Abstract
In birds, as in mammals, one pair of chromosomes differs between the sexes. In birds, males are ZZ and females ZW. In mammals, males are XY and females XX. Like the mammalian XY pair, the avian ZW pair is believed to have evolved from autosomes, with most change occurring in the chromosomes found in only one sex – the W and Y chromosomes1–5. By contrast, the sex chromosomes found in both sexes – the Z and X chromosomes – are assumed to have diverged little from their autosomal progenitors2. Here we report findings that overturn this assumption for both the chicken Z and human X chromosomes. The chicken Z chromosome, which we sequenced essentially to completion, is less gene-dense than chicken autosomes but contains a massive tandem array containing hundreds of duplicated genes expressed in testes. A comprehensive comparison of the chicken Z chromosome to the finished sequence of the human X chromosome demonstrates that each evolved independently from different portions of the ancestral genome. Despite this independence, the chicken Z and human X chromosomes share features that distinguish them from autosomes: the acquisition and amplification of testis-expressed genes, as well as a low gene density resulting from an expansion of intergenic regions. These features were not present on the autosomes from which the Z and X chromosomes originated but were instead acquired during the evolution of the Z and X as sex chromosomes. We conclude that the avian Z and mammalian X chromosomes followed convergent evolutionary trajectories, despite their evolving with opposite (female vs. male) systems of heterogamety. More broadly, in birds and mammals, sex chromosome evolution involved not only gene loss in sex-specific chromosomes, but also marked expansion and gene acquisition in sex chromosomes common to males and females.
A century ago, Herman Muller proposed the first theory of sex chromosome evolution --that the X and Y chromosomes of Drosophila evolved from an ordinary pair of autosomes, and that genes on the Y chromosome had gradually deteriorated while their counterparts on the X were preserved1. In the 1960’s, Susumu Ohno applied Muller’s theory to the sex chromosomes of vertebrates, arguing that while the sex-specific W and Y chromosomes of birds and mammals had degenerated, the content of the Z and X chromosomes remained intact2. Four decades on, comparisons of the human X and Y chromosomes have underscored the dramatic evolutionary changes on the Y chromosome,3–5 but the assumption that the X chromosome has been evolutionarily stable remains unexamined.
The evolutionary relationship between the mammalian X chromosome and the avian Z chromosome has been the subject of much speculation, but it also remains unresolved. Ohno conjectured that the X chromosomes of mammals were orthologous to the Z chromosomes of birds2. However, comparative mapping of 30 Z-linked genes indicated that the chicken Z chromosome was orthologous to human chromosomes 5, 8, 9, and 18, not to the human X chromosome6,7. These findings were supported by the draft sequence of the chicken genome, but only about one third of the sequence of the Z chromosome was present in the assembly, leaving open the possibility that regions of orthology between the avian Z and mammalian X had yet to be detected8. The recent discovery that a subset of the five platypus X chromosomes contain orthologs of genes on the chicken Z chromosome renewed speculation that the avian Z and mammalian X share a common origin9–12. To accommodate the results of comparative gene mapping experiments, some have proposed that the chicken Z and human X were derived from different portions of an ancestral proto sex chromosome which broke apart, leaving Z orthologs autosomal in mammals and X orthologs autosomal in birds11,12.
To reconstruct and compare the evolutionary trajectories of the avian Z and mammalian X chromosomes, we have produced the finished sequence of the chicken Z chromosome, the first for any Z chromosome (Supplementary Figures 1–3). The resulting sequence spans roughly 80 megabases (Mb), is complete apart from four gaps, and is accurate to about one nucleotide per megabase. The chicken Z chromosome contains ~1000 genes (Supplementary Table 1). This makes the Z chromosome less gene-dense than any chicken autosome, with 11 genes per megabase, which is less than half of the chicken autosomal average of 25 genes per megabase (Table 1)8. Conversely, the density of interspersed repeats is 60% higher in the Z chromosome than in chicken autosomes (Supplementary Figure 1, Table 1). Most of these repeats are LINE elements, whose abundance in the Z chromosome is 70% higher than in autosomes (Supplementary Figure 1, Table 1). As a result, the Z chromosome is structurally distinct from the rest of the chicken genome.
Table 1.
Comparison of Z chromosome and X chromosome structural features
| Table 1a: Comparison of structural features of Z and X with autosomes | ||||
|---|---|---|---|---|
| Chicken Z | Chicken autosomes | Human X | Human autosomes | |
| Genes/Mb | 12 | 25 | 7 | 12 |
| Interspersed Repeats | 15% | 9.4% | 56% | 45% |
| LINEs | 11% | 6.4% | 32% | 21% |
| Gene Size (kb) | 21 | 27 | 49 | 57 |
| Table 1b: Comparison of structural features of Z-orthologous and X-orthologous autosomal regions with all autosomes | ||||
|---|---|---|---|---|
| Z-orthologous regions of human chromosomes 5, 9, and 18 | Human autosomes | X-orthologous regions of chicken chromosomes 1 and 4 | Chicken autosomes | |
| Genes/Mb | 10 | 12 | 23 | 25 |
| Interspersed Repeats | 48% | 45% | 8.9% | 9.4% |
| LINEs | 23% | 21% | 6.0% | 6.4% |
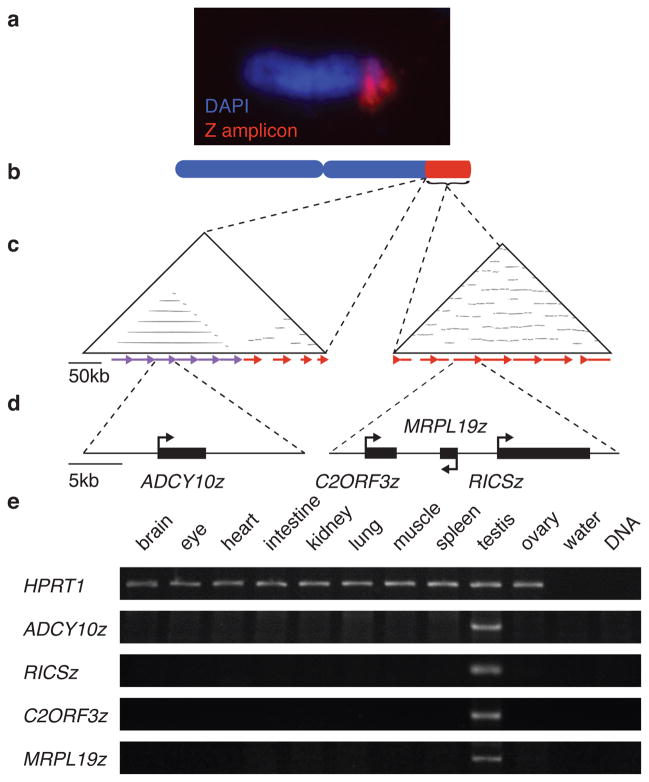
The Z chromosome’s most prominent feature is a previously unrecognized tandem array of testis-expressed genes, extending over 11 megabases at the distal end of the long arm (Fig. 1, Supplementary Figure 4). This array constitutes nearly 15% of the Z chromosome, one-fifth of all chicken segmental duplications, and 1% of the entire chicken genome (Fig. 1A & B)8. This sequence was initially reported as heterochromatin13, but we find three genes are present in each repeat unit, and a smaller flanking array contains a fourth (Fig. 1C & D, Supplementary Figure 5, Supplementary Table 2). Together, these four gene families total hundreds of copies and comprise almost one third of the protein-coding genes on the Z chromosome (Fig. 1D, Supplementary Table 2). All four gene families are expressed predominantly in the testis (Fig. 1E). We have termed this massive array of testis-expressed genes the Z amplicon.
Figure 1. The Z Amplicon.
a. Fluorescence in situ hybridization of Z-amplicon BAC CH261-77N6 (red) to distal long arm of Z chromosome (blue).
b. Z amplicon (red) comprises most distal 11 Mb of Z chromosome.
c. Triangular dot plots each comparing the sequence of a Z-chromosome BAC to itself. Within the plot, each dot represents a perfect match of 50 base pairs. Direct repeats appear as horizontal lines; scale bar represents 50kb. On left, BAC CH261-73L15 contains six tandem repeats covering 120 kb immediately proximal to Z amplicon. On right, BAC CH261-137P21, a representative Z amplicon clone. Each 25–30kb repeat unit is ~95% similar to any other, though some units have been disrupted by insertions and deletions.
d. Genes in repeat units of Z amplicon; scale bar represents 5kb. Each 20kb repeat unit of small array in CH261-73L15 contains one copy of ADCY10z. Each 25–30kb repeat unit of Z amplicon contains one copy each of C2ORF3z, MRPL19z, and RICSz.
e. RT-PCR analysis of Z-amplicon gene expression in adult tissues. HPRT1 is widely expressed in adult tissues and serves as positive control for reverse transcriptase reaction. All Z-amplicon genes are expressed in testis, but not other tissues.
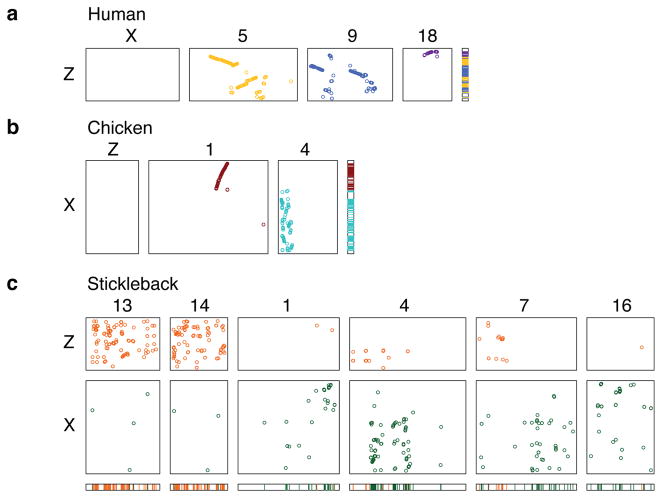
With the finished sequence of the Z chromosome in hand, we set out to test Ohno’s hypothesis that the avian Z and mammalian X chromosomes are orthologous. To reconstruct and visualize evolutionary relationships between chicken and human chromosomes, we systematically plotted the locations of orthologous gene pairs (Supplementary Fig. 6 & 7). We find that none of the ~1000 genes on the chicken Z chromosome has an ortholog on the human X chromosome (Fig. 2A & B, Supplementary Table 1). The Z chromosome is orthologous only to portions of human autosomes 5, 9, and 18 (Fig. 2A). Contrary to initial reports7, the Z chromosome is not orthologous to human chromosome 8 (Supplementary Fig. 6). In reciprocal fashion, the human X chromosome is orthologous only to portions of chicken autosomes 1 and 4, not to the Z chromosome (Fig. 2B)5. Based on this comprehensive analysis, we conclude that genes that are sex-linked in chickens are autosomal in humans, and vice versa, in broad agreement with earlier comparative mapping experiments6,7.
Figure 2. Independent origin of chicken Z and human X chromosomes.
Rectangular dot plots show chromosomal locations of Z-orthologous or X-orthologous genes in other species.
a. Chicken Z chromosome versus selected human chromosomes. Chicken Z chromosome is not orthologous to human X, but is orthologous to portions of human autosomes 5 (yellow), 9 (blue), and 18 (purple). At right: three-color projection of dot plots onto a unified schematic of chicken Z, showing that orthology to human chromosomes 5, 9, and 18 accounts for most of Z chromosome, with exception of Z amplicon on distal long arm.
b. Human X chromosome versus selected chicken chromosomes. Human X chromosome is not orthologous to chicken Z, but is orthologous to portions of chicken autosomes 1 (red) and 4 (cyan). At right: two-color projection of dot plots onto unified schematic of human X, showing that orthology to chicken chromosomes 1 and 4 spans X (colored bar).
c. Chicken Z chromosome (orange) and human X chromosome (green) versus selected stickleback chromosomes. Chicken Z and human X orthologs occupy separate and distinct locations within stickleback genome. Chicken Z orthologs are present on stickleback chromosomes 13 and 14, while human X orthologs are present on stickleback chromosomes 1, 4, 7, and 16.
Although the Z and X chromosomes show no signs of orthology, it is conceivable that they were recruited from different portions of a proto sex chromosome in the common ancestor of birds and mammals11. Some investigators have raised this possibility based on comparative gene mapping in the platypus11. However, the platypus does not form an outgroup to birds and mammals, and cannot resolve which is the ancestral state: a platypus-like linkage of Z-orthologous genes and X-orthologous genes, or the separation we observe in chicken and human. Others have attempted to resolve this question with comparisons to an outgroup genome that is far from complete12. Instead, we compared the Z and X chromosomes to the genomes of the four closest out-group species whose genomes are sequenced and assembled. Each species represents a different order of teleost fish, which diverged from land vertebrates over 450 million years ago14. After they diverged from birds and mammals, but before they diverged from each other, these fish species experienced a whole genome duplication, complicating the identification of 1:1 orthologs14. Nevertheless, we observe that most orthologs of Z and X-linked genes occupy separate portions of each fish genome (Supplementary Fig. 8–11). For example, three-spine stickleback linkage groups 13 and 14 carry the bulk of Z-orthologous genes, while X-orthologous genes mostly reside on stickleback linkage groups 1, 4, 7, and 16 (Fig. 2C). Since we observe that Z-orthologous genes are separated from X-orthologous genes in birds, mammals, and each of these four fish, we conclude that the Z and the X chromosomes have evolved independently from distinct portions of the ancestral vertebrate genome.
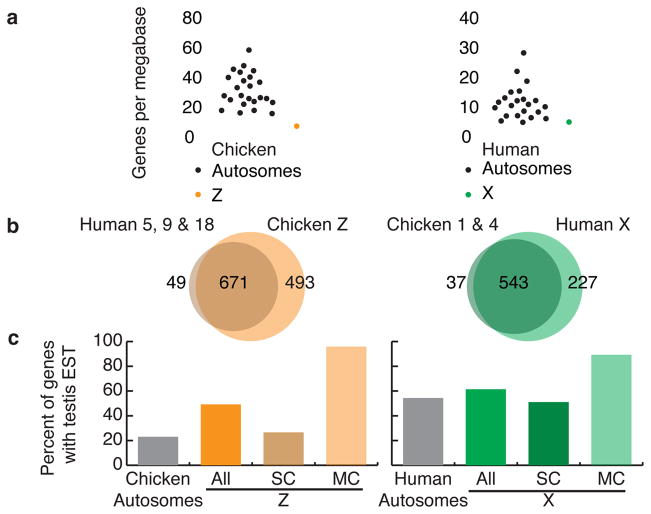
Although we rejected the hypothesis that the avian Z and mammalian X chromosomes share a common origin, we discovered that the chicken Z and human X chromosomes share common features. Like the chicken Z chromosome, the human X chromosome has a low gene density; there are half as many genes per megabase on the X chromosome as on the average human autosome (Fig. 3A, Table 1A)5. Other investigators have observed that low gene density is often associated with increased interspersed repeat content, specifically LINEs5,15. We also observe this association on the Z and X chromosomes (Supplementary Fig. 1, Table 1A).
Figure 3. Convergent gene gain on the chicken Z and human X chromosomes.
a. Gene density of Z and X chromosomes compared to autosomes. Both are unusually gene poor, with about half the gene density of a typical autosome.
b. Venn diagrams comparing gene content of chicken Z and human X chromosomes to orthologous autosomes. Most genes on orthologous autosomes remain on the sex chromosomes; few have been lost. Both chicken Z and human X gained hundreds of genes not present on orthologous autosomes.
c. Percentage of protein coding genes with testis ESTs in Unigene. Left panel: Compared to chicken autosomes, Z chromosome is enriched for testis-expressed genes. Single-copy Z chromosome genes (SC) show no enrichment for testis ESTs compared to autosomal gene, but nearly all multi-copy (MC) genes are expressed in testis. Right panel: Similar results on human X chromosome.
Two scenarios could account for these features of the chicken Z and human X chromosomes. Either the Z and X chromosomes arose from autosomes pre-adapted for the role of sex chromosomes, or they arose from ordinary autosomes that convergently evolved into specialized sex chromosomes. If the Z and X chromosomes arose from pre-adapted autosomes, then the structural features shared by the Z and X chromosomes should also be found on the orthologous autosomal regions. We tested this theory by comparing each sex chromosome to the orthologous autosomes in the other species (Fig. 2, Table 1, Supplementary Tables 3 & 4). As a group, the autosomal regions that correspond to the Z and the X chromosomes are typical of their respective genomes (Table 1B). Although these regions show a slight deficit in gene density relative to the average within their respective genomes, the difference is too small to account for the extremely low gene density of the Z and X chromosomes. Because the orthologous autosomes in the other species do not share the structural features common to the Z and X chromosomes, we infer that these convergent features arose during the process of sex chromosome evolution, and not before.
To explain the paucity of genes on the Z and X chromosomes, we looked for evidence that both chromosomes lost genes during sex chromosome evolution. Instead, we observed that both the Z and the X chromosomes gained protein-coding genes. We compared the gene content of the Z and the X chromosomes to the orthologous autosomes from the other species as a surrogate for the ancestral gene content of the Z and X chromosomes (Fig. 2, Fig. 3B, Table 1, Supplementary Tables 3 & 4). We found that only a few dozen genes present on the orthologous autosomes are absent from the Z and X chromosomes (Fig. 3B). In contrast, hundreds of genes present on the Z and X chromosomes are absent from the orthologous autosomes (Fig. 3B). We conclude that both the Z and X chromosomes experienced substantial net gene gain.
The majority of genes gained by the Z and X chromosomes are members of multi-copy families (Fig. 3B, Supplementary Tables 3 & 4). On the chicken Z chromosome, these are the genes of the Z amplicon. The human X chromosome has gained thirteen different cancer-testis antigen gene families5. All of the Z amplicon genes are expressed predominantly in testis (Fig. 1E), as are the cancer-testis antigen genes of the human X chromosome16. The addition of these multi-copy gene families has biased the Z and X chromosomes toward testis-expressed genes (Fig. 3C). Both the Z and the X chromosomes have an elevated proportion of genes expressed in testis tissue compared to autosomes as measured by the number of genes with a testis EST in Unigene17 datasets (Fig. 3C). However, when multi-copy genes are removed, the remaining conserved single-copy genes show no bias (Fig. 3C). Others have observed a bias towards sex and reproduction related genes on the human X chromosome18. Our comparison suggests that the Z chromosome shares this bias. This bias was not a feature of the autosomes that gave rise to the sex chromosomes of birds and mammals; it arose by gene acquisition and amplification during sex chromosome evolution in each lineage.
In light of this convergent gene gain, we looked for factors other than gene loss that could account for the low gene density of the Z and X chromosomes. Low gene density could result from Z-linked and X-linked genes that are larger than those on autosomes, resulting in fewer genes in the same amount of sequence. However, we find that genes on both the Z and X chromosomes are smaller, on average, than autosomal genes (Table 1A). The only remaining explanation for the unusually low gene density of the Z and X chromosomes is a massive expansion of non-coding intergenic sequences which spread the genes further apart. We estimate that intergenic regions were expanded by about 40 Mb in the case of the Z chromosome and 80 Mb in the case of the X chromosome – nearly half the present lengths of these chromosomes. No single class of non-coding sequence can account for this change, but the two-fold enrichment for LINEs on both the Z and X chromosomes (Table 1A) suggests that the doubling of intergenic sequence may have been driven by recurrent insertion and divergence of transposable elements. In mammalian genomes, high LINE density is associated with reduced rates of crossing over19, and suppression of crossing over is a key step in the evolution of differentiated sex chromosomes. However, the Z and X chromosomes are enriched for LINE elements compared to autosomal regions with similarly low rates of crossing over (Supplementary Figure 12, Supplementary Note 1).
Our comparison of the finished sequences of the chicken Z and human X chromosomes reveals that each evolved independently from different portions of the ancestral genome, from separate pairs of ordinary autosomes. In each lineage, different portions of the ancestral genome were substantially remodelled to become specialized sex chromosomes. The Z and X chromosomes have converged on a set of structural features that distinguish them from autosomes: a high density of interspersed repeats, and long intergenic distances resulting in low gene density. Furthermore, the Z and X chromosomes have both gained multi-copy gene families that are expressed in testis, biasing the gene content of both chromosomes toward male reproductive functions.
This convergent specialization of Z and X chromosomes for male reproduction is surprising given that the Z chromosome evolved with female heterogamety and the X chromosome evolved in opposite circumstances, with male heterogamety. One might have anticipated that the Z and X chromosome would exhibit opposing rather than convergent biases in gene content. While strong selective pressures drive the evolution of genes related to male reproduction20,21, these selective pressures influence autosomes as well as sex chromosomes. But, unlike autosomes, sex chromosomes are uniquely susceptible to selection for traits that benefit one sex more than the other22. Our results suggest that, in amniotes, selective pressures to preserve or enhance male reproductive functions have trumped the differences between ZW and XY systems to produce the changes in gene content that we observe.
For nearly 100 years, it has been thought that sex chromosome evolution involved dramatic modification of sex-specific chromosomes but only modest change in chromosomes shared by the sexes1,2. In the past decade, this understanding was reinforced by comprehensive molecular comparisons between the human X and Y chromosomes3–5, and by more limited comparison of sex chromosome pairs in other plants and animals23–25. These X–Y or Z-W comparisons revealed extensive genetic decay in the sex-specific Y or W chromosome, while assuming that Z and X chromosomes faithfully represent their autosomal progenitors. By contrast, the Z to autosome and X to autosome comparisons in this study reveal that the chicken Z chromosome and the human X chromosome have undergone dramatic evolutionary changes that were not anticipated and that previous studies could not detect. In birds and mammals, sex chromosome evolution was not limited to gene loss from sex-specific chromosomes, but extended to expansion and gene acquisition on the chromosomes shared between the sexes.
Methods Summary
Mapping and sequencing
All Z-chromosome BAC and fosmid clones that we selected for sequencing (see Supplementary Table 5 for GenBank accession numbers) were from six libraries generated from the same female of the inbred line of red jungle fowl (UCD001) as was used for the whole genome shotgun sequence of the chicken8,26. As a result, the sequence we obtained is that of a single Z chromosome haplotype. We made use of available BAC fingerprint maps to select tiling paths across the Z chromosome. Contigs were ordered and oriented by radiation hybrid mapping27, and confirmed by FISH28 (Supplementary Figures 2 & 3).
Sequence Analysis
We used RepeatMasker (http://www.repeatmasker.org) to identify and mask interspersed repeats. We used BLAT29 to detect intrachromosomal similarity and custom Perl scripts to construct triangular dot plots (http://jura.wi.mit.edu/page/Y/azfc/self_dot_plot.pl).
Comparative Genomics
We detected orthology by using BLAT to align peptide sequences (Ensembl version 52)30 and identifying the best reciprocal hit between species. We constructed the inter-species dot plots using the chromosomal coordinates extracted from Ensembl (or in the case of the Z chromosome, the coordinates from this study). For counts of gene gain and loss, we used the list of orthologs of human and chicken genes compiled by Ensembl, which we then manually reviewed to ensure accuracy. In cases where genes on the sex chromosomes did not have a one-to-one ortholog on a corresponding autosome in the other species (or vice versa), we looked to outgroup species (fish and amphibians) to determine the lineage (chicken or human) on which a gene was gained or lost.
Methods
Mapping and sequencing
All Z-chromosome BAC and fosmid clones selected for sequencing (Supplementary Table 5) were from six libraries (CH261, TAM31, TAM32, TAM33, J_AD, and J_AE) generated from the same female of the inbred line of red jungle fowl (UCD001) as was used for the whole genome shotgun sequence of the chicken8,26. As a result, the Z chromosome we obtained is that of a single haplotype. We made use of publicly available BAC fingerprint maps and BAC-end sequences as a source of mapping information and markers. Individual BAC fingerprint contigs were ordered and oriented by radiation hybrid mapping using ChickRH627. Unfortunately, no cell line is available from any bird of the UCD001 line, so we used chicken embryonic fibroblasts derived from White Leghorn (available from Charles River Labs) for FISH experiments to provide independent confirmation of the order and orientation of the sequence (Supplemental Figures 9 – 11).
Chromosomal FISH
One- or two-colour FISH to chicken chromosomes was performed as previously described28.
Z chromosome sequence similarity
Analyses of intrachromosomal similarity were performed using BLAT29 to compare all 5-kb sequence segments, in 1-kb steps, to the entire remainder of the Z chromosome sequence. For each segment, we recorded the highest percent identity to a non-overlapping segment.
Genes and transcription units
We identified potential transcripts in three ways:
We used human (Ensembl version 52, NCBI 36)30 as the informant genome and chicken EST sequences as additional evidence to identify potential transcripts on the repeat-masked chicken Z chromosome, using Twinscan31,32. We compared the output with the Ensembl 52 annotations for chicken and human to identify previously unrecognized genes in our prediction. We considered previously unrecognized chicken genes valid if they were spliced in chicken and conserved to human.
For the novel genes in the Z-amplicon region we relied on BLAST33 matches to cDNA sequences to identify copies of ADCY10z, C2ORF3z, MRPL19z, and RICSz that showed evidence of splicing. We then tested for transcription by RT–PCR across a panel of adult tissues.
We used a combination of methods to locate non-coding transcripts on the chicken Z. We used tRNAscan-SE34 for tRNA predictions. For other non-coding RNAs, we used BLAST to compare our sequence with those of known chicken non-coding RNAs in GenBank35 and mirBase36
Interspersed repeats
We electronically identified interspersed repeats with RepeatMasker37.
Triangular dot plots
We performed dot-plot analysis using custom Perl script38.
Expressed Sequence Tags
We used EST sequences from the BBSRC ChickEST database39, supplemented by our own 454 EST runs on ovary and testis (SRA# SRP000097).
Z amplicon size
To estimate the amount of Z ampliconic sequence missing from our assembly, we compared the average depth of chicken fosmid end sequences on the single copy region of the Z to the depth in the ampliconic region, reasoning that the excess depth in the ampliconic region could be attributed to sequence we could not obtain because the similarity between individual repeat units precluded either cloning or the assembly of BACs. We used BLAT to map 23977 fosmid end sequences to 72.2 Mb of single-copy Z sequence, giving an average of 331 ends per megabase. In the 5.6 Mb of the Z amplicon we found 3787 fosmid end hits, for 666 ends per megabase, roughly a two-fold enrichment. Therefore, we concluded that the Z amplicon comprises roughly 11.4 Mb.
RT-PCR
We used chicken total RNA (Zyagen) and the RETROscript Kit (Ambion). We amplified 1μl of the RT product through 30 cycles of PCR with an annealing temperature of 55°C.
Primers are as follows:
HPRT1 (116 bp product):
F: GGATTTGAAGTGCCAGACAAA
R: GCTTTGTACTTCTGCTTCCCC
ADCY10z (145 bp product):
F: GTTTGTCAGGTCTCTGTGGGA
R: GTAGAGGTCCTCGAGCAAGGC
RICSz (144 bp product):
F: GACAGAGATCAGGGACATGGA
R: AAACAGGAACACCAACTGCAT
C2ORF3z (131 bp product):
F: TGTTCAAAATTCCAAGGCAGA
R: AGGTAACGATTCAGCAGCTTG
MRPL19z (242 and 60 bp products):
F: CAAGCAGAAGCAGAGAGAGGA
R: TGACCATGGTTGAGGTTTCA
Orthologous chromosomes
To identify orthologous chromosomes in inter-species comparisons, we relied on a gene-based approach. We conducted reciprocal BLAT searches using Ensembl 52 peptide sequence databases from Gallus gallus, Homo sapiens, Danio rerio, Gasterosteus aculeatus, Oryzias latipes, and Tetraodon nigroviridis. Considering only the longest peptide sequences for each Ensembl gene, we flagged best reciprocal BLAT hits between two genomes as orthologous genes. We constructed the inter-species dot plots using the chromosomal coordinates extracted from Ensembl (or in the case of the Z chromosome, the coordinates from this study). Individual dots represent a pair of orthologous genes.
Gene gain and loss
We relied on the assignments of chicken and human orthologs in the Ensembl database (Ensembl version 52). However, we manually reviewed genes on chicken chromosomes 1, 4, and Z as well as human chromosomes 5, 9, 18, and X which did not have simple 1:1 orthologs in the Ensembl database to find pairs of orthologous genes that were missing from the database or not properly identified.
To study gene gain and loss on the chicken Z chromosome, we divided genes into several categories on the basis of their location in chickens, humans, and outgroup species:
(A) Z-linked genes with orthologs on human autosomes 5, 9, and 18
(B i) Z-linked genes present only in birds, but not outgroups or human
(B ii) Z-linked genes present in birds and outgroups but not human
(B iii a) Z-linked genes with human orthologs not on autosomes 5, 9, 18 that were not syntenic with neighbors in outgroups or human
(B iii b) Z-linked genes with human orthologs not on autosomes 5, 9, 18 that were syntenic with neighbors in outgroups
(C i) Genes on human autosomes 5, 9, and 18 with orthologs only in mammals, not in outgroups or chicken
(C ii) Genes on human autosomes 5, 9, and 18 with orthologs only in mammals and outgroups, but not chicken
(C iii a) Genes on human autosomes 5, 9, and 18 with chicken orthologs not on the Z chromosome that were not syntenic with neighbors in outgroups or chicken
(C iii b) Genes on human autosomes 5, 9, and 18 with chicken orthologs not on the Z chromosome that were syntenic with neighbors in outgroups
Category (A) was counted as shared, Categories (B i) and (B iii a) were counted as gains to the Z, while Categories (C ii) and (C iii b) were counted as losses from the Z. Category (B ii) and (B iii b) (representing losses from the human autosomes) and categories (C i and C iii a) (representing gains to the human autosomes) were excluded. We carried out an analogous analysis on the human X chromosome and chicken autosomes 1 and 4.
Biased gene content
We searched for Unigene17 EST clusters from normal chicken and human testis that corresponded with chicken and human genes to identify genes expressed in the testis. We determined the percentage of genes with at least one testis EST for each category (Autosomes, Z or X chromosome total, Z or X chromosome single-copy, Z or X chromosome multi-copy). Multi-copy genes on the Z chromosome include those of the Z amplicon, while multi-copy genes on the X chromosome include those identified as cancer-testis antigen genes in the finished sequence of the X chromosome5.
Supplementary Material
Acknowledgments
We thank E. Rapoport, for technical assistance; S. Repping, and S. van Daalen for experimental advice; and E. Anderson, T Endo, M. Gill, A. Hochwagen, C. Hongay, Y. Hu. J. Hughes, J. Marszalek J. Mueller, and Y. Soh for comments on the manuscript. We thank the Broad Institute Genome Sequencing Platform and Genome Sequencing and Analysis Program, Federica Di Palma, and Kerstin Lindblad-Toh for making the unpublished data for Gasterosteus aculeatus available. Supported by the National Institutes of Health and the Howard Hughes Medical Institute.
Footnotes
Supplementary Information accompanies this paper. Predicted Z amplicon transcript sequences and the complete assembled sequence of the Z chromosome are available at: http://jura.wi.mit.edu/page/HIVODCRVRAFKKPXNUW.
Author Contributions DWB, HS, WW, SR, RKW, and DCP planned the project. DWB and LB performed BAC mapping. DWB performed RT-PCR analysis. TAG and CK were responsible for finished BAC sequencing. DWB and HS performed comparative sequence analyses. TP performed FISH analysis. ERM performed 454 sequencing. DWB and DCP wrote the paper.
The authors declare no competing financial interests.
References
- 1.Muller HJ. A gene for the fourth chromosome of Drosophila. Journal of Experimental Zoology. 1914;17:325–336. [Google Scholar]
- 2.Ohno S. Sex chromosomes and sex-linked genes. Springer-Verlag; New York: 1967. [Google Scholar]
- 3.Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–7. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- 4.Skaletsky H, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–37. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 5.Ross MT, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–37. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nanda I, et al. 300 million years of conserved synteny between chicken Z and human chromosome 9. Nature Genetetics. 1999;21:258–9. doi: 10.1038/6769. [DOI] [PubMed] [Google Scholar]
- 7.Nanda I, Haaf T, Schartl M, Schmid M, Burt DW. Comparative mapping of Z-orthologous genes in vertebrates: implications for the evolution of avian sex chromosomes. Cytogenetics and Genome Research. 2002;99:178–84. doi: 10.1159/000071591. [DOI] [PubMed] [Google Scholar]
- 8.Hillier LW. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 9.Grutzner F, et al. In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird Z and mammal X chromosomes. Nature. 2004;432:913–7. doi: 10.1038/nature03021. [DOI] [PubMed] [Google Scholar]
- 10.Rens W, et al. Resolution and evolution of the duck-billed platypus karyotype with an X1Y1X2Y2X3Y3X4Y4X5Y5 male sex chromosome constitution. Proceedings of the Natlional Acadademy of Sciences of the United States of America. 2004;101:16257–61. doi: 10.1073/pnas.0405702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ezaz T, Stiglec R, Veyrunes F, Marshall Graves JA. Relationships between vertebrate ZW and XY sex chromosome systems. Current Biology. 2006;16:R736–43. doi: 10.1016/j.cub.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 12.Smith JJ, Voss SR. Bird and mammal sex-chromosome orthologs map to the same autosomal region in a salamander (ambystoma) Genetics. 2007;177:607–13. doi: 10.1534/genetics.107.072033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hori T, et al. Characterization of DNA sequences constituting the terminal heterochromatin of the chicken Z chromosome. Chromosome Research. 1996;4:411–426. doi: 10.1007/BF02265048. [DOI] [PubMed] [Google Scholar]
- 14.Kasahara M, et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–9. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- 15.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 16.Scanlan MJ, Simpson AJ, Old LJ. The cancer/testis genes: review, standardization, and commentary. Cancer Immunity. 2004;4:1. [PubMed] [Google Scholar]
- 17.Wheeler DL, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Research. 2005;33:D39–45. doi: 10.1093/nar/gki062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saifi GM, Chandra HS. An apparent excess of sex- and reproduction-related genes on the human X chromosome. Proceedings of the Royal Society B: Biological Sciences. 1999;266:203–9. doi: 10.1098/rspb.1999.0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wichman HA, Bussche RA, Hamilton MJ, Baker RJ. Transposable elements and the evolution of genome organization in mammals. Genetica. 1992;86:287–293. doi: 10.1007/BF00133727. [DOI] [PubMed] [Google Scholar]
- 20.Wyckoff GJ, Wang W, Wu CI. Rapid evolution of male reproductive genes in the descent of man. Nature. 2000;403:304–9. doi: 10.1038/35002070. [DOI] [PubMed] [Google Scholar]
- 21.Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nature Reviews Genetics. 2002;3:137–44. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- 22.Rice WR. Sex Chromosomes and the Evolution of Sexual Dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 23.Filatov DA. Evolutionary history of Silene latifolia sex chromosomes revealed by genetic mapping of four genes. Genetics. 2005;170:975. doi: 10.1534/genetics.104.037069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Handley LJL, Ceplitis H, Ellegren H. Evolutionary strata on the chicken Z chromosome: implications for sex chromosome evolution. Genetics. 2004;167:367. doi: 10.1534/genetics.167.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicolas M, et al. A gradual process of recombination restriction in the evolutionary history of the sex chromosomes in dioecious plants. PLoS Biology. 2005;3:e4. doi: 10.1371/journal.pbio.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wallis JW, et al. A physical map of the chicken genome. Nature. 2004;432:761–4. doi: 10.1038/nature03030. [DOI] [PubMed] [Google Scholar]
- 27.Morisson M, et al. ChickRH6: a chicken whole-genome radiation hybrid panel. Genetics Selection Evolution. 2002;34:521–33. doi: 10.1186/1297-9686-34-4-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saxena R, et al. Four DAZ genes in two clusters found in the AZFc region of the human Y chromosome. Genomics. 2000;67:256–67. doi: 10.1006/geno.2000.6260. [DOI] [PubMed] [Google Scholar]
- 29.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Research. 2002;12:656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hubbard TJ, et al. Ensembl 2009. Nucleic Acids Research. 2009;37:D690–7. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korf I, Flicek P, Duan D, Brent MR. Integrating genomic homology into gene structure prediction. Bioinformatics. 2001;17 (Suppl 1):S140–8. doi: 10.1093/bioinformatics/17.suppl_1.s140. [DOI] [PubMed] [Google Scholar]
- 32.Flicek P, Keibler E, Hu P, Korf I, Brent MR. Leveraging the mouse genome for gene prediction in human: from whole-genome shotgun reads to a global synteny map. Genome Research. 2003;13:46–54. doi: 10.1101/gr.830003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 34.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–64. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Research. 2009;37:D26–31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Research. 2008;36:D154–8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. 1996–2004. http://www.repeatmasker.org.
- 38.Kuroda-Kawaguchi T, et al. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nature Genetics. 2001;29:279–86. doi: 10.1038/ng757. [DOI] [PubMed] [Google Scholar]
- 39.Boardman PE, et al. A comprehensive collection of chicken cDNAs. Current Biology. 2002;12:1965–9. doi: 10.1016/s0960-9822(02)01296-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.