Abstract
Somites are segmental units of the mesoderm in vertebrate embryos that give rise to the axial skeleton, muscle and dermis. Somitogenesis occurs in a periodic manner and is governed by a segmentation clock that causes cells to undergo repeated oscillations of gene expression. Here we present a detailed analysis of cis-regulatory elements that control oscillating expression of the zebrafish her1 gene in the anterior presomitic mesoderm. We identify binding sites for Her proteins and demonstrate that they are necessary for transcriptional repression. This result confirms that direct negative autoregulation of her gene expression constitutes part of the oscillator mechanism. We also characterize binding sites for fused somites/Tbx24 and Suppressor of Hairless proteins and show that they are required for activation of her1 expression. These data provide the foundation for a precise description of the regulatory grammar that defines oscillating gene expression in the zebrafish segmentation clock.
Keywords: somitogenesis, oscillation, her1, Tbx24, fused somites, Su(H), her12
INTRODUCTION
In vertebrates, the segmental structure of the vertebral column is prefigured by the metameric organization of the paraxial mesoderm in the embryo. This tissue forms as two columns flanking the midline (the notochord and neural tube) and is subdivided into a series of reiterated units known as somites. Somitogenesis is a sequential process with new segments forming from a region of unsegmented presomitic mesoderm (PSM) in the tail of the embryo. Groups of cells periodically bud off from the anterior end of the PSM to form a somite, while new cells are added to the PSM at the posterior end as a result of growth of the embryo. Thus, somitogenesis occurs in an anterior to posterior procession that reflects the overall progression of embryonic development. The total number of somites and the rate of somitogenesis are species-specific characteristics. For instance, the zebrafish forms 31 somites at the rate of one approximately every 23 minutes (Kimmel et al., 1995; Schröter et al., 2008), while somitogenesis occurs with a period of two hours in the mouse embryo, generating a total of 65 somites (Tam, 1981). Thus, the frequency of somite formation must be strictly regulated and coupled to overall growth of the embryo to ensure reproducibility in somite size and number.
Over the last 12 years, a substantial amount of molecular evidence has accumulated in support of the “clock and wavefront” model of somitogenesis, which was first proposed more than 30 years ago (Cooke and Zeeman, 1976). According to this model, the frequency of somite formation is defined by the period of the clock, which causes cells in the PSM to undergo repeated oscillations of gene expression. Cells are only competent to form a somite during a particular phase of oscillation. The position of a new somite boundary is determined by the wavefront, which moves posteriorly in association with growth of the embryo. A somite forms only when cells in the anterior PSM that have escaped the wavefront also experience the appropriate phase of the clock. Thus, the size of a somite is determined by the distance traveled by the wavefront during one cycle of the clock.
The Fgf and Wnt signaling pathways are important components of the wavefront. For both pathways, a gradient of activity exists across the PSM, with signaling highest in the posterior and absent in the anterior where segmentation occurs (Dubrulle et al., 2001; Sawada et al., 2001; Dubrulle and Pourquié, 2004; Aulehla et al., 2008; Dunty et al., 2008). Notch signaling has an important function in the segmentation clock and oscillating expression has been documented for several components and targets of the Notch pathway. This oscillation is manifest as reiterated waves of expression, which sweep through the PSM from posterior to anterior. Components of the Notch pathway expressed in this manner include deltaC in zebrafish and Lunatic fringe (Lfng) in mouse and chick (Forsberg et al., 1998; McGrew et al., 1998; Jiang et al., 2000). These genes encode a Notch ligand and a glycosyltransferase that modifies Notch activity, respectively. Particularly prominent among the targets of Notch signaling involved in the segmentation clock are members of the Hairy/Enhancer of Split-related family (abbreviated as Hes in mammals, her in zebrafish). Genes of this type showing oscillating expression are Hes1, Hes5, Hes7 and Hey2 in the mouse, cHairy1, cHairy2 and cHey2 in the chick and her1, her7, her11, her12 and her15 in zebrafish (Palmeirim et al., 1997; Holley et al., 2000; Jouve et al., 2000; Leimeister et al., 2000; Sawada et al., 2000; Bessho et al., 2001; Dunwoodie et al., 2002; Henry et al., 2002; Oates and Ho, 2002; Sieger et al., 2004; Shankaran et al., 2007). These genes encode proteins of the basic-Helix-Loop-Helix (bHLH) family that are thought to function as transcriptional repressors (Fischer and Gessler, 2007).
In zebrafish, a negative feedback loop involving Notch signaling and the her genes is thought to be the core mechanism of the oscillator (Holley et al., 2002; Oates and Ho, 2002; Lewis, 2003). Activation of the Notch receptor by ligand binding leads to cleavage of the Notch intracellular domain (NICD), which translocates to the nucleus and interacts with a DNA binding protein of the CSL family (for CBF-1/RBPJ-k in mammals, Suppressor of Hairless (Su(H) in flies, Lag-1 in nematodes) (Bray, 2006). CSL binds to specific sites in the cis-regulatory regions of targets, presumably including the her genes, and the CSL/NICD complex recruits further cofactors to activate transcription (Bray, 2006). As Her proteins subsequently accumulate, it is supposed that they bind directly to regulatory elements of their own genes and repress transcription. The Her proteins have short half-lives (Giudicelli et al., 2007) and are rapidly degraded, therefore transcriptional repression is quickly relieved and a new cycle of expression can be initiated in response to Notch signaling. In the mouse, transcriptional repression by Hes proteins is also likely to be an important component of the oscillator mechanism (Bessho et al., 2003). However, the situation is substantially more complex as components of the Wnt and Fgf signaling pathways also show cyclical expression (Aulehla et al., 2003; Ishikawa et al., 2004; Dale et al., 2006; Dequeant et al., 2006; Niwa et al., 2007). It is not yet clear how oscillations in these pathways interact with each other and with Notch signaling within the segmentation clock.
In order to obtain a more fundamental understanding of the mechanism of the segmentation clock (and to refine mathematical models of somitogenesis) it will be necessary to acquire detailed information on the cis-regulatory elements of oscillating genes. Specifically, it will be crucial to know how the number and arrangement of binding sites for activators and repressors of transcription defines the proper oscillation of gene expression. However, very little is currently known about this regulatory grammar in clock genes.
In vitro studies have identified binding sites for the mouse Hes7 protein in the promoter regions of Hes7 and Lfng (Chen et al., 2005). Moreover, chromatin immunoprecipitation (ChIP) from mouse PSM extracts demonstrated that Hes7 is bound to DNA upstream of the Hes7 and Lfng genes in vivo (Bessho et al., 2003).Therefore, it seems likely that Hes7 is directly regulating transcription of both genes via the motifs identified by Chen et al. (Chen et al., 2005). However, the role of specific binding sites has not yet been confirmed in vivo. A 2.5kb DNA fragment immediately upstream of the mouse Hes1 gene is sufficient to drive oscillating expression of a luciferase reporter in transgenic animals (Masamizu et al., 2006). Indeed, this construct was used to visualize real-time oscillations in live embryos. Four Hes1 binding sites have been identified close to the Hes1 transcription start site and three of them are required for repression of promoter activity by cotransfected Hes1 in cultured cells (Takebayashi et al., 1994). Furthermore, two CSL binding sites in the Hes1 promoter are required for the induction of reporter expression by Notch signaling in transfection experiments (Jarriault et al., 1995). However, the function of neither the Hes nor the CSL binding sites have yet been determined in respect to Hes1 expression in the segmentation clock.
Analysis of reporter gene expression in transgenic mice demonstrated that as little as 2.3kb of 5′ flanking sequence of the mouse Lfng gene is sufficient to recapitulate the complete cyclic expression pattern (Cole et al., 2002; Morales et al., 2002). A similar fragment (only 2kb) has also been used to image oscillating gene expression in real-time in live embryos (Aulehla et al., 2008). Within the Lfng upstream region are several short elements that are highly conserved between the mouse and human sequences. One of these, referred to as Fringe Clock Element 1 (FCE1) (Cole et al., 2002) or block A (Morales et al., 2002), is necessary for oscillating expression in the posterior PSM. Moreover, deletion of FCE1 from the genomic locus leads to the complete absence of Lfng expression in the posterior PSM and causes significant segmentation defects in the axial skeleton (Shifley et al., 2008). Several putative protein binding sites were identified within FCE1, including two possible targets for bHLH proteins (Cole et al., 2002). Mutation or deletion of these sites completely eliminated expression of reporter genes in the posterior PSM of transgenic embryos. However, binding of specific proteins to these sites has not yet been established and these motifs were not among the Hes7 binding sites identified in vitro (Chen et al., 2005). FCE1 also contains a single consensus binding site for CSL proteins and a second such motif is present in another of the conserved regions, block B (Morales et al., 2002). Mutation of both sites within one construct caused a dramatic reduction in gene expression in the posterior PSM, although the pattern was apparently still cyclical. Subsequently, CSL binding to the site within FCE1/block A was confirmed by an in vitro binding assay (Morales et al., 2002). Currently, this finding represents the only example of a confirmed binding site for a specific protein that has been shown to be important for the regulation of oscillating gene expression in the segmentation clock.
In zebrafish, cis-regulatory elements have only been described for one gene with oscillating expression in the PSM, her1 (Gajewski et al., 2003). The full cyclical pattern of the endogenous gene was recapitulated in transgenic zebrafish using a GFP reporter driven by the 8.6kb of DNA immediately upstream of her1. A smaller 3.3kb fragment did not direct expression in the most posterior region of the PSM but did produce a dynamic pattern of two or three stripes in the intermediate to anterior PSM. However, a further deletion down to −2.8kb led to reporter constructs that displayed only weak expression over a broad region of the PSM with no evidence of oscillation (Gajewski et al., 2003). These experiments defined the 3.3-2.8kb region as essential for her1 expression in the anterior PSM. We therefore refer to this fragment as the Anterior Stripe Element (ASE).
Here, we report a detailed analysis of the ASE. We identify two binding sites for Her proteins and show that they are required for transcriptional repression of her1. We also demonstrate that the ASE contains a single binding site for T-box proteins and that this motif is necessary for expression in the anterior PSM. Furthermore, we show that there are no binding sites for the zebrafish CSL proteins (Su(H)1 and Su(H)2) within the ASE but identify two sites elsewhere within the 3.3kb enhancer that are essential for her1 expression. These experiments provide evidence for direct negative autoregulation of her genes in vivo and confirm that her1 is a direct target of Notch signaling, both important features of the prevailing models of the segmentation clock.
RESULTS
Identification of protein binding sites within the Anterior Stripe Element (ASE) of her1
In order to precisely define the ASE for our investigation, we mapped the primers previously used to generate transgenic constructs (Gajewski et al., 2003) to the genomic sequence. This determined that the exact limits of the ASE are −3282bp and −2879bp, relative to the her1 initiation codon, and are therefore correctly rounded to −3.3kb and −2.9kb (Fig.1A). In our analysis of the ASE, we focused on particular candidate transcription factors that are likely to be direct regulators of her1 expression, based on genetic evidence and their expression patterns. Her proteins, including Her1 itself, are thought to be transcriptional repressors and several members of this family are expressed in the zebrafish PSM. Furthermore, knockdown of these her genes generally affects the expression of oscillating genes in the segmentation clock, including her1 (Holley, 2007). In our binding assays we have used Her1, because autoregulation is thought to be an important feature of the segmentation clock, and Her12 because knockdown of this gene produces a strong perturbation of somitogenesis (Shankaran et al., 2007). Tbx24 is a T-box transcription factor expressed in the anterior PSM and disrupted by the fused somites (fss) mutation (Nikaido et al., 2002). The most anterior stripe of oscillating her1 expression is lost in fss embryos (van Eeden et al., 1998; Holley et al., 2000). Finally, her genes are targets of the Notch signaling pathway, the activity of which is modulated by the CSL proteins (Bray, 2006). There are two paralogous members of this family in zebrafish, Su(H)1 and Su(H)2. These proteins are partially redundant and morpholino-mediated knockdown of either disrupts her1 expression (Sieger et al., 2003; Echeverri and Oates, 2007). We synthesized each of the candidate proteins using an in vitro translation system and tested their ability to bind to oligonucleotide probes derived from the ASE sequence in electrophoretic mobility shift assays (EMSA). The probes represented putative binding sites for the candidate proteins that were identified by visual inspection of the ASE sequence (Fig.1B).
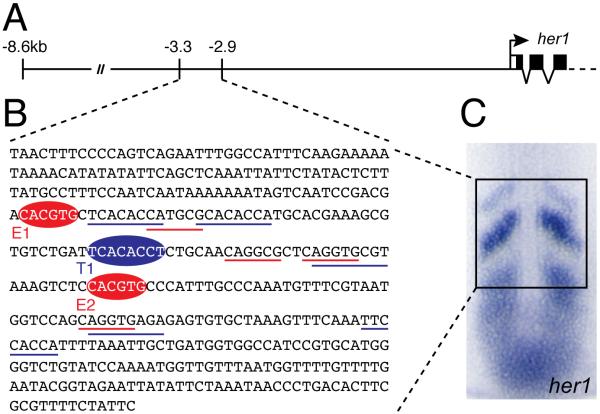
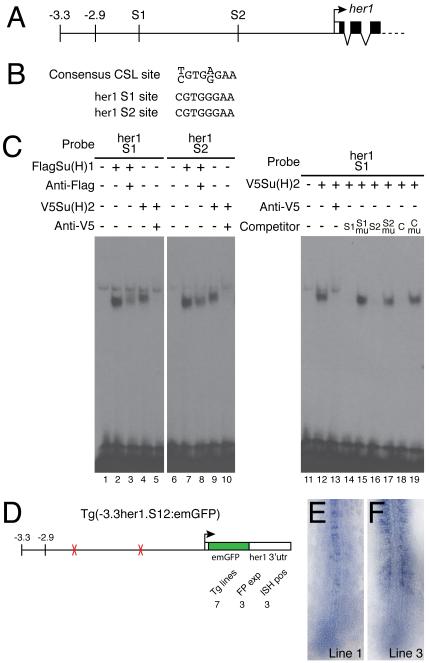
Fig.1. The Anterior Stripe Element (ASE) of the zebrafish her1 gene.
(A) Structure of the her1 genomic locus indicating the limits of known cis-regulatory regions. The ASE is located between 3.3kb and 2.9kb upstream of the her1 coding sequence. (B) Complete sequence of the ASE. Binding sites for Her proteins (E1 and E2) and Tbx proteins (T1) identified in this study are indicated by red and blue ellipses, respectively. Underlining indicates putative sites for Her proteins (red) and Tbx proteins (blue) which were tested but did not bind in our assay. (C) In situ hybridization of a 10 somite stage zebrafish embryo showing expression of her1 in the PSM. The subset of the expression pattern that is controlled by the ASE is boxed.
Her proteins are members of the basic-helix-loop-helix (bHLH) family, which generally bind to DNA motifs known as E-boxes (CANNTG) or N-boxes (CACNAG) (Fischer and Gessler, 2007). More specifically, members of the Hairy/Enhancer of Split class recognize class B or class C E-boxes, with the consensus sequence CACGYG. Both Her1 and Her12 bound a control probe containing a CACGTG site and showed a greater affinity for this sequence over other variations of the E-box or N-box (Fig.2 and data not shown). Consequently, no protein binding was observed with several variant E-boxes (CAGGTG, CAGGCG and CATGCG) located in the ASE (Fig.1B and data not shown). In contrast, two palindromic sites (CACGTG; Fig.1B) were specifically bound by Her12 or Her1 (Fig.2). bHLH proteins bind DNA as dimers, with each monomer making specific base contacts with nucleotides 1 and 2 of the binding site (i.e. CACGTG) on opposing strands (Massari and Murre, 2000). Therefore, we modified four nucleotides in our motifs (GTCGAC) and showed that Her protein binding is completely abolished, using the mutated forms of the control probe and the ASE sites E1 and E2 (Fig. 2).
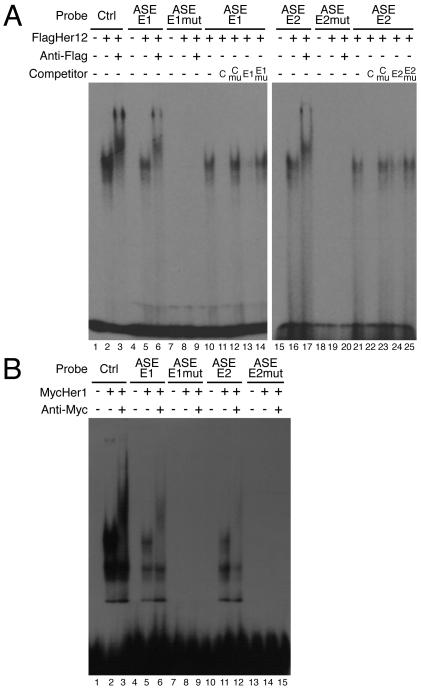
Fig.2. Identification of Her protein binding sites within the ASE.
(A) Two Her protein binding sites were identified in the ASE (lanes 4-6 and 15-17). Control reactions contained unprogrammed lysate (lanes 1, 4 and 15). FlagHer12 forms a single complex with the control E-box, ASE-E1 or ASE-E2 probes (lanes 2, 5 and 16). Specificity of binding was confirmed by supershifting complexes with an anti-Flag antibody (lanes 3, 6 and 17). Mutation of the E-box in the ASE-E1 or ASE-E2 probe abolished Her12 binding (lanes 7-9 and 18-20). Sequence specific binding was confirmed by competition assays (lanes 10-14). Reciprocal competition was observed between the control and ASE-E1 oligonucleotides (lanes 11 and 13). Oligonucleotides containing mutated E-boxes were unable to compete (lanes 12 and 14). An equivalent analysis was performed for the E2 site (lanes 15-25). Unlabelled competitors were added at 100 fold molar excess. (B) Her1 also binds to the E1 and E2 sites. Control reactions contained unprogrammed lysate (lanes 1, 4, 7, 10 and 13). Proteins from MycHer1 synthesis reactions formed three complexes with the control E-box, ASE-E1 or ASE-E2 probes (lanes 2, 5 and 11). However, only one of these complexes could be supershifted using an anti-Myc antibody (lanes 3, 6 and 12) indicating that the other bands are likely experimental artifacts. Mutation of the E-box in the ASE-E1 or ASE-E2 probe eliminated Her1 binding (lanes 7-9 and 13-15).
Although the target sequence has not yet been determined for Tbx24 specifically, binding site selection experiments with several other T-box proteins suggest that all members of this family recognize the consensus sequence TCACACCT (Naiche et al., 2005). Consistent with this data, Tbx24 bound a control probe containing this sequence in EMSA experiments (Fig.3). Note that because full-length Tbx24 was almost impossible to synthesize in vitro under our conditions, we used a truncated version, Tbx24N. This protein consists of the N-terminal 405 amino acids (compared to 874 in the full-length protein) and includes the complete T-box DNA binding domain (residues 62 to 247). EMSA experiments confirmed that this truncated version of Tbx24 was able to bind a control probe containing a proven binding site for T-box proteins (Fig.3) (Goering et al., 2003). We identified five motifs within the ASE that are close matches for the T-box consensus but none were able to bind Tbx24N (Fig.1B and data not shown). However, there is one additional site that perfectly matches the consensus (Fig.1B) and a probe containing this site did specifically bind Tbx24N (Fig.3). A single nucleotide change (TCAGACCT) in the T-box site of the control probe is sufficient to abolish Tbx24N binding (Fig.3), as reported for other T-box proteins (Conlon et al., 2001; Goering et al., 2003). Introducing the equivalent mutation into the T1 probe from the ASE also completely abrogated protein binding (Fig.3).
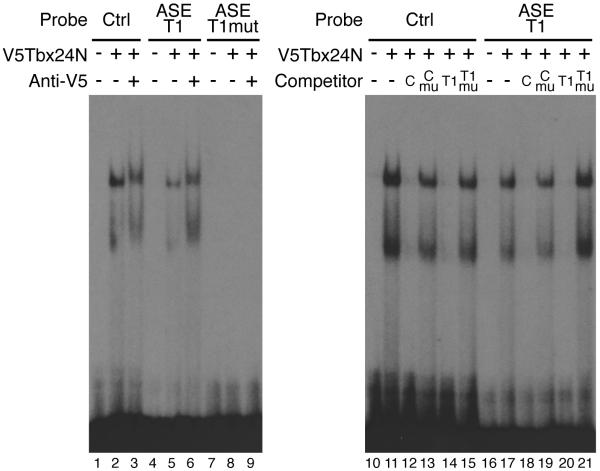
Fig.3. Identification of a Tbx binding site within the ASE.
A single Tbx24 binding site in the ASE was confirmed by EMSA (lanes 4-6). Control reactions contained unprogrammed lysate (lanes 1, 4 and 7). Tbx24N formed two complexes with the control T-box site and ASE-T1 probe (lanes 2 and 5). The faster migrating band is likely due to a truncated protein product in the in vitro translation reaction (data not shown). Specificity of binding was confirmed by supershifting complexes with an antibody to the V5 epitope (lanes 3 and 6). Mutation of the T-box in the ASE-T1 sequence abolished Tbx24N binding (lanes 7-9). Binding specificity was further confirmed by competition assays (lanes 10-21). Reciprocal competition was observed between the control and ASE-T1 oligonucleotides (lanes 12, 14, 18 and 20). Oligonucleotides containing mutated T-box sites were unable to compete (lanes 13, 15, 19 and 21). Unlabelled competitors were added at 100 fold molar excess.
CSL proteins recognize a consensus binding site of YGTGRGAA, which was largely determined by experiments in Drosophila (Nellesen et al., 1999). Although CSL can bind single sites, a common target sequence is the SPS motif, in which two sites are arranged head-to-head and separated by 16-17bp (Nam et al., 2007). Surprisingly, we were unable to identify any bona fide Su(H) binding sites within the ASE despite testing five putative sequences, including an SPS motif previously predicted by bioinformatics (Gajewski and Voolstra, 2002). Lack of binding was not due to technical shortcomings of our assay as we were able to identify Su(H) binding sites in other regions of the her1 upstream sequence (see below). Therefore, it seems likely that the Notch signaling pathway does not directly regulate her1 expression through the ASE.
In summary, our in vitro analysis of the ASE identified two binding sites for Her proteins (E1 and E2) and one for Tbx24 (T1) (Fig.1B). Moreover, we determined specific nucleotide changes that abolish protein binding to each site. In order to test the function of these motifs in the regulation of her1 expression in vivo, we introduced the appropriate mutations into reporter constructs, which were then used to generate transgenic fish.
Activity of the 3.3kb her1 enhancer
The constructs previously used to determine the activity of the 3.3kb region of her1 did not include any her1 coding sequence or the 3′ untranslated region (UTR) (Gajewski et al., 2003). We wanted to replicate this design as closely as possible and, therefore, initially created constructs in which the 3.3kb region of her1 was coupled to the emerald variant of GFP (emGFP) and the SV40 polyadenylation (SV40 polyA) signal. However, in situ hybridization of embryos from four transgenic lines carrying this construct revealed that emGFP transcripts were present in a broad pattern in the anterior PSM, not the expected stripes, and were also maintained throughout the somites (data not shown). The most likely explanation for this pattern is that the SV40 polyA sequence causes excessive stabilization of transcripts, masking the oscillating expression pattern in the PSM and leading to retention of emGFP mRNA in cells well after they have been incorporated into somites. We note that Gajewski et al. (Gajewski et al., 2003) used a transcriptional terminator sequence from the Drosophila hsp70 gene. This sequence obviously did not significantly stabilize transcripts and allowed the authors to detect dynamic expression in the PSM. In order to resolve this issue, we exchanged the SV40 polyA sequence in our constructs for the her1 3′UTR and successfully restricted transgene transcripts to the anterior PSM (see below). This basic design was then used for all reporter constructs, although all modifications of the cis-regulatory sequence were performed in the context of a mCherry reporter instead of emGFP (Fig.4A).
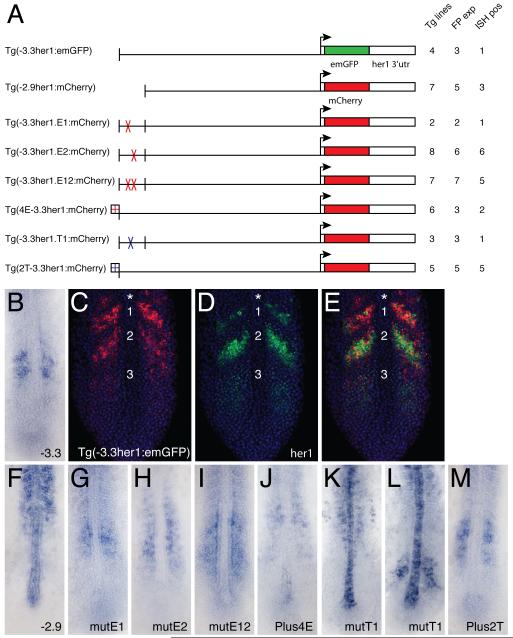
Fig.4. Transgenic analysis of the function of protein binding sites in the ASE.
(A) Schematic representation of the transgenic constructs used to analyze the function of protein binding sites within the ASE. For each construct: Tg lines = the total number of transgenic lines identified, including those only detected by PCR; FP exp = the number of transgenic lines in which expression was observable by fluorescence; ISH pos = the number of transgenic lines that exhibited expression that was detectable by in situ hybridization. (B) Standard in situ hybridization for emGFP mRNA in a transgenic embryo carrying the (−3.3her1:emGFP) construct. (C-E) Double fluorescent in situ hybridization of a Tg(−3.3her1:emGFP) embryo, showing either emGFP (C, red) or her1 (D, green) expression, or both (E). Nuclei stained with propidium iodide are colored blue. The same confocal section is shown in all panels. Numbers indicate specific stripes of gene expression as referenced in the text. Asterisk indicates the additional more anterior stripe seen in transgenic embryos. (F-M) Standard in situ hybridizations showing reporter gene (emGFP or mCherry) expression in transgenic embryos carrying the indicated constructs.
Fluorescent proteins expressed under the control of her1 cis-regulatory elements do not provide an accurate reflection of oscillating expression due to their inherent stability ((Gajewski et al., 2003) and our data not shown). Therefore, fluorescence served as a useful means of screening for transgenesis but analysis of expression was performed by in situ hybridization. However, fluorescence also proved to be a more sensitive assay of expression levels. For almost all of our constructs we obtained transgenic lines for which we observed specific expression patterns by fluorescence but could not detect any signal by in situ hybridization (Fig.4A).
Although we identified three lines for the control construct Tg(−3.3her1:emGFP) by fluorescence, only one of these had a level of mRNA expression that was detectable by in situ hybridization (Fig.4A). As expected, expression was restricted to stripes in the anterior PSM (Fig.4B) in a pattern very similar to that previously attributed to the −3.3kb enhancer (Gajewski et al., 2003). However, we were never able to detect the expected third, most posterior stripe by the standard in situ hybridization method, and the two anterior stripes were generally more poorly defined than previously reported. Fluorescent in situ hybridization resolved the expression pattern of Tg(−3.3her1:emGFP) in greater detail, revealing that there is actually weak expression in the region of the most posterior stripe but confirming that all the stripes are poorly defined (Fig.4C).
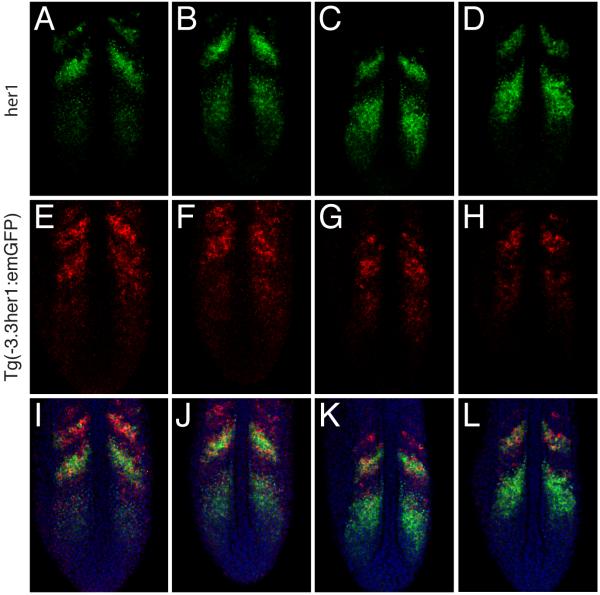
Double in situ hybridization was previously used to demonstrate that the expression pattern driven by the −8.6kb her1 fragment is virtually identical to that of the endogenous gene (Gajewski et al., 2003). However, a similar analysis was not reported for the −3.3kb enhancer. Therefore, we performed double fluorescent in situ hybridization for her1 and emGFP expression in Tg(−3.3her1:emGFP) embryos (Fig.4C-E). This analysis demonstrated that although Tg(−3.3her1:emGFP) expression substantially overlaps with her1, the patterns are not identical. Most notably, the relative intensity of different stripes varies considerably. For Tg(−3.3her1:emGFP), stripe one is more intense than stripe two, while the opposite is true for her1. Moreover, remnants of an even more anterior stripe are detected for Tg(−3.3her1:emGFP) in a region where her1 expression is absent (Fig.4C-E). The expression of endogenous her1 was used to arrange embryos in a series that represents different phases in the progression of the somitogenesis cycle (Fig.5). Comparison of Tg(−3.3her1:emGFP) expression in the same embryos indicated that in the anterior PSM, downregulation of the transgene seems to lag behind that of the endogenous gene leading to the apparent differences in stripe intensity.
Fig.5. The −3.3 kb enhancer does not accurately recapitulate her1 expression in the anterior PSM.
Double fluorescent in situ hybridization of four Tg(−3.3her1:emGFP) embryos. Three images of a single confocal section are shown for each embryo, (A,E,I), (B,F,J), (C,G,K) and (D,H,L), respectively. (A-D) Expression of her1. Embryos are arranged to indicate progression of waves of her1 transcription through the somitogenesis cycle. (E-H) Expression of emGFP mRNA. (I-L) Merged images showing expression of her1 (green) and emGFP (red). Nuclei stained with propidium iodide are colored blue.
Overall, our results suggest that, although Tg(−3.3her1:emGFP) expression resembles that of the endogenous her1 gene in the anterior PSM, it is not as precise and proper temporal control of oscillation is lacking. As the −8.6kb fragment appears to perfectly recapitulate her1 expression, elements in the region between −8.6kb and −3.3kb must be required in conjunction with the ASE to coordinate proper cyclical expression in the anterior PSM.
Activity of the Anterior Stripe Element
Although our data suggest that the ASE is not sufficient to fully recapitulate her1 expression in the anterior PSM, it has been shown to be necessary (Gajewski et al., 2003). We also generated a construct (Tg(−2.9her1:mCherry)) in which reporter gene expression was driven by the −2.9kb her1 fragment (Fig.4A). This construct was intended to serve as a negative control for other constructs containing mutated binding sites by confirming the expression pattern associated with complete absence of the ASE. The 2.9kb fragment was previously reported to direct only weak, diffuse expression in the PSM (Gajewski et al., 2003). In contrast, we consistently saw a complete lack of expression in the PSM, coupled with ectopic transcription in the somites (Fig.4F). The reason for this discrepancy with previous data is not immediately obvious as our construct differs only in the fluorescent protein reporter gene used and the inclusion of the her1 3′utr. We cannot exclude the possibility that elements within the 3′utr drive the ectopic somitic expression because, as described above, constructs that contain the SV40 polyA instead generate excessively stable transcripts that persist throughout the somites anyway (data not shown). Nevertheless, our results suggest that the ASE is not simply necessary for expression of her1 in the PSM but is also required to repress inappropriate transcription within the somites.
The particular transgenic line shown in Fig.4F also displayed significant ectopic expression in the notochord. This is not specific to the Tg(−2.9her1:mCherry) construct as we recovered at least one line for each of our constructs in which the expression pattern included the notochord (for example, Fig.4K,L). Ectopic notochord expression was previously described for transgenic reporters containing the complete 8.6kb her1 enhancer but not the 3.3kb fragment, and attributed to a cryptic notochord enhancer in the 8.6 to 3.3kb region (Gajewski et al., 2003). The notochord expression in our constructs cannot be attributed to an additional enhancer in the her1 3′utr as we also saw this pattern frequently with transgenic reporter constructs that contained the SV40 polyA signal in place of the her1 sequence (data not shown). Therefore, our experiments indicate that the 3.3kb enhancer of her1 does contain sequence capable of directing expression in the notochord and another region at the genomic locus (outside of the −8.6kb fragment) must be responsible for repressing this activity during normal zebrafish development.
Her protein binding sites mediate transcriptional repression of her1 in vivo
Although our results indicate that the −3.3her1:emGFP construct does not fully recapitulate her1 expression in the anterior PSM, it is clear that the ASE is necessary for expression in this region. Therefore, we were still able to use this reporter construct as a context for analyzing the function of the protein binding sites that we identified in vitro. Specific nucleotide changes that abrogate Her protein binding (Fig.2) were introduced in order to generate reporter constructs in which either the E1, E2 or both (E12) binding sites in the ASE were disrupted (Fig.4A). These constructs were then used to generate stable transgenic lines. As these are randomly integrated transgenes, the level of expression obviously depends on both the number of copies and the integration sites. However, as these are reporter constructs, the mutations should only affect the readout from the transgene and should not disrupt the segmentation clock in general. Thus, any effect of the mutations provides a direct measure of the role of these binding sites in her1 regulation and is not complicated by any indirect effect via clock components.
Mutation of the E1 binding site did not have a dramatic effect (Fig.4G). Expression in Tg(−3.3her1.E1:mCherry) embryos was still restricted to the anterior PSM and was not substantially different from that seen with the control construct (Fig.4B), although it could be slightly more disorganized. Mutation of the E2 binding site caused a more pronounced change (Fig.4H). Tg(−3.3her1.E2:mCherry) embryos displayed a splotchy, disorganized pattern with no evidence of stripes. Moreover, the domain of expression was slightly expanded with positive cells detected in more anterior positions than in embryos transgenic for the parent construct (Fig. 4B). In contrast, combined mutation of both E-boxes had a much more dramatic effect (Fig.4I). Relative to the control construct (Fig.4B), expression in Tg(−3.3her1.E12:mCherry) embryos was expanded throughout the intermediate to anterior PSM in an apparently even distribution with no evidence of stripes. Her proteins are thought to function as repressors and the observed pattern is consistent with a failure to repress transcription during appropriate phases of oscillation. Interestingly, the expression pattern in Tg(−3.3her1.E12:mCherry) embryos is very similar to that seen when her1 or her7 function was knocked down in transgenic fish carrying a construct with the unmodified −3.3kb enhancer (Gajewski et al., 2003). This result suggests that the binding sites we have identified are genuine targets for Her protein binding in vivo.
To further investigate the role of Her protein binding sites in the regulation of her1 expression, we determined the effects of increasing the number of Her target sequences. A DNA fragment containing four CACGTG sites, spaced at 10bp intervals, was added at the 5′ end of the −3.3kb her1 enhancer (Fig.4A). Four sites were chosen so that, in combination with the two present in the ASE, the construct was effectively 3x in respect to Her target sites. Injection of this construct, yielded an unusually high proportion (3/6) of non-expressing transgenic lines, suggesting that transcriptional activation is generally downregulated (Fig.4A). Moreover, in those lines with detectable expression, the in situ hybridization signal was very weak and mCherry mRNA was almost completely absent from the PSM, with only a few scattered cells detected in the most anterior unsegmented region (Fig.4J). Combined with the results of the binding site mutations, these data indicate an important role for E-box motifs in mediating transcriptional repression of her1 in the segmentation clock, likely through specific binding of Her proteins.
T-box protein binding sites mediate activation of her1 expression
In addition to the Her binding sites, our in vitro analysis identified one Tbx protein target site (T1) within the ASE (Figs.1, 3). A single nucleotide change that abolishes Tbx24 binding to the T1 site (Fig.3) was introduced into a reporter construct that was subsequently used to generate transgenic zebrafish (Fig.4A). Tbx24 is thought to be an activator of her1 expression (Nikaido et al., 2002), so mutation of the T1 binding site might be expected to abolish expression of the reporter gene. However, all the transgenic lines that we identified by PCR also showed specific patterns of expression by fluorescence, although only one of these lines had a level of expression that was detectable by in situ hybridization. These Tg(−3.3her1.T1:mCherry) embryos displayed ectopic expression in the notochord plus some splotchy expression in the somites. As noted above for the Tg(−2.9her1:mCherry) construct, the notochord is a frequent site of ectopic expression and therefore this is unlikely to be a specific effect of mutating the Tbx site. The additional expression in the somites is reminiscent of the effect of deleting the entire ASE (Fig.4F) and could imply a role for Tbx proteins in repressing transcription of her1 in this region. However, additional transgenic lines will be required to confirm or refute that hypothesis. Strikingly, no expression was detected in the anterior PSM of Tg(−3.3her1.T1:mCherry) embryos (Fig.4K,L). However, a proportion of the embryos (15/38) showed additional patches of expression in central regions of the PSM (Fig.4L). It seems likely that this represents the most posterior stripe (stripe 3) driven by the 3.3kb enhancer and that the variable appearance of this band in Tg(−3.3her1.T1:mCherry) embryos reflects the dynamic expression pattern. Nevertheless, it is clear that the T-box binding site in the ASE is required for activation of her1 expression in the anterior PSM.
As we had done for Her binding sites, we sought to gain further insight into the regulation of her1 by Tbx proteins by increasing the number of available binding motifs. Employing the same rationale, we introduced two extra T-box sites so that, in addition to the one present in the ASE, the construct was 3x in respect to T-box protein targets. Therefore, a DNA fragment containing two TCACACCT sites, 10bp apart, was cloned upstream of the −3.3her1:mCherry reporter (Fig4.A). Tg(2T-3.3her1:mCherry) embryos showed only a modest increase in expression relative to the control construct (Fig.4B,M). Furthermore, although expression is expanded to a larger area of the PSM, some evidence of a striped pattern remains, indicating that the normal cyclical expression has not been completely over-ridden. This result suggests that these two extra T-box sites are not sufficient to mediate robust upregulation of the transgene.
Su(H) binding sites located outside the ASE are necessary for her1 expression in the PSM
As noted above, we were surprised by the lack of Su(H) binding sites in the ASE because her1 is known to be a target of the Notch signaling pathway. Therefore, we searched the rest of the −3.3kb fragment for potential Su(H) target sites and identified two motifs (S1 and S2) that perfectly matched the consensus CSL sequence (Fig.6A, B). Both of these were individual sites, rather than SPS motifs, and were located 2337bp (S1) and 1155bp (S2) upstream of the her1 initiation codon. EMSA experiments confirmed that both sites would bind either zebrafish Su(H) paralogue (Fig.6C). The specificity of binding was confirmed by competition assays. An oligonucleotide containing an SPS motif from the mouse Hes1 gene (Jarriault et al., 1995; Nam et al., 2007) effectively competed for Su(H) binding to either the S1 or S2 site (Fig.6C and data not shown). Furthermore, specific nucleotide changes that have been shown to prevent CSL binding (Nam et al., 2007), completely abolished the ability of the Hes1 SPS oligonucleotide to compete. Similarly, removing the putative 8bp Su(H) sites from the S1 and S2 probes renders them unable to compete for protein binding (Fig.6C). Data is shown for the S1 probe with Su(H)2 protein but identical results were obtained with both probes using either zebrafish Su(H) paralogue (data not shown).
Fig.6. Su(H) binding sites within the −2.9kb region are necessary for her1 expression.
(A) Representation of part of the her1 genomic locus indicating the position of the two Su(H) binding sites (S1 and S2). (B) The S1 and S2 sequences are perfect matches for the consensus CSL binding site. (C) EMSA experiments confirm that the S1 and S2 sites bind both Su(H)1 and Su(H)2 (lanes 1-10). Control reactions contained unprogrammed lysate (lanes 1 and 6). A weak non-specific complex is seen in every lane. Specificity of binding was confirmed by supershifting or blocking complexes with antibodies to the appropriate epitope tag (lanes 3, 5, 8 and 10). Recognition of specific sequences was confirmed by competition assays (lanes 11-19). Unlabeled competitor oligonucleotides were added at 100-fold molar excess. C indicates the control oligonucleotide including the SPS motif from the mouse Hes1 gene; mu indicates competitors in which the CSL site was deleted (lanes 15 and 17) or mutated (lane 19). (D) Schematic representation of the construct used to analyze the function of the two Su(H) binding sites. The total number of transgenic lines are indicated (Tg lines), as are the number of lines in which expression was detected by fluorescence (FP exp) and in situ hybridization (ISH pos). (E,F) In situ hybridization of Tg(-3.3her1.S12:emGFP) embryos from two different lines showing expression of emGFP mRNA.
We engineered two 8bp deletions in our basic reporter construct that removed both of the Su(H) binding sites and generated transgenic fish carrying the modified construct (Fig.6D). An unusually high proportion (4/7) of the lines that we recovered showed no fluorescence and could be identified by PCR only (Fig.6D), indicating that deletion of the Su(H) binding sites leads to a substantial downregulation of reporter gene activity. Moreover, of the three lines in which expression was detectable by in situ hybridization, two showed no expression of emGFP mRNA in the PSM (Fig.6E). In the one line that exhibited PSM expression, the pattern was clearly abnormal with only a few scattered positive cells and no evidence of stripes (Fig.6F). Thus, even at the maximum level of expression observed with this construct, oscillating expression in the anterior PSM is severely disrupted. Therefore, our data demonstrates that Su(H) sites located outside the ASE are required for normal activation of her1 transcription in the anterior PSM.
DISCUSSION
In the present study, we have defined binding sites for Her proteins that are essential in vivo for oscillating gene expression in the segmentation clock. These sites are important for transcriptional repression, thus confirming an important component of the oscillator model: direct negative autoregulation of her gene expression. Mathematical modeling predicts that the number of binding sites for Her/Hes repressors will be an important influence on the behavior of the oscillator (Zeiser et al., 2006). We identified two Her target sites within the ASE (E1 and E2) and observed that mutating either of them individually has a much less pronounced effect than mutating both. However, the level of resolution of our experiments does not permit us to draw any inferences about the role of Her binding site number in controlling the dynamics of her1 expression. Furthermore, a third CACGTG motif was present in all our constructs. This site is located outside the ASE at approximately 2.5kb upstream of the her1 coding region. However, we have not confirmed that this site binds Her proteins, although that can be expected based on our current data, or investigated whether it is important for the regulation of her1 expression.
The affinity of Her protein binding sites will likely also have a significant effect on the behavior of the oscillator (Zeiser et al., 2006). Our in vitro binding experiments indicate that the E1 motif is a higher affinity site than the E2, yet mutation of E2 alone produced a more obvious effect on gene expression than disruption of E1. However, we do not know definitively which members of the Her protein family bind these sites in vivo. Several her genes are expressed in the zebrafish PSM, and the potential exists for multiple homo- and hetero-dimer species. Modeling has shown that ascribing different properties to different dimer species can help to more accurately recapitulate the biological data (Cinquin, 2007). Our current data indicates that there are no differences in target site specificity between different Her proteins but it will be important to determine the relative affinities of different proteins for the binding sites in the ASE in order to fully understand the regulation of her1 expression. Finally, cooperative binding of Her proteins to multiple sites is also predicted to effect clock dynamics (Zeiser et al., 2006). We have not tested this prediction explicitly for E1 and E2 but the substantial space between them suggests that this may not be a factor in the regulation of her1.
Issues of affinity and cooperativity may underlie the slightly counterintuitive result obtained with our construct that included additional Her binding sites. As the transgene was expressed in the context of a functional endogenous segmentation clock, Her proteins should still only be available to repress transcription in a cyclical manner. Thus, we expected to see some residual transcription of the transgene rather than the complete loss of expression that was observed. However, the additional Her binding sites were all of the type present in the control probe used in our EMSA experiments. These sites contain optimized flanking residues (Jennings et al., 1999) and are apparently higher affinity sites than the endogenous sites in the ASE (Fig.2). Moreover, the extra sites were arranged in a closely spaced array (with 10bp between each site), while the two sites in the ASE are separated by 79bp. It seems plausible that this arrangement could also facilitate cooperative binding of Her proteins to adjacent motifs, further enhancing the affinity of these sites. Thus, this array of four E-boxes might mediate the assembly of a robust transcriptional repression complex that is not appropriately degraded or displaced during progression of the clock. Ultimately, analysis of Tg(4E-3.3her1:mCherry) embryos provided additional confirmation of the role of Her protein binding sites in mediating transcriptional repression but did not offer further significant insight into the mechanism of the oscillator. More instructive data may be obtained in the future by adding fewer and/or lower affinity Her protein binding sites or by modifying the affinity of the endogenous sites in the ASE. Furthermore, if cooperative binding of Her proteins does occur on adjacent sites, then the spacing between them may also prove to be relevant. It is also important to note that we have not formally excluded the possibility that the array of Her binding sites in this transgene also impacts the endogenous clock. If the pool of available Her proteins is limiting, the high affinity sites in the Tg(4E-3.3her1:mCherry) construct might sequester them away from regulating endogenous oscillating genes. However, we did not observe any segmentation defects in these embryos (or any of our transgenic lines), indicating that the clock is indeed functioning normally.
The correct balance between the number of binding sites for activators and repressors is likely to be critical for proper behavior of the oscillator. We identified a single Tbx protein binding site in the ASE, specifically confirming an interaction with Tbx24. Mutating this site completely abolishes reporter expression in the anterior PSM supporting a role for Tbx proteins in the activation of her1 transcription. In contrast, loss of tbx24 function in fss mutants results only in the loss of the most anterior stripe of her1 expression (van Eeden et al., 1998; Holley et al., 2000). This discrepancy might indicate that other Tbx proteins are regulating her1 through the same binding site. Indeed, recent studies suggest that her1 could be regulated by two other Tbx proteins, No tail (Ntl) and Spadetail (Spt/Tbx16) (Garnett et al., 2009; Morley et al., 2009). However, the expression domains of ntl and spt preclude a role in controlling her1 expression in the anterior PSM. Another family member, tbx6, is expressed in the posterior to intermediate PSM, overlapping slightly with tbx24 (Nikaido et al., 2002), but not in the more anterior region in which reporter expression is lost after mutation of our binding site. Thus, although it is possible that any or all of these other Tbx proteins regulate her1 expression via the site we have identified in other regions of the embryo, it is not yet clear why mutation of the Tbx site in the ASE causes a more pronounced effect than loss of tbx24 function.
Tbx24 is expressed throughout the anterior PSM but obviously only activates oscillating genes periodically. One explanation for this would be that binding of Her proteins and Tbx24 to cis-regulatory elements is mutually exclusive. We have not explicitly tested this hypothesis for the ASE but given the spacing of the sites we believe it is unlikely. It seems more plausible that Tbx24 can bind to target sites in cells throughout the anterior PSM but is only capable of activating transcription in synergy with periodic loss of Her repression. Accordingly, introducing two additional T-box binding sites to our reporter caused a very modest increase in expression at most and apparently did not override the normal oscillation in the anterior PSM.
Our Tbx24 experiments were also constrained by our relative ignorance of what constitutes a target sequence for Tbx24 binding. Although it seems that all Tbx proteins recognize sites very similar to the TCACACCT consensus, it is not yet clear what variations in that sequence are tolerated by specific Tbx proteins in vivo. Moreover, preferences for orientation and spacing of binding sites have been documented for some Tbx proteins (Conlon et al., 2001; Farin et al., 2007). Interestingly, the zebrafish Tbx24 protein binds motifs in the medaka mespb promoter that contain two adjacent sites in opposite orientations (Yasuhiko et al., 2008). Precisely defining the preferred binding motif for Tbx24 should allow us to more effectively manipulate oscillating expression in the future.
Notch signaling does not seem to regulate her1 expression directly through the ASE as we could not identify Su(H) binding sites within that region. Instead we confirmed two such motifs within the −2.9kb region. These sites are essential for transcriptional activation as expression is almost completely abolished in the anterior PSM when the sites are mutated. In contrast, all previous zebrafish studies in which Notch signaling was disrupted have demonstrated that her1 expression is not eliminated but present in a salt and pepper pattern in the anterior PSM (Holley, 2007). Since our transgene lacking Su(H) binding sites was analyzed in the context of a normal segmental pattern, we would not expect to see the salt and pepper expression pattern. Nevertheless, as some her1 expression is maintained in the absence of Notch signaling, we might expect to see some residual reporter gene transcription, perhaps even stripes. Furthermore, the salt and pepper expression of her1 in the anterior PSM after loss of Notch activity is dependent on fss/tbx24 (van Eeden et al., 1998; Holley et al., 2000; Sieger et al., 2003). We have shown that our constructs contain a site for Tbx24 binding, yet they show no expression after deletion of the Su(H) binding sites. Together, our result suggests that the 3.3kb enhancer of her1 lacks some other cis elements necessary for residual expression in the absence of input from the Notch pathway. These elements would likely also contribute to normal regulation of oscillating expression.
Our experiments suggest that the ASE as currently defined does not accurately recapitulate the expression of her1 in the anterior PSM. It remains possible that a more extensive deletion series through the 8.6kb of upstream sequence would define an element genuinely capable of driving precise oscillating expression in this region. However, it may also be true that the elements driving expression in the posterior and anterior PSM cannot actually be uncoupled. In this regard, it has not yet been shown that the 8.6kb to 3.3kb region, or any fragment of it, is sufficient for oscillating expression in the posterior PSM (although the whole region is clearly necessary). The activator and repressor binding sites required for oscillation may be spread throughout the full 8.6kb enhancer and might contribute to expression in both regions of the PSM. Resolving these issues will be key to determining the full significance of the binding sites that we have identified in this study. This is particularly evident in light of experiments demonstrating that temporary disruption of oscillating gene expression does not affect formation of somites by cells already in the anterior PSM (Giudicelli et al., 2007). However, it does affect cells in the posterior PSM, leading to delayed segmentation defects. Thus, the function of the segmentation clock is likely more dependent on proper regulation of oscillations in the posterior rather than the anterior PSM. Moreover, we do not yet know whether any of these cis-regulatory elements are shared with the her7 gene, which lies just 12kb away from her1, or if the two genes are regulated independently. Finally, our results suggest that the ASE is also responsible for suppressing expression of her1 in the somites.
In summary, we have performed a detailed characterization of the protein-DNA interactions that regulate expression of her1 in the anterior PSM. Further investigation of the her1 upstream sequence will reveal the full compliment of binding sites necessary for the complete her1 expression pattern throughout the PSM. Moreover, similar detailed analyses of the cis-regulatory regions of additional oscillating genes should lead to a comprehensive understanding of the regulatory logic that defines oscillating gene expression in the segmentation clock.
EXPERIMENTAL PROCEDURES
In vitro transcription/translation
Constructs for protein synthesis were derived from the pAMB-CAT plasmid supplied with the ActivePro In Vitro Translation kit (Ambion). Initially, specific pairs of complementary, overlapping oligonucleotides, designed to encode standard epitope tags, were cloned into the pAMB vector, replacing the chloramphenicol acetyltransferase (CAT) gene. The genes encoding the proteins of interest were then cloned into the modified vectors to produce N-terminally tagged constructs. The truncated version of V5tbx24 was generated by digesting the full length construct with StuI and XhoI to remove the 3′ 1406 bp of coding sequence, blunting the ends and religating. Proteins were synthesized using the ActivePro In Vitro Translation kit (Ambion), according to the manufacturers instructions. Synthesis of proteins was confirmed by SDS-PAGE and western blotting. Epitope tags were detected with the following primary antibodies: anti-Flag M2 monoclonal (Sigma); anti c-myc 9E10 monoclonal (Covance); anti-V5 V5-10 monoclonal (Sigma). Blots were developed using an anti-mouse IgG peroxidase conjugated secondary antibody (Sigma) and ECL Plus Detection Reagents (GE Healthcare). Note: the ActivePro kit has been discontinued but we have successfully used our vectors with the Easy Xpress Protein Synthesis kit from Qiagen.
Electrophoretic Mobility Shift Assay
Complementary pairs of oligonucleotides were end-labeled by standard procedures using γ32P-dATP (370MBq/ml; MP Biomedicals) and T4 polynuceotide kinase (NEB). Labeled probes were purified using Mini Quick Spin Oligo columns (Roche). 20 μl binding reactions (see below for details of binding buffers) containing 1 μg poly[dI.dC] and 1-5 μl of synthesized protein (plus antibodies and/or competitor oligonucleotides as required) were incubated on ice for 20 minutes. 10 fmol of labeled probe was then added and reactions incubated for a further 20 minutes on ice. Reactions were run on 5% native polyacrylamide gels in 0.5x TBE at 200V at room temperature. Dried gels were exposed to Biomax MR film (Kodak) in the presence of a Biomax intensifying screen (Kodak). Her binding buffer (1x): 25 mM Tris-HCl (pH7.5), 200 mM KCl, 1 mM DTT, 10% glycerol. Tbx binding buffer (1x): 25 mM Tris-HCl (pH7.5), 100 mM KCl, 1 mM DTT, 10% glycerol. Su(H) binding buffer (1x): 10 mM Tris-HCl (pH8.0), 0.1 mM EDTA, 10 mM MgCl2, 2 mM DTT, 17.5% glycerol, 2 mg/ml BSA. The following oligonucleotides were used as probes or unlabelled competitors (the sequence of only one strand is shown for each complementary pair): Her control GGTGGCACGTGCCATT; Her control mutant GGTGGGTCGACCCATT; ASE-E1 GACGACACGTGCTCAC; ASE-E1 mutant GACGAGTCGACCTCAC; ASE-E2 GTCTCCACGTGCCCAT; ASE-E2 mutant GTCTCGTCGACCCCAT; Tbx control CCCCAATTTCACACCTGGGCTGCAG; Tbx control mutant CCCCAATTTCAGACCTGGGCTGAG; ASE-T1 TGTCTGATTCACACCTCTGCAACAG; ASE-T1 mutant TGTCTGATTCAGACCTCTGCAACAG; Her1-S1 TTATACTTTCCCACGGGATTTT; Her1-S1 mutant AACATTATACT—GGATTTTTCTT; Her1-S2 AATATTGTTCCCACGAGAAAAA; Her1-S2 mutant CCACAATATTG—AGAAAAATCCT; Hes1-SPS GGTTACTTGTGGGAAAGAAAGTTTGGGAAGTTTCACACGAGCCGTTCC; Hes1-SPS mutant GGTTACTGACGCTAAGAAAGTTTGGGAAGTTAGAGTCGAGCCGTTCC. The nucleotides altered in the mutant probes are underlined. For the S1 and S2 mutants, the position of the deleted binding site is indicated by a dash.
Reporter constructs
A custom DNA fragment was designed containing the rabbit β-globin 5′utr, the emGFP coding sequence (codon optimized for expression in zebrafish) and the SV40 polyadenylation signal, flanked by the minimal Tol2 transposon elements (Urasaki et al., 2006). This 2.1kb fragment was synthesized commercially (GeneArt) and cloned into a pBluescript backbone to create the pTol2übervector. An alternative version was created by cloning the mCherry gene in place of emGFP. PCR products representing the 3.3kb and 2.9kb enhancer regions of her1 were inserted upstream of the emGFP or mCherry genes, replacing the rabbit β-globin sequence. The her1 3′utr sequence was amplified by PCR and cloned into the required vectors, immediately downstream of fluorescent protein coding sequence and replacing the SV40polyA signal. Specific mutations of binding sites were generated by PCR. Additional protein binding sites were introduced by cloning specifically designed oligonucleotides into restriction sites immediately upstream of the 3.3kb enhancer sequence. The accuracy and integrity of all constructs was confirmed by sequencing.
Generation and identification of transgenic zebrafish
Transposase mRNA was synthesized from NotI-linearised pCS-TP plasmid (a gift from Koichi Kawakami (Urasaki et al., 2006)) using the Sp6 mMESSAGEmMACHINE kit (Ambion). Plasmid DNA was purified using Qiagen Midiprep kits and injected into early one-cell stage embryos as circular DNA at 15 to 25 ng/μl, mixed with transposase mRNA at 25 ng/μl. Injected embryos were examined around 15hpf and healthy embryos exhibiting fluorescence were retained and raised to adulthood. Potentially transgenic fish were crossed to each other and their progeny screened for fluorescence. Positive embryos were collected and fixed at the 10-12 somite stage for analysis by in situ hybridization. Transgenic founder fish were isolated and assigned a specific line number. Founders were mated regularly and fluorescent embryos collected for in situ hybridization. During screening, clutches containing no fluorescent embryos were additionally analyzed by PCR. Extraction of genomic DNA was performed essentially as described (Gilmour et al., 2002), except that the volume of buffer was adjusted to the number of embryos in each sample (10 μl/embryo) and samples were not pooled for DNA precipitation. 200μl of each sample was precipitated and the pellets resuspended in 50 μl TE. 1 μl of genomic DNA was added to 20 μl PCR reactions using the Mastermix system (5 Prime). Specific primers were designed to amplify a 236 bp fragment of emGFP or a 275 bp fragment of mCherry (details below). Each reaction contained an additional pair of primers that amplify a 484 bp fragment of the keratin4 (krt4) promoter that serves as a positive control for the presence of genomic DNA. Cycling parameters were: 95°C 4min; 30 cycles of 95°C 30s, 59°C 30s, 72°C 30s; 72°C 5min. Primers: emGFPTg-F GTGAACGGACACAAGTTCAGC; emGFPTg-R AAGATGGTTCTCTCCTGCACG; mCherryTg-F GTTCATGCGCTTCAAGGTGC; mCherryTg-R CGTCCTCGAAGTTCATCACG; krt4-F ACACAAGACGGCACAAGACG; krt4-R GCATTTAGTTATTGCCCAGCAG. Fish identified as positive by PCR were carefully re-screened for transgene expression but only one line (of 42 total) was subsequently found to show weak fluorescence.
In situ hybridization
The mCherry sequence was cloned into pCS2+ and the resulting plasmid was linearized with BamHI for probe synthesis. The emGFP probe was synthesized from a purified 1kb linear fragment produced by SacII digestion of pCS2+itga5emGFP (provided by Dörthe Jülich). The her1 probe has been described previously (Holley et al., 2000). Probe synthesis and standard and fluorescent in situ hybridization were performed as previously described (Jülich et al., 2005; Brend and Holley, 2009). Images of fluorescent in situs were captured using a Zeiss LSM 510 confocal microscope.
ACKNOWLEDGEMENTS
We thank Andrew Collard for assistance in screening transgenic fish and Koichi Kawakami and Dörthe Jülich for providing plasmids. This work was supported by a grant from the National Institute of Child Health and Human Development (NIHCHD: R01 HD045738) to S.A.H.
Funded by: NICHD; Grant number: R01 HD045738
REFERENCES
- Aulehla A, Wehrle C, Brand-Saberi B, Kemler R, Gossler A, Kanzler B, Herrmann BG. Wnt3a Plays a Major Role in the Segmentation Clock Controling Somitogenesis. Dev Cell. 2003;4:395–406. doi: 10.1016/s1534-5807(03)00055-8. [DOI] [PubMed] [Google Scholar]
- Aulehla A, Wiegraebe W, Baubet V, Wahl MB, Deng C, Taketo M, Lewandoski M, Pourquie O. A beta-catenin gradient links the clock and wavefront systems in mouse embryo segmentation. Nat Cell Biol. 2008;10:186–193. doi: 10.1038/ncb1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho Y, Hirata H, Masamizu Y, Kageyama R. Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock. Genes Dev. 2003;17:1451–1456. doi: 10.1101/gad.1092303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho Y, Sakata R, Komatsu S, Shiota K, Yamada S, Kageyama R. Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev. 2001;15:2642–2647. doi: 10.1101/gad.930601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Brend T, Holley SA. Zebrafish whole mount high-resolution double fluorescent in situ hybridization. J Vis Exp. 2009 doi: 10.3791/1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kang L, Zhang N. Negative feedback loop formed by Lunatic fringe and Hes7 controls their oscillatory expression during somitogenesis. Genesis. 2005;43:196–204. doi: 10.1002/gene.20171. [DOI] [PubMed] [Google Scholar]
- Cinquin O. Repressor Dimerization in the Zebrafish Somitogenesis Clock. PLoS Comput Biol. 2007;3:e32. doi: 10.1371/journal.pcbi.0030032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SE, Levorse JM, Tilghman SM, Voght TF. Clock Regulatory Elements Control Cyclic Expression of Lunatic fringe during Somitogenesis. Dev Cell. 2002;3:75–84. doi: 10.1016/s1534-5807(02)00212-5. [DOI] [PubMed] [Google Scholar]
- Conlon FL, Fairclough L, Price BM, Casey ES, Smith JC. Determinants of T box protein specificity. Development. 2001;128:3749–3758. doi: 10.1242/dev.128.19.3749. [DOI] [PubMed] [Google Scholar]
- Cooke J, Zeeman EC. A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J Theor Biol. 1976;58:455–476. doi: 10.1016/s0022-5193(76)80131-2. [DOI] [PubMed] [Google Scholar]
- Dale JK, Malapert P, Chal J, Vilhais-Neto G, Maroto M, Johnson T, Jayasinghe S, Trainor P, Herrmann B, Pourquie O. Oscillations of the snail genes in the presomitic mesoderm coordinate segmental patterning and morphogenesis in vertebrate somitogenesis. Dev Cell. 2006;10:355–366. doi: 10.1016/j.devcel.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Dequeant ML, Glynn E, Gaudenz K, Wahl M, Chen J, Mushegian A, Pourquie O. A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science. 2006;314:1595–1598. doi: 10.1126/science.1133141. [DOI] [PubMed] [Google Scholar]
- Dubrulle J, McGrew MJ, Pourquié O. FGF Signaling Controls Somite Boundary Position and Regulates Segmentation Clock Control of Spatiotemporal Hox Gene Activation. Cell. 2001;106:219–232. doi: 10.1016/s0092-8674(01)00437-8. [DOI] [PubMed] [Google Scholar]
- Dubrulle J, Pourquié O. fgf8 mRNA decay establishes a gradient that couples axial elongation to patterning in the vertebrate embryo. Nature. 2004;427:419–422. doi: 10.1038/nature02216. [DOI] [PubMed] [Google Scholar]
- Dunty WC, Jr., Biris KK, Chalamalasetty RB, Taketo MM, Lewandoski M, Yamaguchi TP. Wnt3a/ -catenin signaling controls posterior body development by coordinating mesoderm formation and segmentation. Development. 2008;135:85–94. doi: 10.1242/dev.009266. [DOI] [PubMed] [Google Scholar]
- Dunwoodie SL, Clements M, Sparrow DB, Sa X, Conlon RA, Beddington RS. Axial skeletal defects caused by mutation in the spondylocostal dysplasia/pudgy gene Dll3 are associated with disruption of the segmentation clock within the presomitic mesoderm. Development. 2002;129:1795–1806. doi: 10.1242/dev.129.7.1795. [DOI] [PubMed] [Google Scholar]
- Echeverri K, Oates AC. Coordination of symmetric cyclic gene expression during somitogenesis by Suppressor of Hairless involves regulation of retinoic acid catabolism. Dev Biol. 2007;301:388–403. doi: 10.1016/j.ydbio.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Farin HF, Bussen M, Schmidt MK, Singh MK, Schuster-Gossler K, Kispert A. Transcriptional repression by the T-box proteins Tbx18 and Tbx15 depends on Groucho corepressors. J Biol Chem. 2007;282:25748–25759. doi: 10.1074/jbc.M703724200. [DOI] [PubMed] [Google Scholar]
- Fischer A, Gessler M. Delta-Notch--and then? Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Res. 2007;35:4583–4596. doi: 10.1093/nar/gkm477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg H, Crozet F, Brown NA. Waves of mouse Lunatic fringe expression, in four-hour cycles at two-hour intervals, precede somite boundary formation. Curr Biol. 1998;8:1027–1030. doi: 10.1016/s0960-9822(07)00424-1. [DOI] [PubMed] [Google Scholar]
- Gajewski M, Sieger D, Alt B, Leve C, Hans S, Wolff C, Rohr KB, Tautz D. Anterior and posterior waves of cyclic her1 gene expression are differentially regulated in the presomitic mesoderm of zebrafish. Development. 2003;130:4269–4278. doi: 10.1242/dev.00627. [DOI] [PubMed] [Google Scholar]
- Gajewski M, Voolstra C. Comparative analysis of somitogenesis related genes of the hairy/Enhancer of split class in Fugu and zebrafish. BMC Genomics. 2002;3:21. doi: 10.1186/1471-2164-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett AT, Han TM, Gilchrist MJ, Smith JC, Eisen MB, Wardle FC, Amacher SL. Identification of direct T-box target genes in the developing zebrafish mesoderm. Development. 2009;136:749–760. doi: 10.1242/dev.024703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DT, Jessen JR, Lin S. Manipulating gene expression in the zebrafish. In: Nüsslein-Volhard C, Dahm R, editors. Zebrafish. Oxford University Press; New York: 2002. pp. 121–144. [Google Scholar]
- Giudicelli F, Ozbudak EM, Wright GJ, Lewis J. Setting the Tempo in Development: An Investigation of the Zebrafish Somite Clock Mechanism. PLoS Biol. 2007;5:e150. doi: 10.1371/journal.pbio.0050150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goering LM, Hoshijima K, Hug B, Bisgrove B, Kispert A, Grunwald DJ. An interacting network of T-box genes directs gene expression and fate in the zebrafish mesoderm. Proc Natl Acad Sci U S A. 2003;100:9410–9415. doi: 10.1073/pnas.1633548100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry CA, Urban MK, Dill KK, Merlie JP, Page MF, Kimmel CB, Amacher SL. Two linked hairy/Enhancer of split-related zebrafish genes, her1 and her7, function together to refine alternating somite boundaries. Development. 2002;129:3693–3704. doi: 10.1242/dev.129.15.3693. [DOI] [PubMed] [Google Scholar]
- Holley SA. The genetics and embryology of zebrafish metamerism. Dev Dyn. 2007;236:1422–1449. doi: 10.1002/dvdy.21162. [DOI] [PubMed] [Google Scholar]
- Holley SA, Geisler R, Nüsslein-Volhard C. Control of her1 expression during zebrafish somitogenesis by a Delta-dependent oscillator and an independent wave-front activity. Genes & Dev. 2000;14:1678–1690. [PMC free article] [PubMed] [Google Scholar]
- Holley SA, Jülich D, Rauch GJ, Geisler R, Nüsslein-Volhard C. her1 and the notch pathway function within the oscillator mechanism that regulates zebrafish somitogenesis. Development. 2002;129:1175–1183. doi: 10.1242/dev.129.5.1175. [DOI] [PubMed] [Google Scholar]
- Ishikawa A, Kitajima S, Takahashi Y, Kokubo H, Kanno J, Inoue T, Saga Y. Mouse Nkd1, a Wnt antagonist, exhibits oscillatory gene expression in the PSM under the control of Notch signaling. Mech Dev. 2004;121:1443–1453. doi: 10.1016/j.mod.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Jennings BH, Tyler DM, Bray SJ. Target specificities of Drosophila enhancer of split basic helix-loop-helix proteins. Mol Cell Biol. 1999;19:4600–4610. doi: 10.1128/mcb.19.7.4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y-J, Aerne BL, Smithers L, Haddon C, Ish-Horowicz D, Lewis J. Notch signaling and the synchronization of the somite segmentation clock. Nature. 2000;408:475–479. doi: 10.1038/35044091. [DOI] [PubMed] [Google Scholar]
- Jouve C, Palmeirim I, Henrique D, Beckers J, Gossler A, Ish Horowicz D, Pourquié O. Notch signalling is required for cyclic expression of the hairy-like gene HES1 in the presomitic mesoderm. Development. 2000;127:1421–1429. doi: 10.1242/dev.127.7.1421. [DOI] [PubMed] [Google Scholar]
- Jülich D, Lim C-H, Round J, Nicolaije C, Davies A, Schroeder J, Geisler R, Consortium TS. Lewis J, Jiang Y-J, Holley SA. beamter/deltaC and the role of Notch ligands in the zebrafish somite segmentation, hindbrain neurogenesis and hypochord differentiation. Dev Biol. 2005;286:391–404. doi: 10.1016/j.ydbio.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WM, R KS, Ullmann B, Schilling TF. Stages of Embryonic Development of the Zebrafish. Dev. Dynam. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Leimeister C, Dale K, Fischer A, Klamt B, Hrabe de Angelis M, Radtke F, McGrew MJ, Pourquié O, Gessler M. Oscillating expression of c-Hey2 in the presomitic mesoderm suggests that the segmentation clock may use combinatorial signaling through multiple interacting bHLH factors. Dev Biol. 2000;227:91–103. doi: 10.1006/dbio.2000.9884. [DOI] [PubMed] [Google Scholar]
- Lewis J. Autoinhibition with Transcriptional Delay: A simple Mechanism for the Zebrafish Somitogenesis Oscillator. Curr Biol. 2003;13:1398–1408. doi: 10.1016/s0960-9822(03)00534-7. [DOI] [PubMed] [Google Scholar]
- Masamizu Y, Ohtsuka T, Takashima Y, Nagahara H, Takenaka Y, Yoshikawa K, Okamura H, Kageyama R. Real-time imaging of the somite segmentation clock: revelation of unstable oscillators in the individual presomitic mesoderm cells. Proc Natl Acad Sci U S A. 2006;103:1313–1318. doi: 10.1073/pnas.0508658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrew MJ, Dale JK, Fraboulet S, Pourquié O. The Lunatic Fringe gene is a target of the molecular clock linked to segmentation in avian embryos. Curr Biol. 1998;8:979–982. doi: 10.1016/s0960-9822(98)70401-4. [DOI] [PubMed] [Google Scholar]
- Morales AV, Yasuda Y, Ish-Horowicz D. Periodic Lunatic fringe Expression is Controlled during Segmentation by a Cyclic Transcriptional Enhancer Responsive to Notch Signaling. Dev Cell. 2002;3:63–74. doi: 10.1016/s1534-5807(02)00211-3. [DOI] [PubMed] [Google Scholar]
- Morley RH, Lachani K, Keefe D, Gilchrist MJ, Flicek P, Smith JC, Wardle FC. A gene regulatory network directed by zebrafish No tail accounts for its roles in mesoderm formation. Proc Natl Acad Sci U S A. 2009;106:3829–3834. doi: 10.1073/pnas.0808382106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE. T-box genes in vertebrate development. Annu Rev Genet. 2005;39:219–239. doi: 10.1146/annurev.genet.39.073003.105925. [DOI] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Pear WS, Aster JC, Blacklow SC. Cooperative assembly of higher-order Notch complexes functions as a switch to induce transcription. Proc Natl Acad Sci U S A. 2007;104:2103–2108. doi: 10.1073/pnas.0611092104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellesen DT, Lai EC, Posakony JW. Discrete enhancer elements mediate selective responsiveness of enhancer of split complex genes to common transcriptional activators. Dev Biol. 1999;213:33–53. doi: 10.1006/dbio.1999.9324. [DOI] [PubMed] [Google Scholar]
- Nikaido M, Kawakami A, Sawada A, Furutani-Seiki M, Takeda H, Araki K. Tbx24, encoding a T-box protein, is mutated in the zebrafish somite-segmentation mutant fused somites. Nat Genet. 2002;31:195–199. doi: 10.1038/ng899. [DOI] [PubMed] [Google Scholar]
- Niwa Y, Masamizu Y, Liu T, Nakayama R, Deng CX, Kageyama R. The initiation and propagation of Hes7 oscillation are cooperatively regulated by Fgf and notch signaling in the somite segmentation clock. Dev Cell. 2007;13:298–304. doi: 10.1016/j.devcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Oates AC, Ho RK. Hairy/E(spl)-related (Her) genes are central components of the segmentation oscillator and display redundancy with the Delta/Notch signaling pathway in the formatoin of anterior segmental boundaries in the zebrafish. Development. 2002;129:2929–2946. doi: 10.1242/dev.129.12.2929. [DOI] [PubMed] [Google Scholar]
- Palmeirim I, Henrique D, Ish-Horowicz D, Pourquié O. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell. 1997;91:639–648. doi: 10.1016/s0092-8674(00)80451-1. [DOI] [PubMed] [Google Scholar]
- Sawada A, Fritz A, Jiang Y-J, Yamamoto A, Yamasu K, Kuroiwa A, Saga Y, Takeda H. Zebrafish Mesp family genes, mesp a and mesp b are segmentally expressed in the presomitic mesoderm, Mesp b confers the anterior identity to the developing somites. Development. 2000;127:1691–1702. doi: 10.1242/dev.127.8.1691. [DOI] [PubMed] [Google Scholar]
- Sawada A, Shinya M, Jiang YJ, Kawakami A, Kuroiwa A, Takeda H. Fgf/MAPK signalling is a crucial positional cue in somite boundary formation. Development. 2001;128:4873–4880. doi: 10.1242/dev.128.23.4873. [DOI] [PubMed] [Google Scholar]
- Schröter C, Herrgen L, Cardona A, Brouhard GJ, Feldman B, Oates AC. Dynamics of zebrafish somitogenesis. Dev Dyn. 2008;237:545–553. doi: 10.1002/dvdy.21458. [DOI] [PubMed] [Google Scholar]
- Shankaran SS, Sieger D, Schroter C, Czepe C, Pauly MC, Laplante MA, Becker TS, Oates AC, Gajewski M. Completing the set of h/E(spl) cyclic genes in zebrafish: her12 and her15 reveal novel modes of expression and contribute to the segmentation clock. Dev Biol. 2007;304:615–632. doi: 10.1016/j.ydbio.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Shifley ET, Vanhorn KM, Perez-Balaguer A, Franklin JD, Weinstein M, Cole SE. Oscillatory lunatic fringe activity is crucial for segmentation of the anterior but not posterior skeleton. Development. 2008 doi: 10.1242/dev.006742. [DOI] [PubMed] [Google Scholar]
- Sieger D, Tautz D, Gajewski M. The role of Suppressor of Hairless in Notch mediated signalling during zebrafish somitogenesis. Mech Dev. 2003;120:1083–1094. doi: 10.1016/s0925-4773(03)00154-0. [DOI] [PubMed] [Google Scholar]
- Sieger D, Tautz D, Gajewski M. her11 is involved in the somitogenesis clock in zebrafish. Dev Genes Evol. 2004;214:393–406. doi: 10.1007/s00427-004-0427-z. [DOI] [PubMed] [Google Scholar]
- Takebayashi K, Sasai Y, Sakai Y, Watanabe T, Nakanishi S, Kageyama R. Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1. Negative autoregulation through the multiple N box elements. J Biol Chem. 1994;269:5150–5156. [PubMed] [Google Scholar]
- Tam PP. The control of somitogenesis in mouse embryos. J Embryol Exp Morphol. 1981;65(Suppl):103–128. [PubMed] [Google Scholar]
- Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174:639–649. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eeden FJM, Holley SA, Haffter P, Nüsslein-Volhard C. Zebrafish segmentation and pair-rule patterning. Dev Genet. 1998;23:65–76. doi: 10.1002/(SICI)1520-6408(1998)23:1<65::AID-DVG7>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Yasuhiko Y, Kitajima S, Takahashi Y, Oginuma M, Kagiwada H, Kanno J, Saga Y. Functional importance of evolutionally conserved Tbx6 binding sites in the presomitic mesoderm-specific enhancer of Mesp2. Development. 2008;135:3511–3519. doi: 10.1242/dev.027144. [DOI] [PubMed] [Google Scholar]
- Zeiser S, Liebscher HV, Tiedemann H, Rubio-Aliaga I, Przemeck GK, de Angelis MH, Winkler G. Number of active transcription factor binding sites is essential for the Hes7 oscillator. Theor Biol Med Model. 2006;3:11. doi: 10.1186/1742-4682-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]