Abstract
Background
NPM1 gene at chromosome 5q35 is involved in recurrent translocations in leukemia and lymphoma. It also undergoes mutations in 60% of adult acute myeloid leukemia (AML) cases with normal karyotype. The incidence and significance of NPM1 deletion in human leukemia have not been elucidated.
Methodology and Principal Findings
Bone marrow samples from 145 patients with myelodysplastic syndromes (MDS) and AML were included in this study. Cytogenetically 43 cases had isolated 5q-, 84 cases had 5q- plus other changes and 18 cases had complex karyotype without 5q deletion. FISH and direct sequencing investigated the NPM1 gene. NPM1 deletion was an uncommon event in the “5q- syndrome” but occurred in over 40% of cases with high risk MDS/AML with complex karyotypes and 5q loss. It originated from large 5q chromosome deletions. Simultaneous exon 12 mutations were never found. NPM1 gene status was related to the pattern of complex cytogenetic aberrations. NPM1 haploinsufficiency was significantly associated with monosomies (p<0.001) and gross chromosomal rearrangements, i.e., markers, rings, and double minutes (p<0.001), while NPM1 disomy was associated with structural changes (p = 0.013). Interestingly, in complex karyotypes with 5q- TP53 deletion and/or mutations are not specifically associated with NPM1 deletion.
Conclusions and Significance
NPM1/5q35 deletion is a consistent event in MDS/AML with a 5q-/-5 in complex karyotypes. NPM1 deletion and NPM1 exon 12 mutations appear to be mutually exclusive and are associated with two distinct cytogenetic subsets of MDS and AML.
Introduction
In humans, nucleophosmin (NPM1) is implicated in the genesis of different cancers [1]. We previously found NPM1 exon 12 somatic mutations in approximately 60% of adult acute myeloid leukemia (AML) and normal karyotype [2]. NPM1 fusion proteins are also specifically involved: t(5;17)(q35;q21)/NPM1-RARA causes acute promyelocytic leukemia; t(2;5)(p23;q35)/NPM1-ALK underlies anaplastic large cell lymphoma; t(3;5)(q26;q35)/NPM1-MLF1 is associated with myelodysplastic syndromes (MDS) and AML with trilineage dysplasia and poor prognosis (http://AtlasGeneticsOncology.org). In the last cytogenetic subgroup of MDS/AML a cryptic loss of NPM1 gene may occur at the 5q35 breakpoint of the translocation [3]. Despite of this sporadic observation the role of NPM1 deletion in hematological malignancies remains to be elucidated. In human leukemia an interstitial isolated deletion at chromosome 5q identifies a myelodysplastic syndrome with a benign clinical course (the “5q- syndrome”) [4]. Moreover a partial or complete 5q loss (5q-/-5) may occur as part of a complex karyotype in high risk MDS/AML, including cases arising after radio- and/or chemo-therapy [5]. Chromosomes 5q- are heterogeneous in size as deleted segments vary in extension and have different centromeric and telomeric breakpoints [6], [7]. Experimental evidences indicate that the 5q- syndrome originates from haploinsufficiency of more than one critical pathogenetic gene included in the hemizygous chromosome deletion [8]–[12]. Thus identification of genes that go lost in different 5q- chromosomes of MDS and AML is very important to understand the pathophysiology of the associated malignant disorders. Focusing on NPM1 gene we investigated a large series of MDS and AML with and without a 5q deletion.
Materials and Methods
Patient inclusion criteria
AML or MDS and one of the following abnormal karyotypes, according to the ISCN [13]: isolated 5q-; 5q-/-5 plus one or more additional changes (non-isolated); complex karyotypes without 5q-/-5. 145 patients (60 males; 85 females; age range 9–94 years; 71 with MDS and 74 with AML) were recruited from the Hematological Institutions, University of Perugia, Italy, Sant Pau Hospital, Barcelona, Spain, and the Demokritos Cancer Centre, Athens, Greece.
FISH
Protocols have already been described [14]. Commercial FISH probes were provided by Abbott/Vysis (Downers Grove, IL, USA): LSI EGR1/D5S721, D5S23 dual color probe; LSI CSF1R (5q33–q34) spectrum orange/D5S721, D5S23 spectrum green; LSI c-MYC dual color break apart rearrangement probe; LSI 13 (RB1) 13q14 spectrum orange probe. Caltech DNA clones CTC-286C20 (FGFR4/5q35.2), CTC-549A4 (NSD1/5q35.3), CTD-2131H15 (3′CTNNA1/5q31), CTD-2324F6 (5′CTNNA1/5q31), CTD-2342K5 (BRCA2/13q12), and CTD-3199J23 (BRCA1/17q21) were purchased from Invitrogen (Carlsbad, CA, USA). Other DNA clones were selected from the RPCI libraries (http://www.ncbi.nlm.gov/project/mapview, Build 37.1), provided by Mariano Rocchi, Department of Genetics and Microbiology, University of Bari, Italy and grown, labelled, and validated at the Cytogenetics and Molecular Genetics Laboratory, Hematology Department, University of Perugia, Italy. Clones mapping at 5q11.2–5q14.1 listed from centromere to telomere: RP11-266N13, RP11-489L13, RP11-298P6, RP11-79C20, RP11-170N5, RP11-195G20, RP11-633M1, RP11-195E2, RP11-771B3, RP11-79P5, RP5-910M8, RP11-229C3, RP11-469J18, RP11-996M9, RP11-168A11, RP11-80K5, RP11-1089B2, and RP11-885P10. Gene-specific RPCI clones: RP11-89G4 (IRF1/5q31), RP11-946D14 (RPS14/5q33), RP11-204L7 (3′SPARC/5q33), RP11-642K17 (5′SPARC/5q33), RP11-117L6 (NPM1/5q35.1), RP1-240G13 (5qter subtelomeric probe), RP11-480O8 (BUB1A/2q14), RP11-383G6 (3′ATR/3q23), RP11-427D1 (5′ATR/3q23), RP11-669K4 (FBW7/4q31), RP11-149I2 (CDKN2A/B/9p21), RP11-380G5 (PTEN/10q23), RP11-241D13 (ATM/11q22), RP11-880O16 (CHEK1/11q24), RP11-1137N1 (MDM2/12q15), RP11-248I17 (BUB1B/15q15), RP11-436C9 (3′CHEK2/22q12), RP11-444G7 (5′CHEK2/22q12).
NPM1 and TP53 Mutational Analysis
Exon 12 NPM1 mutations were studied by direct sequencing of PCR products from genomic DNA using the following primers: NPM1_ex12for 5′-ATGTCTATGAAGTGTTGTGGTTCC-3′ and NPM1_ex12rev 5′- CAGGCATTTTGGACAACACA-3′. TP53 gene (NC_000017.10, 7590863.7571720) mutations on exons 2-12 ( Table 1 ) were studied using PCR-based denaturing HPLC using a WAVE-MD™ System (Transgenomic, Omaha, NE) equipped with a DNASep Cartridge. PCR assays were performed in a volume of 25 µl, containing 100 ng of genomic DNA, 6 pmol of forward and reverse primer, 200 mM dNTPs, 0.3 U of AmpliTaq Gold (Life Technologies Corp., Carlsbad, CA, USA). Gradient elution and melting temperature conditions were established by Wave-Maker Navigator version 1.7 software (Transgenomic). Bidirectional sequencing was performed on samples with abnormal chromatographs using ABI prism 3130 (Life Technologies). Missense and frameshift mutations were detected using Finch TV version 1.4.0 and described according to CCDS 11118.1 (NM_0000546.4).
Table 1. Primers used to amplify TP53 coding exons (NC_000017.10).
| TP53 Exon | Primer | Sequence |
| 2 | P53_ex2FP53_ex2TGFP53_ex2R | 5′-TTTTCCTCTTGCAGCAGCCA-3′ 5′-CCAGGTGACCCAGGGTTGGAAG-3′ 5′-CAAGAGCAGAAAGTCAGTCC-3′ |
| 3 | P53_ex3FP53_ex3R | 5′-AGACCTGTGGGAAGCGAAAA-3′ 5′-GGGACAGCATCAAATCATCC-3′ |
| 2–3 | P53_ex2-3FP53_ex2-3R | 5′-TGCCTTCCGGGTCACTGCC-3′ 5′-AGCCCTCCAGGTCCCCAGCC-3′ |
| 4 | P53_ex4FP53_ex4AFP53_ex4RP53_ex4AR | 5′-ATCTACAGTCCCCCTTGCCG-3′ 5′-ACCTGGTCCTCTGACTGCTC-3′ 5′-GCAACTGACCGTGCAAGTCA-3′ 5′-GCCAAAGGGTGAAGAGGAAT-3′ |
| 5 | P53_ex5FP53_ex5AFP53_ex5RP53_ex5AR | 5′-GCTGCCGTGTTCCAGTTG-3′ 5′-GCTTTATCTGTTCACTTGTGCC-3′ 5′-ACCAGCCCTGTCGTCTCTC-3′ 5′-AGCCCTGTCGTCTCTCCAG-3′ |
| 6 | P53_ex6FP53_ex6R | 5′-CCAGGCCTCTGATTCCTCAC-3′ 5′-GCCCCCCTACTGCTCACC-3′ |
| 7 | P53_ex7FP53_ex7RP53_ex7AR | 5′-GCCACAGGTCTCCCCAAG-3′ 5′-TGTGCAGGGTGGCAAGTG-3′ 5′-TGCAGGGTGGCAAGTGGCTC-3′ |
| 8 | P53_ex8FP53_ex8R | 5′-TGCCTCTTGCTTCTCTTTTCC-3′ 5′-GGCATAACTGCACCCTTGG-3′ |
| 9 | P53_ex9FP53_ex9R | 5′-GCGGTGGAGGAGACCAAG-3′ 5′-GCTACAACCAGGAGCCATTG-3′ |
| 10 | P53_ex10FP53_ex10R | 5′-GGCAGTGATGCCTCAAAGAC-3′ 5′-CTAGGCTAAGCTATGATGTTCC-3′ |
| 11 | P53_ex11FP53_ex11AFP53_ex11DFP53_ex11RP53_ex11ARP53_ex11BR | 5′-CCCCCTCCTCTGTTGCTGC-3′ 5′-TACTTCTCCCCCTCCTCTG-3′ 5′-GAACCATCTTTTAACTCAGGTAC-3′ 5′-GGCAGGGGAGTAGGGCCAG-3′ 5′-GAAGGCAGGATGAGAATGGA-3′ 5′-AGCTGCCTTTGACCATGAAG-3′ |
| 12 | P53_ex12FP53_ex12AFP53_ex12BFP53_ex12RP53_ex12AR | 5′-CACTCATGTGATGTCATCTCTC-3′ 5′-CTCACTCATGTGATGTCATCT-3′ 5′-CTCTGAGGTGCTCAGTAAAC-3′ 5′-GCTGTCAGTGGGGAACAAGA-3′ 5′-GCAAGCAAGGGTTCAAAGAC-3′ |
NPM1 expression by Real Time Quantitative PCR
RT-qPCR was performed on cryopreserved bone marrow RNA samples from 9 patients with NPM1+/- and 5q-, 10 patients with NPM1+/+ and 5q-, 5 patients with NPM1+/+ without 5q- and 11 healthy controls. Total RNA was extracted using Trizol (Invitrogen) according to the manufacturer's protocol with minor modifications. Two micrograms of total RNA were treated with 4 Units of Dnase I (Deoxyribonuclease I Amplification Grade, Sigma) in a total volume of 11 µl. One microgram of treated RNA was reverse transcribed to single strand cDNA according to the EAC protocol [15] and 1 µg was used in a control reaction (-RT) using the same procedure without the M-MLV reverse transcriptase enzyme (Perkin Elmer Applied Biosystems, Foster City, USA). The RT-qPCR mixture reaction contained 12.5 µl Taq Man Universal PCR Master Mix (Life Technologies), 300 nM primers, 200 nM probe and 5 µl cDNA (1/10 of RT product), in a total volume of 25 µl. Amplification conditions were: 2 minutes at 50°C, 10 minutes at 95°C followed by 40 cycles at 95°C for 15 seconds and at 60°C for 1 minute [16]. RT-qPCR reactions and fluorescence measurements were performed on an ABI PRISM 7700 Sequence Detection System (Life Technologies). Gene expression levels of NPM1 and ABL1 were detected using specific assays on Demand (Life Technologies) following the manufacturer's instructions: ID: Hs02339479_g1 (exon 4–5) for NPM1 and ID: Hs00245445_m1 (exon 3–4) for ABL1. RT-qPCR reactions were performed in quadruplicate for NPM1 and in duplicate for ABL1. NPM1 transcript levels were quantified relative to endogenous ABL1 and expressed as 2−ΔΔCt [17]. Universal Human Reference RNA (Stratagene, La Jolla, CA, USA) was used as calibrator in all experiments. A threshold value of 0.1 was used throughout the study.
Statistical analysis
Inter-group differences were analyzed by non parametric tests using first analysis of variance (ANOVA) with the Kruskal-Wallis test to compare three groups and the Mann-Whitney U test to compare two. If significant, multiple comparisons were carried out using the Mann-Whitney U test with the Bonferroni correction (i.e. 0.05/numbers of comparisons). The multivariable Poisson regression model analyzed the adjusted effect of NPM1 and TP53 on gross chromosomal changes and monosomies.
Contingency tables studied associations among categorical variables which were analyzed by Fisher's Exact test for 2X2 tables or the chi-square test. Unless otherwise indicated, significance was set at <0.05. SPSS version 17.0 software (Chicago, IL) was used for statistical analysis.
Ethics statement
This study was carried out solely on encoded archival samples that were originally taken for diagnostic purposes. The residual of these samples was used after all diagnostic procedures had been completed. Our specific study was approved by the Institutional Review Board of the Hematology Department of the University of Perugia (IRB 00003450) which guidelines allow the use of anonymous residual diagnostic samples for research purposes. Written informed consent was obtained. The “Demokritos” Cancer Research Centre Ethics Committee (Athens, Greece) allows the use of anonymous or encoded residual diagnostic samples for research purposes, unless the patient expresses its refusal to participate in research studies. The authors declare that all samples used in this study were encoded anonymously and that none of the patients had explicitly refused to participate in research.
Results and Discussion
One hundred forty five patients with MDS/AML (60 males and 85 females; median age = 68 years, range 9–94) were included in the study (Table S1).
The mono-allelic NPM1 deletion was a very infrequent lesion in the “5q- syndrome” because it was not detected in 42/43 cases. It identified two molecular sub-groups, here named NPM1+/− and NPM1+/+, since it was present in 38/84 (45%) cases with complex karyotypes and 5q-. Interestingly Lessard et al. [18] demonstrated loss of the 5q35 chromosome band in 30% of de novo and therapy related AML. NPM1 loss may have occurred as an early step in MDS/AML as it remained stably mono-allelic in 4 NPM1+/− patients and did not develop in 6 NPM1+/+ over a median of 9 months follow-up (range: 2–156 months) ( Table 2 ).
Table 2. FISH: NPM1 status over time (median 9 months; range 2–156) in 10 adult patients with MDS/AML.
| No. | Diagnosis | Karyotype | Follow-up (months)/Disease status | Karyotype/NPM1 |
| Non-isolated 5q- NPM1 +/+ | ||||
| 45 | RA | 47,XX,del(5q),+21 | +156/AML | 46,idem,-7,t(12;22(p13;q11))/U |
| 48 | RAEB | 46,XX,add(11)(q),del(5q)/46,XX | +24/RAEB | U/U |
| 57 | AML | 46,XX,del(5q),t(2;3)(p21;q26) | +4/AML | 45,idem,−7/U |
| 72 | AML | 45–46,XX,t(3;11)(p21;q23),del(5)(q13q31),−7,+8,del(12)(p13),add(16)(p13),add(17)(q),−18,+mar/46,XX | +13/2nd relapse | U/U |
| 73 | RAEB | 46,XX,del(5)(q13q33),del(7)(q22q32),del(7)(q22q32),der(20)/46,idem,add(7)(q36) | +13/MDS-U | U/U |
| 76 | AML | 41–49,XY,del(2p),der(3)(p21),−5,del(8)(q22),der(13;15)(q10;q10),−17,−18,+1−6 mar/46,XY | +5/2nd relapse | U/U |
| Non-isolated 5q- NPM1 +/− | ||||
| 93 | AML | 42–44,XX,−5,add(6)(p21),+8,1–3mar | +9/1st relapse | n.d./U |
| 110 | AML | 44,XY,−2,−3,−5,−7,−13,−17,+mar1,+mar2,+mar3/46,XY | +9/1st relapse | n.d./U |
| 121 | RAEB | 40–48,XY,t(1;3)(p32;p21),−5,−7,−13,−18,−20,−22,+1−6 mar/46,XY | +2/RAEB | n.d./U |
| 122 | AML | 40−43,XY,add(1)(q),−4,del(5)(q13q33),−7,−9,−12,−17,−20,+2-6mar | +9/resistant | n.d./U |
Patient no. (see Supplementary Table 1); AML, acute myeloid leukemia; RAEB, refractory anemia with excess of blasts; RA, refractory anemia; NPM1+/+ no monoallelic NPM1 deletion; NPM1+/− monoallelic NPM1 deletion; U, unchanged; n.d., not done.
The NPM1 loss was not cryptic since it was absent in 18 patients with complex karyotypes without 5q- (Table S1). Chromosome 5q breakpoints were distributed over a wide chromosomal area within genomic regions which extended beyond the isolated 5q deletion breakpoints. In NPM1+/− patients telomeric breakpoints clustered between NPM1 and FGFR4 (approximately 5.7 megabases) in 4/16 cases and included subtelomeric sequences in 12 (75%) ( Figure 1A,B,C , and Table S1). Centromeric breakpoints involved chromosome bands from q11.2 to q14.1 in more than 50% and were proximal to RP11-80K5 ( Figure 1A,B,C , and Table S1). Notably in cases with isolated 5q deletions centromeric breakpoints always fell distally to RP11-80K5 ( Figure 1A and Table S1).
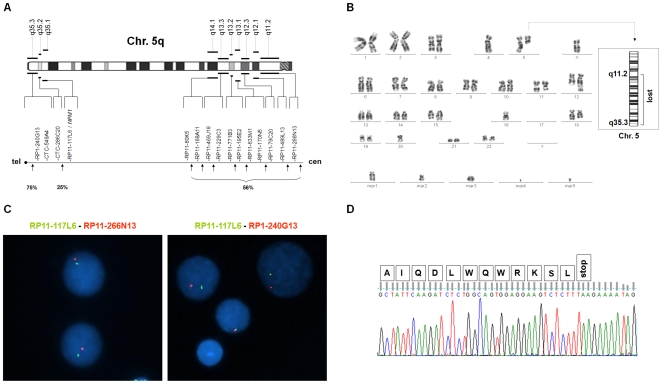
Figure 1. FISH, cytogenetics and mutational analysis.
A) Results of FISH in 38 patients with MDS/AML and NPM1 monoallelic deletion (NPM1+/−): schema of 5q breakpoints. Upper: ideogram of the long arm of chromosome 5 in G banding. Lower: genomic clones at proximal (q11.2– q14.1 sub-bands) and distal (q35.1–q35.3 sub-bands) breakpoints with percentages of cases. B) G-banded karyotype of a representative case (n.103, Supporting Table S1) showed a complex karyotype including monosomies, unbalanced translocations, five markers, and a very small deleted chromosome 5 corresponding to the largest 5q deletion including NPM1 with breakpoints at q11.2 and q35.3 (Right ideogram). C) FISH of case n.103: concomitant deletion of RP11-117L6/NPM1 (green) and RP11-266N13 (red) indicate centromeric breakpoint (left) and RP1-240G13/subtelomeric sequences (red) indicate telomeric breakpoint (right). Only one copy of each clone is present. D) Gene sequencing of case n.103 shows no NPM1 exon 12 mutation in the non-deleted chromosome 5. The last 12 amino acids encoded by exon 12 of wild type NPM1 (NM_002520) are annotated on top of the sequence.
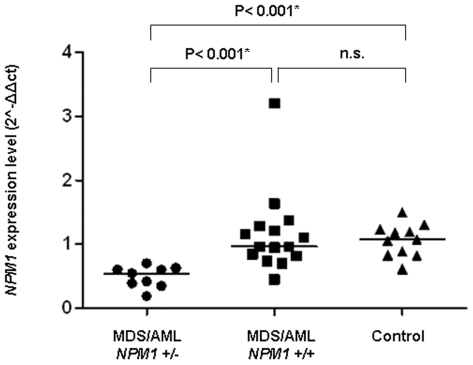
Exon 12 mutations were not found in this study indicating that monoallelic NPM1 deletion and heterozygous mutation can not occur at the same time ( Figure 1D and Table S1). Furthermore, the NPM1 deletion corresponded to reduced NPM1 expression in MDS/AML cells which was significantly lower in NPM1+/− cases than in the NPM1+/+ (P<0.001) and normal controls (P<0.001) ( Figure 2 ). Notably, NPM1 is a multifunctional protein which regulates ribosome biogenesis, TP53 protein activity and stability, ARF protein stability and localization, and DNA integrity [1], [19], [20]. In keeping with evidence from murine models [21], in which NPM1 loss induced morphological signs of MDS, favored c-MYC lymphomagenesis and underlay chromosomal instability, chromosome rearrangements constituting complex karyotypes were significantly different in NPM1+/− and NPM1+/+ patients. In NPM1+/+ structural chromosome rearrangements (translocations; deletions; insertions; inversions) were significantly more frequent (median 3, range 0–7 vs median 1.5, range 0–5; P = 0.013) (Table S2) but in the NPM1+/− group gross chromosomal changes (markers i.e. derivative chromosomes unclassified by karyotyping, rings, and double minutes) were significantly more common (median 3, range 0–9 vs median 0, range 0–6; P<0.001) (Table S2). Aneuploidy was present in 47/64 NPM1+/+ patients (73%) and in 38/38 NPM1+/− patients (100%) (P<0.001). Significantly more monosomies were found in the NPM1+/− subgroup (median 2, range 0–8 vs median 1, range 0–6; P<0.001) (Table S2), with the most frequent, monosomy 7, being detected in 18/38 (47%) of NPM1+/− and in 11/64 (17%) of NPM1+/+ (P = 0.002).
Figure 2. RT-qPCR and NPM1 gene expression.
NPM1 expression in 9 patients with MDS/AML NPM1+/− (median: 0.5443; range: 0.18÷0.70), in 15 with MDS/AML NPM1+/+ (median: 0.96; range: 0.44÷3.20) and in 11 healthy controls (median 1.07; range 0.61÷1.50). Horizontal lines indicate median NPM1 expression in each group. Y axis = 2∧−ΔΔCt value (NPM1 gene expression level normalized to endogenous ABL1 gene) ΔCt = Ct NPM1 −Ct ABL1, ΔΔCt = ΔCt sample−ΔCtcalibrator. (P values according to the Mann-Whitney U test and Bonferroni correction); n.s. = non significant.
Since NPM1 is related to TP53 [19], , a master gene of chromosome stability that is frequently involved in MDS/AML with complex karyotype [24], 57 cases from this series were investigated ( Table 3 and TableS1). Univariate analysis on TP53 and chromosome rearrangements showed a significant association between the number of monosomies and TP53 deletion and/or mutation (median 2 range 0−8 vs median 0 range 0−3; P = 0.006) (Table S3). However, both NPM1 and TP53 gene status maintained significant association with monosomies in a multivariate analysis (P = 0.014 and P = 0.013, respectively) (Poisson regression model), suggesting each gene plays an independent role, but may interact synergistically, in determining chromosomal abnormalities.
Table 3. TP53 monoallelic deletion and/or mutations in 49/57 patients with MDS/AML and non-isolated 5q-.
| TP53 del | TP53 mut | TP53 del/TP53 mut | |
| NPM1 +/+ | 11 | 7 | 5 |
| NPM1 +/− | 11 | 11 | 4 |
del, monoallelic deletion; mut, mutation.
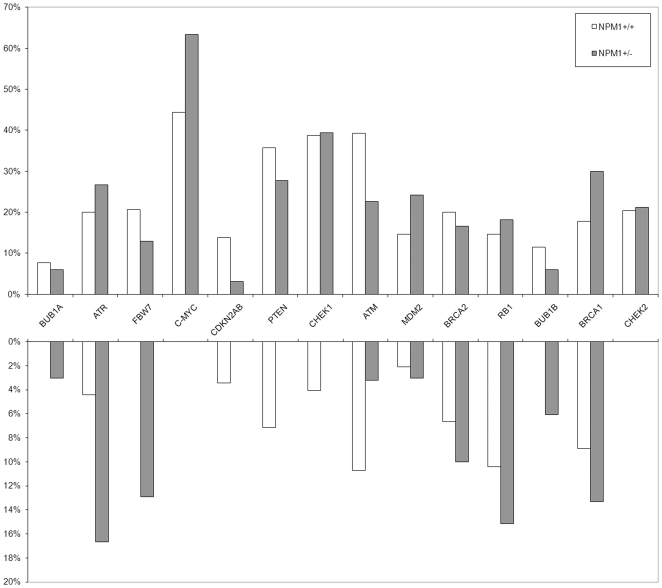
NPM1 loss was not associated with copy number variations of 14 genes other than TP53 that are putatively involved in the complex regulatory network of genetic stability. Gains and losses of clones containing BUB1A, ATR, FBW7, c-MYC, CDKN2A-2B, PTEN, CHEK1, ATM, MDM2, BRCA2, RB1, BUB1B, BRCA, CHEK2 were equally distributed in NPM1+/+ and NPM1+/− cases ( Figure 3 ), suggesting that whatever roles these genes play in MDS/AML with complex karyotypes, they are independent of the NPM1 deletion.
Figure 3. FISH investigations on clones encompassing 14 genes that are putatively involved in genomic stability.
Gains (upper) and losses (lower) in NPM1+/+ patients (white columns) and NPM1+/− (grey columns).
In conclusion these findings prove that haploinsufficiency of the NPM1 gene, is present in more than 40% of patients with high risk MDS/AML characterised by chromosome 5q- and complex karyotypes. NPM1 haploinsufficiency and mutation appear to exert very different effects ( Figure 4 ) as patients with NPM1 mutated-AML have normal karyotype, rare TP53 involvement, good prognosis if FLT3-ITD mutations are absent, and a predominance of females [2], [25], [26]. NPM1 haploinsufficiency was not linked to female gender (Table S4). It significantly reduced NPM1 expression and, in synergy but independently of TP53 deletion and/or mutation, was associated with a strong tendency to perturb chromosomal stability. Consequently NPM1 gene status appears to indicate diverse genetic backgrounds in two different myeloid leukemias.
Figure 4. Mutually exclusive NPM1 abnormalities indicate different leukemogenic pathways.
Supporting Information
Demographics, clinical findings, results of cytogenetics, FISH and mutational analysis in 145 patients with MDS/AML. Footnotes: M, male; F, female; RAEB, refractory anemia with excess of blasts; RA, refractory anemia; AML, acute myeloid leukemia; RCMD, refractory cytopenia with multilineage dysplasia; MDS-U, myelodysplastic syndrome, unclassifiable; RARS, refractory anemia with ringed sideroblasts; ex, exon; DEL, monoallelic deletion; NL, normal hybridization pattern; GAIN, ≥3 hybridization signals; (a) cases with NPM1 RT-qPCR analysis; (b) copy number >5.
(0.12 MB XLS)
Distribution of markers, monosomies, structural aberrations, and trisomies in NPM1+/+ (0) and NPM1+/− (1) cases (Mann-Whitney U Test).
(0.47 MB DOC)
Distribution of markers, monosomies, structural aberrations, and trisomies according to the TP53 status (0 = no deletion or mutation; 1 = deletion and/or mutation) (Mann-Whitney U Test).
(0.47 MB DOC)
Sex distribution between in NPM1+/+ and NPM1+/− groups. Sex (males = 0, females = 1) distribution between in NPM1+/+ (0) and NPM1+/− (1) groups.
(0.47 MB DOC)
Acknowledgments
Authors wish to thank Dr. Geraldine Boyd for assistance in preparing the manuscript.
Footnotes
Competing Interests: Brunangelo Falini and Cristina Mecucci applied for a patent on clinical use of NPM1 mutants. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: This work was supported by Ministero per l'Istruzione, l'Universita e la Ricerca Scientifica - Fondo per gli Investimenti per la Ricerca di Base (MIUR-FIRB, http://firb.miur.it/) and Associazione Italiana Ricerca sul Cancro (AIRC, http://www.airc.it/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- 2.Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 3.Berger R, Busson M, Baranger L, Hélias C, et al. Loss of the NPM1 gene in myeloid disorders with chromosome 5 rearrangements. Leukemia. 2006;20:319–321. doi: 10.1038/sj.leu.2404063. [DOI] [PubMed] [Google Scholar]
- 4.Van den Berghe H, Cassiman JJ, David G, Fryns JP, Michaux JL, et al. Distinct haematological disorder with deletion of long arm of no. 5 chromosome. Nature. 1974;251:437–438. doi: 10.1038/251437a0. [DOI] [PubMed] [Google Scholar]
- 5.Pedersen-Bjergaard J, Christiansen DH, Desta F, Andersen MK. Alternative genetic pathways and cooperating genetic abnormalities in the pathogenesis of therapy-related myelodysplasia and acute myeloid leukemia. Leukemia. 2006;20:1943–1949. doi: 10.1038/sj.leu.2404381. [DOI] [PubMed] [Google Scholar]
- 6.Boultwood J, Fidler C, Strickson AJ, Watkins F, Gama S, et al. Narrowing and genomic annotation of the commonly deleted region of the 5q- syndrome. Blood. 2002;99:4638–4641. doi: 10.1182/blood.v99.12.4638. [DOI] [PubMed] [Google Scholar]
- 7.Royer-Pokora B, Trost D, Müller N, Hildebrandt B, Germing U, et al. Delineation by molecular cytogenetics of 5q deletion breakpoints in myelodyplastic syndromes and acute myeloid leukemia. Cancer Genet Cytogenet. 2006;167:66–69. doi: 10.1016/j.cancergencyto.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Ebert BL. Deletion 5q in myelodysplastic syndrome: a paradigm for the study of hemizygous deletions in cancer. Leukemia. 2009;23:1252–1256. doi: 10.1038/leu.2009.53. [DOI] [PubMed] [Google Scholar]
- 9.Barlow JL, Drynan LF, Hewett DR, Holmes LR, Lorenzo-Abalde S, et al. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q- syndrome. Nat Med. 2010;16:59–66. doi: 10.1038/nm.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisenmann KM, Dykema KJ, Matheson SF, Kent NF, DeWard AD, et al. 5q- myelodysplastic syndromes: chromosome 5q genes direct a tumor-suppression network sensing actin dynamics. Oncogene. 2009;28:3429–41. doi: 10.1038/onc.2009.207. [DOI] [PubMed] [Google Scholar]
- 11.Starczynowski DT, Kuchenbauer F, Argiropoulos B, Sung S, Morin R, et al. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med. 2010;16:49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 12.Graubert TA, Payton MA, Shao J, Walgren RA, Monahan RS, et al. Integrated Genomic Analysis Implicates Haploinsufficiency of Multiple Chromosome 5q31.2 Genes in De Novo Myelodysplastic Syndromes Pathogenesis. PLoS ONE. 2009;4:e4583. doi: 10.1371/journal.pone.0004583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.ISCN Shaffer LG, Tommerup N, editors. An International System for Human Cytogenetic Nomenclature. 2005. Basel∷ Karger, 2005.
- 14.Crescenzi B, La Starza R, Romoli S, Beacci D, Matteucci C, et al. Submicroscopic deletions in 5q- associated malignancies. Haematologica. 2004;89:281–285. [PubMed] [Google Scholar]
- 15.van Dongen JJ, Macintyre EA, Gabert JA, Delabesse E, Rossi V, et al. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Report of the BIOMED-1 Concerted Action: investigation of minimal residual disease in acute leukemia. Leukemia. 1999;13:1901–28. doi: 10.1038/sj.leu.2401592. [DOI] [PubMed] [Google Scholar]
- 16.Gabert J, Beillard E, van der Velden VH, Bi W, Grimwade D, et al. Standardization and quality control studies of ‘real-time’ quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia - a Europe Against Cancer program. Leukemia. 2003;17:2318–2357. doi: 10.1038/sj.leu.2403135. [DOI] [PubMed] [Google Scholar]
- 17.Beillard E, Pallisgaard N, van der Velden VH, Bi W, Dee R, et al. Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using ‘real-time’ quantitative reverse-transcriptase polymerase chain reaction (RT-qPCR) - a Europe against cancer program. Leukemia. 2003;17:2474–2486. doi: 10.1038/sj.leu.2403136. [DOI] [PubMed] [Google Scholar]
- 18.Lessard M, Hélias C, Struski S, Perrusson N, Uettwiller F, et al. Fluorescence in situ hybridization analysis of 110 hematopoietic disorders with chromosome 5 abnormalities: do de novo and therapy-related myelodysplastic syndrome-acute myeloid leukemia actually differ? Cancer Genet Cytogenet. 2007;176:1–21. doi: 10.1016/j.cancergencyto.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 19.Colombo E, Bonetti P, Lazzerini Denchi E, Martinelli P, et al. Nucleophosmin is required for DNA integrity and p19Arf protein stability. Mol Cell Biol. 2005;25:8874–8886. doi: 10.1128/MCB.25.20.8874-8886.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gjerset RA. DNA damage, p14ARF, nucleophosmin (NPM/B23), and cancer. J Mol Histol. 2006;37:239–251. doi: 10.1007/s10735-006-9040-y. [DOI] [PubMed] [Google Scholar]
- 21.Grisendi S, Bernardi R, Rossi M, Cheng K, Khandker L, et al. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437:147–153. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- 22.Kurki S, Peltonen K, Laiho M. Nucleophosmin, HDM2 and p53: players in UV damage incited nucleolar stress response. cell Cycle. 2004;3:976–9. [PubMed] [Google Scholar]
- 23.Latonen L, Laiho M. Cellular UV damage responses—functions of tumor suppressor p53. Biochim Biophys Acta. 2005;1755:71–89. doi: 10.1016/j.bbcan.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 24.Bowen D, Groves MJ, Burnett AK, Patel Y, Allen C, et al. TP53 gene mutation is frequent in patients with acute myeloid leukemia and complex karyotype, and is associated with very poor prognosis. Leukemia. 2009;23:203–206. doi: 10.1038/leu.2008.173. [DOI] [PubMed] [Google Scholar]
- 25.Thiede C, Koch S, Creutzig E, Steudel C, Illmer T, et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML). Blood. 2006;107:4011–4020. doi: 10.1182/blood-2005-08-3167. [DOI] [PubMed] [Google Scholar]
- 26.Döhner K, Döhner H. Molecular characterization of acute myeloid leukemia. Haematologica. 2008;93:976–982. doi: 10.3324/haematol.13345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Demographics, clinical findings, results of cytogenetics, FISH and mutational analysis in 145 patients with MDS/AML. Footnotes: M, male; F, female; RAEB, refractory anemia with excess of blasts; RA, refractory anemia; AML, acute myeloid leukemia; RCMD, refractory cytopenia with multilineage dysplasia; MDS-U, myelodysplastic syndrome, unclassifiable; RARS, refractory anemia with ringed sideroblasts; ex, exon; DEL, monoallelic deletion; NL, normal hybridization pattern; GAIN, ≥3 hybridization signals; (a) cases with NPM1 RT-qPCR analysis; (b) copy number >5.
(0.12 MB XLS)
Distribution of markers, monosomies, structural aberrations, and trisomies in NPM1+/+ (0) and NPM1+/− (1) cases (Mann-Whitney U Test).
(0.47 MB DOC)
Distribution of markers, monosomies, structural aberrations, and trisomies according to the TP53 status (0 = no deletion or mutation; 1 = deletion and/or mutation) (Mann-Whitney U Test).
(0.47 MB DOC)
Sex distribution between in NPM1+/+ and NPM1+/− groups. Sex (males = 0, females = 1) distribution between in NPM1+/+ (0) and NPM1+/− (1) groups.
(0.47 MB DOC)