD). A, minor HA binding (Δf = −2 Hz) is retained on (HABD)2-Fc after incubation with BRIC-235 mAb for 60 min. B, binding was fully abolished on His-HABD-covered surfaces after incubation with 20 μg/ml BRIC-235 mAb for 120 min. Surfaces were prepared as shown in Figs. 2 and 3, respectively.
Estimation of CD44 Surface Density
Having characterized the thickness, stability, and morphological features of the functionalized lipid bilayer, we next quantified the amounts of each component adsorbed, in particular the effective density of CD44 and HA, by in situ ellipsometry (Fig. 5).
A maximal surface density of 4.0 ± 0.2 pmol/cm2, or a surface area per molecule of 42 ± 2 nm2, was obtained for his-HABD (Table 2). This corresponds to a mean distance between neighboring anchor sites of 7.0 ± 0.2 nm, which is about double the size of the peptidic core of the CD44 HABD (3.5 nm). It is likely that the heavy glycosylation of HABD increases the effective size of the receptor and thereby limits the packing density. It is also notable that the mean surface area per HABD monomer at saturation for (HABD)2-Fc (72 ± 8 nm2) is about 70% higher than for His-HABD. The surface density of AnxA5-Z (5.0 pmol/cm2) exceeds that of (HABD)2-Fc by severalfold and hence is unlikely to limit binding. Instead, we propose that it is the elongated and rather bulky Fc domain that hinders further adsorption sterically. The pronounced slowdown of binding with increasing coverage that we observed for (HABD)2-Fc (Figs. 2 and 5) would be consistent with such a scenario. We stress that, in line with results from QCM-D, all protein layers were found to be stable upon rinsing in buffer, illustrating the stability of the assembled multilayer systems.
TABLE 2.
Stably bound amounts of HABDs and HA and stoichiometry of the binding interaction
Values were determined by ellipsometry, after rinsing in buffer, in measurements analogous to those displayed in Fig. 5; S.E. values correspond to variations between two separate measurements and experimental noise.
| HABD |
HA |
HABD monomers available per HA | |||||
|---|---|---|---|---|---|---|---|
| Mass | Adsorbed amount | Mass | Adsorbed amount | ||||
| kDa | ng/cm2 | pmol/cm2 | kDa | ng/cm2 | fmol/cm2 | ||
| His-HABD | 39 | 156 ± 6 | 4.0 ± 0.2 | 262 | 15 ± 1.0 | 57 ± 4 | 70 ± 5 |
| (HABD)2-Fc | 116 | 146 ± 15 | 1.26 ± 0.13 | ≤11 | <0.5 | ||
| 146 ± 15 | 1.26 ± 0.13 | 30 | 6.1 ± 2.0 | 200 ± 70 | 12 ± 5 | ||
| 122 ± 15 | 1.05 ± 0.13 | 262 | 11.5 ± 2.0 | 44 ± 8 | 48 ± 15 | ||
| 140 ± 15 | 1.21 ± 0.13 | 1156 | 13.5 ± 1.5 | 12 ± 1.3 | 207 ± 45 | ||
| 133 ± 15 | 1.15 ± 0.13 | 2400 | 11.0 ± 2.0 | 4.6 ± 0.8 | 500 ± 150 | ||
Quantification of HA Binding and Stoichiometry of the Interaction
HA250 (262 kDa) adsorbed to a maximum level of 11.5 ± 2 ng/cm2, or approximately 40 fmol/cm2, on (HABD)2-Fc and to a maximum of 15 ± 1 ng/cm2, or ∼57 fmol/cm2, on His-HABD-coated SLBs (Fig. 5 and Table 2). The bound masses of HA are hence an order of magnitude below those observed for either of the adsorbed protein species yet are still well above the detection limit (∼0.5 ng/cm2).
Assuming a minimal footprint of HA10 per receptor (7, 13, 44), we estimate that a single HA chain of 262 kDa could potentially accommodate 138 receptors. Furthermore, based on the surface density of CD44 determined by ellipsometry, we estimate the mean number of HABD monomers that are available per bound HA chain as 50 for (HABD)2-Fc and 70 for His-HABD. Of course, such estimates do not necessarily imply that all of these receptors are engaged in binding at any given time.
Influence of HA Concentration on Binding
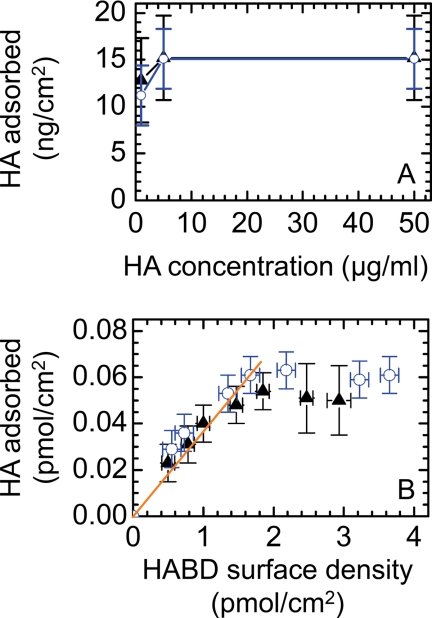
The HA concentration in solution did not strongly influence binding over the range of concentrations tested (see Fig. 7A). More than half-maximal binding was already reached at 1 μg/ml, and saturation was readily attained at 10 μg/ml. The responses for both CD44 constructs were identical. Below 1 μg/ml, adsorption kinetics became exceedingly slow, and undesired sample depletion by nonspecific adsorption to chamber walls became predominant.
FIGURE 7.
A, effect of HA concentration on HA binding. Shown is adsorption of 262-kDa HA to SLBs that were densely coated with His-HABD (blue circles) and (HABD)2-Fc (black triangles) as a function of HA concentration. B, effect of receptor surface density on HA binding. 10 μg/ml 262-kDa HA were exposed to SLBs that were coated with His-HABD (blue circles) and (HABD)2-Fc (black triangles) at varying surface densities. A linear fit through the origin for receptor surface densities <1.5 pmol/cm2 is also shown (solid orange line). Adsorbed amounts were determined by QCM-D; frequency shifts were converted into adsorbed masses or molar surface densities with the aid of calibration curves that were acquired from combined QCM-D/ellipsometry measurements (supplemental Fig. S1). Error bars correspond to experimental noise and uncertainties in the calculation of masses or surface densities from the frequency shifts.
Influence of CD44 Surface Density on HA Binding
To investigate how receptor density affects HA binding, we performed a systematic study with 262-kDa HA (Fig. 7B and supplemental Fig. S1). Although binding did not depend significantly on the presentation of CD44 (i.e. monomeric versus dimeric), two adsorption regimes were found. At high receptor densities (>1.5 pmol/cm2), HA binding attained a plateau, whereas between 0.5 and 1.5 pmol/cm2, binding scaled approximately linearly with receptor density (i.e. with 27 ± 4 receptors being on average available per HA chain). Remarkably, HA binding remained stable, even for the lowest receptor surface densities (0.5 pmol/cm2) investigated (not shown). Binding was also stable with 30-kDa HA at the lowest receptor densities tested (1.0 pmol/cm2).
Influence of HA Size on Binding to CD44-coated Bilayers
Next, we investigated how the size of HA affects binding to CD44. Given the similarity in binding characteristics observed with both CD44 model systems, we restricted this systematic analysis to (HABD)2-Fc-covered surfaces.
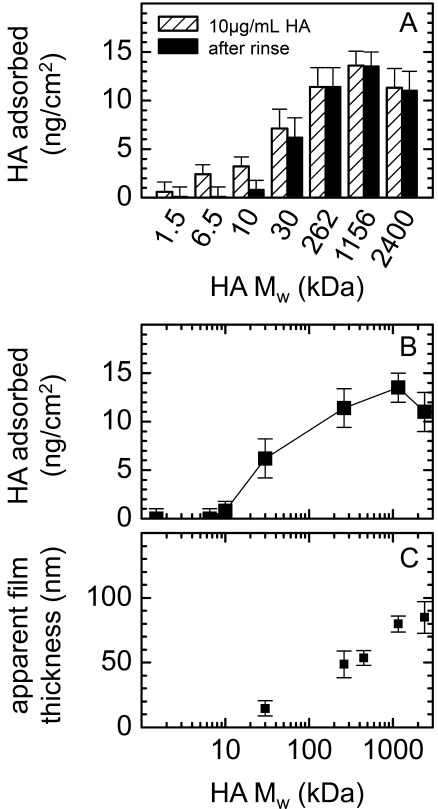
As shown in Fig. 6, A and B, the amount of HA binding increased in a sigmoidal fashion with HA size and saturated for molecular weights of ≥262 kDa (see also Table 2). Approximately half maximal binding was observed with 30-kDa HA, for which the average number of receptors available per stably bound HA molecule (12 ± 5; Table 2) corresponds quite closely to the maximal number of 16 available binding sites on the sugar chain.
FIGURE 6.
Effect of HA size on the extent and reversibility of binding to CD44. A, absolute amounts of HA with different molecular weights bound to (HABD)2-Fc-coated SLBs are shown following application at concentrations of 10 μg/ml (hatched bars) and subsequent rinsing (black filled bars). Error bars correspond to variations between typically two separate measurements. B, absolute amounts of stably bound HA, from A, displayed as a function of molecular weight. C, apparent thickness of HA films formed by molecular weight preparations of 30, 262, 450, 1156, or 2400 kDa, determined by colloidal probe RICM. Displayed values represent the increase in the total measured film thickness upon the addition of (HABD)2-Fc and HA of different molecular weight.
Significantly, the reversibility of HA binding was strongly dependent on the size of the bound HA molecule. Specifically, HA with molecular weights up to and including 10 kDa bound reversibly, whereas binding of HA with a molecular weight of 30 kDa or more was essentially irreversible (Fig. 6A).
Thickness and Morphology of HA Films
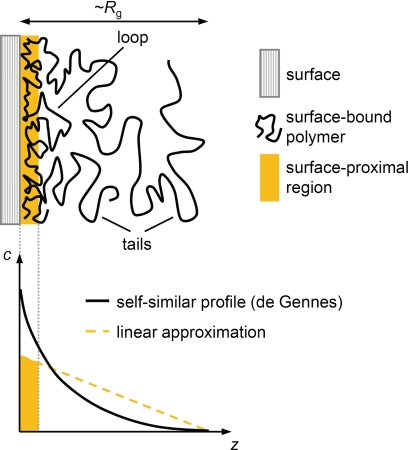
We employed colloidal probe reflection interference contrast microscopy (RICM) to estimate the thickness of stably bound HA films (Fig. 6C and supplemental Fig. S2). Again, (HABD)2-Fc-covered surfaces were chosen for a systematic investigation. Compared with the CD44 HABD layer alone, the film thickness increased only marginally in the presence of 30-kDa HA. In contrast, we found an apparent thickness of around 50 nm for 262-kDa HA. The thickness increased further with the molecular weight of HA, albeit to a smaller extent. The film thickness for HA molecular weights of ≥262 kDa hence exceeds the size of the CD44 HABD by an order of magnitude, implying that long HA chains engage in binding only along a fraction of their contour. Extended parts of the polymer chains dangle in the solution in the form of flexible tails and loops (see Fig. 8).
FIGURE 8.
Theoretical properties of flexible polymers, such as HA, adsorbing to a planar adhesive surface. A, schematic presentation of the morphology of the polymer film. Some parts of the polymer attach to the surface, whereas others dangle in solution, in the form of flexible loops and tails. The film thickness is similar to the polymer's radius of gyration, Rg. B, the polymer concentration, c, is highest at the surface and decreases with increasing distance, z, from it. A simple theoretical model by de Gennes (49) predicts a power law dependence (solid black line). For simplicity, we have used a linear dependence (dotted orange line) to estimate the amount of polymer in the surface-proximal region (orange area under the line).
From the amount of bound HA (Fig. 6B and Table 2) and the film thickness (Fig. 6C), we estimate that the HA chains occupy only a small fraction (1–3%) of the film's volume (i.e. ample space remains available for the incorporation of other molecules). With its low carbohydrate density and the dangling tails and loops, the HA film would be expected to be soft, in agreement with our observations by QCM-D (see above).
Estimation of the Occupancy of Surface-bound Receptors
At HA molecular weights of ≥30 kDa, we found a rather large number of receptors to be available on average per HA chain (Table 2). Are all of these receptors indeed engaged in binding at any given time? Expressing the adsorbed amounts of HA in units of HA10, the minimal footprint that is required for binding to individual CD44 molecules at the cell surface (7, 44), we derive maximal surface densities of ∼3 pmol/cm2 for 30-kDa HA and ∼6 pmol/cm2 for higher molecular weight HA. These values are indeed similar to or exceed the maximal surface density of membrane-bound CD44 HABD monomers (2.4 pmol/cm2 for (HABD)2-Fc and 4 pmol/cm2 for His-HABD).
For films made of HA with molecular weights of ≥262 kDa, the receptors can access only those portions of the HA chains within the 5–10-nm surface proximal region (Fig. 8). A local concentration of 2.4–4.8 mm HA10 would be estimated for this surface proximal region, when assuming that the concentration of HA in a film of 50-nm thickness decreases linearly with the distance from the surface (Fig. 8). Based on the law of mass action, KD = [HABD] × [HA10]/[HABD-HA10], a rather large occupancy of CD44 HABDs of 50–85% would result, assuming a KD of ∼65 μm.
It should, however, be born in mind that successful binding relies not only on the mere availability of HA at the receptor site but also on the accurate spatial alignment of the HA chain and the HA binding groove of CD44. The rotational mobility of the surface-bound receptors and the flexibility of HA should facilitate this alignment to some extent. However, due to the stereospecificity of HA binding, not all spatial constraints can be released by rotational or lateral motion of the receptors. The exact number of individual interactions that are engaged, on average, per HA chain thus remains difficult to assess. However, using this methodology, we are able to determine the maximum number of such interactions that would be possible for a particular length of HA and surface density of CD44.
DISCUSSION
We have developed in vitro model surfaces that reproduce the presentation of HA receptors on the plasma membrane in the sense that they expose the HA binding domain of CD44 in a natively oriented and stable manner and in a two-dimensional configuration. The surface density and type of receptors can be controlled, and their rotational and lateral mobility is likely to be retained. This is particularly so for the His-HABD monomer, which is anchored by a single (or at most very few) bis-NTA lipid to the fluid lipid membrane. In the case of the double-headed (HABD)2-Fc construct, the AnxA5 anchors are also likely to be laterally mobile under the conditions employed here (26, 45), although their dense packing may slow down large scale lateral motions.
In conjunction with a toolbox of surface sensitive characterization techniques, these model surfaces allowed for the binding of HA to be studied in a highly controlled, quantitative, and systematic manner.
Our approach differs from conventional binding assays in that the multivalent interactions between multiple binding sites on each HA chain and multiple receptors on the surface can be investigated directly. The affinity of the employed individual receptors for HA is rather low (KD of ∼65 μm (21)), and the lifetimes of individual interactions are thus short, as observed for HA8 and molecular weights up to 10 kDa (Fig. 6A). For high molecular weight (≥30 kDa) HA, we found irreversible binding. This drastic increase in stability is a direct consequence of the avidity increase incurred by multivalent interactions.
We recall that the binding strength of a multivalent interaction can potentially increase very quickly with the valency. Neglecting entropic contributions and assuming that the enthalpic contributions to the free energy of binding are additive, the binding affinities of the individual interactions would be multiplicative (46). Based on a KD of ∼65 μm for the interaction between CD44 and high molecular weight HA, this would imply that a valency of 3 would already result in a subpicomolar avidity. In comparison, the numbers of ≥27 receptors that we found to be available for HA chains of ≥262 kDa (Fig. 7B and Table 2) appear remarkably large.
HA has been shown to exhibit many features that are characteristic for a flexible polymer (47, 48). In particular, the conformation of HA in dilute aqueous solution is well described by a swollen random coil, the size of which is given by the radius of gyration, Rg, and depends on the molecular weight as follows, Rg = 1.3 nm × (molecular weight (kDa))0.6 (47). To interact with several membrane-bound receptors simultaneously, a given HA chain will have to undergo conformational changes. The energetic gain upon binding to several receptors will hence be offset by the entropic cost associated with a reduction in conformational freedom, and for a flexible chain, this penalty will be particularly high. We propose that this may be the reason why HA needs a rather large number of receptors for stable binding and hence that the mechanical properties of HA play an important role in determining the stability of HA binding to a receptor-covered cell surface.
Polymer physics has developed a theoretical framework that successfully describes the conformation of flexible polymers upon adsorption to surfaces (49, 50), and it is instructive to compare these theoretical predictions (Fig. 8) with the adsorption behavior of HA.
For the adsorption at sufficiently high coverage (the so called pseudoplateau regime), a film thickness comparable with the polymer's radius of gyration would be expected (50). For HA250, the Rg is 37 nm (47), which is in reasonable agreement with our experimental data revealing an apparent film thickness of ∼50 nm (Fig. 6C). However, the film thicknesses, as measured by colloidal probe RICM, increased more slowly than Rg with molecular weight. This discrepancy is not unexpected because the colloidal probe will compress very soft films to some extent, resulting in underestimation of the thickness values (see supplemental material for details).
Theory also predicts that the adsorbed amount becomes independent of molecular weight for high molecular weights (50) and that in this regime, saturation is reached at very low polymer concentrations in solution. Furthermore, adsorption is expected to decrease drastically below a threshold molecular weight, provided that the attraction between individual binding sites on the polymer and the surface is weak. Experimentally, we could distinguish two different regimes for CD44 HA binding. For high molecular weight species (≥262 kDa), binding was indeed irreversible and independent of molecular weight and HA concentration in solution, with adsorbed amounts of about 12 ng/cm2. With decreasing molecular weight, adsorbed amounts decreased strongly, and binding became progressively weaker or reversible and HA concentration-dependent. Stable half-maximal binding occurred at MW50 of ∼30 kDa. Taken together, these observations indicate that binding of HA can be described well in terms of adsorption of a flexible polymer to an adhesive surface.
Stoichiometry of HA Binding at Low Receptor Surface Density
At sufficiently low receptor coverage, the amount of adsorbed HA scaled linearly with the CD44 surface density, with ∼27 receptors being available per 262-kDa HA chain (Fig. 7B). It is instructive to compare the surface area per receptor at the lowest investigated receptor coverage (330 nm2) with the dimensions of HA in solution. The essentially spherical swollen random coil that the HA chains adopt in solution would have a projected area of approximately π × Rg2 ≈ 4300 nm2. An unperturbed HA chain that approaches the surface would hence cover a surface area that corresponds to ∼13 receptors, which is 2 times less than the number of available receptors.
This apparent discrepancy may be explained by one of or a combination of the following scenarios. The receptor density may increase locally in the vicinity of a bound HA chain. The design of our model systems allowed lateral displacement of the bound receptors, as is the case on the cell surface, and clustering or oligomerization of CD44 was indeed proposed as one possible mechanism to enhance HA binding to cell surfaces (7, 51, 52). Further experiments with model surfaces carrying immobile receptors will be useful to investigate whether ligand-induced reorganization of receptors can enhance ligand binding. In addition, the conformation of HA chains may change upon binding, adopting a flattened, pancake-like conformation on the surface, thereby increasing their projected area and the number of accessible receptors. Finally, there is systematic uncertainty in the determination of receptor and HA surface densities (supplemental Fig. S1).
Stability of Binding and Signaling
It has repeatedly been observed that the biological effect of HA depends on its molecular weight (18, 53, 54). Specifically, CD44 binding of a fraction of HA defined operationally as “low molecular weight” (20–500 kDa) has been reported to stimulate proliferation, whereas higher molecular weight fractions were inhibitory (18), but the molecular mechanism behind the size dependence remains unknown. In our in vitro assay, we find a strong dependence between HA size and stability of binding. One may speculate that the stability of binding could, in the cellular/tissue context, be a trigger for signaling, which would relate molecular weight and signaling in a very simple functional way.
We note that the threshold molecular weight for stable binding of HA may well depend on a number of parameters, such as the affinity, mobility, and (local) surface density of active receptors, or modifications of the HA chains in ways that remain to be elucidated. Large HA complexes, such as the recently reported cable-like structures or cross-linked HA (10), for example, should bind even more stably to the cell surface than the longest individual HA chains.
Stability of Interaction and Mechanical Cues
We have assessed the stability of binding under conditions in which external mechanical forces on the HA chains or the receptors were negligible. Given that stable binding is the result of an ensemble of multiple weak HA-receptor interactions, binding can quickly become labile under even minor mechanical stress. Leukocytes, for example, may readily start to roll along an HA-covered surface upon application of shear flow, because under such conditions, the individual interactions between CD44 on the leukocyte surface and HA could be ruptured one-by-one, in a zipper-like fashion.
Comparison between Monomeric and Dimeric HABD
HA of 262 kDa bound stably, in similar amounts and with similar conformation to bilayers coated with either monomeric or dimeric CD44 HABD, suggesting that the strength of binding is not strongly affected by the dimeric presentation of HABD in the form of the Fc fusion. We have speculated previously that dimerization of CD44 can lead to the activation of its HA binding function via promotion of a conformational change in the HABD (13). Clearly, the dimeric presentation that occurs in the context of the Fc fusion protein does not cause an increase in affinity and most likely does not represent the dimers that can be formed on the cell surface.
Comparison with the Cell Surface
The model surfaces that we have investigated here presented a rather high surface density of receptors, between 10- and 100-fold higher than the average densities that are typically found on CD44-expressing cells (55, 56). The total amount of adsorbed HA decreased with decreasing receptor density (Fig. 7B), although within the investigated range of receptor densities and for laterally mobile receptors, the molecular weight that is required for stable binding of HA did not depend strongly on the receptor density.
The receptor distribution on the cell surface is likely to be heterogeneous, and local enrichment (e.g. in membrane domains (57) or by dimerization via the cytoplasmic domains of CD44 (22)) has indeed been proposed. Our model surfaces may thus well represent local receptor concentrations and hence provide quantitative insight into the mechanism of local accumulation of HA on the cell surface.
We note that our receptor-bound HA films remain rather thin when compared with the typical thickness of hyaluronan-rich cell coats that can be found on the endothelial cell surface (58) or around chondrocytes (59, 60), for example. The thickness and HA surface density is also considerably lower than what we have reported previously for HA films in which the molecules were grafted by one of their ends to the surface (31), in agreement with theoretical predictions for adsorbed and end-grafted polymers, respectively. These observations imply that binding of HA to a cell surface that is covered with CD44 or other HA receptors alone is not sufficient to generate the thick coats of several hundred nm or more that are found around cells in vivo. Contributions from additional HA binding molecules, such as the pericellular matrix proteoglycans versican or aggrecan (59–63), and physical factors, such as membrane protrusions (64), are almost certainly responsible for these observed differences. Nevertheless, it should be possible to incorporate some of these components during formation of in vitro HA films in future experiments to more closely reproduce the pericellular HA coats found in vivo.
Conclusions and Perspectives
We have developed model systems that provide direct and quantitative information about the binding of HA to an assembly of HA receptors in a biologically relevant arrangement. We showed that the multivalent interaction of a given HA chain with a receptor-covered surface confers stable binding, provided that the HA chains are long enough, whereas binding decreased rapidly for HA smaller than 30 kDa. The amount of bound HA decreased with decreasing receptor surface density, whereas the stability of binding was not affected. Our results suggest a biophysical explanation for the different binding behaviors of low and high Mr HA that may underlie their distinct biological functions. The nature of HA as a flexible polymer needs to be considered to understand its binding and functional activities.
Supplementary Material
Acknowledgments
We thank Oleg Borisov (Pau, France) for fruitful discussions and Jacob Piehler (Osnabrück, Germany) for providing bis-NTA-functionalized lipids.
This work was supported by German Federal Ministry of Education and Research Grant 0315157, Spanish Ministry of Science and Innovation Grants MAT2008-04192 and RYC2009-04275, the Department of Industry of the Basque Government, and the Max-Planck-Society (to R. P. R. and P. M. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
I. Carton, A. R. Brisson, and R. P. Richter, submitted for publication.
- HA
- hyaluronan
- HABD
- hyaluronan binding domain
- Fc
- immunoglobulin Fc domain
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine
- DOPS
- 1,2-dioleoyl-sn-glycero-3-phosphoserine
- OEOA
- octadec-9-enyl-octadecyl-amine
- SUV
- small unilamellar vesicle
- QCM-D
- quartz crystal microbalance with dissipation monitoring
- RICM
- reflection interference contrast microscopy
- SLB
- supported lipid bilayer
- TEV
- tobacco etch virus
- His-HABD
- His10-tagged monomer of HABD.
REFERENCES
- 1.Teriete P., Banerji S., Noble M., Blundell C. D., Wright A. J., Pickford A. R., Lowe E., Mahoney D. J., Tammi M. I., Kahmann J. D., Campbell I. D., Day A. J., Jackson D. G. (2004) Mol. Cell 13, 483–496 [DOI] [PubMed] [Google Scholar]
- 2.Banerji S., Day A. J., Kahmann J. D., Jackson D. G. (1998) Protein Expr. Purif. 14, 371–381 [DOI] [PubMed] [Google Scholar]
- 3.Haynes B. F., Liao H. X., Patton K. L. (1991) Cancer Cells 3, 347–350 [PubMed] [Google Scholar]
- 4.Puré E., Cuff C. A. (2001) Trends Mol. Med. 7, 213–221 [DOI] [PubMed] [Google Scholar]
- 5.Clark R. A., Alon R., Springer T. A. (1996) J. Cell Biol. 134, 1075–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeGrendele H. C., Estess P., Siegelman M. H. (1997) Science 278, 672–675 [DOI] [PubMed] [Google Scholar]
- 7.Lesley J., Hascall V. C., Tammi M., Hyman R. (2000) J. Biol. Chem. 275, 26967–26975 [DOI] [PubMed] [Google Scholar]
- 8.Day A. J., Sheehan J. K. (2001) Curr. Opin. Struct. Biol. 11, 617–622 [DOI] [PubMed] [Google Scholar]
- 9.Lesley J., English N. M., Gál I., Mikecz K., Day A. J., Hyman R. (2002) J. Biol. Chem. 277, 26600–26608 [DOI] [PubMed] [Google Scholar]
- 10.Day A. J., de la Motte C. A. (2005) Trends Immunol. 26, 637–643 [DOI] [PubMed] [Google Scholar]
- 11.English N. M., Lesley J. F., Hyman R. (1998) Cancer Res. 59, 3736–3742 [PubMed] [Google Scholar]
- 12.Skelton T. P., Zeng C., Nocks A., Stamenkovic I. (1998) J. Cell Biol. 140, 431–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banerji S., Wright A. J., Noble M., Mahoney D. J., Campbell I. D., Day A. J., Jackson D. G. (2007) Nat. Struct. Mol. Biol. 14, 234–239 [DOI] [PubMed] [Google Scholar]
- 14.de la Motte C. A., Hascall V. C., Drazba J., Bandyopadhyay S. K., Strong S. A. (2003) Am. J. Pathol. 163, 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bollyky P. L., Falk B. A., Long S. A., Preisinger A., Braun K. R., Wu R. P., Evanko S. P., Buckner J. H., Wight T. N., Nepom G. T. (2009) J. Immunol. 183, 2232–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bollyky P. L., Falk B. A., Wu R. P., Buckner J. H., Wight T. N., Nepom G. T. (2009) J. Leukoc. Biol. 86, 567–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bollyky P. L., Lord J. D., Masewicz S. A., Evanko S. P., Buckner J. H., Wight T. N., Nepom G. T. (2007) J. Immunol. 179, 744–747 [DOI] [PubMed] [Google Scholar]
- 18.Kothapalli D., Flowers J., Xu T., Puré E., Assoian R. K. (2008) J. Biol. Chem. 283, 31823–31829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singleton P. A., Dudek S. M., Ma S. F., Garcia J. G. (2006) J. Biol. Chem. 281, 34381–34393 [DOI] [PubMed] [Google Scholar]
- 20.Puré E., Assoian R. K. (2009) Cell. Signal. 21, 651–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banerji S., Hide B. R., James J. R., Noble M. E., Jackson D. G. (2010) J. Biol. Chem. 285, 10724–10735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perschl A., Lesley J., English N., Trowbridge I., Hyman R. (1995) Eur. J. Immunol. 25, 495–501 [DOI] [PubMed] [Google Scholar]
- 23.Mahoney D. J., Aplin R. T., Calabro A., Hascall V. C., Day A. J. (2001) Glycobiology 11, 1025–1033 [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 25.Löwenadler B., Jansson B., Paleus S., Holmgren E., Nilsson B., Moks T., Palm G., Josephson S., Philipson L., Uhlén M. (1987) Gene 58, 87–97 [DOI] [PubMed] [Google Scholar]
- 26.Richter R. P., Him J. L., Tessier B., Tessier C., Brisson A. R. (2005) Biophys. J. 89, 3372–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lata S., Gavutis M., Piehler J. (2006) J. Am. Chem. Soc. 128, 6–7 [DOI] [PubMed] [Google Scholar]
- 28.Lata S., Reichel A., Brock R., Tampé R., Piehler J. (2005) J. Am. Chem. Soc. 127, 10205–10215 [DOI] [PubMed] [Google Scholar]
- 29.Richter R., Mukhopadhyay A., Brisson A. (2003) Biophys. J. 85, 3035–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eisele N. B., Frey S., Piehler J., Görlich D., Richter R. P. (2010) EMBO Rep. 11, 366–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richter R. P., Hock K. K., Burkhartsmeyer J., Boehm H., Bingen P., Wang G., Steinmetz N. F., Evans D. J., Spatz J. P. (2007) J. Am. Chem. Soc. 127, 5306–5307 [DOI] [PubMed] [Google Scholar]
- 32.Wolny P. M., Spatz J. P., Richter R. P. (2010) Langmuir 26, 1029–1034 [DOI] [PubMed] [Google Scholar]
- 33.Bingen P., Wang G., Steinmetz N. F., Rodahl M., Richter R. P. (2008) Anal. Chem. 80, 8880–8890 [DOI] [PubMed] [Google Scholar]
- 34.Benes M., Billy D., Benda A., Speijer H., Hof M., Hermens W. T. (2004) Langmuir 20, 10129–10137 [DOI] [PubMed] [Google Scholar]
- 35.Richter R. P., Brisson A. (2004) Langmuir 20, 4609–4613 [DOI] [PubMed] [Google Scholar]
- 36.Keller C. A., Kasemo B. (1998) Biophys. J. 75, 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schilling J., Sengupta K., Goennenwein S., Bausch A. R., Sackmann E. (2004) Phys. Rev. E 69, 021901 [DOI] [PubMed] [Google Scholar]
- 38.Kühner M., Sackmann E. (1996) Langmuir 12, 4866–4876 [Google Scholar]
- 39.Richter R. P., Bérat R., Brisson A. R. (2006) Langmuir 22, 3497–3505 [DOI] [PubMed] [Google Scholar]
- 40.Bérat R. (2007) Assemblages 2D de l'Annexine A5: Applications Biotechnologiques & Aspects Fonctionnels, Ph.D. thesis, University of Bordeaux I [Google Scholar]
- 41.Johannsmann D., Reviakine I., Richter R. P. (2009) Anal. Chem. 81, 8167–8176 [DOI] [PubMed] [Google Scholar]
- 42.Deisenhofer J. (1981) Biochemistry 20, 2361–2370 [PubMed] [Google Scholar]
- 43.Anstee D. J., Gardner B., Spring F. A., Holmes C. H., Simpson K. L., Parsons S. F., Mallinson G., Yousaf S. M., Judson P. A. (1991) Immunology 74, 197–205 [PMC free article] [PubMed] [Google Scholar]
- 44.Tammi R., MacCallum D., Hascall V. C., Pienimäki J. P., Hyttinen M., Tammi M. (1998) J. Biol. Chem. 273, 28878–28888 [DOI] [PubMed] [Google Scholar]
- 45.Vats K., Knutson K., Hinderliter A., Sheets E. D. (2010) ACS Chem. Biol. 5, 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mammen M., Choi S.-K., Whitesides G. M. (1998) Angew. Chem. Int. Ed. 37, 2754–2794 [DOI] [PubMed] [Google Scholar]
- 47.Takahashi R., Kubota K., Kawada M., Okamoto A. (1999) Biopolymers 50, 87–98 [Google Scholar]
- 48.Gribbon P., Heng B. C., Hardingham T. E. (2000) Biochem. J. 350, 329–335 [PMC free article] [PubMed] [Google Scholar]
- 49.de Gennes P. G. (1979) Scaling Concepts in Polymer Physics, Cornell University Press, Ithaca, NY [Google Scholar]
- 50.Fleer G. J., Cohen Stuart M. A., Scheutjens J. M., Cosgrove T., Vincent B. (1993) Polymers at Interfaces, Chapman & Hall, London [Google Scholar]
- 51.Sleeman J., Rudy W., Hofmann M., Moll J., Herrlich P., Ponta H. (1996) J. Cell Biol. 135, 1139–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lesley J., English N., Charles C., Hyman R. (2000) Eur. J. Immunol. 30, 245–253 [DOI] [PubMed] [Google Scholar]
- 53.Camenisch T. D., McDonald J. A. (2000) Am. J. Respir. Cell Mol. Biol. 23, 431–433 [DOI] [PubMed] [Google Scholar]
- 54.Stern R., Asari A. A., Sugahara K. N. (2006) Eur. J. Cell Biol. 85, 699–715 [DOI] [PubMed] [Google Scholar]
- 55.Visintin A., Mazzoni A., Spitzer J. H., Wyllie D. H., Dower S. K., Segal D. M. (2001) J. Immunol. 166, 249–255 [DOI] [PubMed] [Google Scholar]
- 56.Alves C. S., Burdick M. M., Thomas S. N., Pawar P., Konstantopoulos K. (2008) Am. J. Physiol. Cell Physiol. 294, C907–C916 [DOI] [PubMed] [Google Scholar]
- 57.Chen J., Pei Y., Chen Z., Cai J. (2010) Micron 41, 198–202 [DOI] [PubMed] [Google Scholar]
- 58.Nijenhuis N., Mizuno D., Spaan J. A., Schmidt C. F. (2009) Phys. Biol. 6, 025014 [DOI] [PubMed] [Google Scholar]
- 59.Boehm H., Mundinger T. A., Boehm C. H. J., Hagel V., Rauch U., Spatz J. P., Curtis J. E. (2009) Soft Matter 5, 4331–4337 [Google Scholar]
- 60.Lee G. M., Johnstone B., Jacobson K., Caterson B. (1993) J. Cell Biol. 123, 1899–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knudson W., Knudson C. B. (1991) J. Cell Sci. 99, 227–235 [DOI] [PubMed] [Google Scholar]
- 62.Evanko S. P., Angello J. C., Wight T. N. (1999) Arterioscler. Thromb. Vasc. Biol. 19, 1004–1013 [DOI] [PubMed] [Google Scholar]
- 63.Yamaguchi Y. (2000) Cell. Mol. Life Sci. 57, 276–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rilla K., Tiihonen R., Kultti A., Tammi M., Tammi R. (2008) J. Histochem. Cytochem. 56, 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.