Abstract
Idiosyncratic drug hepatotoxicity is a major problem in pharmaceutical development due to poor prediction capability of standard preclinical toxicity assessments and limited knowledge of its underlying mechanisms. Findings in animal models have shown that adverse effects of numerous drugs with idiosyncratic hepatotoxicity in humans can be reproduced in the presence of coincident inflammatory cytokine signaling. Following these observations, we have recently developed an in vitro drug/inflammatory cytokine co-treatment approach that can reproduce clinical drug hepatotoxicity signatures -- particularly for idiosyncratic drugs -- in cultured primary human hepatocytes. These observations have suggested that drug-induced stresses may interact with cytokine signaling to induce hepatic cytotoxicity, but the hepatocyte signaling mechanisms governing these interactions are poorly understood. Here, we collect high-throughput phosphoprotein signaling and cytotoxicity measurements in cultured hepatocytes, from multiple human donors, treated with combinations of hepatotoxic drugs (e.g. trovafloxacin, clarithromycin) and cytokines (tumor necrosis factor-α, interferon-γ, interleukin-1α, and interleukin-6). We demonstrate, through orthogonal partial least-squares regression (OPLSR) modeling of these signal-response data, that drug/cytokine hepatic cytotoxicity is integratively controlled by four key signaling pathways: Akt, p70 S6 kinase, MEK–ERK, and p38–HSP27. This modeling predicted, and experimental studies confirmed, that the MEK–ERK and p38–HSP27 pathways contribute pro-death signaling influences in drug/cytokine hepatic cytotoxicity synergy. Further, our four-pathway OPLSR model produced successful prediction of drug/cytokine hepatic cytotoxicities across different human donors, even though signaling and cytotoxicity responses were both highly donor-specific. Our findings highlight the critical role of kinase signaling in drug/cytokine hepatic cytotoxicity synergies and reveal that hepatic cytotoxicity responses are governed by multi-pathway signaling network balance.
Keywords: partial least-squares regression (PLSR), inflammation, xenobiotic, adverse drug reactions, signaling network, predictive data models
Introduction
Hepatotoxicity† is a major cause of failures in the pre-clinical, clinical and post-approval stages of drug development and thus represents a major challenge for the pharmaceutical industry1, 2. Idiosyncratic drug hepatotoxicity -- a hepatotoxicity subset that occurs in a very small fraction of human patients (~1 in 10,000) and accounts for ~10% of acute liver failure cases -- is poorly predicted by standard preclinical models and in clinical trials and frequently leads to post-approval drug failure3. Following the establishment of bacterial liposaccharide (LPS)-administered rodent models to assess inflammation-associated idiosyncratic drug hepatotoxicity3, 4, we have previously developed an drug/inflammatory cytokine co-treatment cell culture approach for studying inflammation-associated drug hepatic cytotoxicity in vitro5. Our approach demonstrated that numerous drugs with clinical findings of idiosyncratic chronic hepatotoxicity, but not their non-toxic control compounds, exhibit synergistic induction of acute hepatocellular death in multiple hepatocyte cell culture models co-dosed with drugs and cytokines at concentrations near their upper limits of clinical liver exposure. In that work, we showed that the drugs exhibiting inflammation-associated toxicities induce mild hepatocellular stresses that synergize with cytokine signaling to elicit hepatocellular death3. A novel concept speculated upon therein was that signaling network dysregulation underlies cooperative drug/cytokine hepatic cytotoxicity, and we now undertake direct examination of that hypothesis in this study.
The factors identified as potentiating inflammation-associated drug hepatic cytotoxicity (LPS and the cytokines tumor necrosis factor-α [TNF], interferon-γ [IFN-γ], interleukin-1α [IL-1α], and interleukin-6 [IL-6]) . in these animal and cell culture models stimulate a diversity of intracellular signaling pathways related to hepatocyte survival, stress, and apoptosis6-11 (Fig. 1). For example, TNF activates the MEK–ERK, IKK–NF-κB, JNK, and p38–HSP27 signaling pathways and the caspase cascade, whose integrated activities specify hepatocyte death responses to TNF6, 12. Further, physiologically relevant growth factor co-stimuli such as insulin can provide activation of survival signaling pathways that antagonize cytokine-induced apoptosis13, 14 and modulate hepatocyte metabolism of xenobiotic compounds15. How these pathways regulate drug/cytokine toxicity outcomes in hepatocytes is not clear, but there is evidence that, in a LPS-administered mouse model, hepatotoxicity synergy between LPS-induced cytokine release and the idiosyncratic hepatotoxicant ranitidine is dependent on p38-mediated TNF release in the resident liver macrophages, Kupffer cells11, 16-18. Drug-induced hepatocellular stresses can induce activation of many of the signaling pathways stimulated by cytokines and/or growth factors (Fig. 1). Glutathione depletion can perturb cellular nutrient levels, possibly leading to attenuation of mTOR signaling19, 20, and can alter mitochondrial redox potentials, increasing susceptibility to mitochondria-mediated apoptosis21. Reactive oxygen species accumulation can activate the JNK and p38–HSP27 stress-response signaling pathways6. Consequently, we hypothesize that further identification of the signaling mechanisms governing hepatocyte cell death responses to hepatotoxicants and inflammatory cytokines would benefit from a network-level examination of multiple intracellular signaling pathways.
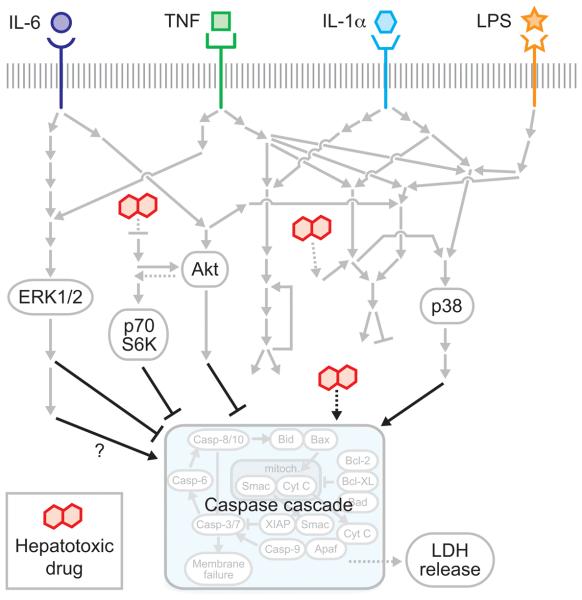
Figure 1.
A schematic of the drug- and cytokine-induced hepatocellular death signaling network, curated from literature references6-14, 19-21. Inflammatory cytokines bind to their cognate receptors and activate shared downstream phosphoprotein signaling pathways regulating cell death. Hepatotoxic drugs (hepatotoxicants) lead to various cellular stresses (e.g. ROS accumulation) that are integrated into stress-related phosphoprotein signaling (e.g. JNK pathway) and mitochondrial control of apoptosis. Phosphoproteins and cell death phenotypes experimentally assayed in this study are indicated.
Here, we use orthogonal partial least-squares regression (OPLSR) data modeling to analyze a hepatic cytotoxicity data compendium, spanning diverse drug/cytokine stimulation combinations and containing measurements of 17 phosphoproteins from multiple signaling pathways and a cell toxicity readout, in cultured human hepatocytes. OPLSR modeling indicated that the activation profiles of four signaling pathways -- Akt, p70 S6K, MEK–ERK, and p38–HSP27 -- are sufficient to accurately predict toxicity responses to drug/cytokine co-treatments. Model predictions of pro-death contributions from p38–HSP27 and, counter-intuitively, MEK–ERK signaling were confirmed using small molecule kinase inhibitors. This work highlights the importance of a multi-pathway signaling network state for characterizing hepatic cytotoxicity arising from drug/cytokine synergies.
Results
Drug/cytokine co-treatments elicit shared regulation of multiple phosphoprotein signaling pathways
To examine the signaling mechanisms regulating inflammatory cytokine-associated drug hepatic cytotoxicity, we collected a ‘cue-signal-response’ (CSR) drug- and cytokine-induced hepatic cytotoxicity data compendium in primary human hepatocytes (Fig. 2). Human hepatocytes (donor #1) were treated with 66 different combinations of 11 ‘drug’ conditions (six compounds with inflammation-associated hepatic cytotoxicity, four corresponding ‘comparison’‡ compounds, and a DMSO control) and six ‘cytokine’ conditions (no cytokine, TNF, IL-1α, IL-6, LPS, and a mix containing all three cytokines and LPS). The hepatotoxicants (e.g. trovafloxacin, nefazodone, nimesulide, clarithromycin) and cytokines were selected based on identification of their role in inducing synergistic hepatic cytotoxicity synergy in our previous large-scale drug/cytokine co-treatment screen5. This study focused on single drug and cytokine dosing concentrations (drugs at 100*plasma Cmax5, 22) -- rather than examining drug and/or cytokine dose-response effects -- in order to enable a diversity of combinatorial drug/cytokine treatments within the constraints of limited human primary hepatocyte samples. Bead-based phosphoprotein and LDH release assays were used to quantitatively assess signaling pathway activation and cell death, respectively. Seventeen phosphoproteins, including signaling components of the MEK–ERK, mTOR–p70 S6K, PI3K–Akt, IKK–NF-κB, JNK, p38–HSP27, STAT3, STAT6, cell cycle regulatory, and DNA damage signaling pathways, were measured at both early (0 and 20 min) and delayed time-points (4, 24, and 48 hr) following drug and/or cytokine stimulation to capture a diversity of intracellular signaling dynamics. These pathways are downstream of the cytokine and growth factor stimuli examined, but to our knowledge have not been reported to be directly regulated by the drugs used in this study (Fig. 1).
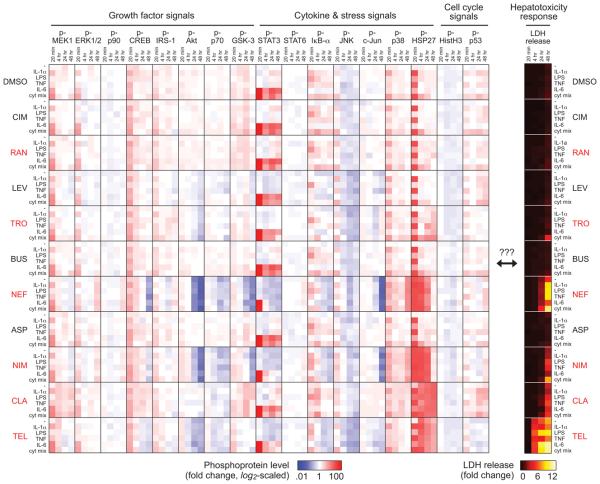
Figure 2.
A cue-signal-response (CSR) drug/cytokine-induced hepatic cytotoxicity data compendium for model training. Cultured primary human hepatocytes (donor #1) were treated with 66 combinations of 11 ‘drug’ conditions (six idiosyncratic hepatotoxicants, four corresponding ‘comparison’ compounds, and a DMSO control) and six ‘cytokine’ conditions (no cytokine, 20 ng/ml IL-1α, 10 μg/ml LPS, 100 ng/ml TNF, 20 ng/ml IL-6, and a mix containing all three cytokines plus LPS). All drugs were dosed at 100*Cmax concentrations (see Experimental): 1.5 mM cimetidine (CIM), 142 μM ranitidine (RAN), 1.6 mM levofloxacin (LEV), 770 μM trovafloxacin (TRO), 0.46 μM buspirone (BUS), 86 μM nefazodone (NEF), 552 μM aspirin (ASP), 2.1 mM nimesulide (NIM), 334 μM clarithromycin (CLA), and 277 μM telithromycin (TEL). Idiosyncratic hepatotoxicants (red font) and corresponding ‘comparison’ compounds are presented in vertical juxtaposition. Seventeen phosphoproteins (left) and LDH release (right) were quantitatively assayed at both early (0 and 20 minutes) and delayed time-points (4, 24, and 48 hours) following drug and/or cytokine stimulation. Phosphoprotein levels were fold-change normalized to untreated samples at 0 minutes and are plotted on a log2-scaled colormap to capture both up- and down-regulated signaling levels. LDH release levels were fold-change normalized to untreated samples at 48 hours post-stimulation and are plotted on a linearly scaled colormap. Single biological replicates were used for both phosphoprotein and LDH assays.
Across this CSR compendium, it was evident that cytokine-only treatments induced early-phase (20 min-4 hr) activation of multiple signals, including p-MEK1, p-STAT3, p-IκB-α, and p-p38. Many toxic drug-only treatments induced a late-phase (24-48 hr) down-regulation of certain pro-survival signals (e.g. p-Akt, p-p70 S6K; see also Fig. 3D-E). This late-phase down-regulation was not evident for all phosphoproteins, as some survival (e.g. p-ERK1/2) and stress (e.g. p-HSP27) signals had sustained activation under toxic conditions (see also Fig. 4C,F). Multiple drug/cytokine co-treatments (e.g. trovafloxacin ± the three-cytokine/LPS mix) synergistically induced both sustained p38–HSP27 signaling and cell death (Fig. 1 [donor #1] and Fig. 4B,F [donor #2]). This paired synergy raised the possibility that a single phosphoprotein signal might unilaterally yield a predictive readout of cell death across the entire data compendium. However, we calculated the correlation (Pearson coefficients) between observed cell death at 48 hours post-drug/cytokine co-treatment and each 17 phosphoprotein signal levels, and even the most positively (p-HSP27 at t = 4 hr, R = 0.78) and negatively (p-Akt at t = 24 hr, R = −0.74) correlated single phosphoprotein was poorly predictive of measured cell death (Fig. S3). This demonstrated need for multivariate analysis of signal-response relationships present in drug- and cytokine-induced hepatic cytotoxicity.
Figure 3.
OPLSR modeling identifies key molecular signals regulating drug/cytokine-induced hepatic cytotoxicity. An OPLSR model was trained on the 66-condition CSR data compendium from human hepatocyte donor #1 to relate the 102 time-dependent phosphoprotein signaling metrics (17 phosphoproteins × 6 metrics) to the cell death measurement (LDH release at t = 48 hr). (A) A correlation plot relating the observed and OPLSR model-predicted LDH release responses for all 66 drug/cytokine co-treatment conditions for the 17-phosphoprotein and a reduced 4-phosphoprotein (p-ERK1/2, p-Akt, p-p70 S6K, and p-HSP27) model both demonstrate good model fitness (R2)28, 29. (B-C) OPLSR model loadings and VIP scores from the 17-phosphoprotein model. Positive loadings contribute to and negative loadings antagonize the model's cell death prediction. VIP scores identify the relative importance of individual phosphoprotein signaling metrics, with the threshold value of 1 for informative VIP scores28, 31 used as the lower-limit of the color scale in (B) and is plotted in (C). In (D), VIP scores from individual metrics and averages for each phosphoprotein signal are plotted, with the 20-min metrics were omitted, as these metrics were uniformly uninformative. (D) Model fitness sensitivity to the removal of phosphoprotein signaling metrics. Model complexity was reduced using: (i) step-wise removal of the lowest average VIP phosphoprotein; (ii) by selecting four (p-ERK1/2, p-Akt, p-p70 S6K, and p-HSP27) or six (those four plus p-MEK1 and p-p38) phosphoproteins; or (iii) using an equivalent number of signaling metrics as the four- and six-phosphoprotein models (24 and 36 metrics, respectively) but selected from the top VIP scores across all phosphoprotein metrics.
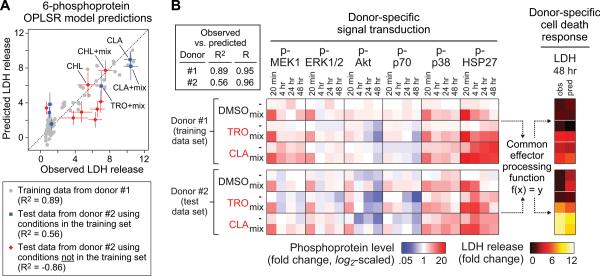
Figure 4.
A CSR drug/cytokine-induced hepatic cytotoxicity data compendium for model training. (A) Cultured primary human hepatocytes (donor #2) were treated with 14 combinations of seven ‘drug’ conditions (two idiosyncratic hepatotoxicants used in initial compendium, four idiosyncratic hepatotoxicants not used in the initial compendium, and a DMSO control) and two ‘cytokine’ conditions (no cytokine and the three-cytokine/LPS mix). Idiosyncratic hepatotoxicants (red font) were dosed at 100*Cmax concentrations: 770 μM trovafloxacin (TRO), 334 μM clarithromycin (CLA), 111 μM chlorpromazine (CHL), 10 μM nortriptyline (NOR), 13 μM clomipramine (CLO), and 107 μM riluzole (RIL). Six phosphoproteins (p-MEK1, p-ERK1/2, p-Akt, p-70 S6K, p-p38, p-HSP27) and LDH release were assayed at both early (0 and 20 min) and delayed time-points (4, 24, and 48 hr) following stimulation. Some conditions were also assayed at 1 and 12 hr post-stimulation (see (C-F)). Biological triplicates were used for both phosphoprotein and LDH assays. Phosphoprotein and LDH release levels were normalized and plotted as in Fig. 2. (B) Trovafloxacin, but not clarithromycin, synergizes with cytokine mix co-treatment to induce hepatic cytotoxicity. Differences between drug/cytokine co-treatment and either drug-only or cytokine mix-only conditions are labeled (*) if P < 0.05 by a Student's t test. (C-F) Phosphoprotein signaling time-courses for idiosyncratic drug (TRO or CLA) and/or cytokine mix treatment demonstrate sustained p-ERK1/2 (C) and p-HSP27 (F) activation and late p-Akt (D) and p-p70 S6K (E) down-regulation under hepatotoxic conditions compared to cytokine mix-only controls. In (C-F), DMSO-only control treatments are not shown but are in (A). In (B-F), data is presented as mean ± s.e.m. of three biological replicates.
Multi-pathway OPLSR modeling identifies key molecular signals associated with drug- and cytokine-induced hepatic cytotoxicity
A spectrum of modeling approaches23, 24 are suited for analysis of signal transduction networks and their relationships to phenotypic cellular responses to diverse stimuli, but many of these approaches require a priori knowledge of how cellular stimuli are connected to signaling pathways and how those pathways mechanistically govern cellular responses of interest. The limited understanding of how idiosyncratic hepatotoxicants and inflammatory cytokine signals interact to synergistically induce hepatocyte cell death motivates the use of data-driven modeling approaches such as partial least-squares regression (PLSR), decision trees, and multiple linear regression, which can identify novel connectivities between measured signaling pathways and cellular responses without requiring an a priori network structure. From these approaches, we chose here to employ a variant of partial least-squares regression, which has been previously successful in establishing novel signal-response relationships from high-throughput signaling and response data13, 23, 25-28. PLSR can make predictions of cellular responses across diverse cell types and experimental conditions and be used to reduce complexity in multi-variate protein signaling data to identify the most critical signaling nodes.
To generate a multi-pathway model relating the observed signaling activities and cell death responses, we subjected the initial CSR data compendium to orthogonal partial-least squares regression (OPLSR). OPLSR modeling relates the measured phosphoprotein signaling data matrix (X) and the observed hepatocellular death responses (Y), each arrayed across all 66 treatment conditions, through a linear regression of the relationship Y = X·B, where B is a vector of regression coefficients that reflect how each phosphoprotein signal contributes to cell death. Following regression, the “fitness”of an OPLSR model can be assessed by comparing how the model “predicts”the cell death responses by multiplying the experimental signaling measurements with the model regression coefficients, and then calculating model fitness28, 29 by comparing the “predicted”and experimentally observed responses. We note that, in contrast to previous studies13, 25, 26, 28 implementing cell signaling-response PLSR modeling in cell line systems, our training data here comprises single biological replicates due to the severe limitations of human donor tissue samples; we chose to trade biological replication for a greater landscape of treatment conditions, within the constraints of this patient-oriented study.
Here, we found that an OPLSR model based of 102 phosphoprotein signaling metrics (17 phosphoproteins × 6 time-dependent metrics; see Experimental) demonstrated very good model fitness (R2 = 0.92) for cross-validated predictions of the observed cell death responses for all 66 drug/cytokine co-treatment conditions (Fig. 3A). Examination of the OPLSR model scores28, 30 projections (Fig. S2D) demonstrated the model's ability to distinguish between toxic and non-toxic treatment conditions by their scores in the first principal component (PC), which represents pro-death model contributions. The second PC of this OPLSR model captures remaining, orthogonal variation in the CSR data compendium irrelevant to response prediction.
OPLSR model loadings28, 30 (w1*c1; Fig. 3B and Fig. S4A) reflect the regression coefficients of the signaling metrics; positive model loadings imply signaling metrics with pro-death contributions, and negative model loadings imply pro-survival contributions. Model variable-importance-of-projection (VIP) scores (Fig. 3B-C and Fig. S4B-C) reflect the relative importance of individual signaling metrics to the model fitness by capturing the loss in model fitness upon removal of their associated signaling metric28, 30, 31. Metrics with VIP score >1 have significant importance in the model whereas metrics with a VIP score <<1 significantly lack unique information in the model. Inspection of model loadings and VIP scores identified four signaling pathways (MEK–ERK, Akt, p70 S6K, and p38–HSP27) with phosphoproteins having informative model contributions from four or more signaling metrics (Fig. 3B-C). p-ERK1/2, p-p38, and p-HSP27 were all identified as having significant pro-death contributions from multiple metrics, and p-Akt and p-p70 S6K were identified as having significant pro-survival contributions from multiple metrics. The identification of MEK–ERK signaling as having pro-death contributions by the model may seem surprising in light of this pathway's more commonly presumed pro-survival role, although this pathway has been recently found to similarly provide pro-death influence under inflammatory cytokine treatment in other human epithelial cell types32; this effect is very clear in our study here given the sustained activation of MEK–ERK signaling evident under certain drug treatments (e.g. clarithromycin; Fig. 2 [donor #1] and Fig. 4C [donor #2]). For all 17 measured phosphoprotein, early time-point (20 min) metrics were uninformative as assessed by VIP scores (Fig. 2B and Fig. S4C), likely because these signaling activities were similarly activated by cytokines in both presence and absence of drug co-stimuli. In contrast, metrics from later time points (4-48 hr) and the average and integral metrics were, on average, more informative (Fig. 2B and Fig. S4C).
The distinct importance of these four signaling pathways suggested that many of the phosphoproteins in the full CSR data compendium were neglectable for model prediction, permitting model reduction to more easily interpretable subset of signals. To test this, phosphoproteins (and all six of their associated signaling metrics) were removed from the model stepwise in order of the lowest average VIP score across all six metrics (Fig. 3D). Model fitness was maintained until the top ~5 phosphoproteins remained (R2 = ~0.87-0.91); further removal of phosphoproteins resulted in the significant losses in model fitness. A similar trend in loss of model fitness was observed by model reduction by removal of each of the 102 individual signaling metrics in order of the lowest VIP scores (Fig. S4D). Thus, an equivalently predictive multi-pathway network model could be generated by focusing on representative phosphoproteins from four pathways -- MEK–ERK, Akt, p70 S6K, and p38–HSP27 -- and representative phosphoproteins (p-ERK1/2, p-Akt, p-p70 S6K, and p-HSP27, with R2 = 0.89; Fig. 3A,D) from these pathways can serve as a signaling ‘network gauge’25 (Fig. S5), whose combined activities specify hepatocellular death responses to drug/cytokine treatment.
Multi-pathway OPLSR model successfully predicts hepatic cytotoxicity signal-response relationships across human hepatocyte donors
In this study, human hepatocyte donors were garnered based on availability of sufficient viable cell quantities for assay demands. Donors with known obesity, alcohol use, or liver disease were excluded; availability limitations precluded matching for more general potential variables such as gender or age (Table S1). To test ability of the OPLSR model to make accurate predictions of hepatic cytotoxicity cell death responses to across human hepatocyte donors, we collected a second CSR hepatic cytotoxicity data compendium (in cells from donor #2) containing drug/cytokine co-treatments that were both present in the CSR from donor #1 and new (Fig. 4A). In the second CSR, human hepatocytes were treated with 14 different combinations of seven drug conditions (two hepatotoxicants used in initial compendium, four hepatotoxicants not used in the initial compendium, and a DMSO control) and two cytokine conditions (no cytokine and the three-cytokine/LPS mix). This second CSR focused on six phosphoprotein signals (p-MEK1, p-ERK1/2, p-Akt, p-p70 S6K, p-p38, and p-HSP27) from the four ‘network gauge’ pathways (Fig. 4A), allowing direct evaluation of reduced six-phosphoprotein OPLSR model generated from the first CSR data compendium. We note that human donor variation is evident in the widespread differences observed in both phosphoprotein activation and cell death observed between donors #1 and #2 in response to the same drug/cytokine co-treatments (Fig. S6).
An OPLSR model trained on data from these six phosphoproteins from donor #1 was evaluated for its ability to make cell death predictions based on signaling data in donor #2 (Fig. 5A). The six-phosphoprotein model produced good quantitative predictions of new donor cell death responses for test data on treatments present in the training compendium. Predictions for the yet more stringent challenge of new donor cell death test data on treatments not present in the training compendium were gratifyingly accurate for some of the novel conditions (chlorpromazine ± cytokine mix) though less so for other of the novel conditions. These results demonstrate that the six-phosphoprotein OPLSR model can generate successful a priori predictions of drug/cytokine responses across different human hepatocyte donors for conditions present in the training data set and even for some conditions beyond. For some of the drugs (e.g., clomipramine, riluzole) that were not present in the training set, it is likely that other signaling pathways play a more substantial role in regulating responses drug/cytokine co-stimulation as these drugs induced cell death without significant activation of the four ‘network gauge’ signaling pathways compared to control treatments (Fig. 4A). We thus expect that building a broader training compendium and a network gauge model including even just a few more signaling pathways could further widen the scope of successful a priori prediction.
Figure 5.
OPLSR model testing on CSR drug/cytokine-induced hepatic cytotoxicity data compendia in different human hepatocyte donors. An OPLSR model accurately predicts cell death responses across human hepatocyte donors for drug/cytokine treatments present in the training data compendium, even though donor-specific signaling network activation profiles and cell death responses were observed. (A) An OPLSR model of LDH release was trained on the time-dependent signaling metrics from six phosphoproteins (p-MEK1, p-ERK1/2, p-Akt, p-70 S6K, p-p38, p-HSP27) using data from donor #1. This trained model was used to predict LDH release responses based on the same signaling metrics measured in hepatocytes from donor #2. A correlation plot relating the observed and predicted LDH release responses for the both training data (donor #1) and the test data (donor #2), which contained six treatments present and eight treatments not present in the training data. Test conditions (CHL ± cytokine mix) not present in the training data but nonetheless predicted accurately are noted. The model-predicted responses are presented as the mean prediction ± cross-validation standard error, calculated by jack-knifing42. The test data is presented as mean ± s.e.m. of three biological replicates. Experimental and prediction uncertainties are not shown for the training data. (B) The predictive accuracy of this OPLSR model suggests that a ‘common effector processing’ mechanism (f(x) = y), encompassing the integration of multiple signaling pathways (x) yields quantitatively concerted cell death responses (y), exists and is shared between hepatocytes from different human donors given similar drug/cytokine treatments. The inset tabulates model fitness (R2) and Pearson correlation coefficient (R) values for the model predictions of these shared conditions. See Fig. S6 for expanded line plots comparing the phosphoprotein and LDH release data in donors #1 and #2.
The ability of our ‘network gauge’ modeling approach to predict hepatic cytotoxicity signaling-response relationships across human hepatocyte donors for similar drug/cytokine conditions suggests that hepatocytes from multiple human donors share a ‘common effector processing’ function26. In cells from two different human donors, particular drug/cytokine treatment conditions (e.g. clarithromycin ± cytokine mix) elicited significantly different signaling network activation profiles and induced different levels of cell death (Fig. S6), but the OPLSR model nonetheless quantitatively captured their common signal-response relationship (Fig. 5A-B).
Multi-pathway OPLSR model accurately predicts experimental observations that MEK and p38 kinase inhibitors antagonize drug- and cytokine-induced hepatic cytotoxicity
The initial 17-phosphoprotein OPLSR model suggested that sustained activation of both the MEK–ERK and p38–HSP27 signaling pathways positively contributes to drug- and cytokine-induced hepatocellular death. Whereas the p38–HSP27 pathway is generally considered pro-apoptotic, due to transcriptional regulation of effector caspases33, the MEK–ERK pathway has been considered generally pro-survival in hepatocytes through activation of anti-apoptotic effectors such as Bad34 (Fig. S7). To test this complementary set of expected (p38) and surprising (ERK) predictions, MEK and p38 inhibitors were selected from panel of candidate small molecules for their ability to potently inhibit signaling while themselves inducing minimal toxicity in human hepatocytes (from donor #3; Fig. S8). The selected MEK (U0126 and PD325901) and p38 (PHA-666859 and PHA-818637) inhibitors were used to evaluate (both through model-prediction and experiment) the control of MEK–ERK and p38–HSP27 signaling pathways in drug- and/or cytokine-induced hepatic cytotoxicity.
First, we tested the ability of the six-phosphoprotein OPLSR model to predict effects of MEK and p38 kinase inhibitor on drug/cytokine-induced hepatic cytotoxicity. We reasoned that a most practically implementable challenge would be to predict effects on new donor hepatocytes from a model constructed on a previously-generated database from other human hepatocyte donors using previously-measured signaling inhibition efficacies. That is, given the ‘common effector processing’ paradigm, a conceivable application could be to use a signal-response model established across multiple previous subjects to predict a priori expected effects of interventions on a newly-arising subject. Accordingly, we calculated signaling time-courses for the DMSO ± three-cytokine/LPS mix and the hepatotoxicant nortriptyline ± cytokine mix conditions using uninhibited signaling data from all conditions examined in donors #1 and/or #2 in concert with data for MEK and p38 inhibitors from donor #3 (Fig. S8). This calculation entailed decreasing the phosphoprotein levels of both the inhibitor's targeted kinase (e.g. p-p38) and its downstream substrate (e.g. p-HSP27) by the experimentally-determined proportions (e.g. 93% decrease for the p38 inhibitor PHA-666859; see Supplementary Experimental Methods). Since these drug/cytokine ± inhibitor conditions produced very low levels of cell death (Fig. 6 and Fig. S9), we logarithmically scaled the response data to remove regression bias towards high-response conditions (see Fig. S9A-B).
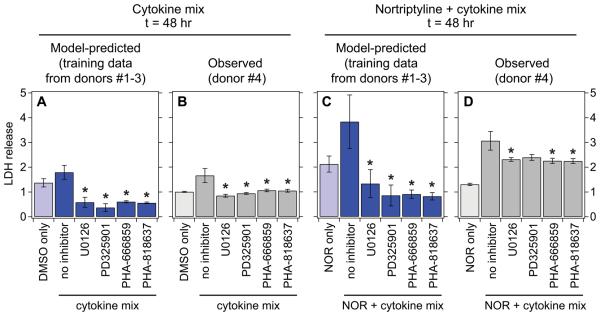
Figure 6.
Kinase inhibitor perturbation of MEK and p38 signaling confirms their model-predicted pro-death signaling regulation of drug/cytokine-induced hepatic cytotoxicity. (A,C) OPLSR model predictions of the effects of MEK and p38 inhibitors in perturbing drug/cytokine-induced hepatic cytotoxicity. To make predictions of the effects of kinase inhibitor perturbation on hepatocellular death responses, uninhibited signaling time-courses measured in cells from donors #1 and/or #2 were ‘computationally inhibited’ based on inhibitor activities measured in cells from donor # 3 (Fig. S8). An OPLSR model trained on data from donors #1 and #2 was used to make cell death response predictions from these ‘computationally inhibited’ time-courses (Fig. S9A-B). Model predictions are presented as the mean ± cross-validation standard error, calculated by jack-knifing42. (B,D) Experimental inhibition of MEK and p38 kinase activities demonstrates their pro-death regulation of cytokine mix- and nortriptyline (NOR) + cytokine mix-induced hepatic cytotoxicity. Cultured human hepatocytes (donor #4) were treated with DMSO control (B) or 10 μM nortriptyline (D) ± a three-cytokine/LPS mix and were assayed for LDH release 48 hr post-treatment. Cells were pretreated with a MEK inhibitor (10 μM U0126 or 1 μM PD325901) or a p38 inhibitor (1 μM PHA-666859 or 1 nM PHA-818637) one hour before stimulation. Data are presented as the mean ± s.e.m. of eight biological replicates. Kinase inhibition did not significantly perturb cell death responses in the absence of cytokine mix treatment (Fig. S9C-D). In (A) to (D), differences between to control and inhibitor conditions are labeled as significant (*) if P < 0.05 by a Student's t test.
Using a log-scale OPLSR model trained on CSR data from donors #1 and #2, we predicted the cell death responses to multiple MEK and p38 inhibitors based on the signaling time-course perturbation method described above (Fig. 6A,C). As expected, the model predicted significant decreases in hepatic cytotoxicity induced by cytokine mix-only and nortriptyline + cytokine mix treatments upon MEK or p38 kinase inhibitor pre-treatment. In human hepatocytes (from donor #4) treated with cytokine mix-only or the idiosyncratic hepatotoxicant nortriptyline + cytokine mix, one or both of MEK and p38 kinase inhibitors reduced the observed hepatotoxicities (LDH release at t = 48 hr; Fig. 6B,D). MEK or p38 inhibition did not perturb hepatic cytotoxicity in DMSO control or nortriptyline only conditions (Fig. S9C-D). Quantitative accuracy of the model predictions (based on data from donors #1-3) was assessed by comparing to these experimental observations in donor #4 (Fig. S9). The log-scaled OPLSR model demonstrated promising model fitness for the training data (R2 = 0.79), but unsatisfactory predictions of the inhibitor test data (R2 = 0.08). However, using a Pearson correlation metric, the model generated well-correlated predictions of both training (R = 0.90) and inhibitor test (R = 0.63) data. Importantly, the model correctly predicted the experimentally observed pro-survival effects of MEK and p38 inhibition under these treatment conditions, confirming that both MEK–ERK and p38–HSP27 signaling indeed both contribute positively to the induction of hepatocellular death by drug/cytokine co-treatment. Moreover, this demonstrates that the reduced six-phosphoprotein OPLSR model is capable of at least qualitatively correct predictions of kinase inhibitor perturbations to drug/cytokine hepatic cytotoxicity in a practically implementable protocol.
Discussion
To date, systems-level analyses of drug-associated hepatotoxicity have largely focused on gene expression35 or metabonomic36 data. Our new work here demonstrates the utility of a systems modeling approach for predicting hepatic cytotoxicity in human hepatocytes based on phosphoprotein signaling data. Using orthogonal partial least-squares regression modeling of multi-pathway phosphoprotein data, we showed that four key signaling pathways are instrumental to the cooperative induction of hepatocellular death by hepatotoxicants and physiologically relevant inflammatory cytokine co-stimuli. This informative subset contained two pathways (Akt and p70 S6K) with model-assigned pro-survival functions, in agreement with their accepted survival signaling roles19, 37 and two pathways (MEK–ERK and p38–HSP27) with model-assigned pro-death functions. These four model-identified signaling pathways represent a useful ‘network gauge’25 capturing the balance between pro-survival and pro-death signaling in drug- and cytokine-stimulated hepatocytes (Fig. S5). Thus, we propose that this kind of hepatic cytotoxicity may be understood in terms of a signaling network state, in which the integrated behavior of multiple pathways characterizes the effects of a candidate drug in the context of diverse, physiologically relevant co-stimuli (Fig. 7).
Figure 7.
Inflammatory cytokine-associated idiosyncratic drug hepatic cytotoxicity as a ‘network toxicity’. The multi-pathway modeling approach presented here suggests that an integration of the activities of multiple intracellular signaling pathways -- namely ERK, Akt, p70 S6K, and p38 -- is necessary for hepatocytes to fully specify death responses to hepatotoxic drug/cytokine co-treatment conditions. This provides motivation of the network-level consideration of multiple survival, stress, and apoptosis signaling pathways in evaluating the hepatic cytotoxicity mechanisms of context-dependent hepatotoxic drugs.
Though p38 and JNK signaling pathways have been individually identified as critical regulators of liver injury associated with the hepatotoxicants ranitidine11, 16, 18 and acetaminophen38, respectively, this work demonstrates that, for diverse hepatotoxicant and cytokine co-stimulatory conditions, specification of hepatic cytotoxicity responses depends on the pro-survival and pro-death signaling contributions from the Akt, p70 S6K, MEK–ERK, and p38–HSP27 pathways. It is likely that other cytokine signaling pathways, including IKK–NF-κB and JNK, are indeed critical to inflammation-associated idiosyncratic drug hepatotoxicity, but they may be more associated with regulation of the inflammatory response in cells other than hepatocytes and/or are crucial for mechanisms of toxicity not shared by the hepatotoxicants in this study. We note that the MEK-ERK and p38-HSP27 pathways highlighted by our present work may influence a variety of downstream effector mechanisms involved in potentially diverse observed hepatocyte injury behaviors5. Future investigation into the mechanisms connecting drug-induced hepatocellular injuries and kinase signaling activation should lead to improved prediction and understanding of inflammatory cytokine-associated idiosyncratic drug hepatic cytotoxicity.
We submit that our findings here motivates complementary future studies -- perhaps using lower drug and cytokine concentrations -- focused on how hepatocyte signaling networks govern sub-lethal hepatic toxicity phenotypes (such as mitochondrial and drug transporter dysfunction22, 39) measureable in high-throughput cell culture that underlie chronic liver toxicity. Moreover, this type of patient-specific data-driven toxicity modeling could be implemented to generate personalized predictions of idiosyncratic hepatotoxicity based on well-trained, multi-donor in vitro signaling-hepatic toxicity models that are further supplemented by focused in vitro studies on patient-specific hepatocytes using patient-tailored drug doses and cytokine levels.
Our data-driven modeling analysis indicated that the MEK–ERK and p38–HSP27 pathways are activated in more sustained manner by hepatotoxic drug and/or cytokine treatments to positively regulate the resulting cell death response. These predicted pro-death signaling contributions were confirmed using small molecule MEK and p38 kinase inhibitors to perturb the cooperative hepatic cytotoxicity induced by nortriptyline and a cytokine mix. While in agreement with the generally appreciated apoptotic signaling role of p38 signaling33, this result is surprisingly counter to the general view of MEK–ERK contributing to survival signaling32, 34. One conceivable mechanism for the surprisingly similar pro-death consequences of MEK–ERK and p38–HSP27 pathway activation is induction of the cellular metalloproteinase TACE. Activated ERK40 and p3816 can both positively regulate TACE activity and its proteolytic release of TNF41. TNF is a crucial pro-death cytokine in inflammation-induced drug hepatotoxicity in vivo10, 11, where its TACE-regulated release by Kupffer cells is p38-dependent16, 18, and in our in vitro drug/cytokine co-treatment system5. We speculate that sustained ERK and p38 activation by the initial drug/cytokine stimuli may lead to enhanced hepatocyte TNF production and autocrine signaling, which could lead to further pro-death signaling (for example, through prolonged p38 activation; see Fig. 4F). This potential mechanism merits dedicated study in future work.
Experimental
Human hepatocyte cell culture and stimulation
Primary human hepatocytes were obtained from CellzDirect (Durham, NC). Human hepatocytes from multiple donors were used in this study, and donor identification is noted for each data set. Donor information is provided in Table S1. Human hepatocytes were seeded on collagen-coated 12- or 96-well plates (BD Biosciences, Franklin Lakes, NJ) at 1.5×105 cells/cm2 in medium containing 5% FBS (Hyclone, Logan, UT) and 5 μg/ml insulin (Sigma, St. Louis, MO), and, one day post-seeding, were overlayed with 0.25 mg/ml Matrigel (BD Biosciences) in medium containing insulin but not FBS, as described previously5. One day following Matrigel overlay, fresh culture medium containing insulin but not FBS was added. For kinase inhibition studies, this medium contained inhibitors at 2× final concentration; one hour later, an equal volume of medium containing drugs and/or cytokines at 2× final concentration was added. All drugs were suspended in 0.25% final DMSO. Drugs with associated idiosyncratic hepatotoxicity and corresponding ‘comparison’ compounds were dosed at a 100*Cmax concentration, a therapeutically appropriate dosing level (see Supplementary Experimental Methods). Recombinant human cytokines were obtained from R&D Systems (Minneapolis, MN) and were used at the following final concentrations: 100 ng/ml TNF, 20 ng/ml IL-1α, and 20 ng/ml IL-6. LPS serotype 1 from E. coli 0111:B4 (Sigma) was used at 10 μg/ml. The MEK inhibitors PD98059, U0126, and MEK inhibitor-1 (MEKi-1) and the p38 inhibitors SB202190 and SB203580 were obtained from EMD Biosciences (San Diego, CA). The MEK inhibitor PD325901 and p38 inhibitors PHA-666859 and PHA-818637 were obtained from Pfizer's chemical sample bank (Groton, CT). All inhibitors were suspended in 0.1% final DMSO. See Supplementary Experimental Methods for details on selection of inhibitor concentrations.
Multiplexed phosphoprotein assays
Cell lysates were collected at 0, 20 min, 4, 24, and 48 hr following drug/cytokine stimulation, clarified, and normalized using a bicinchonicic assay (Pierce). Bio-Plex bead-based assays (Bio-Rad, Hercules, CA) were conducted using a Luminex 200 instrument (Austin, TX), according to manufacturer's instructions, to quantify the following 17 phosphoproteins in cell lysates: p-Akt (Ser473), p-CREB (Ser133), p-c-Jun (Ser63), p-GSK-3α/β (Ser21/Ser9), p-IκB-α (Ser32/Ser36), p-IRS-1 (Ser636/Ser639), p-ERK1/2 (Thr202/Tyr204, Thr185/Tyr187), p-Histone H3 (Ser10), p-HSP27 (Ser78), p-JNK (Thr183/Tyr185), p-MEK1 (Ser217/Ser221), p-STAT3 (Ser727), p-STAT6 (Tyr641), p-p38 (Thr180/Tyr182), p-p53 (Ser15), p-p70 S6 kinase (Thr421/Ser424), and p-p90 RSK (Thr359/Ser363). Phosphoprotein assays were validated for high specificity to known activating conditions (Fig. S1). See Supplementary Experimental Methods for additional details.
Lactate dehydrogenase cell death assay
At 20 min, 4, 24, and 48 hr post-stimulation, conditioned medium samples were collected to assay lactate dehydrogenase (LDH) release using a CytoTox-ONE Homogeneous Membrane Integrity Assay (Promega, Madison, WI) according to manufacturer's recommendations.
Orthogonal partial least-squares modeling of cue-signal-response data compendia
‘Cue-signal-response’ (CSR) data compendia containing phosphoprotein time-course (t = 0-48 hr) and LDH release end-point (t = 48 hr) data were constructed separately for each human hepatocyte donor. Phosphoprotein data were fold-change normalized to untreated samples (at 0 min) for each phosphoprotein assay and separately for each hepatocyte donor. LDH release data were fold-change normalized to untreated samples at 48 hr post-stimulation separately for each hepatocyte donor. These normalized data are available in Supplementary Data. CSR data compendia normalization and fusion was performed using the DataRail toolbox43 for Matlab (The Mathworks, Inc., Natick, MA). For each phosphoprotein signaling time-course, two time-dependent signaling metrics13 were extracted: (i) integral for the entire time-course, and (ii) the average of the late time-points (4-48 hr), reflecting the steady-state signaling level. These were added to the four time-points (20 min, 4, 24, and 48 hr) to yield six signaling metrics for each assayed phosphoprotein. For each compendium, the signaling metrics from all measured phosphoproteins were then fused into a signaling network data matrix (X), and the toxicity response data were cast into a vector (Y), with both X and Y arrayed across all treatment conditions.
The relationship between the signaling data matrix (X) and the toxicity response vector (Y) was modeled using partial least-squares regression (PLSR). Briefly, we implemented PLSR using the NIPALS algorithm in SIMCA-P software (Umetrics, Inc., Kinnelon, NJ) following standard methods13, 25, 26, 28-30, 44, with some modifications. Data matrices and vectors were mean-centered and unit-variance scaled. All models were generated using four principal components under standard optimization criteria30. Model calibration was conducted using leave-one-out cross-validation, and model uncertainties were calculated by jack-knifing42. Calibrated models were subjected to a principal-component space linear transformation by rotating the projection of the single cell death response variable completely into the first principal-component, yielding an orthogonal PLSR (OPLSR) model45 (see Fig. S2), to allow for simplified interpretation of model loadings and scores. Model loadings were calculated using the mean-centered regression coefficients wa*ca from the a-th OPLSR principal-component13. The information content of each signaling metric was assessed by its variable importance of projection (VIP) score, which captures the loss in model fitness upon removal of the individual signaling metric28, 30, 31. The accuracy of model predictions for both training and test data were assessed using a model fitness parameter28, 29 (R2) and Pearson correlation coefficient25 (R). See Supplementary Experimental Methods for additional details.
Statistical analysis
For comparing two means, a Student's t test was used. All tests were performed at a significance level of α = 0.05.
Statement for Graphical Abstract.
Certain pharmaceutical compounds exhibit rare hepatocyte toxicities in humans that may be dependent on coincident inflammatory cytokine signaling but are poorly understood mechanistically. We employ a systems biology approach to identify a set of four kinase signaling pathways that govern these toxicities across human hepatocyte donors.
Supplementary Material
Acknowledgments
We thank David de Graaf, Jinghai J. Xu, Arthur Goldsipe, and Julio Saez-Rodriguez for valuable discussions, and Katerina Chairakaki and Ioannis Melas for technical help. We are grateful for funding support from Pfizer Inc., the Department of Defense Institute for Collaborative Biotechnologies (to D.A.L.), the MIT Center for Cell Decision Processes (NIH grant P50-GM68762; D.A.L., P.K.S.), the MIT Biotechnology Process Engineering Center (L.G.G.), the MIT Center for Environmental Health Sciences (NIH grant U19ES011399; L.G.G.), and a Whitaker Foundation Graduate Fellowship (B.D.C.). We also are appreciative of the anonymous human tissue donors.
Abbreviations
- Akt
serine-threonine protein kinase B/Akt
- ASP
aspirin
- BUS
buspirone
- CHL
chlorpromazine
- CIM
cimetidine
- CLA
clarithromycin
- CLO
clomipramine
- Cmax
average plasma maximum drug concentration
- CSR
cue-signal-response
- DMSO
dimethyl sulfoxide
- ERK
extracellular signal-regulated kinase
- FBS
fetal bovine serum
- GSH
glutathione
- HSP27
heat shock protein 27
- IFN-γ
interferon-γ
- IKK
inhibitor of nuclear factor-κB kinase
- IL-1α
interleukin-1α
- IL-6
interleukin-6
- JNK
c-Jun N-terminal kinase
- LDH
lactate dehydrogenase
- LEV
levofloxacin
- LPS
lipopolysaccharide
- MEK
MAPK-ERK kinase
- mTOR
mammalian target of rapamycin
- NEF
nefazodone
- NF-κB
nuclear factor-κB
- NIM
nimesulide
- NIPALS
non-linear iterative partial least-squares
- NOR
nortriptyline
- OPLSR
orthogonal partial least-squares regression
- p38
p38 mitogen-activated protein kinase
- p70 S6K
p70 S6 protein kinase
- PC
principal component
- PI3K
phosphoinositide 3-kinase
- RAN
ranitidine
- RIL
riluzole
- ROS
reactive oxygen species
- TACE
tumor necrosis factor-α-converting enzyme
- TEL
telithromycin
- TNF
tumor necrosis factor-α
- TRO
trovafloxacin
- VIP
variable-importance-of-projection
Footnotes
We define “hepatotoxicity” as in vivo liver toxicity, largely arising from hepatocyte injury and/or death. We define “hepatic cytotoxicity” as in vitro and in vivo hepatocyte injury and/or death.
‘Comparison’ compounds are defined as paired control compounds to hepatotoxicants with similar molecular target(s), clinical indication, and chemical structure but are non-hepatotoxic.
Author contributions B.D.C. and L.G.A., with assistance from T.H., performed hepatocyte experiments and phosphoprotein and toxicity data collection and compendium construction. B.D.C. performed all data modeling and wrote the manuscript. B.D.C., L.G.A., and D.A.L. designed and interpreted the study and edited the manuscript. B.S.H., P.K.S., and L.G.G. assisted in designing and interpreting the study and editing the manuscript.
References
- 1.Kaplowitz N. Drug Saf. 2001;24:483–490. doi: 10.2165/00002018-200124070-00001. [DOI] [PubMed] [Google Scholar]
- 2.Lee WM. N Engl J Med. 2003;349:474–485. doi: 10.1056/NEJMra021844. [DOI] [PubMed] [Google Scholar]
- 3.Kaplowitz N. Nat Rev Drug Discov. 2005;4:489–499. doi: 10.1038/nrd1750. [DOI] [PubMed] [Google Scholar]
- 4.Ganey PE, Luyendyk JP, Maddox JF, Roth RA. Chem Biol Interact. 2004;150:35–51. doi: 10.1016/j.cbi.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Cosgrove BD, King BM, Hasan MA, Alexopoulos LG, Farazi PA, Hendriks BS, Griffith LG, Sorger PK, Tidor B, Xu JJ, Lauffenburger DA. Toxicol Appl Pharmacol. 2009 doi: 10.1016/j.taap.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwabe RF, Brenner DA. Am J Physiol Gastrointest Liver Physiol. 2006;290:G583–589. doi: 10.1152/ajpgi.00422.2005. [DOI] [PubMed] [Google Scholar]
- 7.Malhi H, Gores GJ. Gastroenterology. 2008;134:1641–1654. doi: 10.1053/j.gastro.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tacke F, Luedde T, Trautwein C. Clin Rev Allergy Immunol. 2008 doi: 10.1007/s12016-008-8091-0. [DOI] [PubMed] [Google Scholar]
- 9.Kolls JK, Lei D, Nelson S, Summer WR, Greenberg S, Beutler B. J Infect Dis. 1995;171:570–575. doi: 10.1093/infdis/171.3.570. [DOI] [PubMed] [Google Scholar]
- 10.Shaw PJ, Hopfensperger MJ, Ganey PE, Roth RA. Toxicol Sci. 2007;100:259–266. doi: 10.1093/toxsci/kfm218. [DOI] [PubMed] [Google Scholar]
- 11.Tukov FF, Luyendyk JP, Ganey PE, Roth RA. Toxicol Sci. 2007;100:267–280. doi: 10.1093/toxsci/kfm209. [DOI] [PubMed] [Google Scholar]
- 12.Wajant H, Pfizenmaier K, Scheurich P. Cell Death Differ. 2003;10:45–65. doi: 10.1038/sj.cdd.4401189. [DOI] [PubMed] [Google Scholar]
- 13.Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. Science. 2005;310:1646–1653. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- 14.Janes KA, Gaudet S, Albeck JG, Nielsen UB, Lauffenburger DA, Sorger PK. Cell. 2006;124:1225–1239. doi: 10.1016/j.cell.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 15.Kim SK, Novak RF. Pharmacol Ther. 2007;113:88–120. doi: 10.1016/j.pharmthera.2006.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng X, Lu J, Lehman-McKeeman LD, Malle E, Crandall DL, Ganey PE, Roth RA. J Pharmacol Exp Ther. 2008;326:144–152. doi: 10.1124/jpet.108.137497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng X, Luyendyk JP, Ganey PE, Roth RA. Pharmacol Rev. 2009;61:262–282. doi: 10.1124/pr.109.001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng X, Luyendyk JP, Zou W, Lu J, Malle E, Ganey PE, Roth RA. J Pharmacol Exp Ther. 2007;322:852–861. doi: 10.1124/jpet.107.122069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarbassov DD, Ali SM, Sabatini DM. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Hotamisligil GS, Erbay E. Nat Rev Immunol. 2008;8:923–934. doi: 10.1038/nri2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan L, Kaplowitz N. Mol Aspects Med. 2008 doi: 10.1016/j.mam.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 22.Xu JJ, Henstock PV, Dunn MC, Smith AR, Chabot JR, de Graaf D. Toxicol. Sci. 2008;105:97–105. doi: 10.1093/toxsci/kfn109. [DOI] [PubMed] [Google Scholar]
- 23.Janes KA, Yaffe MB. Nat Rev Mol Cell Biol. 2006;7:820–828. doi: 10.1038/nrm2041. [DOI] [PubMed] [Google Scholar]
- 24.Cosgrove BD, Griffith LG, Lauffenburger DA. Cell Molec Bioeng. 2008;1:33–41. [Google Scholar]
- 25.Kumar N, Wolf-Yadlin A, White FM, Lauffenburger DA. PLoS Comput Biol. 2007;3:e4. doi: 10.1371/journal.pcbi.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller-Jensen K, Janes KA, Brugge JS, Lauffenburger DA. Nature. 2007;448:604–608. doi: 10.1038/nature06001. [DOI] [PubMed] [Google Scholar]
- 27.Kemp ML, Wille L, Lewis CL, Nicholson LB, Lauffenburger DA. J Immunol. 2007;178:4984–4992. doi: 10.4049/jimmunol.178.8.4984. [DOI] [PubMed] [Google Scholar]
- 28.Gaudet S, Janes KA, Albeck JG, Pace EA, Lauffenburger DA, Sorger PK. Mol Cell Proteomics. 2005;4:1569–1590. doi: 10.1074/mcp.M500158-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Janes KA, Reinhardt HC, Yaffe MB. Cell. 2008;135:343–354. doi: 10.1016/j.cell.2008.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geladi P, Kowalski B. Anal. Chim. Acta. 1986;185:1–17. [Google Scholar]
- 31.Wold S. Chemometrics and intelligent laboratory systems. 1994;23:149–161. [Google Scholar]
- 32.Ballif BA, Blenis J. Cell Growth Differ. 2001;12:397–408. [PubMed] [Google Scholar]
- 33.Ono K, Han J. Cell Signal. 2000;12:1–13. doi: 10.1016/s0898-6568(99)00071-6. [DOI] [PubMed] [Google Scholar]
- 34.McCubrey JA, Milella M, Tafuri A, Martelli AM, Lunghi P, Bonati A, Cervello M, Lee JT, Steelman LS. Curr Opin Investig Drugs. 2008;9:614–630. [PubMed] [Google Scholar]
- 35.Fielden MR, Kolaja KL. Curr Opin Drug Discov Devel. 2006;9:84–91. [PubMed] [Google Scholar]
- 36.Clayton TA, Lindon JC, Cloarec O, Antti H, Charuel C, Hanton G, Provost JP, Le Net JL, Baker D, Walley RJ, Everett JR, Nicholson JK. Nature. 2006;440:1073–1077. doi: 10.1038/nature04648. [DOI] [PubMed] [Google Scholar]
- 37.Hatano E, Brenner DA. Am J Physiol Gastrointest Liver Physiol. 2001;281:G1357–1368. doi: 10.1152/ajpgi.2001.281.6.G1357. [DOI] [PubMed] [Google Scholar]
- 38.Gunawan BK, Liu ZX, Han D, Hanawa N, Gaarde WA, Kaplowitz N. Gastroenterology. 2006;131:165–178. doi: 10.1053/j.gastro.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 39.Xu JJ, Diaz D, O'Brien PJ. Chem Biol Interact. 2004;150:115–128. doi: 10.1016/j.cbi.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 40.Chokki M, Eguchi H, Hamamura I, Mitsuhashi H, Kamimura T. Febs J. 2005;272:6387–6399. doi: 10.1111/j.1742-4658.2005.05035.x. [DOI] [PubMed] [Google Scholar]
- 41.Le GT, Abbenante G. Curr Med Chem. 2005;12:2963–2977. doi: 10.2174/092986705774462851. [DOI] [PubMed] [Google Scholar]
- 42.Efron B, Tibshirani RJ. Chapman and Hall, London. 1993 [Google Scholar]
- 43.Saez-Rodriguez J, Goldsipe A, Muhlich J, Alexopoulos LG, Millard B, Lauffenburger DA, Sorger PK. Bioinformatics. 2008;24:840–847. doi: 10.1093/bioinformatics/btn018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar N, Afeyan R, Kim HD, Lauffenburger DA. Mol Pharmacol. 2008;73:1668–1678. doi: 10.1124/mol.107.043794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bylesjö M, Rantalainen M, Cloarec O, Nicholson JK, Holmes E, Trygg J. J Chemometrics. 2006;20:341–351. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.