ABSTRACT/SUMMARY
The successful isolation and cultivation of prostate stem cells will allow us to study their unique biological properties and their application in therapeutic approaches. Here we provide step-by-step procedures on the basis of previous work in our laboratory for: the harvesting of primary prostate cells from adolescent male mice by a modified enzymatic procedure; the isolation of an enriched population of prostate stem cells through cell sorting; the cultivation of prostate stem cells in vitro; and characterization of these cells and their stem-like activity, including in vivo tubule regeneration. Normally it will take approximately 8 hours to harvest prostate cells, isolate the stem cell enriched population, and set up the in vitro sphere assay. It will take up to 8 weeks to analyze the unique properties of the stem cells, including their regenerative capacity in vivo.
INTRODUCTION
Epithelial stem cells are of interest due to their capacity for organ replenishment and for their potential role in cancer-initiation. During the normal life span of an adult organ, stem cells operate to replace lost or damaged tissue to ensure proper organ function1. More recently, epithelial stem cells have also been demonstrated as a target population for cancer2. Due to their long-lived nature and inherent self-renewal capacity, adult stem cells are a likely cell-of-origin for many cancers3. The isolation of these cells and investigation into their properties will be useful for understanding their basic biological processes and for designing new therapies.
The prostate represents an ideal model system to investigate the properties of adult stem cells due to the seemingly unlimited ability of the rodent prostate gland to undergo cycles of involution after androgen-depletion and subsequent organ regeneration in response to androgen add-back4. Perhaps the most important reason to study prostate stem cells (PrSCs) is that they share the property of androgen-independence (or castration-resistance) with the subset of prostate cancer cells that survive in an androgen-deprived environment and can drive the lethal phase of the disease, termed hormone-refractory or castration-resistant prostate cancer (CRPC)5,6. Identifying critical self-renewal pathways in PrSCs may provide new therapeutic targets for the treatment of CRPC.
Several methods have been described for the isolation of primitive cells from the mouse prostate7,8,9,10,11,12,13. While genetically engineered mouse models can be useful for stem cell isolation, they limit the widespread use of such an approach. We have previously reported the isolation of PrSCs from wild-type mice capable of self-renewal and tri-lineage differentiation in vivo9. PrSCs can be reproducibly isolated by fluorescence activated cell sorting (FACS) using the antigenic profile Lin- Sca-1+ CD49f+ (LSC)10 or Lin- Sca-1+ CD49fhi Trop2hi (LSCT)9. These cells possess a basal phenotype and primarily reside in the region of the gland proximal to the urethra. Primitive cells with basal characteristics and an antigenic profile similar to PrSCs (LSC/LSCT) can be reproducibly isolated from un-fractionated prostate when primary cells are cultured in the prostate sphere assay10,14. Here we will describe our methods to isolate PrSCs from primary prostate tissue, culture PrSCs in vitro, and measure PrSC activity using quantitative in vitro and in vivo assays.
Epithelial cells quickly lose their self-renewal potential when they are cultured in two-dimensions15. We have developed a three-dimensional culture system to maintain and expand primitive prostate cells that retain the capacity for both self-renewal and differentiation14. Cells are suspended in a semi-solid matrix comprised of prostate epithelial growth medium (PrEGM) and Matrigel. Matrigel is comprised primarily of laminin, collagen, fibronectin and other extracellular matrix (ECM) components. This in vitro environment closely mimics the ECM-rich basement membrane where basal cells reside in the gland. More importantly, basal cells express high levels of ECM-binding integrins that promote proper cell signaling and likely keep them in an undifferentiated state16. The culture system is supplemented with selected growth factors and cytokines including EGF and FGF in the PrEGM media.
Three assays (the colony-forming assay, the sphere-forming assay and the in vivo prostate-regeneration assay) have been developed to measure primitive prostate cell activity10. Similar to other epithelial stem cell assays, the colony-forming assay is performed in a two-dimensional culture system and measures both proliferative colony-forming activity and differentiation. Colonies are clonal in origin, express basal and luminal keratins, and can be induced to undergo differentiation upon addition of androgen10,14,15. The sphere-forming assay is performed under three-dimensional conditions, as described above. Spheres are also clonal in origin, are comprised of several hundreds of cells, and can be dissociated and re-plated to measure self-renewal activity10,14,15. Sphere cells undergo spontaneous differentiation with the most primitive cells residing around the outside and the more mature cells oriented towards the lumen, similar to the architecture of the native gland14. Finally, the in vivo prostate-regeneration assay17,18 measures the ability of PrSCs to form prostatic tubules when combined with inductive stroma and implanted under the kidney capsule or skin (subcutaneous) of immunodeficient mice. Regenerated tubules are indistinguishable from primary prostate tubules, with an outer basal layer, an inner luminal layer and rare neuroendocrine cells9. The subrenal regeneration assay can be used to study self-renewal in vivo by implanting prostate cells from transgenic mice harboring a probasin promoter driven luciferase, and performing androgen cycling15. While the in vitro colony-forming and sphere-forming assays take 7–10 days, the in vivo prostate-regeneration assay takes considerably longer (6–10 weeks). We have functionally defined PrSCs based on their ability to generate colonies, spheres and prostatic tubules in these assays.
MATERIALS
1. REAGENTS
Male C57/BL6,β-actin dsRed Tg, or β-actin GFP mice (obtained from Jackson Laboratories and housed and bred at UCLA in accordance with Division of Laboratory Animal Medicine regulations.)
-
CB17SCID/SCID mice (obtained from Jackson Laboratories and housed and bred at UCLA in accordance with Division of Laboratory Animal Medicine regulations.)
! CAUTION All experiments involving live rodents must conform to national and institutional regulations.
Dubelcco’s modified Eagle medium (DMEM)(Invitrogen/Gibcocatalogue #12100-061)
RPMI (Roswell Park Memorial Institute) medium(Invitrogen/Gibcocatalogue #31800-089)
10x PBS(Omega Scientific catalogue #PB-10)
Fetal bovine serum (Omega Scientific catalogue #FB-01)
-
NuSerum (BD Biosciences catalogue #355504)
▲ CRITICAL This serum was found to be optimal for the support of UGSM growth.
-
Dihydrotestosterone (DHT) (Sigma Aldrich)(see REAGENT SETUP)
!CAUTION Handle with care.
Insulin (Sigma Aldrich catalogue #12585-014)
Glutamine (Fisher Scientific catalogue #BP379-100)
Penicillin/Streptomycin (Omega Scientific catalogue #PS-20)
Collagenase (Invitrogen/Gibcocatalogue #17018-029)(see REAGENT SETUP)
Dispase (Invitrogen/Gibcocatalogue #17105-041)(see REAGENT SETUP)
DNase I (Rochecatalogue #10 104 159 00)
Trypan bluestain 0.4% (Invitrogen/Gibco catalogue #15250-061) (see REAGENT SETUP)
Trypsin/0.05% EDTA (Invitrogen/Gibcocatalogue #25300)
10% Trypsin (Invitrogen catalogue #15090046) (see REAGENT SETUP)
-
Antibodies for FACS staining:
-
CD49f-PE (eBioscience cloneeBioGoH3catalogue #12-0495-83)
▲ CRITICAL Some other clones are more diluted and the staining will not look the same.
Sca-1-APC(eBioscience clone D7catalogue #17-5981-82)
Sca-1-PE-Cy7 (BioLegend clone E13-161.7catalogue #122514)
Ter119-FITC(eBioscience clone TER-119catalogue #11-5921-85)
CD31-FITC(eBioscience clone 390catalogue #11-0311-85)
CD45-FITC(eBioscience clone 30-F11catalogue #11-0451-85)
Trop2-biotin(R+D Systems catalogue #BAF1122)
SA-APC(BD Biosciences catalogue #554067)
-
-
PrEGM(Clonetics/Lonzacatalogue #CC-3165)(see REAGENT SETUP)
▲CRITICAL STEP All of the published data was obtained using this medium to grow prostate spheres and colonies. Other media were tested and had lower success rate.
-
Matrigel (BD Biosciences catalogue #354234)
▲CRITICAL STEP All of the published sphere data were obtained using this Matrigel.
Collagen(BD Biosciences catalogue #354236)
-
1N NaOH (Sigma catalogue #S2770)
!CAUTION Corrosive agent – handle using appropriate safety gear.
-
Acetone(Fisher Scientific catalogue #A18-500)
!CAUTION Flammable and vapors may cause irritation. Handle with care.
-
Crystal Violet (Fisher Scientific catalogue #C581-25)
!CAUTION May be harmful of inhaled; handle with care.
-
Acetic Acid, Glacial (Fisher Scientific catalogue #A38S-212)
!CAUTION Corrosive agent – may cause burns, vapors are harmful. Handle using appropriate safety gear.
Histogel (Richard-Allan Scientific catalogue #92957065)
Tween 20 (Fisher Scientific catalogue #BP337-100)
Normal goat serum(Calbiochem catalogue #566380)
-
10% Buffered Formalin(Fisher Scientific catalogue #SF100-4)
!CAUTION Contains formaldehyde, which can cause cancer.
Vectashield mounting medium (Vector catalogue #H-1200)
-
Ethanol
!CAUTION Flammable liquid and vapor. Handle with care.
2. EQUIPMENT
Cell culture disposables: Petri dishes (BD Flacon), centrifuge tubes (Eppendorf), pipettes, pipette tips, filter units(Millipore), FACS tubes with filter tops(BD Falcon)
12-well tissue culture plates (BD Falcon)
18-gauge, 20-gauge, and 28-gauge needles(Kendall)
3cc syringes(BD Biosciences)
10 or 20cc syringes(BD Biosciences)
Syringe attachable 22μmpore size filters(Millipore)
Razor blades(Personna)
Dissecting tools: dissecting scissors, dissecting forceps (Roboz)
Dissecting microscope (Olympus)
Nylon mesh 40 and 70μmpore size(BD Biosciences)
Cell culture centrifuge(Beckman Coulter)
Tissue culture hood(SteriGuard)
CO2 incubator set to 8% CO2 and 37°C (SteriCult)
Inverted microscope with fluorescence, phase contrast objectives (phase x4, x10, x20) from Zeiss
Shaker in 37 degree room or area
FACS Aria cell sorter (Becton Dickinson or similar (see EQUIPMENT SETUP)
Glass slides and cover slips
Concave plate (Fisher)
Alcohol and Iodine prepswabs
6-0 Coated Vicryl Sutures (Ethicon)
Wound clip applier, clips, and remover (Fisher Catalogue #01-804, 01-804-5, and 01-804-15)
3. REAGENT SETUP
Dissecting medium: DMEM supplemented with 10% (vol/vol) FBS, 1% (vol/vol) 100x glutamine and 1% (vol/vol) 100x penicillin/streptomycin solution. Store at 4°C for up to 1 month.
Collagenase solution: Dissolve the nonsterile lyophilized collagenase type I in RPMI medium to a concentration of 10 mg ml−1. Filter through a .22μm filter. Prepare 1ml aliquots and store at −20°C for up to 3 months. Thaw stock solution in refrigerator immediately before use and dilute into Dissecting media to a final concentration of 1mg ml−1.
Dispase solution: Dissolve the nonsterile lyophilized Dispase in PrEGM medium to a concentration of 1 mg ml−1. Filter through a .22μm filter. Prepare 1ml aliquots and freeze at −20°C for longer term storage, or refrigerate at 4°Cfor up to 2 weeks. Thaw immediately before use and add to Matrigel.
100x Glutamine: Weigh out 23.36g glutamine powder, and dissolve in 350ml water. Stir the solution, and when the powder starts to go into solution, add 12M HCl until all of the glutamine has been dissolved, and the solution is clear. Add water to a final volume of 400ml, and filter through a .22μmfilter. Aliquot and store at −20°C for up to 3 months.
Trop2-biotin antibody: Reconstitute the lyophilized antibody (from R+D Systems) with sterile Tris-buffered saline pH 7.3 containing 0.1% BSA to a final concentration of 100ug ml−1. Store at 4°C until expiration date.
FACS Collection medium: DMEM supplemented with 50% (vol/vol) FBS, 1x glutamine and 1x penicillin/streptomycin soulution. Prepare small volumes fresh before each sort, do not store.
PrEGM medium: Thaw supplements directly before preparing medium. Add all 9 supplements to the basal medium. Store at 4°C for up to 3 weeks, or aliquot into 50 ml aliquots and freeze at −20°C for storage. Aliquots can be stored for up to 2 months. Thaw in 37°C water bath before use.
3T3 Medium: DMEM supplemented with 10% (vol/vol) FBS and 1x glutamine. Store at 4°C for up to 1 month.
Dihydrotestosterone stock: Dissolve in ethanol to a final concentration of 10−3 M. Freeze and store at −20°Cfor up to 1 year.
1% Trypsin: Dilute the 10% Trypsin (Invitrogen) to 1% (vol/vol) in sterile 1x PBS. Make a small aliquot fresh every time; do not store.
BFS growth medium for UGSM cells: DMEM supplemented with 5% (vol/vol) FBS, 5% (vol/vol) NuSerum IV, 1x glutamine, 1x penicillin/streptomycin solution, 10−8 M (0.01μM) final concentration of Dihydrotestosterone, 25ug ml−1 insulin. Store at 4°C for up to 1 month.
Colony differentiation medium: PrEGM complete medium supplemented with DHT to a final concentration of 10−8 M (0.01μM). Keep refrigerated at 4°C for up to 2 weeks.
Turk’s solution: Mix together 10mg Crystal Violet powder, 3ml acetic acid (Glacial), and fill up volume to 100ml with water. Solution can be stored at RT for at least 1 year.
Collagen: Mix together 11.5μl 1N NaOH, 56.8μl 10x PBS, and 500μl collagen. Leave on ice for at least 15 minutes before use. Make fresh each time grafts are made.
4. EQUIPMENT SETUP
FACS sorting for PrSC enrichment: The Special Order BD FACSAria II cell sorter is equipped with four solid-state lasers (laser outputs at 488, 640, 561, and355 nm). FITC is excited by a 488-nm laser and measured through a 530/30nm BP 505nm LP filter. PE is excited by a 561-nm laser and measured through a 585/15nm BP 570nm LP filter. PE-Cy7 is excited by a 561nm laser and measured through a 780/60nm BP 755nm LP filter. APC is excited by a 640-nm laser and measured through a 660/20nm BP filter. Wild type prostate cells are used as control to set the cutoff value for background fluorescence. Single color stained wild-type prostate cells are used for compensating between the different color channels. Samples are sorted at 4°C using a 100μmnozzle and 23 psi pressure, and collected into FACS Collection Media (see REAGENT SETUP) chilled at 4°C. BD FACSDiva v6.1.1 software is used for collection, storage and analysis of the digital data.
PROCEDURE
Isolation of prostate cells • TIMING 3 to 4 h
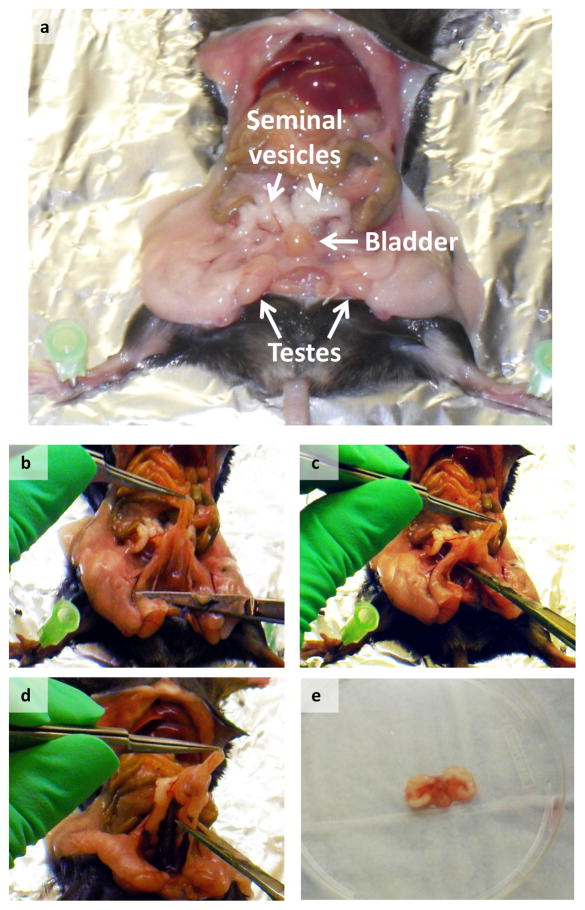
1] Harvest and dissect prostate as described below. Sacrifice a male mouse (8–12 weeks old), remove the majority of the urogenital system by pulling up on the bladder and cutting the connective tissue below the UGS (Figure 2). Place UGS in a 10cm Petri dish containing Dissecting Media (see REAGENT SETUP). You should have the bladder, seminal vesicles, prostate, urethra, and part of the ureters in the dish at this point.
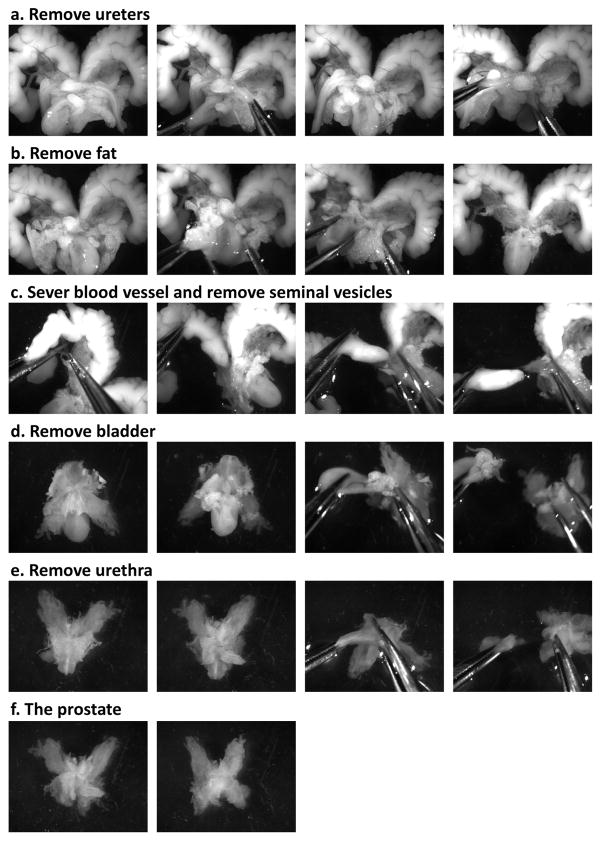
2] The following steps are shown in Figure 3. Using two dissecting tweezers hold onto the bladder and remove the two ureters. Using the two tweezers, separate the anterior lobes of the prostate from the seminal vesicles by severing the connecting blood vessel. Remove the seminal vesicles one at a time by holding onto the urethra/prostate with one pair of tweezers and gently pulling the seminal vesicle away at the base. Pull away any chunks of fat still attached to the UGS. Holding on to the prostate, gently pull the bladder away, leaving behind the prostate and urethra. Unfurl the prostate tubules and pull the urethra away. Transfer the prostate into a new 10cm Petri dish containing fresh Dissecting Media.
3] Place each prostate onto a clean surface and mince tissue using a razor blade. Transfer minced prostate tissue into a 15ml Falcon tube into 9mls of Dissecting media. Add 1ml 10x Collagenase solution, and incubate on a shaker in 37°C for 2 hours.
4] Spin down the tissue chunks at 1300rpm (400xg) for 5 minutes at 25°C. Remove the supernatant and add 2mls warm Trypsin/0.05% EDTA to the cell pellet, mix using a pipette, and place in 37°C incubator for 5 minutes.
-
5] Mix the cell/tissue suspension with a P1000 pipette and add 2–3mls of dissecting media containing 500U DNase I to inactivate the trypsin and break up any DNA released from dead cells. Pass through an 18G syringe 5 times, and then through a 20G syringe 5 times. Repeat steps 4 and 5 for the highest cellular yield.
? TROUBLESHOOTING
-
6] Filter mixture through a nylon mesh filter with a 40μm pore size into a 50ml Falcon tube. Wash out 15ml Falcon tube and filter with fresh dissecting media. Pellet the collected cells at 1300rpm(400xg)for 5 minutes at 25°C, and resuspend in 1ml fresh dissecting media. Count cells using a hemocytometer.
? TROUBLESHOOTING
Figure 2. Harvest of the Urogenital System.
(a) Image depicts the location of the urogenital sinus inside the mouse abdomen, (b–d). Images show how the urogenital sinus in separated and excised in one piece, (e) Image shows the entire urogenital sinus, which is obtained after the procedure.
Figure 3. Prostate micro-dissection.
Images show the step by step micro-dissection procedure used to dissect the prostate away from the rest of the urogenital system. Specifically, images show the (a) removal of the ureters, (b) removal of fat, (c) removal of the seminal vesicles, (d) removal of the bladder, (e) removal of the urethra, and (f) the final product, the prostate.
Enrichment of prostate stem cells • TIMING 2 to 3 h
7] To enrich for the LSC fraction of cells using FACS sorting, follow Option A. To enrich for the LSCT+ cell population, follow Option B. To have a rough enrichment for the PrSC-like cells through in vitro selection, refer to steps 9–13under CULTURE OF PrSCs.
(A) PrSC enrichment through sorting for the LSC population
Aliquot out 500μl of dissecting media into 4 eppendorf tubes and add 5×10e4 cells to each tube. These will be used to set up the compensation for FACS sorting. Divide the remaining cells into new eppendorf tubes up to 3×10e6 cells per tube in a 1ml total volume. These will be stained for sorting.
To set up the compensation cells and sorting cells, refer to Table 1 and as described below. To set up the compensation, leave one tube of cells unstained. Add 1μl CD49f-PE to the second tube, 1μl Sca-1-APC to the third tube, and 1μleach of CD31-FITC, CD45-FITC, and Ter119-FITC to the fourth tube. When setting up the gates for the first time, FMO (All Minus One) compensation should also be performed (Supplementary Table 1).
To the sample tubes add 3μl CD49f-PE, 2μl Sca-1-APC, and 4μl each of CD31-FITC, CD45-FITC, and Ter119-FITC. Wrap all of the tubes in aluminum foil and place on a shaker at 4°C for 20 minutes.
Pellet the cells in a bench top centrifuge at 2300rpm (500xg) for 2 minutes at 25°C. Aspirate the media off and resuspend in 500μl of fresh dissecting media for the compensation cells and 1ml for the sorting sample cells. Transfer cells into 40μmfilter top FACS tubes.
-
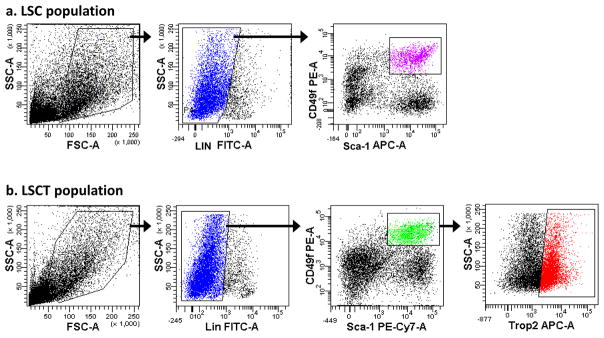
After completing the compensation (see Supplementary Figure 1a), sort the FITC−PEhiAPC+ cell population (Figure 4a) using the FACSAria cell sorter (see EQUIPMENT SETUP). Collect cells into 15ml Falcon tubes containing FACS Collection media (see REAGENT SETUP).
? TROUBLESHOOTING
Table 1.
Antibodies used for FACS staining
(see REAGENTS for company and catalogue number information)
| Step | Condition | Antibody | Dilution | Volume |
|---|---|---|---|---|
| Compensation tubes | ||||
| 7A(ii) | no stain | N/A | N/A | N/A |
| PE | CD49f-PE | 1:500 | 1ul | |
| APC | Sca-1-APC | 1:500 | 1ul | |
| FITC | CD31-FITC | 1:500 | 1ul | |
| CD45-FITC | 1:500 | 1ul | ||
| Ter119-FITC | 1:500 | 1ul | ||
| 7B(ii) | PE | CD49f-PE | 1:500 | 1ul |
| PE-Cy7 | Sca-1-PE-Cy7 | 1:500 | 1ul | |
| FITC | CD31-FITC | 1:500 | 1ul | |
| CD45-FITC | 1:500 | 1ul | ||
| Ter119-FITC | 1:500 | 1ul | ||
| APC | Trop-2-Biotin | 1:250 | 2ul | |
| 7B(v) | SA-APC | 1:500 | 1ul | |
| Sample tubes | ||||
| 7A(iii) | LSC sort | CD49f-PE | 1:333 | 3ul |
| Sca-1-APC | 1:500 | 2ul | ||
| CD31-FITC | 1:250 | 4ul | ||
| CD45-FITC | 1:250 | 4ul | ||
| Ter119-FITC | 1:250 | 4ul | ||
| 7B(iii) | LSCT+/LSCT-sort | CD49f-PE | 1:333 | 3ul |
| Sca-1-PE-Cy7 | 1:500 | 2ul | ||
| CD31-FITC | 1:250 | 4ul | ||
| CD45-FITC | 1:250 | 4ul | ||
| Ter119-FITC | 1:250 | 4ul | ||
| Trop-2-Biotin | 1:100 | 10ul | ||
| 7B(v) | SA-APC | 1:200 | 5ul | |
Figure 4. FACS plots of the LSC and LSCT populations.
(a) FACS plots show the side and forward scatter (area), Lin-FITC, and Sca-1-APC/CD49f-PE plots and the gates used to sort the final LSC population of prostate cells, (b) FACS plots show the side and forward scatter (area), Lin-FITC, Sca-1-PE-Cy7/CD49f-PE, and Trop-2-APC plots in addition to the gates used to sort the LSCT population of prostate cells.
(B) PrSC and basal cell enrichment through sorting for the LSCT+ population
Aliquot out 500μl of dissecting media into 5 eppendorf tubes and add 5×10e4 cells to each tube. These will be used to set up the compensation for FACS sorting. Divide the remaining cells into new eppendorf tubes up to 3×10e6 cells per tube in a 1ml total volume. These will be stained for sorting.
To set up the compensation cells and sorting cells, refer to Table 1 and as described below. To set up the compensation, leave one tube of cells unstained. Add 1μl CD49f-PE to the second tube, 1μl Sca-1-PE-Cy7 to the third tube, 2μl Trop2-biotin into the fourth tube, and 1μl each of CD31-FITC, CD45-FITC, and Ter119-FITC to the fifth tube. When setting up the gates for the first time, FMO (All Minus One) compensation should also be performed (Supplementary Table 1).
To the sample tubes add 3μl CD49f-PE, 2μl Sca-1-PE-Cy7, 10μl Trop2-biotin, and 4μl each of CD31-FITC, CD45-FITC, and Ter119-FITC. Wrap all of the tubes in aluminum foil and place on a shaker at 4°C for 20 minutes.
Pellet the cells in a bench top centrifuge at 2300rpm (500×g) for 2 minutes at 25°C. Aspirate the media off and resuspend in 500μl of fresh dissecting media for the compensation cells and 1ml for the sorting sample cells. Keep samples on ice.
Add 1μl SA-APC secondary antibody to the APC compensation tube, and 5μl SA-APC antibody to each of the sample tubes. Wrap tubes in aluminum foil and place on a shaker at 4°C for 15 minutes. Pellet cells in a bench top centrifuge at 2300rpm (500×g) for 2 minutes at 25°C. Aspirate the media off and resuspend in 500μl of fresh media for the compensation cells and 1ml for the sorting sample cells.
-
After completing the compensation (see Supplementary Figure 1b), sort the FITC−PEhiPE-Cy7+APChi cell population (Figure 4b) using the FACSAria cell sorter (see EQUIPMENT SETUP). Collect cells into 15ml Falcon tubes containing FACS Collection media (see REAGENT SETUP).
? TROUBLESHOOTING
8] Pellet cells at 1300rpm (400xg) for 5 minutes at 25°C, resuspend in 500μl PrEGM (see REAGENT SETUP), and count cells using a hemocytometer. Cells at this point can be cultured in vitro (described in the ‘Culture of PrSCs’ section), or implanted in the renal capsule regeneration assay to directly analyze tubule regenerating capacity (Box1).
Box 1. Sub-renal prostate regeneration assay.
-
This procedure can be done using freshly isolated prostate stem cells (LSC cells or LSCT cells). The subrenal dissociated prostate cell regeneration assay has previously been described in detail17, and is described briefly below. Images of the procedure can be seen in Supplementary Figure 4.
! CAUTION All experiments involving live rodents must conform to national and institutional regulations.
-
For the LSC or LSCT cell population, aliquot out 5×10e3 to 1×10e5 cells into an eppendorf tube. Add 1–2×10e5 UGSM cells (see Box 1). Mix the cells with a pipette, and spin them down in a benchtop centrifuge at 2300rpm (500×g) for 2 minutes at 25°C. Aspirate off as much of the media as possible without disturbing the pellet. Resuspend the pellet in 15μl collagen mix (see REAGENT SETUP).
▲CRITICAL STEP Be careful not to introduce any bubbles to the mixture!
-
Set a pipette to 18μl, collect the cell/collagen mix, and pipette into a 6cm tissue culture dish. Place the plate in a 37°C incubator for 20–40 minutes to allow the collagen to solidify. Add warm BFS media to the plate, and put back into the incubator until surgery.
■ PAUSE POINT The collagen grafts could be left in the incubator overnight, if necessary. Do not wait more than 24 hours before implanting grafts under the kidney capsule.
Surgery: Anesthetize a male SCID mouse using a protocol approved by national and institutional regulations. Place the mouse prone, and shave a 3cm × 3cm area in the mid back region. Sterilize the area by alternating alcohol and Betadine swabs three times (Supplementary Figure 4a).
Cut a small incision in the skin around the middle of the back, approximately 1cm lateral to the spine (Supplementary Figure 4b). Cut a second small hole in the peritoneum exposed (Supplementary Figure 4c).
Hold the peritoneum open using forceps, and search for the kidney. Once located, use the small fat pad located at the tip of the kidney to pull it to the surface, and out above the skin. (Supplementary Figure 4d,e).
-
Gently grab the thin membrane on the outside of the kidney with two forceps, and tear a small hole. Lift the edge of the capsule; push the collagen graft underneath the membrane. (Supplementary Figure 4f–h).
▲CRITICAL STEP Make sure the collagen plug is securely underneath the kidney capsule.
Grab the peritoneum with forceps, and gently allow the kidney to slip back into the body. Make sure the graft doesn’t slip out (Supplementary Figure 4i).
-
Suture the peritoneum together, careful not to puncture any organs (Supplementary Figure 4j). Insert a testosterone pellet subcutaneously. Finally, use metal clips to staple the skin closed (Supplementary Figure 4k,l).
! CAUTION Make sure to monitor the mouse until the anesthetics wear off and the mouse is ambulatory. Monitor and medicate the mice in accordance to national and institutional animal care protocols.
Remove staples 7 days after surgery.
Grafts can be harvested 6–12 weeks after surgery.
Culture of PrSCs • TIMING Set up 2 h, then culturing for 7–10d
9] This step could be performed with sorted cells to culture PrSCs, as described below. It could also be used with unsorted cells as a way to enrich for a more stem-like cell population, as only these cells can form spheres. If using unsorted primary prostate cells, dilute cells to 2. 5×10e5 cells ml−1 in PrEGM and follow the protocol below.
10] Dilute sorted cells to 7.5×10e4 cells ml−1 in PrEGM. Mix 40μl of cells mixture with 60μl of cold Matrigel, and pipette the mixture around the rim of a well of a 12-well plate. Swirl the plate so that the mixture is evenly distributed around the rim, and place in a 37°C CO2 incubator for 30 minutes to allow the Matrigel to solidify.
11] Add 800μl warm PrEGM to each well, aiming the pipette to the center of the well as to not disturb the Matrigel ring. ▲CRITICAL STEP Media must be warm when added or the solidified Matrigel might dissolve or lose its integrity.
12] Perform a half media change every 3 days. Tilt the plate to a 45 degree angle, aspirate off approximately half of the media, and add 400μl of fresh warm PrEGM.
-
13] Culture and monitor the spheres for 7–10 days.
? TROUBLESHOOTING
Counting prostate spheres: • TIMING 30 minutes to 1 hour
14] The spheres can be counted easily by using 50x magnification on an inverted microscope, and following the rim of Matrigel around the wells. If there are too many spheres, the wells can be divided into 4 quadrants, out of which 1–2 could be counted and averaged. ? TROUBLESHOOTING
Maintenance and Characterization of PrSCs
15] After 7–10 days in culture the Matrigel can be digested and the spheres collect for sectioning, or the spheres could be dissociated into a single cell suspension to maintain the culture and prepare cells for other characterization.
16] Aspirate the PrEGM from each well, and add 1ml of Dispase (see REAGENT SETUP). Incubate the plate at 37°C for 1 hour.
17] Collect spheres into a 15ml Falcon tube and pellet at 1000rpm (233xg) for 5 min at 25°C. ▲CRITICAL STEP For sphere sectioning, you must leave the spheres intact (See step 20 Option A: ‘Sectioning and Immunostaining’ below). For all other characterization and maintenance, continue to dissociate spheres into single cells.
18] Resuspend the sphere pellet in 1ml warm Trypsin/0.05% EDTA, and transfer into a 1.5ml Eppendorf tube. Incubate the tube at 37°C for 5 minutes. Pass spheres through a 1ml syringe and a 27G needle 5 times to dissociate. Spin down cells in a bench top centrifuge at 2300rpm (500×g) for 2 minutes at 25°C.
-
19] Aspirate all but 50μl of the Trypsin/0.05% EDTA off, and resuspend pellet in fresh PrEGM. Pellet again in a bench top centrifuge at 2300rpm (500×g) for 2 minutes at 25°C. Resuspend in 500μl fresh PrEGM, and filter through a 40μm pore size filter. Pellet again in a bench top centrifuge at 2300rpm (500×g) for 2 minutes at 25°C. Resuspend in 500μl fresh PrEGM count cells using a hemocytometer.
■ PAUSE POINT Cells can be frozen down and stored in liquid Nitrogen.
20] The following assays can be used to maintain and characterize the dissociated sphere cells and their unique properties. Each of the options A–D described below should be carried out with separate samples of cells.
(A) Sectioning and Immunostaining of spheres
After collecting and pelleting the intact spheres (see steps 16 and 17), wash spheres with 1 ml PBS and spin down again. Aspirate as much of the PBS off as possible without disturbing the pellet.
To fix the spheres, resuspend the spheres in 1ml 10% buffered Formalin, and leave at room temperature for 1 hour. Spin down the spheres at 1000rpm (233×g) for 5 minutes at 25°C.
Aspirate the formalin off and resuspend the spheres in 70% vol/vol ethanol, and leave at room temperature for 1 hour. Spin down the spheres at 1000rpm (233×g) for 5 minutes at 25°C. Aspirate as much of the ethanol off as possible without disturbing the pellet.
Resuspend the sphere pellet in 20μl of 60°C Histogel. Mix by pipetting up and down 3 times. Collect the Histogel/sphere mixture, and pipette it onto a parafilm lined petridish. Allow the Histogel plug to solidify at room temperature, or you could speed up the solidification process by placing the dish at 4°C.
Embed in paraffin and section.
A standard immunofluorescence or immunohistochemical staining protocol can be carried out at this stage. (See Table 2 for antibody clones and dilutions typically used).
Table 2.
List of antibodies for colony and sphere characterization
| Antibody | Clone/Company | Dilution | Cell type stained |
|---|---|---|---|
| CK5 | PRB-160P/Covance | 1:1000 | Single positive: basal and stem cells Double positive with CK8: trans-amplifying cells |
| CK8 | MMS-162P/Covance | 1:1000 | Single positive: luminal cells Double positive with CK5: trans-amplifying cells |
| p63 | sc-8431/Santa Cruz | 1:200 | Basal cells |
| AR | sc-816/Santa Cruz | 1:200 | Luminal cells |
| SMA | ab5694-100/AbCam | 1:400 | Stroma/smooth muscle cells |
| Vimentin | ab39376-100/AbCam | 1:400 | Stroma |
| Synaptophysin | MAB5258/Chemicon | 1:200 | Neuroendocrine cells |
(B) In vitro passaging (to maintain cells or to study self-renewal)
Dilute dissociated sphere cells to 1.25×10e5 cells ml−1 in PrEGM. Mix 40ul of cells mixture with 60μl of cold Matrigel, and pipette the mixture around the rim of a well of a 12-well plate. Swirl the plate so that the mixture is evenly distributed around the rim, and place in a 37°C CO2 incubator for 30 minutes to allow the Matrigel to solidify.
Add 800μl warm PrEGM to each well, aiming the pipette to the center of the well as to not disturb the Matrigel ring. ▲CRITICAL STEP Media must be warm when added or the solidified Matrigel might dissolve or lose its integrity.
Perform a half media change every 3 days. Tilt the plate to a 45 degree angle, aspirate off approximately half of the media, and add 400μl of fresh warm PrEGM.
After 7 days in culture you can count the spheres and/or dissociate the spheres and set up a fresh sphere assay.
(C) In vitro differentiation
(i) Dissociated sphere cells can be plated into a 2 dimensional colony assay in order to drive the cells to terminal differentiation. Two alternate feeder layers can be used: irradiated 3T3 cells (described in ii.a) or Matrigel (described in ii.b).
(ii)a Plate 1×10e5 3T3 cells in 60mm tissue culture plates the night before the experiment. The next morning, irradiate the cells with 500rads of γ-irradiation. Change the media after 1 hour to fresh 3T3 media, and keep in the incubator until the sphere cells are ready for plating. Immediately before plating the dissociated sphere cells, change the media again to PrEGM.
(ii)b Prepare 6 well tissue culture plates by pipetting 20μl cold Matrigel into each well and distribute it evenly using a cell scraper. Place plates in a 37°C incubator for 30 minutes. Wash with 1x PBS, and aspirate off. Add 2.5ml warm PrEGM into each well. Keep in incubator until the cells are ready.
(iii) Dilute dissociated sphere cells to 1.25×10e5 cells ml−1 in PrEGM. Add 40μl of the cell/PrEGM mixture into each well, and shake gently to distribute cells evenly.
(iv) Allow colonies to form for 7 days, changing media every 3days. After 7 days, remove the media and add PrEGM supplemented with DHT to a final concentration of 10−8 M (0.01μM). Change media every 2 days, and allow colonies to grow for another 5–7 days.
(v) To analyze the colonies, they can be counted or characterized using immunohistochemistry.
(vi) To visualize the colonies for overview images, aspirate PrEGM, and wash the plate with 1x PBS. Add Turk’s solution (see REAGENT SETUP), and leave for 10–15 minutes. When the colonies are stained, aspirate the Turk’s solution off and wash plate with 1x PBS twice.
(vii) To stain the colonies, wash the wells twice with 1x PBS. Fix the colonies with cold acetone for 2 minutes. Wash with 1x PBS.
(viii) A standard Immunofluorescence protocol can then be carried out to stain the colonies. Typical markers to use for analyzing the differentiation status are cytokeratin 5 and 8, p63, AR, and synaptophysin. (See Table 2 for antibody clones and dilutions.)
(D) In vivo regeneration -subcutaneous injection
-
(i) This procedure can be done using freshly isolated prostate stem cells or dissociated sphere cells. Cells can be combined with UGSM(see Box 2)and Matrigel, and then injected subcutaneously into SCID mice as described below. Alternatively, the cells could be mixed with UGSM and collagen, and implanted under the kidney capsule of SCID mice, as previously described18 and briefly described in Box 1.
! CAUTION All experiments involving live rodents must conform to national and institutional regulations.
(ii) To test the tubule regenerative capacity of the primary prostate stem cells or the dissociated sphere cells, start this procedure with the cell preparation from Step 12/18, or Step 26 respectively.
(iii)a. For the LSC or LSCT cell population, aliquot out 5×10e3 to 1×10e5 cells into an eppendorf tube. Add 1-2×10e5 UGSM cells (see Box2). Mix the cells with a pipette, and spin them down in a bench top centrifuge at 2300rpm (500xg) for 2 minutes at 25°C. Aspirate off as much of the media as possible without disturbing the pellet. Resuspend in 10μl of PrEGM. Place in ice.
(iii)b. For sphere cells, aliquot 5×10e4-5×10e5 cells into an eppendorf tube. Add 2×10e5 UGSM cells (see Box1). Mix the cells with a pipette, and spin them down in a bench top centrifuge at 2300rpm (500×g) for 2 minutes at 25°C. Aspirate off as much of the media as possible without disturbing the pellet. Resuspend in 5μl of PrEGM. Place on ice.
(iv) Prepare an insulin syringe for each injection, and place them on ice. Prepare a 500μl aliquot of Matrigel, and place on ice. Prepare all of the surgical tools and equipment for sterile, aseptic surgery.
(v) Anesthetize an 8 week-old SCID mouse with Ketamine/Xylazine or Isofluorine gas. Once the animal is anesthetized, shave the target injection site. Sterilize with 3 rounds of iodine and alcohol swabs.
(vi) Immediately before injection, mix the cell mixture with a pipette. Add in 30μl cold Matrigel, mix, and place back on ice. Pull up the cell/Matrigel mixture into the cold syringe, making sure no bubbles are formed in the process.
(vii) Using a clean pair of forceps, pull up on the freshly shaved and cleaned skin of the mouse, creating a tent between the skin and musculature of the flank. Inject the cell/Matrigel mixture, and pull out the needle slowly. Hold onto the skin with the forceps for a minute to make sure the Matrigel has settled.
(viii) Monitor mice in accordance to institution’s animal care protocols.
(ix) Grafts can be harvested and analyzed 8 weeks after injection.
Box 2. Urogenital Sinus Mesenchyme (UGSM) Cell Collection.
-
Prepare UGSM cells as previously described17. Briefly, set up timed pregnancies and sacrifice pregnant females at day 16 of the pregnancy.
! CAUTION All experiments involving live rodents must conform to national and institutional regulations.
Remove the uterus with the embryos, and place in Dissecting Media (see REAGENT SETUP).
Cut the uterus laterally, separate the embryos from the placenta, and place them in a new 10cm Petri dish containing Dissecting Media.
Cut embryos in half, below the liver. Place the bottom half of the embryos into a new dish containing sterile 1× PBS.
Place the bottom half of the embryos in a supine position and cut the abdomen open while holding the hind legs apart with forceps.
The urogenital sinus is connected to the bladder. As in the adult, the urogenital sinus could be removed intact by gently pulling up on the bladder. Dissect the pelvic UGS and clean off the attached tubular structures, and finally cut off the bladder.
Place the pelvic UGS onto a concave glass slide (placed in 10 cm Petri dish) containing 250μl Dissecting Media.
When all of the UGS are excised, wash them 3 times with 1× PBS. Aspirate the last wash of PBS carefully, and add 1ml 1% Trypsin. Keep the plate in 4°C for 90 minutes to allow for digestion.
Carefully remove the Trypsin with a pipette, and add Dissecting Media. Carefully pipette the media off, and add 1m Dissecting Media containing 500U DNase I. Let sit for 5 minutes.
Wash the UGS three more times with fresh Dissecting Media.
After the third wash, take 2 28-gauge needles and separate the mesenchyme away from the epithelium. The epithelium can be identified as the opaque cylinder shaped object inside the more translucent and vascular mesenchyme.
Collect all of the mesenchyme fragments into a 15ml Falcon tube containing 9mls of Dissecting Media, and add 1ml 10× collagenase (see REAGENT SETUP). Digest at 37°C for 2 hours.
After digestion, filter the cells through a nylon mesh filter with 40μm pore size, and wash the filter with 10mls of Dissecting Media to collect all of the cells from the mesh. Spin down at 1300rpm (400×g) at 25°C for 5 minutes. Aspirate off the media, resuspend in 1ml BFS media (see REAGENT SETUP), and plate in a 10cm Petri dish containing 9mls of BFS media.
Culture for 5–7 days, and monitor cell growth. ▲CRITICAL STEP Make sure there are no epithelial colonies, as this means the UGSM is contaminated with UGSE and cannot be used for successful in vivo regeneration.
-
Passage and expand the UGSM cells when they get to 80% confluency.
■ PAUSE POINT UGSM cells could be frozen down and stored at −80°C for 4 weeks, or in liquid nitrogen for up to 6 months.
-
Cells could be used at this time for in vivo regeneration experiments (in both subcutaneous injection and subrenal prostate regeneration assays).
▲CRITICAL STEP Do not passage UGSM cells more than 5 times, as they lose their inductive capacity in later passages.
• TIMING
Steps 1–6, isolation of prostate cells: ~3–4 hours
Step 7A or 7B, enrichment of PrSCs: 2–3 hours
Steps 8–13, culturing PrSCs: 1 hour set-up, followed by a 7–10day incubation
Step 14, counting prostate spheres: ~ 30 minutes – 1 hour
Steps 15–20, dissociating prostate spheres: 2 hours
Step 20A, Immunostaining: up to 1 week
Step 20B, Sphere passaging: 2 hours, followed by a 7–10day incubation
Step20C, Colony assay: 15 minute set-up, followed by a 10 day incubation
Step20D, Subcutaneous and subrenal regeneration assay: 2 hour set-up followed by 8-week incubation
Complete characterization of PrSCs: 8 weeks for performing all of the experiments
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 3.
Table 3.
Troubleshooting
| Step | Problem | Possible Reasons | Solution |
|---|---|---|---|
| 5 | Large clumps of cells present in the media | Cells may have stuck together due to DNA in the mixture, and they did not go through filter. | Add more DnaseI into the solution before filtering the cells, and wait 10 minutes. Pipette the cells thoroughly before filtering again. |
| 6 | Low cell yield | Digestion may have been incomplete. | Digest longer in Collagenase solution (up to 4 hours), and add an extra round of Trypsin/EDTA digestion. |
| 7A(v)and 7B(vi) | Low LSC percentage or low LSCT+/LSCT− percentage | Incomplete digestion of the prostate leads to incomplete release of the basal cells and therefore a lower percentage of LSC and LSCT populations. | Leave prostates in collagenase longer and add an extra Trypsin digestion step. Add more DnaseI into the media before filtering the cells, and wait 10 minutes. Pipette the cells thoroughly before filtering again. |
| 13 | Floating chunks of Matrigel | Too much PrEGM was mixed with the Matrigel | The ration of Matrigel:Media should be at least 1:1, but preferably 2:1. Anything lower will compromise the integrity of the Matrigel. Increase the amount of Matrigel mixed in with the cells. |
| Matrigel may be expired | Get a fresh batch of Matrigel. Matrigel is only guaranteed for 3 months. | ||
| Cold PrEGM was added after solidification | Once the Matrigel solidifies, make sure to only add warm PrEGM to the wells. Cold media will compromise the integrity of the Matrigel. | ||
| 14 | Low sphere count | Matrigel may be expired | Get a fresh batch of Matrigel. Matrigel is only guaranteed for 3 months. |
| Not enough cells were plated | Increase the cell number plated to 1e4 per well. | ||
| Cells died in the sorting process | Make sure to keep the cells at 4°C during and after sorting. | ||
| 20a | Can’t see many spheres in the slides | Too few spheres must have been collected | Collect at least 3 wells of spheres for each sample. Resuspend in as little Histogel as possible (minimum = 20ul). Check the Histogel plug under the microscope to make sure lots of spheres are present. |
| 20b | No or low secondary sphere growth | Number or quality of dissociated sphere cells may have been low | Do not use spheres that are more than 8 days old. Make sure the spheres look healthy and are not overcorwded prior to digestion. Make sure spheres are completely digested by Trypsin, otherwise the majority will be lost in the filtration. |
| Reagents may be too old | Check the Matrigel to make sure it’s not expired. Make sure to use fresh PrEGM (maximum 2–3 weeks old). | ||
| 20c | No or low colony growth | Number or quality of dissociated sphere cells may have been low | See advice above (27a) |
| Feeders: 3T3s may have been overcrowded, or Matrigel expired | Plate fewer 3T3 cells the day before, or plate them later in the afternoon the day before the experiment. Make sure the Matrigel is not expired, and that it is evenly coating the plate. |
||
| 20d | No or low efficiency tubule regeneration | Number or quality of dissociated sphere cells may have been low | See advice above (27a) |
| Part of the sample may have gotten stuck in the syringe | Make sure to keep the syringes and needles on ice up until the injection to prevent the Matrigel from solidifying. |
ANTICIPATED RESULTS
In our hands we can get ~1.0 – 1.5×106 prostate cells from an 8 to 10 week old C57/Bl6 mouse. We get around 20–23% LSC staining and 8–10% LSCT staining (when gating on the Lin- fraction). From one 8- to 10-week old prostate we can obtain approximately 8–10×104 LSC cells by FACS count, which correlates to approximately 3–5×104 cells by hand count. We get 2–4×104 LSCT cells by FACS count, which correlates to approximately 1–2×104 cells by hand count.
One in 35 LSC cells and 1 in 11 LSCT cells can form spheres in the first generation, as reported in our most recent publications9,10. It is important to remember that basal cells express higher levels of matrix binding integrins, which may give these cells a growth advantage in the extracellular matrix-rich Matrigel. The additional growth factors in the Matrigel and PrEGM may also drive more progenitor-like cells to give rise to spheres in addition to the stem cells.
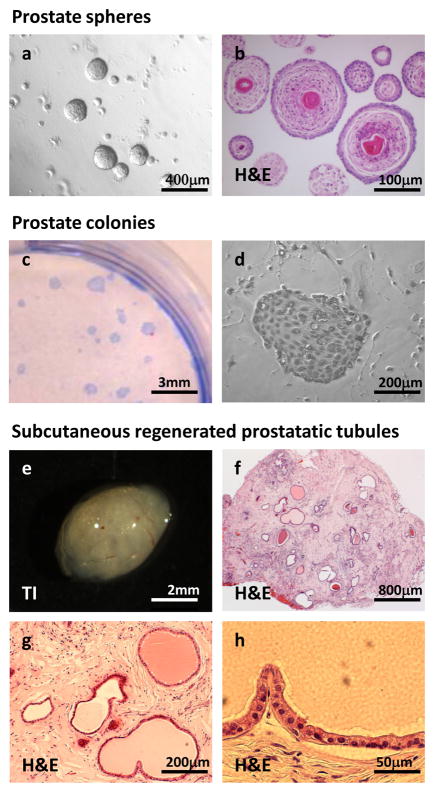
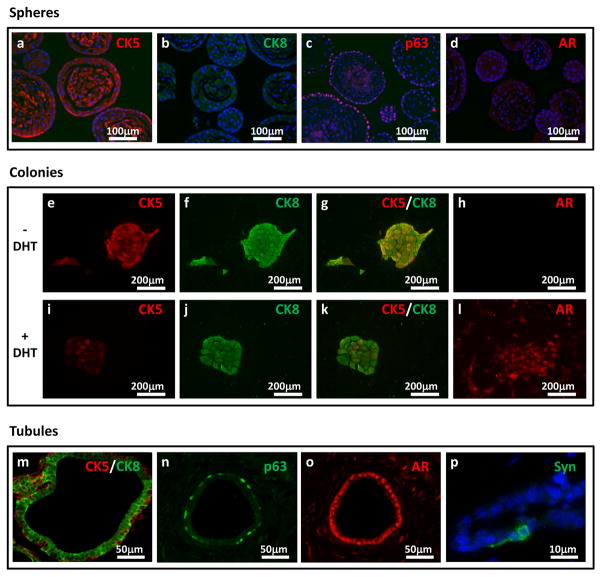
The majority of primary prostate spheres are solid in appearance (Figure 5a), and a subset have secretions in the center (Figure 5b). The spheres contain mostly basal cells that express high levels of cytokeratin 5 and low levels of cytokeratin 8 (Figure 6a and b). The cells located on the outer rim are the most primitive basal cells that express p63 (Figure 6c). Full length AR protein cannot be detected in the majority of the spheres (Figure 6d). Western blot analysis has revealed that the sphere cells do express AR protein, but in the absence of testosterone, the protein is quickly degraded. Only small bands representing the degraded protein are seen on Western blots, and not the full length receptor14.
Figure 5. Representative overview images of spheres, colonies, and prostatic tubules.
(a) A phase contrast image of prostate spheres in Matrigel, taken at a 50× magnification, and (b) an H&E stained sphere section taken at 200× magnification (right), (c) Image shows a 6cm tissue culture dish with prostate colonies stained with crystal violet and (d) a phase contrast image of a colony taken at 100× magnification (right), (e) Transillumination image (TI) of a Matrigel plug after 8 weeks in vivo, (f) an H&E stained section of a regenerated tubules in a Matrigel plug taken at 25× magnification, (g) taken at 100× magnification, and (h) taken at 400× magnification. All institutional regulations were followed when performing experiments using live mice.
Figure 6. Differentiation marker stains of spheres, colonies and prostatic tubules.
(a–d) Images show prostate sphere sections stained with cytokeratin 5 (red; a), cytokeratin 8 (green; b), p63 (red, c), and AR (red; d). Sections were co-stained with DAPI (blue) and taken at 200× magnification. Images are similar to previously published images from our lab14. Images of prostate colonies growth in the absence (e–h), or the presence (i–l) of DHT, and stained with CK5 (red; e, i), cytokeratin 8 (green; f, j), CK5/CK8 merged (red/green, g, k), and AR (red; h, I). Images were taken at 100× magnification. Images are similar to previously published images14 (m–p) Images of regenerated prostate tubules stained with cytokeratin 5 and 8 (red and green; m), p63 (green; n), AR (red, o), and Synaptophysin (green; p). First three images were taken at 400× magnification. Last image was co-stained with DAPI (blue) and taken at 1000× magnification for better visualization. All institutional regulations were followed when performing experiments using live mice.
In vitro passaging: One in 20 dissociated first generation sphere cells will form secondary spheres. In later generation, the sphere forming activity becomes closer to 1 in 10.
In vitro colony assay and differentiation: Approximately 1 in 25 primary sphere cells will form colonies in the colony assay (Figure 5c). The colonies appear epithelial, with a cobblestone morphology (Figure 5d). In the absence of DHT, 95% of the colonies are CK5/CK8 double positive, which is indicative of an intermediate/transit-amplifying phenotype (Figure 6e–g). None of these colonies express full length AR (Figure 6h). There are occasional CK8 single positive colonies, which are probably derived from a luminal-restricted colony-forming cell. Another small percentage of the colonies is CK5 single positive. With the addition of DHT, the majority of the colonies become CK8 high and CK5 low to negative(Figure 6i–k). Most of the colonies also express AR in the presence of DHT(Figure 6l).
In vivo prostate regeneration: Primary isolated LSC and LSCT cells will grow in vivo in both the subrenal and subcutaneous regeneration assays. Cells tend to have more robust growth under the kidney capsule, most likely due to the rich blood supply and inductive microenvironment existing in that area. Stem cells cultured in vitro in the prostate sphere assay will grow better in the subcutaneous regeneration assay. Subcutaneous grafts will likely have a largely mesenchymal or fibroblastic appearance due to UGSM outgrowth, with scattered tubules (Figure 5e–g). One in a few hundred cells from an enriched preparation of stem cells (LSCT) will be expected to generate prostatic tubules. Regenerated tubules should contain a typical double-layered epithelial appearance (Figure 5h). Immunostaining will show an outer layer of cells expressing the basal keratins (K5, K14) and the transcription factor p63 (Figure 6m and n). The inner epithelial layer is larger and columnar in shape, expressing high levels of the luminal keratins (K8, K18) and AR(Figure 6m and o). Occasional cells within the basal layer or between layers will express the neuroendocrine marker synaptophysin(Figure 6p). In the subrenal regeneration experiment, the tubule forming activity of the LSC and LSCT cells are similar to the subcutaneous assay, but the tubules appear larger (Supplementary Figure 3a and 3b). As in the subcutaneous regeneration assay, cells in the outer layer express the basal markers CK5 and p63 (Supplementary Figures 3c and 3d), while the inner layer cells express the luminal markers CK8 and AR (Supplementary Figures 3eand 3f). The subrenal regeneration assay is also preferred for carcinogenesis studies. Use of SCID mice for the regeneration assays allows testing of the tubule regenerating capabilities of both primary mouse and human prostate cells in a parallel manner. Studies to purify and characterize stem and progenitor cells from the human prostate using the in vivo model are underway in our laboratory.
Supplementary Material
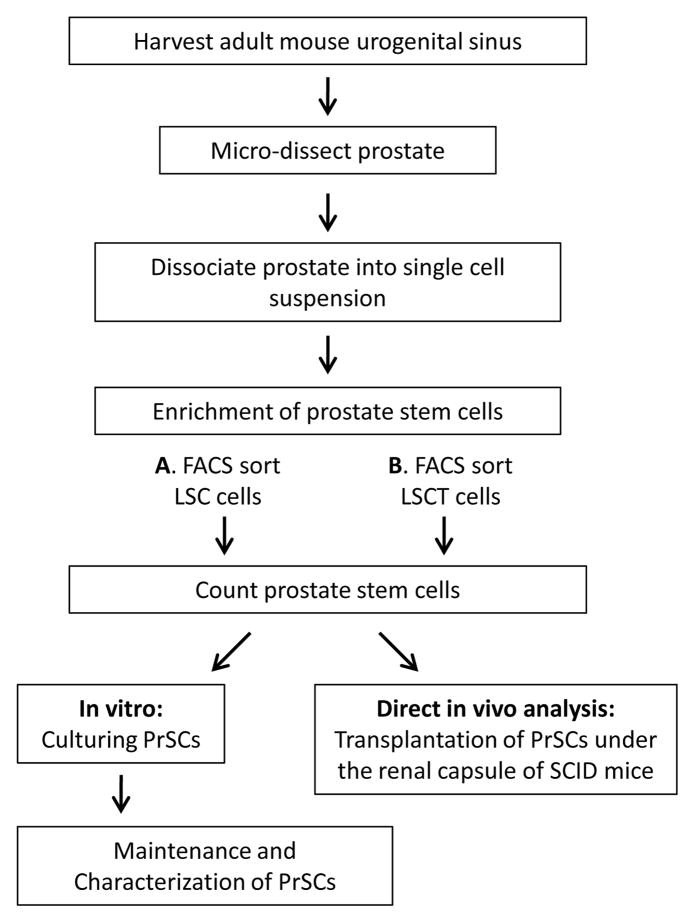
Figure 1. A schematic overview of the major steps in the procedure.
A. Sectioning and Immunostaining
B. In vitro passaging/self-renewal
C In vitro differentiation
D. In vivo subcutaneous regeneration
Acknowledgments
We thank Houjian Cai and Yang Zong for their helpful comments on the protocols and for demonstrating the procedure in Supplementary Figure 4. R.U.L. is supported by the California Institute for Regenerative Medicine training grant (T1-00005 and TG2-01169). A.S.G. is supported by Ruth L. Kirschstein National Research Service Award GM07185. This work has also been supported by the Prostate Cancer Foundation Challenge Award for Defining Targets and Biomarkers in Prostate Cancer Stem Cells. O.N.W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTION STATEMENT
All authors contributed to the development of the methodology. RUL, ASG, and ONW contributed to the description of the protocols.
The authors declare that they have no competing financial interests.
References
- 1.Blanpain C, Horsley V, Fuchs E. Epithelial stem cells: turning over new leaves. Cell. 2007;128:445–458. doi: 10.1016/j.cell.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 3.Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124:1111–1115. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 4.Tsujimura A, et al. Proximal location of mouse prostate epithelial stem cells: a model of prostatic homeostasis. The Journal of cell biology. 2002;157:1257–1265. doi: 10.1083/jcb.200202067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scher HI, Sawyers CL. Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. J Clin Oncol. 2005;23:8253–8261. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 6.Taplin ME, Balk SP. Androgen receptor: a key molecule in the progression of prostate cancer to hormone independence. Journal of cellular biochemistry. 2004;91:483–490. doi: 10.1002/jcb.10653. [DOI] [PubMed] [Google Scholar]
- 7.Burger PE, et al. High aldehyde dehydrogenase activity: a novel functional marker of murine prostate stem/progenitor cells. Stem cells (Dayton, Ohio) 2009;27:2220–2228. doi: 10.1002/stem.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burger PE, et al. Sca-1 expression identifies stem cells in the proximal region of prostatic ducts with high capacity to reconstitute prostatic tissue. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7180–7185. doi: 10.1073/pnas.0502761102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstein AS, et al. Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20882–20887. doi: 10.1073/pnas.0811411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawson DA, Xin L, Lukacs RU, Cheng D, Witte ON. Isolation and functional characterization of murine prostate stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:181–186. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leong KG, Wang BE, Johnson L, Gao WQ. Generation of a prostate from a single adult stem cell. Nature. 2008;456:804–808. doi: 10.1038/nature07427. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xin L, Lawson DA, Witte ON. The Sca-1 cell surface marker enriches for a prostate-regenerating cell subpopulation that can initiate prostate tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:6942–6947. doi: 10.1073/pnas.0502320102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xin L, Lukacs RU, Lawson DA, Cheng D, Witte ON. Self-renewal and multilineage differentiation in vitro from murine prostate stem cells. Stem cells (Dayton, Ohio) 2007;25:2760–2769. doi: 10.1634/stemcells.2007-0355. [DOI] [PubMed] [Google Scholar]
- 15.Lukacs RU, et al. Epithelial stem cells of the prostate and their role in cancer progression. Cold Spring Harbor symposia on quantitative biology. 2008;73:491–502. doi: 10.1101/sqb.2008.73.012. [DOI] [PubMed] [Google Scholar]
- 16.Pontier SM, Muller WJ. Integrins in mammary-stem-cell biology and breast-cancer progression--a role in cancer stem cells? Journal of cell science. 2009;122:207–214. doi: 10.1242/jcs.040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunha GR, Lung B. The possible influence of temporal factors in androgenic responsiveness of urogenital tissue recombinants from wild-type and androgen-insensitive (Tfm) mice. J Exp Zool. 1978;205:181–193. doi: 10.1002/jez.1402050203. [DOI] [PubMed] [Google Scholar]
- 18.Xin L, Ide H, Kim Y, Dubey P, Witte ON. In vivo regeneration of murine prostate from dissociated cell populations of postnatal epithelia and urogenital sinus mesenchyme. Proceedings of the National Academy of Sciences of the United States of America. 2003;100 (Suppl 1):11896–11903. doi: 10.1073/pnas.1734139100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.