A new human influenza A virus (IAV) strain of swine-like origin emerged in Mexico and the United States in March and April of 2009 [1]. The virus disseminated globally with sufficient celerity that on June 11, 2009 the World Health Organization (WHO) raised the Flu Pandemic Alert Level to Phase 6, officially designating a new global pandemic. At this time there were 30,000 confirmed cases in 74 countries. Less than a month later (as of July 1, 2009), the WHO confirmed over 77,000 cases with 332 deaths.
The collaborative nature of the global response to the outbreak has been remarkable. Within days of the initial reporting of the virus, researchers completely sequenced the genome of several viral isolates and posted the sequences on the internet. Sequences of new isolates continue to be uploaded daily on websites such as the Global Initiative on Sharing Avian Influenza Data (http:platform.gisaid.org) and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/genomes/FLU/SwineFlu/html). Nearly in real time, phylogeneticists identified the complex origins of the virus. Within weeks, a vaccine seed stock was isolated, and the private and public sector began intense collaborative efforts to produce a vaccine post-haste [2].
This is the first major introduction of a pandemic influenza strain since 1977, and the first to benefit from the biotechnology revolution. Those looking to defend the investment of world governments into biomedical research have another shining example of the enormous payoff in both lives and money gained from achieving a better understanding of biology. Retrospectively, concerns over the pandemic potential of avian IAVs prepared society well for the current pandemic, as recently upgraded resources have played critical roles in monitoring and responding rapidly. Scientists, physicians, administrators, and politicians who fought for these resources deserve the gratitude of their nations.
Clearly, appropriate actions have been taken to limit disease and death associated with the new virus. The contrast with the 1918 pandemic, whose causative agent was not known for 13 years, could not be starker. Despite our advances, it is critical to recognize that today, as in 1918, humanity can neither control nor predict the genetic and antigenic evolution of this virus. Here we discuss factors in the evolution of 2009 swine-origin H1N1 IAV (hereafter referred to as S-IAV-2009).
Overview of IAV Proteins
IAVs are negative single stranded RNA viruses with 8 gene segments encoding at least 11 functional proteins. Virions consist of a matrix (M1) shell covered by a lipid membrane. Within the shell, the 8 segments are coated with nucleoprotein (NP) and small quantities of the viral polymerase complex (PB1, PB2, and PA), and NS2 (also known as nuclear export protein). Two viral glycoproteins are embedded in the membrane, hemagglutinin (HA) and neuraminidase (NA). HA mediates binding to cellular receptors, which are sialic acid moieties terminally linked to oligosaccharides on cellular proteins and lipids. NA cleaves sialic acid, playing a critical role in progeny virus release from host cells and in preventing viral aggregation. Another small viral non-glycosylated membrane protein, M2, is an ion channel involved in viral entry and exit. The virus encodes two proteins excluded from virions, NS1 and PB1-F2. NS1 blocks innate immune anti-viral responses and contributes to viral gene expression. While the functions of PB1-F2 remain to be firmly established, it appears to play an important role in pathogenicity [3]. PB1-F2 is encoded by an overlapping reading frame in the PB1 gene, and contemporary H1 subtype viruses express a truncated form of less than 60 amino acids (compared to ~ 90 amino acids in other strains). Intriguingly, while PB1-F2 is encoded by nearly all human and avian isolates, swine viruses, including S-IAV-2009, encode extremely truncated forms (less than 20 residues) that are likely to lack functionality. Additional novel IAV gene products probably play important roles in viral replication and pathogenicity [4,5]; it is to be expected that IAV makes the most of its extremely limited coding capacity.
In Came S-IAV-2009
When multiple IAVs infect a single cell, reassortment of gene segments between viruses in progeny is common. Pandemic viruses typically arise from the recombination of animal and human viruses, generating a virus with animal glycoprotein genes on a background of mostly, or entirely human genes.
The genetic makeup of S-IAV-2009 is complex, containing genes derived from human (PB1), avian (PA, PB2), classical swine (HA, NP, NS), and Eurasian swine (NA, M) IAVs [6–8]. Despite their disparate origins, each of the genes appears to have circulated in swine for at least 10 years [7]. While the virus most likely jumped directly from pigs to man, it is impossible to rule out a bridging host. Due to the relatively limited surveillance of swine IAVs, it is possible that the reassorted virus circulated in swine for years.
Swine IAVs routinely infect humans, although prior to this year, the Center for Disease Control and Prevention only reported 1–2 annual cases. Most cases of human infection with swine IAVs are likely unreported since symptoms are indistinguishable from the spectrum of symptoms caused by human IAV infections [9]. The precise moment when S-IAV-2009 crossed the species will never be known. One of the earliest cases was reported in La Gloria, Mexico in February 2009, and further genetic analysis of individual viral isolates predicts a common ancestor virus that dates back to January 2009 [10].
Immunity to S-IAV-2009
The immune response to IAV is comprised of multiple elements of innate and adaptive immunity. While the innate immune system plays an essential role in limiting viral replication in the first days of infection, it is relatively non-specific, and has yet to be rationally manipulated to provide a protective effect against infection. Rather, vaccines are based on the induction of B and T cell responses. CD8+ T cells may contribute to decreasing human IAV mortality (as has been extensively documented in mice [11], and even chickens [12]), but as yet are unproven vaccine targets. Rather, vaccines are designed to elicit anti-IAV antibodies (Abs).
Three IAV proteins elicit robust Ab responses during infection: NP, NA, and HA. Although NP is expressed on cell surfaces, anti-NP Abs have never been shown to play an important role in immunity to IAV. NA-specific Abs prevent virus release from cells and limit transmission. HA-specific Abs block entry into cells by blocking virus attachment or fusion with cellular membranes, and these Abs are the only effector function of the immune system that prevents IAV infection. Consequently, HA is by far the most important IAV protein from the standpoint of epidemiology and vaccination. M2 is less immunogenic, but is a promising target for cross-protective vaccination, since its short extracellular target domain is highly conserved, and Abs can protect against lethal challenge [13]. This contrasts with HA and NA, which rapidly evolve antigenically under Ab pressure. The process of antigenic evolution that results from the accumulation of amino acid substitutions is known as “antigenic drift”. More radical changes in HA (or NA) antigenicity, “antigenic shift”, occurs when an entirely new gene is introduced into the human population from the animal reservoir.
Due to the importance of IAV as a human pathogen, and the central role of HA in IAV ecology in humans, HA has recorded many “firsts”. These include: first viral protein to have its structure determined by crystallography, first protein to be mapped antigenically with monoclonal Abs (mAbs), and first to have its membrane fusion mechanism dissected.
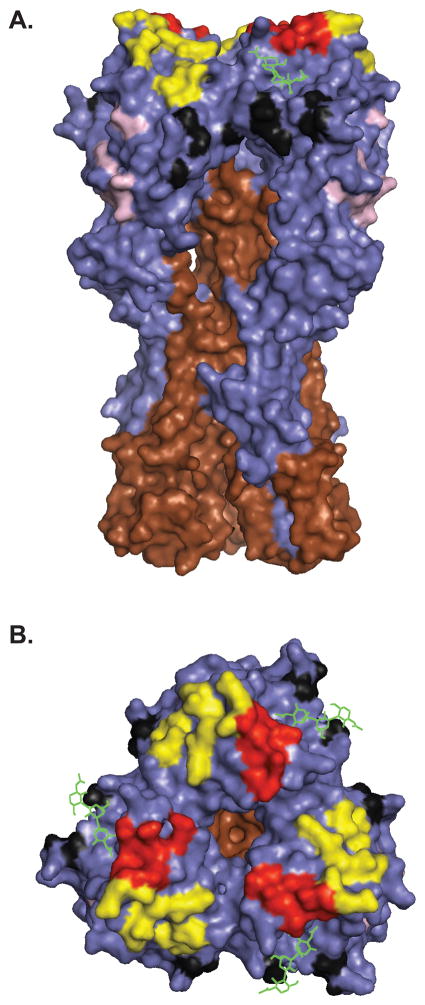
HA is a homotrimer, with each monomer consisting of two disulfide linked subunits, HA1 and HA2, that are generated by cleavage in the late secretory compartment during HA biogenesis (Figure 1). Cleavage is required to activate HA fusion activity, which is based on the low pH induced exposure of a highly hydrophobic peptide at the HA2 NH2-terminus. Abs with potent neutralizing activity are specific for the globular domain formed by loops of the HA1 chain at the tip of the HA spikes. Most anti-HA Abs neutralize by preventing viral binding to sialic acid bearing cellular receptors [14]. A subset of Abs inhibit the fusion step of virus entry by binding either HA2 or membrane proximal regions of HA1. Fusion inhibiting Abs are potentially important in providing protection between drifted, or even shifted strains, since the targeted domains are more highly conserved that the globular regions [15,16]. At the same time, the conservation of these domains, suggests that such Abs exert little pressure in nature, and indeed, fusion inhibiting Abs typically are far less efficient than receptor blocking Abs at neutralizing virus.
Fig 1.
The antigenic sites of H1. (A) Side and (B) top views of A/PR/8/34 HA (PDB: 1RVZ). HA1 and HA2 are shown in blue and brown, respectively and sialic acid is shown in green. Antigenic sites are shown in different colors: Sa (yellow), Sb (red), Ca (black), and Cb (pink). Images were generated using Pymol software.
H1: HA Antigenicity in Hi-Definition
As luck would have it, thanks to Walter Gerhard and his disciples at the Wistar Institute, the H1 HA is the probably most extensively characterized antigen in existence. The “first” mAbs mentioned above were generated by Walter & co. against the A/PR/8/34 HA, and were followed by hundreds more over the course of his career. These mAbs were used to invent “epitope mapping”, a three step process. Step one: propagate virus in the presence of sufficient Ab to neutralize all wt virus, resulting in the selection of escape mutants, which demonstrate at least 100-fold decreased affinity for the selecting mAb [17]. Step two: sequence mutants by nascent nucleic acid sequencing technology [18,19]. Step three: use mutants to map the epitopes of a large mAb panel by direct binding assay.
Epitope mapping revealed that the H1 HA possesses 4 physically distinct, antigenic sites designated Sa, Sb, Ca, and Cb [19]. The ‘S’ in Sa and Sb refers to ‘strain-specific’, as these sites are highly variable between different stains. The ‘C’ in Ca and Cb refers to ‘cross-reactive’, as these sites are positioned more proximal to the viral membrane and are more conserved between different viral strains. The ‘a’ and ‘b’ designate physically distinct sites.
Operationally, the antigenic sites are distinct, based on large number of mAbs whose bindings is diminished by mutations in a single site. Physically (Figure 1), Sa and Sb sites are situated at the top of the globular domain, on the upper ridge of the sialic acid binding pocket. The Ca site is just below the receptor binding site and sits astride the trimer interface, subdividing the site into Ca1 and Ca2 sub-sites. The Cb site resides further down the spike. Each of the sites are formed by residues from different loops of the HA and are exquisitely sensitive to denaturation. Consequently, despite the initial hoopla [20,21], it is apparently not possible to use short synthetic peptides corresponding to linear HA sequences to elicit Abs that bind to the globular domain with sufficient affinities to enable effective vaccination.
Drift: A Well Defined Mystery
The typical response to HA probably contains thousands of different Abs with neutralization activity [22]. How then can HA escape neutralization? Though different Abs interact with a common antigen in a unique manner, they share many common features of recognition, and they are similarly negatively affected by amino acid substitutions. Retrospective analysis of Gerhard et.al [18], reveals that single amino acid substitutions in escape mutants on average block binding of ~ half of the Abs specific for a given site (Table 1). If individuals make a balanced neutralizing response against each of the 4 sites, then 8 (or more) mutations would be needed to generate a mutant capable of escaping all Abs. Since the frequency of escape mutants with single amino substitutions is on the order of 10−5 [23], anything beyond a double substitution (frequency, 10−10) is essentially impossible during the course of infection of one individual, given the amount of virus replication required (at least 1015 virions, which is a lot considering each cell produces ~ 103 progeny virions, and humans only have ~ 1013 cells).
Table 1.
| Antigenic Site | Number Mutants Tested | Number Abs Tested | Avg # Abs Eliminated by Single Substitution | % Abs Affected |
|---|---|---|---|---|
| Sa | 11 | 15 | 8 ± 2.3 | 53 |
| Sb | 9 | 9 | 5.1 ± 1.5 | 57 |
| Ca | 7 | 9 | 3.6 ± 1.6 | 40 |
| Cb | 7 | 16 | 8.9 ± 2.5 | 55 |
This leads to two distinct possibilities. First, some humans may mount dominant Ab responses against individual antigenic sites, leading to sequential selection of variants and gradual antigenic drift [24,25]. Second, some amino acid substitutions may confer resistance to neutralization by a mechanism independently of their antigenic modifications. Indeed, growing PR8 in the presence of a mixture of mAbs chosen to provide equal neutralization pressure on all 4 sites resulted in the generation of absorptive variants, which are mutants with higher affinity for cellular sialic acid receptors with extremely limited antigenic alterations [26].
Could such absorptive mutants be selected in vivo by individuals with intermediate levels of neutralizing Abs? This might explain why antigenic evolution in humans is punctuated rather than gradual [27,28]. It might also help explain why ~30% of HA substitutions are located outside of defined antigenic sites [28], if such mutations modulate HA binding affinity. Clearly, a great deal remains to be learned about antigenic drift, including why IAV drifts much more rapidly in humans than other species, and why other human viruses with similar mutation rates and receptors (such as parainfluenza viruses) essentially are antigenically stable.
A Silver Lining to the 1976 Swine Vaccine Debacle?
In the last century, pandemic influenza has always been associated with antigenic shifts. S-IAV-2009 is a H1N1 virus, and H1N1 viruses have been circulating since their reintroduction into human population in 1977 (probably from a laboratory freezer, since the 1977 H1N1 virus was nearly identical to a 1950’s H1N1 strain). S-IAV-2009 however, might break the “shift rule”, since swine viruses are quite distinct antigenically from other H1N1 viruses. S-IAV-2009 is no exception, as at least 13 amino acid substitutions in the HA and 4 in the NA have not been observed in previous human H1N1 viruses [29]. Comparison of the S-IAV-2009 to the H1N1 component of the current vaccine (A/Brisbane/59/07) revealed only 56% and 38% sequence homology within the antigenic sites of HA and NA, respectively [29]. As vaccine strains sometimes need to be modified after only 1 or 2 HA amino acid substitutions, it is unlikely that the current vaccine will afford any protection against S-IAV-2009 infection. Indeed, ferret antisera against a 2009 seasonal H1N1 human IAV did not react with S-IAV-2009 in HA-inhibition (HI) assays [6]. Further support for S-IAV-2009’s pandemic potential comes from 1947, when a significant alteration in H1 antigenicity resulted in a complete vaccine failure that probably would have led to a pandemic had the novel virus not exhibited low pathogenicity [30].
The transmission of S-IAV-2009 in the USA may be mitigated, however, by the 1976 swine vaccination campaign that vaccinated 40,000,000 US citizens before being terminated due to the occurrence of Guillian-Barré syndrome in ~ 1 in 100,000 vaccinees (see http://www.iom.edu/CMS/AboutIOM/4081/65926.aspx to download the fascinating history The Swine Flu Affair). Plasma cells are extremely long lived, as spectacularly documented by the recovery of neutralizing Abs specific for 1918 HA from nonagenarian survivors of the epidemic [31]. It will be of great interest to correlate 1976 vaccination with the presence of anti-swine HA antibodies and protection against S-IAV-2009.
Bioinformatics: No Crystal Ball
It is very likely that due its antigenic novelty, S-IAV-2009 will become established in the human population, and that massive quantities of vaccine will be necessary for the coming flu seasons. There is every reason to believe that S-IAV-2009 will behave like every other known human IAV and begin to drift antigenically, necessitating constant vaccine reformulation. Given the enormous progress in sequencing and bioinformatics, is it possible to predict the direction of drift, and maintain a lead on generating vaccines? In a nutshell: no.
Approximately 50 amino acids contribute to HA antigenicity, and point mutations in the corresponding codons can introduce multiple potential substitutions (limited by the genetic code and also the structural flexibility of HA in maintaining function with a given substitution), leading to a daunting number of potential structures. Attempts to empirically predict drift by in vitro selection with antiserum have failed miserably [32]. The reintroduction of the 1950’s H1N1 virus in 1977 led to a unique natural experiment: how would drift compare during two independent trials in humans? The answer is not at all [33].
There is some good news, at least in the near term. First, drift in S-IAV-2009 seems to be on the languid side compared to other human IAVs [27,34]. Second, S-IAV-2009 lacks several molecular motifs associated with high transmission and pathogenesis. Unlike virtually all human isolates, S-IAV-2009 encodes Lys in place of the normal Glu at position 627 of PB2. It was recently reported that K627 is important for efficient viral transmission, by allowing the virus to replicate at lower temperatures in the upper airway [35]. Third, PB1-F2 protein, a defined pathogenesis factor in mouse models [36–38], is truncated in S-IAV-2009 to the point of functional irrelevance. Fourth, S-IAV-2009 is sensitive to the NA inhibitors oseltamivir and zanamivir.
The bad news is that S-IAV-2009 is probably just a handful of mutations (or a recombination event or two) from becoming highly pathogenic. Should this happen, mass vaccination will become an emergency, as the absence of pre-existing immunity could result in rapid and extensive transmission. The financial price, alone, of this possibility, will dwarf the costs of maximizing investments in monitoring, vaccine development, and IAV research.
References
- 1.Swine influenza A (H1N1) infection in two children--Southern California, March-April 2009. Mmwr. 2009;58(15):400–402. [PubMed] [Google Scholar]
- 2.Clark T, Stephenson I. Influenza A/H1N1 in 2009: a pandemic in evolution. Expert review of vaccines. 2009;8(7):819–822. doi: 10.1586/erv.09.54. [DOI] [PubMed] [Google Scholar]
- 3.Conenello GM, Palese P. Influenza A virus PB1-F2: A small protein with a big punch. Cell Host & Microbe. 2007;2(4):207–209. doi: 10.1016/j.chom.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Wise HM, Foeglein A, Sun J, et al. A complicated message: identification of a novel PB1-related protein translated from influenza A segment 2 mRNA. J Virol. 2009 doi: 10.1128/JVI.00826-09. JVI.00826-00809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhirnov OP, Poyarkov SV, Vorob’eva IV, Safonova OA, Malyshev NA, Klenk HD. Segment NS of influenza a virus contains an additional gene NSP in positive-sense orientation. Dokl Biochem Biophys. 2007;414(1):127–133. doi: 10.1134/s1607672907030106. [DOI] [PubMed] [Google Scholar]
- 6.Garten RJ, Davis CT, Russell CA, et al. Antigenic and Genetic Characteristics of Swine-Origin 2009 A(H1N1) Influenza Viruses Circulating in Humans. Science (New York, NY. 2009 doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith GJ, Vijaykrishna D, Bahl J, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459(7250):1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 8.Dawood FS, Jain S, Finelli L, et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. The New England journal of medicine. 2009;360(25):2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 9.Myers KP, Olsen CW, Gray GC. Cases of swine influenza in humans: a review of the literature. Clin Infect Dis. 2007;44(8):1084–1088. doi: 10.1086/512813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser C, Donnelly CA, Cauchemez S, et al. Pandemic potential of a strain of influenza A (H1N1): early findings. Science (New York, N Y. 2009;324(5934):1557–1561. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stambas J, Guillonneau C, Kedzierska K, Mintern JD, Doherty PC, La Gruta NL. Killer T cells in influenza. Pharmacology & Therapeutics. 2008;120(2):186–196. doi: 10.1016/j.pharmthera.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Seo SH, Webster RG. Cross-Reactive, Cell-Mediated Immunity and Protection of Chickens from Lethal H5N1 Influenza Virus Infection in Hong Kong Poultry Markets. J Virol. 2001;75(6):2516–2525. doi: 10.1128/JVI.75.6.2516-2525.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerhard W, Mozdzanowska K, Zharikova D. Prospects for universal influenza virus vaccine. Emerg Infect Dis. 2006;12(4):569–574. doi: 10.3201/eid1204.051020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knossow M, Skehel JJ. Variation and infectivity neutralization in influenza. Immunology. 2006;119(1):1–7. doi: 10.1111/j.1365-2567.2006.02421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekiert DC, Bhabha G, Elsliger MA, et al. Antibody recognition of a highly conserved influenza virus epitope. Science (New York, N Y. 2009;324(5924):246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sui J, Hwang WC, Perez S, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nature structural & molecular biology. 2009;16(3):265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yewdell JW, Gerhard W. Antigenic characterization of viruses by monoclonal antibodies. Annual Review of Microbiology. 1981;35:185–206. doi: 10.1146/annurev.mi.35.100181.001153. [DOI] [PubMed] [Google Scholar]
- 18.Gerhard W, Yewdell J, Frankel ME, Webster R. Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature. 1981;290(5808):713–717. doi: 10.1038/290713a0. [DOI] [PubMed] [Google Scholar]
- 19.Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31(2 Pt 1):417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 20.Green N, Alexander H, Olson A, et al. IMMUNOGENIC STRUCTURE OF THE INFLUENZA-VIRUS HEMAGGLUTININ. Cell. 1982;28(3):477–487. doi: 10.1016/0092-8674(82)90202-1. [DOI] [PubMed] [Google Scholar]
- 21.Muller GM, Shapira M, Arnon R. ANTI-INFLUENZA RESPONSE ACHIEVED BY IMMUNIZATION WITH A SYNTHETIC CONJUGATE. Proceedings of the National Academy of Sciences of the United States of America-Biological Sciences. 1982;79(2):569–573. doi: 10.1073/pnas.79.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Staudt LM, Gerhard W. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. I. Significant variation in repertoire expression between individual mice. The Journal of experimental medicine. 1983;157(2):687–704. doi: 10.1084/jem.157.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yewdell JW, Webster RG, Gerhard WU. Antigenic variation in three distinct determinants of an influenza type A haemagglutinin molecule. Nature. 1979;279(5710):246–248. doi: 10.1038/279246a0. [DOI] [PubMed] [Google Scholar]
- 24.Wilson IA, Cox NJ. Structural basis of immune recognition of influenza virus hemagglutinin. Annual review of immunology. 1990;8:737–771. doi: 10.1146/annurev.iy.08.040190.003513. [DOI] [PubMed] [Google Scholar]
- 25.Wang ML, Skehel JJ, Wiley DC. Comparative analyses of the specificities of anti-influenza hemagglutinin antibodies in human sera. Journal of virology. 1986;57(1):124–128. doi: 10.1128/jvi.57.1.124-128.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yewdell JW, Caton AJ, Gerhard W. Selection of influenza A virus adsorptive mutants by growth in the presence of a mixture of monoclonal antihemagglutinin antibodies. J Virol. 1986;57(2):623–628. doi: 10.1128/jvi.57.2.623-628.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith DJ, Lapedes AS, de Jong JC, et al. Mapping the antigenic and genetic evolution of influenza virus. Science (New York, N Y. 2004;305(5682):371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 28.Nelson MI, Simonsen L, Viboud C, et al. Stochastic processes are key determinants of short-term evolution in influenza a virus. PLoS pathogens. 2006;2(12):e125. doi: 10.1371/journal.ppat.0020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soundararajan V, Tharakaraman K, Raman R, et al. Extrapolating from sequence--the 2009 H1N1 ‘swine’ influenza virus. Nature biotechnology. 2009;27(6):510–513. doi: 10.1038/nbt0609-510. [DOI] [PubMed] [Google Scholar]
- 30.Kilbourne ED, Smith C, Brett I, Pokorny BA, Johansson B, Cox N. The total influenza vaccine failure of 1947 revisited: Major intrasubtypic antigenic change can explain failure of vaccine in a post-World War II epidemic. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(16):10748–10752. doi: 10.1073/pnas.162366899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu X, Tsibane T, McGraw PA, et al. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature. 2008;455(7212):532–536. doi: 10.1038/nature07231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fazekasd S. EVOLUTION AND HIERARCHY OF INFLUENZA VIRUSES. Archives of Environmental Health. 1970;21(3):293. doi: 10.1080/00039896.1970.10667241. [DOI] [PubMed] [Google Scholar]
- 33.Raymond FL, Caton AJ, Cox NJ, Kendal AP, Brownlee GG. The antigenicity and evolution of influenza H1 haemagglutinin, from 1950–1957 and 1977–1983: two pathways from one gene. Virology. 1986;148(2):275–287. doi: 10.1016/0042-6822(86)90325-9. [DOI] [PubMed] [Google Scholar]
- 34.Russell CA, Jones TC, Barr IG, et al. The global circulation of seasonal influenza A (H3N2) viruses. Science (New York, N Y. 2008;320(5874):340–346. doi: 10.1126/science.1154137. [DOI] [PubMed] [Google Scholar]
- 35.Van Hoeven N, Pappas C, Belser JA, et al. Human HA and polymerase subunit PB2 proteins confer transmission of an avian influenza virus through the air. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(9):3366–3371. doi: 10.1073/pnas.0813172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS pathogens. 2007;3(10):1414–1421. doi: 10.1371/journal.ppat.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zamarin D, Ortigoza MB, Palese P. Influenza A virus PB1-F2 protein contributes to viral pathogenesis in mice. Journal of virology. 2006;80(16):7976–7983. doi: 10.1128/JVI.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steel J, Lowen AC, Mubareka S, Palese P. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS pathogens. 2009;5(1):e1000252. doi: 10.1371/journal.ppat.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]