Summary
Understanding the functional complexity of protein interactions requires mapping biomolecular complexes within the cellular environment over biologically-relevant time scales. Herein we describe a novel set of reversible, multicolored heteroprotein complementation fragments based on various firefly and click beetle luciferases that utilize the same substrate, D-luciferin. Luciferase heteroprotein fragment complementation systems enabled dual-color quantification of two discreet pairs of interacting proteins simultaneously or two distinct proteins interacting with a third shared protein in live cells. Using real-time analysis of click beetle green and click beetle red luciferase heteroprotein fragment complementation applied to β-TrCP, an E3-ligase common to the regulation of both β-catenin and IκBα, GSK3β was identified as a novel candidate kinase regulating IκBα processing. These dual-color protein interaction switches may enable directed dynamic analysis of a variety of protein interactions in living cells.
Introduction
Protein-protein interactions are fundamental to living systems, mediating many cellular functions, including signal transduction, cell cycle progression, apoptosis and metabolic control. Traditional techniques in biochemistry for isolation and purification of protein complexes tend to select for only the most robust of interactions, whereas weak and transient protein interactions may escape detection (Sali et al., 2003). Furthermore, many protein interactions arise from cell-to-cell-mediated signaling in a tissue-restricted manner that cannot be investigated fully with in vitro systems and thus, there is considerable interest in imaging signal transduction and protein-protein interactions noninvasively in their normal physiological context within living organisms (reviewed in Villalobos et al., 2007). Previous strategies have employed transcriptional modification, activated signal transduction, or modification of enzymatic activity to generate readily observable biological or physical reporter readouts (Luker et al., 2003; Toby and Golemis, 2001). Of the many strategies for detecting protein interactions in cellulo, several features of the protein fragment complementation assay (PCA) render it attractive for monitoring protein interactions in general, and in live cells in particular (Galarneau et al., 2002; Hu and Kerppola, 2003; Luker and Piwnica-Worms, 2004; Luker et al., 2004; Michnick et al., 2007; Ozawa and Umezawa, 2001; Remy and Michnick, 1999; Remy and Michnick, 2004; Remy et al., 1999; Villalobos et al., 2007; von Degenfeld et al., 2007; Wehrman et al., 2002). Protein fragment complementation assays conventionally depend on division of a monomeric enzyme into two separate inactive components which are fused to interacting proteins of interest (Michnick et al., 2007; Villalobos et al., 2007). Upon association of the interacting proteins, the reporter fragments reassemble thereby generating reporter activity. Thus, a fundamental advantage of the PCA strategy is that the hybrid proteins rapidly and directly reconstitute an enzymatic reporter activity within the cell, offering the potential benefit of real-time analysis and enhanced sensitivity for detecting weakly interacting proteins. In principle, protein interactions may be detected in any sub-cellular compartment, and assembly of protein complexes with a broad range of affinities may be monitored. The innovation of luciferase protein fragment complementation has further allowed for rapid, readily observable signals that can be interrogated in real-time in living animals in vivo (Villalobos et al., 2007).
Several luciferase protein fragment complementation variants have been designed based upon firefly (P. pyralis), Renilla and Gaussia luciferases (Kaihara et al., 2003; Kim et al., 2007; Kim et al., 2004; Luker and Piwnica-Worms, 2004; Luker et al., 2004; Paulmurugan and Gambhir, 2003; Paulmurugan and Gambhir, 2005; Remy and Michnick, 2006; Stefan et al., 2007). As with the intact luciferases, the primary differences between firefly and Renilla/Gaussia PCA reporters are the cognate substrates, spectral properties and quality of emission (Villalobos et al., 2007; Wilson and Hastings, 1998). Luciferase PCA reporters are useful for real-time imaging of inducible and reversible interactions within cells or animals and can also be efficiently utilized in high-throughput systems with a high degree of sensitivity for protein interaction kinetics. A particularly favorable firefly luciferase (FLuc) PCA system was initially identified using an incremental truncation library of large N- and C-terminal luciferase sequences, and optimal fragments were found to contain a common overlap sequence (residues 398–416 of firefly luciferase) (Luker et al., 2004). This reporter system has been shown to be effective in monitoring protein interactions in lysates, in live cells and in living animals with low background and minimal effect on the interacting proteins of interest (Luker et al., 2004; Villalobos et al., 2007; Porter et al., 2008).
To further explore cellular and biochemical mechanisms of macromolecular assemblies, we have now developed advanced constructs derived from D-luciferin-based click beetle green (CBG) and click beetle red (CBR) luciferases (Pyrophorus plagiophthalamus). Because secondary structure mapping indicated a significant degree of conserved topology, especially within a region assigned to the overlapping fragments of our original firefly luciferase PCA system (Luker et al., 2004), we hypothesized that the firefly luciferase PCA system could provide a template for optimal bisection sites of any D-luciferin-based enzyme into functional complementation fragments, including CBR and CBG. This strategy was then employed to develop a plethora of novel luciferase PCA pairs, comprising both homologous and heterologous combinations of various firefly and click beetle luciferase fragments. Selected luciferase PCA reporters showed increased photon output and fold-inducibility as well as the ability to stably emit spectrally distinguishable colors, thereby allowing simultaneous quantification of multiple protein interactions within the same live cell in real-time using a single bioluminescence substrate.
Results
Development of color variations of luciferase PCA reporters
To further develop efficient protein fragment complementation pairs with useful spectral emission characteristics, CBG and CBR were chosen due to their use of the same substrate, D-luciferin. Although firefly luciferase and click beetle luciferases only retain approximately 48% amino acid homology, secondary structure predictions were remarkably similar (Supplemental Figure S1). Click beetle luciferases exhibit a large separation in spectral emission with CBR emitting at λmax = 615 nm and CBG emitting at λmax = 540 nm. These luciferases also show little color sensitivity to pH and temperature changes as opposed to firefly luciferase, in which the maximal emission color can vary by as much as 40 nm under different reaction conditions.
Using Clustal V protein domain mapping strategies, fragments were identified within the click beetle proteins that corresponded to residues 2–416 and 398–550 of our previously published firefly luciferase PCA constructs (Luker et al., 2004). These novel PCA constructs, comprising 2–413 and 395–542 of the click beetle luciferases, contained an overlap region consisting of nineteen amino acid residues (395–413) present on each click beetle luciferase fragment.
The rapamycin-binding domain of human mTOR (residues 2024–2113; FRB) and human FK506-binding protein-12 (FKBP) provided a well-characterized macromolecular interaction platform to validate these new protein fragment complementation pairs (Chen et al., 1995; Luker et al., 2004). The N-termini fragments of all luciferase PCA constructs were each fused to a flexible G-S linker and the C-terminus of FRB, while the C-termini fragments of all constructs were each fused to a flexible G-S linker and the N-terminus of FKBP (Figure 1A). Using HEK293T cells, protein interactions interrogated by luciferase protein fragment complementation were analyzed by live cell bioluminescence imaging.
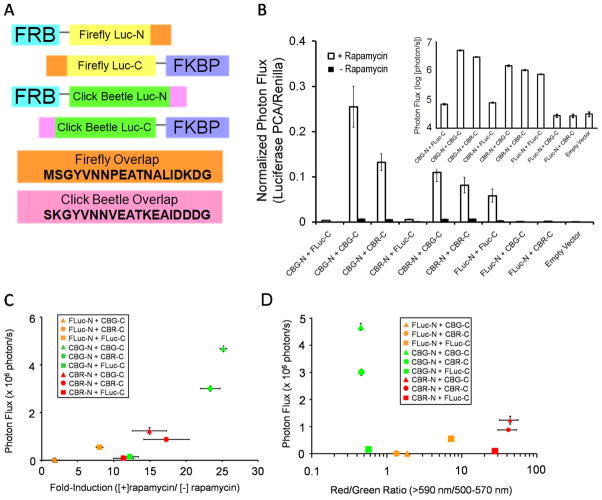
Figure 1.
Characterization of various luciferase PCA reporter pairs. A) The basic design of all luciferase PCA reporter constructs is shown. All references to Luc-N and Luc-C include fusion to the proteins FRB and FKBP, respectively, unless otherwise stated. The overlap sequence found on both N and C termini are exactly the same on all click beetle luciferase fragments. Firefly luciferase fragments contain a homologous overlap region. The Gly-Ser linker region (black line) is the same for all constructs. B) Photon flux in the presence or absence of rapamycin from all possible iterations of luciferase PCA reporter pairs expressed in HEK293T cells. D-luciferin PCA signals were normalized for transfection efficiency using coelenterazine to capture Renilla luciferase expression. Firefly N-terminus (FLuc-N) only produced photon output when combined with firefly C-terminus (FLuc-C). Click beetle green and red N-termini (CBG-N and CBR-N, respectively) were active with any C-terminal fragment, but only marginally with FLuc-C. The inset shows photon flux of rapamycin-treated cells in log scale to better illustrate values of heterologous protein fragment complementation activities. C) CBG-N + CBG-C fragments exhibited the best fold-induction and total photon output of all possible combinations, while CBR-N + CBG-C also showed improvement over our previous firefly luciferase PCA reporter. D) The color of emission was primarily determined by the N-terminus in all combinations. The red/green ratio of photon flux in the presence of rapamycin is calculated as light output from the 590 nm long pass filter divided by the 500–570 nm band pass filter. Higher values correspond to enhanced red output. See also Figures S1 and S2.
The optimal luciferase pairs based on total photon flux and fold-induction by rapamycin (100 nM) utilized the C-terminal fragment of CBG99 (CBG-C) and the N-terminal fragment of CBG99 (CBG-N) (Figure 1B, C; Figure S2). Using the FRB-FKBP protein interaction system to drive productive CBG-C and CBG-N complementation and comparing the output to our original firefly luciferase pair, 100 nM rapamycin induced a 5-fold enhancement in total photon flux and a greater than 3-fold enhanced inducibility of CBG over firefly luciferase in live cells. When combining the CBR N-terminus (CBR-N) with CBG-C, photon flux was comparable to that of our original firefly luciferase PCA pair, but fold-inducibility more than doubled. The dominant emission color for all fragment pairs tracked with the identity of the N-terminus (Figure 1D), as might be predicted from the crystal structure showing inclusion of the major D-luciferin reaction center within this fragment (Conti et al., 1996). All unpaired fragments, when expressed alone, yielded no detectable emissions (data not shown), demonstrating the lack of nonspecific bioluminescence with this system.
Specificity of Complementation
To further demonstrate the specificity of the system, we replaced wild-type FRB with ΔFRB in the luciferase PCA systems (referred to as ΔCBG-N, ΔCBR-N and ΔFLuc-N). The ΔFRB protein contains a S2035I mutation, which is known to effectively abrogate binding of the FKBP:rapamycin complex to FRB (Chen et al., 1995), thus acting as a negative control. Figure 2A shows HEK293T cells that were equally transfected with wild-type- or Δ-N fusions plus homologous C fusion PCA pairs along with intact Renilla luciferase to act as a transfection control. With all fusion pairs, upon treatment with rapamycin for 6 hrs, Δ-N exhibited no detectable induction, while wild-type N fusion constructs consistently exhibited 20- to 75-fold induced signal (Figure 2B).
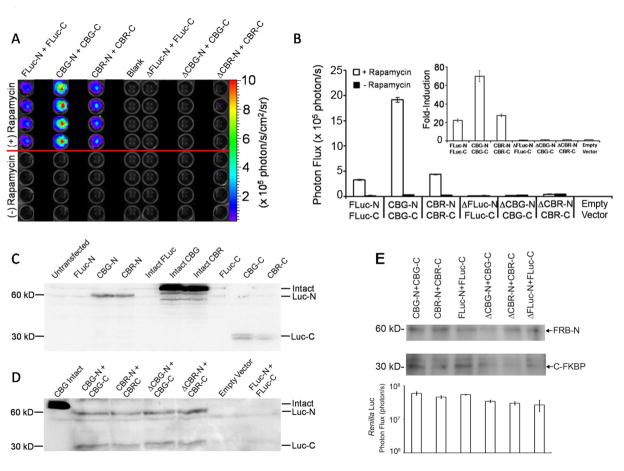
Figure 2.
Biochemical specificity of luciferase PCA. A S2035I mutation in FRB (Δ) which prevented FRB binding to the FKBP:rapamycin complex was introduced into the Luc-N fusion fragment of all reporter constructs. Each column represents cells that were batch co-transfected with PCA reporters to ensure equal light output and co-transfected with RLuc to document equal plating and transfection efficiency during the experiment (data not shown). A) Image of HEK293T cells in a 96-well plate. The top 4 rows were treated with 100 nM rapamycin for 6 hrs, while the bottom rows were treated with vehicle. B) Photon flux of treated versus untreated cells. Inset shows fold-induction of treated over untreated cells. C) Western blot using a novel anti-click beetle luciferase (CB) primary antibody shows high avidity binding to intact click beetle luciferases and significant binding to each click beetle luciferase fusion fragment. FRB-Luc-N and Luc-C-FKBP fusion fragments have molecular weights of approximately 30 kD and 60 kD, respectively. There is no cross-reactivity to any firefly luciferase protein or fragment. D) Western blot (anti-CB) of cell extracts from the experiment shown in panels A and B shows comparable expression of both wild-type CB and ΔCB fusion fragments. E) Western blots (anti-FRB, top panel; anti-FKBP, middle panel) of cell extracts from identical experiments show comparable expression of all wild-type and Δ fusion fragments. Bioluminescence of co-transfected Renilla luciferase confirms comparable levels of transfection efficiency and total cell viability following treatment with rapamycin (bottom panel).
While the optimal response was again attributed to the CBG-N/CBG-C PCA pair (Figure 2B), because both overall photon output and fold-inducibilty could be a function of the expression levels of the various fusion fragments (Villalobos et al., 2007), it was necessary to confirm equal protein content. First, to facilitate direct determination of protein expression levels of all homologous and heterologous click beetle luciferase pairs without antigen bias, we generated a novel polyclonal antibody raised against the common overlap sequence of all click beetle fragments (anti-CB; Supplemental Material). Control Western blots, performed with the intent to characterize our new polyclonal anti-CB antibody, validated the detection of all click beetle luciferase fragments and intact proteins (Figure 2C). The N-terminal fusion fragments migrated at ~60 kD, while the C-terminal fusion fragments migrated at ~30 kD, and as expected, anti-CB antibody did not recognize firefly luciferase. Then, lysates of cells used in bioluminescence assays expressing wild-type-N and Δ-N click beetle luciferase fusion pairs were analyzed (Figure 2D), confirming that all CB fusion proteins were indeed expressed equally. Furthermore, anti-FRB and anti-FKBP immunoblotting also confirmed that wild-type-N and Δ-N (~60 kD) as well as C-terminal (~30 kD) fusion pairs of both CB and FLuc were expressed at comparible levels (Figure 2E). Finally, imaging co-transfected Renilla luciferase with coelenterazine confirmed comparable levels of transfection efficiency and total cell viability within each set of luciferase pairs following treatment with rapamycin (Figure 2E). Thus, we concluded that the non-productive interactions of Δ-N fusion fragments as well as the enhanced activity of the CBG fusion pair could not be attributed to significant differences in protein expression levels.
Quantitation of rapamycin induction and complementation reversibility
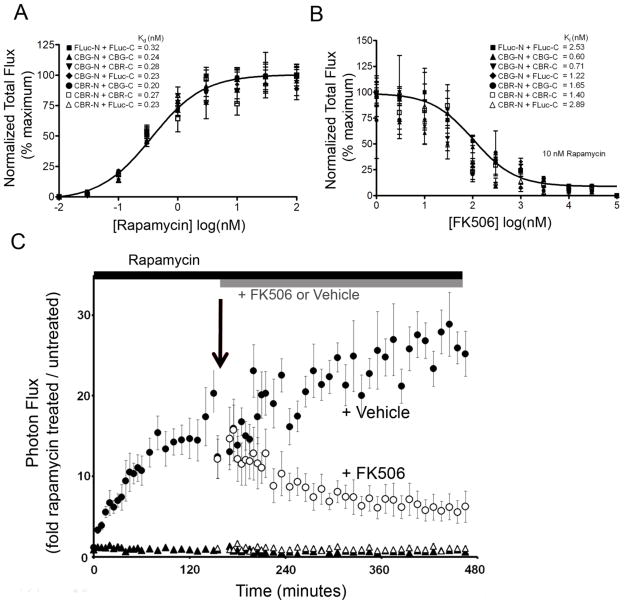
In live HEK293T cells, homologous and heterologous luciferase PCA fusions with FRB and FKBP were analyzed following a 6 hr incubation with rapamycin and competitively inhibited with FK506, each over a range of concentrations. Each complementation pair was individually curve fitted and apparent Kd values calculated (Figure 3A). Interestingly, individual Kd values and the mean apparent Kd value (0.25 ± 0.05 nM), irrespective of the composition of the paired luciferases, spectral characteristics, total photon flux, or fold-inducibility, matched previously reported Kd values for rapamycin (0.2 nM) (Bierer et al., 1990), suggesting that expression of the various luciferase PCA fusions in cells had no appreciable impact on the equilibrium state of the interacting proteins, in this case, FRB-FKBP. In some cases, there was greater than a 100-fold difference in total photon flux (e.g., CBG-N + CBG-C versus CBR-N + FLuc-C), but no significant difference in the apparent Kd (Kd, 0.24 nM versus 0.23 nM, respectively). Furthermore, the individual apparent Ki values for competitive inhibition by FK506 with all the different luciferase PCA pairs were remarkably close when tested in the presence of 10 nM rapamycin (Figure 3B). FK506 displacement curves yielded a mean apparent Ki value (1.6 ± 0.9 nM) comparable to previously reported literature values (0.4 nM) (Bierer et al., 1990). Importantly, because affinities of macromolecular interactions are dominated by dissociation rates, the surprising precision of these quantitative pharmacological titration curves from live cells (apparent Kd and Ki values) strongly suggested that the various homologous and heterologous complementation pairs, regardless of their overall photon output and spectral properties, represented reversible systems. Furthermore, because these luciferase PCA fusion pairs with FRB-FKBP reported apparent affinities highly comparable to purified FRB-FKBP protein systems (Bierer et al., 1990), this implied that the overall free energies of association/dissociation of these luciferase fragments were, as desired, effectively near zero.
Figure 3.
Titration of rapamycin induction and FK506 inhibition of FRB-FKBP complex formation by luciferase heteroprotein and homoprotein PCA. A) HEK293T cells were incubated in increasing concentrations of rapamycin and photon output determined. Each curve was individually fitted to yield the calculated Kd values as indicated; the solid line denotes a curve fit of CBG-N and CBG-C. The mean Kd was 0.25 ± 0.05 nM. B) Cells were incubated in the presence of 10 nM rapamycin and increasing concentrations of the inhibitor FK506. Each curve was individually fitted to yield the calculated Ki values as indicated; the solid line denotes a curve fit of CBG-N and CBG-C. The mean Ki was 1.6 ± 0.9 nM. C) Reversibility of the CBG-N and CBG-C PCA system. HEK293T cells were transfected with plasmids encoding CBG-C and either CBG-N (●, ○), or ΔCBG-N (▲, △). First, cells were treated with rapamycin (10 nM; ●, ▲) to induce FRB-FKBP interactions or vehicle and imaged sequentially for 150 min. Then, FK506 (10 μM final concentration; ○, △) or vehicle were added to the wells and cells imaged sequentially for an additional 325 min. Error bars represent propagated SEM. See also Figures S3 and S4.
While equilibrium analysis in live cells was promising, to directly demonstrate reversible kinetics of the CBG-N + CBG-C PCA pair, we performed FK506 displacement experiments following rapamycin pre-treatment while imaging live cells continuously in real time. HEK293T cells were transfected with plasmids encoding CBG-N and CBG-C PCA fusions and first treated with rapamycin (10 nM) to induce FRB-FKBP interactions (Figure 3C). Luciferase activity increased within minutes of rapamycin induction and continued to drift upward. As expected, the ΔCBG-N fusion paired with the CBG-C fusion showed no inducibility when monitored continuously by live cell real-time imaging (Figure 3C). At 2.5 hours post induction, FK506 (10 μM final concentration) or vehicle (DMSO) were added to the same media and cells sequentially imaged in real-time. Over this longer time-course, a brisk loss of bioluminescence occurred only with FK506, consistent with competitive FK506-mediated disruption of rapamycin-induced FRB-FKBP interactions and importantly, expectations for reversible kinetics in live cells. In DMSO treated cells, bioluminescence signal continually drifted higher over the time course of the experiment. Furthermore, when the translation inhibitor cycloheximide (100 μM) was added to cells that had previously been induced with rapamycin, luciferase activity remained stable for up to 8 hours (Supplemental Figure S3), suggesting that the upward drift in signal with vehicle was due to additional synthesis of new protein. However, upon addition of FK506 to the rapamycin- and cycloheximide-pre-treated cells, an identical rate of signal loss was observed as in the absence of cycloheximide (Supplemental Figure S3). Thus, the loss of PCA signal could not be attributed to re-synthesis and turnover of new non-interacting fusion proteins, but favored a model whereby the existing pool of interacting partners was reversibly dissociating in the presence of FK506. Note that the observed time constants of the changes in bioluminescence signal from live cells in these kinetic experiments were limited by the membrane permeabilities of both rapamycin and FK506, which are known to be somewhat low (Inoue et al., 2005), and not necessarily by inherently slow association/dissociation rates of the PCA fusion proteins within the cytosol. With these compounds, live cell kinetic analysis cannot be directly compared to live cell equilibrium analysis. Additional control experiments showed that luciferase PCA reporters were not affected by the D-luciferin concentration (Supplemental Figure S4).
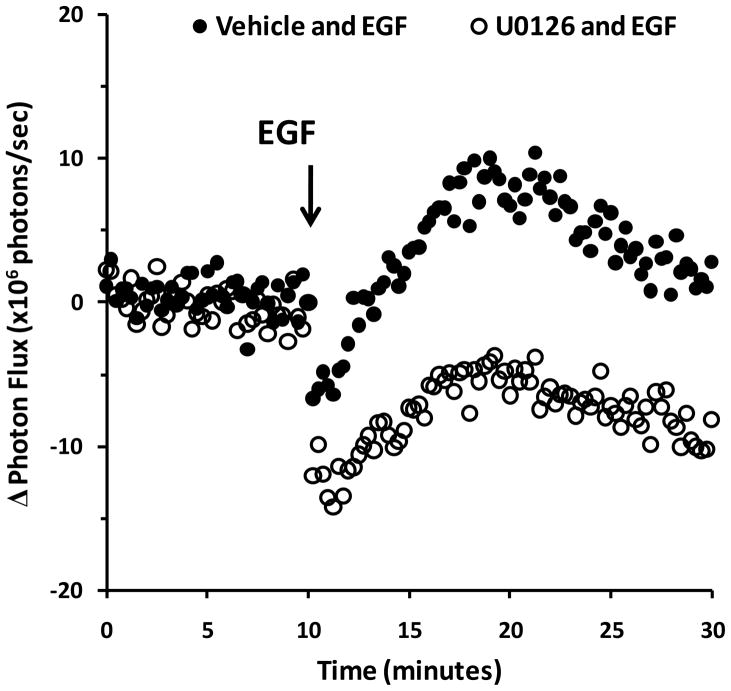
To further demonstrate reversible kinetics of our CBG fragments in a temporally faster system independent of rapamycin permeabilty, both the CBG N- and C-termini fragments were fused to C-termini of EGFR to monitor ligand-induced EGFR protein conformation changes (Yang et al., 2009). CHO TetOn cells were co-transfected with plasmids expressing the EGFR-CBG PCA fusion pairs and treated with vehicle or EGF ligand to induce receptor activation. With live cell imaging before ligand, a steady signal was observed, consistent with the presence of pre-existing EGFR dimers in close proximity within the plasma membrane. Addition of EGF resulted in a rapid biphasic signal response, characterized by an immediate drop in luciferase activity within the first 15–30 sec after EGF addition followed by a recovery of signal over the subsequent 20 minutes (Figure 4; Figure S5). The initial signal loss correlated highly with known EGFR autophosphorylation events during the first 15–30 sec after ligand binding, asymmetric movement of the cytosolic C-termini tails of EGFR dimers, and activation of tyrosine kinase activity as previously reported (Yang et al., 2009). Our data were consistent with adoption of a constrained conformation by EGFR after autophosphorylation, which prevented complementation. The subsequent recovery in luciferase PCA activity corresponded temporally to the known MAPK-mediated phosphorylation of Thr-669, which places the relaxed C-termini tails once again in close proximity, leading to desensitization of the receptor (Yang et al., 2009). As further proof-of-principle, addition of the MAPK inhibitor U0126 (10 μM final concentration) resulted in a blunted recovery of luciferase PCA activity (Figure 4), consistent with the requirement for MAPK-dependent conformational changes to induce post-activation rearrangements of the cytoplamic domain of EGFR. Note that the known ligand-induced internalization of EGFR and trafficking to endosomal/lysosomal compartments occurs on a slower time scale, beginning 10–12 minutes after addition of ligand (Huang et al., 2006; Kesarwala et al., 2009), when compared to these protein conformation changes. Thus, sensing the rapid ligand-induced conformational changes of the cytoplasmic tails of EGFR dimers provided evidence for the rapid reversibility of the CBG luciferase PCA system.
Figure 4.
Luciferase PCA reversibility assessed using CBG complementation fragments fused to the C-terminus of EGFR. CHO TetON cells in 96-well plates were transfected for 24 hours with plasmids expressing the EGFR-CBG PCA reporters. This was followed by serum starvation for 3 hours prior to imaging to ensure responsiveness to EGF stimulation. Cells were pre-incubated for 20 minutes in the absence (untreated) or presence of either vehicle (DMSO; ●) or 10 μM U0126 (○) for thermal equilibration. Cells were then imaged sequentially every 15 sec for 10 min to establish a baseline signal, treated with 25 nM recombinant mouse EGF to initiate activation of EGFR, and again imaged sequentially for 20 min. Photonic signals from untreated wells (not shown) were subtracted from EGF-treated wells to isolate EGF-induced signal changes (Δ). See Supplemental Figure S4 for raw photonic data with error bars. Each data point represents the mean of 8 replicate wells. See also Figure S5.
Imaging a two-color PCA protein interaction switch in live cells
Color separation using a bioluminescence spectral deconvolution algorithm enables accurate quantification of multi-colored luciferase reporters (blue vs. green vs. red) within the same region of interest (Gammon et al., 2006). As validation, deconvolution analysis readily separated rapamycin-induced FRB-FKBP interactions tagged with CBR-N/CBG-C (red) from those tagged with CBG-N/CBG-C (green) within the same HEK293T cells (Supplemental Figure S6). In effect, this demonstrated the quantitative imaging of a red/green protein interaction switch in live cells.
To directly demonstrate use of our interaction switch to interrogate a node within a cellular protein network, we examined in detail two signaling cascades regulated by the ubiquitin-proteasome pathway, the central mediator of proteolysis (Deshaies, 1999). Signaling through several important regulators of transcription, such as β-catenin (β-cat), a key activator of Wnt signaling (Aberle et al., 1997), and IκBα, an inhibitor of nuclear factor κB (NFκB) (Karin and Ben-Neriah, 2000), depends on regulated proteolysis for mediation of downstream transcriptional activation. Phosphorylation of key Ser/Thr residues of each of these proteins renders them substrates for rapid polyubiquitination by β-TrCP, an E3-ligase common to both pathways (Fuchs et al., 2004). In the resting state, β-cat interacts continuously with β-TrCP (Hart et al., 1999; Kitagawa et al., 1999), due to binding induced by site-specific phosphorylation of β-cat by GSK3β (Siegfried et al., 1992). As a result, low levels of β-cat are maintained in the cytosol and downstream Wnt targets are suppressed. Conversely, in an unstimulated state, IκBα resides in a cytoplasmic complex with NFκB (Ghosh and Karin, 2002; Karin et al., 2002; Li and Verma, 2002). Upon stimulation with canonical upstream receptor ligands, such as tumor necrosis factor (TNFα), IκBα is phosphorylated on Ser-32 and Ser-36, subsequently engaged by β-TrCP, polyubiquitinated and rapidly degraded, leading to release and nuclear translocation of free NFκB for activation of target genes. Thus, β-TrCP represents a common interaction node in two salient signaling cascades, providing opportunity to characterize novel modes of cross-talk between these two pathways in real-time.
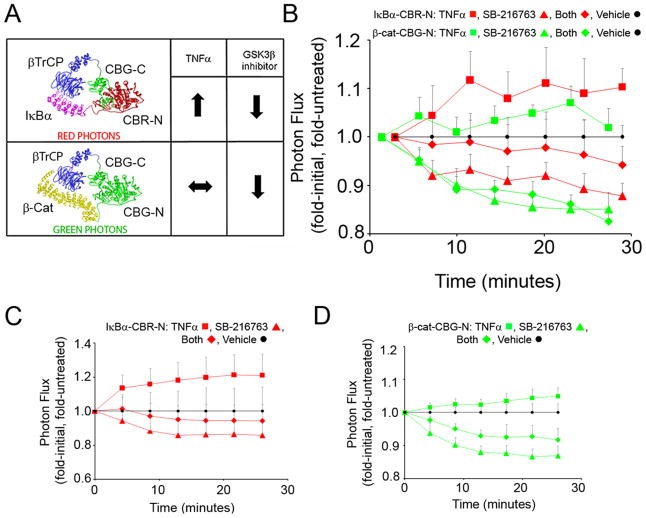
Therefore, we generated a PCA color switch using CBG-N fused to the C-terminus of β-cat, CBR-N fused to the C-terminus of IκBα, and CBG-C fused to the N-terminus of β-TrCP, the common partner (Figure 5A). HEK293T cells were co-transfected with all three fusion constructs and, after 48 hours, live cell imaging was performed every 3 minutes following addition of TNFα alone, the GSK3β inhibitor SB-216763 (Frame and Cohen, 2001) alone, both TNFα and SB-216763, or vehicle (DMSO). Red and green photon fluxes were monitored in rapid succession, deconvoluted, and presented as fold-initial, fold-untreated photon flux (Figure 5B; non-normalized data are also presented in Supplemental Figure S7). Upon stimulation with TNFα, IκBα and β-TrCP fusions exhibited an increase in red photon flux corresponding to the anticipated ligand-induced increase in IκBα/β-TrCP interactions, while β-cat and β-TrCP fusions showed no substantial change in basal green photon flux as expected. However, while addition of the GSK3β inhibitor (SB-216763; 50 μM) attenuated basal β-cat/β-TrCP interactions as would be predicted, both basal and TNFα-induced IκBα/β-TrCP interactions were similarly and unexpectedly inhibited. As deconvolution controls, HEK293T cells were either co-transfected with IκBα (red) and β-TrCP PCA fusions (Figure 5C) or separately with β-cat (green) and β-TrCP PCA fusions (Figure 5D), and imaged identically. Herein, red and green photon fluxes could be monitored separately, again yielding the same results as cells co-transfected with all three fusion reporters (Figure 5B). Control experiments also showed that SB-216763 had no effect on the PCA luciferase enzyme activity per se (data not shown).
Figure 5.
Imaging a two-color protein interaction switch in HEK293T cells. A) β-TrCP fused to CBG-C interacted with IκBα or β-cat fused to CBR-N and CBG-N, respectively, to generate either red light (IκBα) or green light (β-cat). The effects of TNFα and/or SB-216763 (a GSK3β inhibitor) on processing of IκBα and β-cat, which in turn would modulate interactions with β-TrCP, were characterized. B) Cells were co-transfected with plasmids encoding IκBα-CBR-N (red lines), β-cat-CBG-N (green lines) and CBG-C-β-TrCP for 48 hours, and then imaged in rapid succession with open, green and red filters at the times indicated immediately following addition of 20 ng/ml TNFα (squares), 50 μM SB-216763 (triangles), TNFα plus SB-216763 (diamonds) or vehicle (circles). Photon flux images were subsequently deconvoluted and shown as fold-initial, fold-untreated. Small time shifts are evident after normalization of the data due to the sequential nature of photon acquisition with the different filter sets. Error bars represent propogated SEM (n=4, each). Five independent experiments showed identical trends. C) In separate wells, cells were transfected with only IκBα-CBR-N and CBG-C-β-TrCP (red lines) and imaged with an open filter following addition of the same compounds. D) In separate wells, cells were transfected with only β-cat-CBG-N and CBG-C-β-TrCP (green lines) and imaged with an open filter following addition of the same compounds. See also Figures S6, S7 and S8.
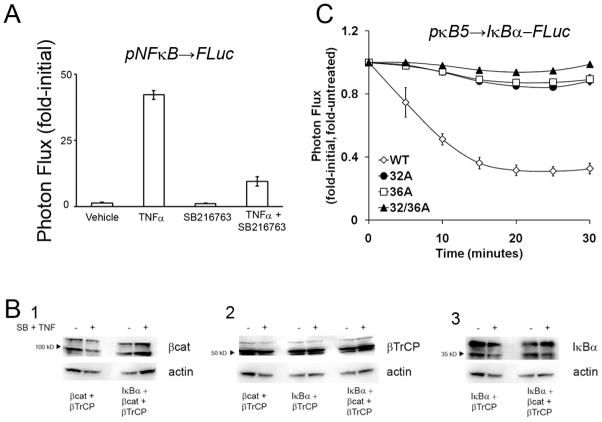
To independently validate this unanticipated interaction, the effect of GSK3β inhibition was assessed with an NFκB-dependent transcriptional reporter, wherein NFκB binding to κB response elements drives FLuc expression (Gross and Piwnica-Worms, 2005). While robust transcriptional stimulation was confirmed upon TNFα treatment, the effect was dramatically attenuated when TNFα treatment was combined with SB216763 (Figure 6A), thus corroborating our finding that GSK3β activity was important for NFκB pathway activiation. Furthermore, Western blot analysis indicated both the lack of gross overexpression of PCA fusion proteins compared to endogenous protein levels of β-cat, β-TrCP and IκBα (Figure 6B), as well as the lack of drug-induced protein degradation, providing direct evidence that the sensitivity of the bioluminescence technique did not require artifactual overexpression of reporter fusions for analysis.
Figure 6.
Validation of GSK3β-mediated regulation. A) Cells were transfected with an pNFκB→FLuc reporter plasmid for 48 hours and imaged pre- and 2 hours post-treatment with vehicle, TNFα, SB-216763 or TNFα together with SB-216763. Photon flux values are represented as mean fold-initial +/− S.E.M. (n=3 per treatment condition). B) For comparison of endogenous and fusion protein levels, Western blot analysis was performed on HEK293T cells co-transfected with plasmids encoding either β-cat-CBG-N and CBG-C-β-TrCP, IκBα-CBR-N and CBG-C-β-TrCP, or β-cat-CBG-N, IκBα-CBR-N and CBG-C-β-TrCP together, as indicated. Cells were treated with either vehicle or GSK3β inhibitor (SB216763) plus TNFα prior to processing. Bands for both endogenous (lower band) and fusion (upper band) proteins can be seen in the top panels in each lane: (1) β-cat and β-cat-CBG-N visualized with anti-β-cat Ab, (2) β-TrCP and CBG-C-β-TrCP visualized with anti-β-TrCP Ab, and (3) IκBα and IκBα-CBR-N visualized with anti-IκBα Ab; bands for actin loading controls are in the lower panels. C) For analysis of IκBα processing, HepG2 cells were transiently transfected with pκB5-IκBα-FLuc plasmids (wild type, S32A, S36A, or S32/36A), and treated with TNFα (20 ng/mL) or vehicle. Photon flux data over time are presented as fold-initial, fold-untreated (mean +/− S.E.M. when larger than the symbol; n=3 independent experiments each performed in triplicate wells).
In support of the physiological significance of this finding, protein databases revealed that IκBα (NCBI; NP 065390) contains two S/T-X-X-X-S/T(p) motifs, the canonical GSK3β substrate sequence (Frame and Cohen, 2001), at residues IκBα(32–36) and IκBα(164–168). Interestingly, upon TNFα activation of the pathway, Ser-32 and Ser-36 are known targets for inhibitor of κB kinase (IKK)-mediated phosphorylation of IκBα, marking IκBα for β-TrCP-mediated polyubiquitination and degradation. Mechanistically, phosphorylation of Ser-36 would concurrently establish a primed GSK3β motif, raising the possibility of synergistic regulation of IκBα degradation by both IKK and GSK3β, a novel level of regulation not previously characterized. To begin to test this model, we modified a previously published transcriptionally-coupled IκBα-FLuc fusion reporter (Moss et al., 2008) to generate Ala mutations at Ser-32, Ser-36, or both in IκBα. We then characterized the dynamic processing of the IκBα mutants compared to wild type IκBα by real-time bioluminescence imaging of HepG2 cells transiently transfected with the reporter constructs. Upon treatment with TNFα, loss of signal from the canonical TNFα-induced degradation of IκBα was observed with the wild type IκBα-FLuc reporter (Figure 6C). However, the S32A and S36A mutants, both singly and combined, completely abrogated ligand-induced signal loss, consistent with the model. While the detailed contributions of IKK and GSK3β remain to be resolved, these observations further supported the PCA color switch data that identified GSK3β as a novel candidate kinase regulating IκBα processing.
Discussion
Protein fragment complementation offers a straightforward, potentially quantitative approach to monitor protein interactions in living cells and organisms (Michnick et al., 2007). Other common strategies, such as two-hybrid transactivation (Chien et al., 1991) and canonical designs of the split ubiquitin system (Johnsson and Varshavsky, 1994; Stagljar et al., 1998), require transcription and translation of the reporter protein in response to stimuli before generating an active signal. A substantial delay from the moment when target protein interaction occurs to detection of a positive reporter signal limits their use as true temporal readouts of protein interactions. Furthermore, luciferase PCA systems allow excitation-free imaging of these reversible protein interaction systems in live cells and animals (Luker et al., 2004; Villalobos et al., 2007). The temporal resolution of luciferase PCA systems are amenable to accurate kinetic analysis, providing the capacity to monitor medium term changes in protein interactions, even those regulated by intracellular trafficking, protein degradation and post-translational modifications. Herein, the development of new multicolored heteroprotein fragment complementation systems may allow for more stringent analysis and deconvolution of complex protein interaction schemes in real-time within living cells. Additionally, development of the CBG luciferase PCA fragments introduces a new, reversible, and highly effective protein complementation system for the measurement of single protein interaction pairs with high signal-to-noise ratios. Further characterization of steric limitations and distance requirements between each protein fragment may serve to additionally clarify the optimal utility of these fragments.
Due to the importance of reversibility in assessing dynamic processes and associations in cellulo, we have employed several strategies to understand reversibility within the context of these CBR/CBG luciferase PCA pairs. First, under equilibrium conditions in live cells, the Kd for FRB-FKBP interactions and the Ki for FK506 were faithfully reproduced when compared to literature values. Because reversible rate equations are fundamental to the derivation of Kd and Ki values, experimentally titrating accurate values implied reversibility for our luciferase PCA systems (Michnick et al., 2007). Furthermore, because our original PCA fragments were discovered by a cell-based screen of an incremental truncation library and validated in live mammalian cells (Luker et al., 2004), the fragments were required to survive the proteasome. Because intracellular chaperones bind unfolded proteins for targeted degradation in the proteasome (Kubota, 2009), we favor a modification of the canonical PCA model wherein our luciferase fragments reside in a partially folded transition state (rather than unfolded) independent of driver protein-protein interactions, which would protect resting fragments from the proteasome. A partially folded transition state also would be consistent with a low activation energy and low ΔG contribution from the complementation fragments to the overall binding energy of the protein interaction system under study, thereby enabling readout of accurate Kd and Ki values in live cells. Second, on the time scale of the experiments exploring FK506-induced FRB-FKBP dissociation in live cells, it could be argued that protein turnover would have appeared as a decline in luciferase activity, i.e., as a subpopulation of rapamycin-FKBP-FRB complexes degraded in the presence of excess FK506, newly synthesized proteins could have been blocked by FK506 from forming FRB-FKBP complexes. However, cycloheximide data indicated that this was not the case. Last, a completely independent readout was used to assess complementation on a faster time scale. Building on our previously published studies on bioluminescense sensing of EGFR protein conformation changes induced by EGF (Yang et al., 2009), complementation reversibility was apparent from the rapid biphasic response of the CBG luciferase PCA sensor. When taken together, these data supported the rapid reversibiliy of our luciferase PCA pairs.
The considerable variation in spectral emission of the novel luciferase constructs described in this study may allow for the interrogation of different proteins of interest tagged with various luciferase fragments. Due to the differences in color, the signals can be quantified and followed in real-time, thereby expanding the repertoire of useful information derived from co-transfected cells using one luciferase substrate (D-luciferin). The luciferase PCA systems described herein allow at least two different methods of measuring multiple interactions simultaneously. First, because protein interactions that force proximity of incompatible fragments of firefly luciferase and click beetle luciferase (e.g., FLuc-N + CBG-C or CBG-N + FLuc-C; Figure 1B) do not emit significant amounts of light, signals from these fragments will be exceedingly low when compared to the photon flux of other matched homologous pairs. This feature, generalizable from the rapamycin-induced protein interaction data (Figure S6), allows for the simultaneous measurement of two discrete protein interactions (A + B and X + Y) by pairing CBG-N + CBG-C and FLuc-N + FLuc-C fusion constructs, respectively, within the same cell (Figure S8A). Importantly, the distinct property of the N-termini of both CBG and CBR luciferases to reconstitute activity with the C-terminus of CBG allows use of CBG-C as the contact point for a PCA color switch in the analysis of a multi-interaction protein network. Thus, the CBG-C fragment, when fused to a protein that interacts with two different proteins tagged with CBG-N and CBR-N, may enable the ratio of competing protein complexes (A + C or B + C) to be monitored as a color switch between states (Figure S8B). This principle of heteroprotein fragment complementation was applied successfully to real-time analysis of β-TrCP (Figure 5), an E3-ligase common to the regulation of both β-cat and IκBα, to identify GSK3β as a novel candidate kinase regulating IκBα processing. This was supported by pharmacological and mutational analysis.
Using the complementation fragment sites identified in our original incremental truncation screen with FLuc (Luker et al., 2004), a recent report generated analogous N- and C-terminal fragments of Brazilian C. distinctus luciferase (Emerald Luc, ELuc(1–413; 394–542) and CBR(1–414; 395–542) as PCA reporters (Hida et al., 2009). Further mutagenesis of the C-terminal fragment of CBR identified a mutant CBR-C(F420I, G421A, E453S; termed McLuc1) that performed similarly to our CBG-C in its ability to complement productively with both CBR-N (red) and ELuc-N (green). However, unlike our CBG-C, McLuc1 could also complement with FLuc-N (red). Signals 12-fold and 40-fold over native FLuc and ELuc PCA systems, respectively, as well as strong inducibility and reversibility were reported. However, all quantitative analysis in the report were from lysates (Promega reagent) measured in a luminometer rather than real-time live cell imaging in an IVIS as described in the present report. While analysis of lysates traverses membrane permeability issues with compounds such as rapamycin and FK-506, commercial luminescence reagents contain signal enhancers and stabilizers not found in living cells. Thus, in addition to the use of different cell types, the results are difficult to compare quantitatively. Nonetheless, both McLuc1 and CBG-C would appear to afford robust multi-color luminescent PCA systems. Thus, multi-color luciferase heteroprotein fragment complementation systems may prove useful for assessing multi-protein interactions in live cells in real-time. The CBR/CBG system may also enable imaging within the physiological context of animals in vivo (Figure S9).
Because the CBR/CBG constructs provide convenient cloning sites, the platforms illustrated herein could generalize to a broad array of protein interaction complexes. Heteroprotein fragment complementation using a single bioluminescence substrate provides a promising tool for high throughput screening, direct and indirect quantification of drug and ligand binding to specific protein complexes, and the noninvasive characterization of mechanisms of therapeutic response. This strategy may permit real-time examination of reversible protein interaction switches previously difficult to kinetically discern in living organisms.
Significance
Full understanding of the functional complexity of the protein interactome requires mapping of biomolecular complexes within the cellular environment over biologically-relevant time scales. Herein we described a novel set of reversible, multicolored heteroprotein fragment complementation systems based on various firefly and click beetle luciferases that utilize the same substrate, D-luciferin. Compared to fluorescent proteins, luciferase strategies provide minimal or no background activity, reversibility and excitation-free readouts. The click beetle green luciferase complementation fragments provide an effective protein complementation system for the measurement of single protein interaction pairs, while mixed click beetle red and green luciferase fragments enable simultaneous dual-color quantification of two discreet pairs of interacting proteins simultaneously or two distinct proteins interacting with a third shared protein. These multicolored heteroprotein fragment complementation systems may allow for more stringent analysis and detailed study of multi-protein interactions, provide a strategy for higher order high-throughput screens, and help deconvolute interaction schemes in real-time within living cells.
Methods and Materials
Plasmids
Click beetle luciferase (Pyrophorus plagiophthalamus) complementation fragments were generated using the plasmids pCBG99 and pCBR (Promega). PCR was performed on pCBG99 and pCBR to introduce convenient restriction sites (BsiWI 5′ and XhoI 3′) for direct insertion of pCBG99 or pCBR open reading frames into our previously described firefly luciferase complementation fragment constructs (Luker et al., 2004). The amino acid sequences expressed by the click beetle inserts corresponded to 2–413 for the N-terminus and 395–542 for the C-terminus. The primers used were as follows: forward primer for pCBR-N: AGAGCGTACGCGTCCCGGGGCGGTGGCTCATCTGGCGGAGGTGTAAAGCGTGAGAAAAATGTCATC; forward primer for pCBG-N: AGAGCGTACGCGTCCCGGGGCGGTGGCTCATCTGGCGGAGGTGTGAAGCGTGAGAAAAATGTCATC; reverse primer for both click beetle N-termini: AGAGGGATCCGCCACCATGAAGGGTTATGTCAATAACGTTGAAGC; forward primer for both click beetle C-termini: AGAGGGATCCGCCACCATGAAGGGTTATGTCAATAACGTTGAAGC; reverse primer for pCBG-C: AGAGCGTACGAGATCTGACCTCCGCCAGATGAGCCACCTCCACCGCCGGCCTTCTCCAACAATTGTTT; reverse primer for pCBR-C: AGAGCGTACGAGATCTGACCTCCGCCAGATGAGCCACCTCCACCGCCGGCCT TCACCAACAATTGTTT. The PCR products were subcloned into a topoisomerase plasmid (pZeroBlunt PCR4Blunt, Invitrogen) for sequencing and subsequently transferred using BsiWI and XhoI, or BsiWI and BamHI into pFRB-FLuc-N and pFLuc-C-FKBP (Luker et al., 2004), respectively. All the vectors were then transferred identically into pTriEx3Neo (Novagen) triple expression vector to assure equivalent expression levels in transient transfection experiments and to minimize error.
To construct pEGFR-CBG-N, the NLuc fragment was removed from pEGFR-NLuc (a kind gift of L. Pike, Washingtin University in St Louis (Yang et al., 2009)) and replaced with CBG-N from pFRB-CBG-N using BsiWI and AvrII sites available in both plasmids. pEGFR-CBG-C was created in two steps. First, CBG-C was amplified by PCR and cloned by replacement of CBG-N in pFRB-CBG-N, using BsiWI and XbaI sites. The primers used in amplification of CBG-C from pCBG-C-FKBP were as follows: forward: GCCGCGTACGCGCAGATCGCGTCCCGGGGCGGTGGCTCA TCTGGCGGAGGTTCCGGTAAGGGTTATGTCAATAACGTTGAAGC; reverse: CGGCTCTAGACTAACCGCCGGCCTTCTCCA. This resulted in pFRB-CBG-C. FRB was then removed from pFRB-CBG-C by digestion with HindIII and BsiWI and replaced with EGFR from pEGFR-NLuc using the same enzymes.
pCBG-C-β-TrCP was constructed by PCR amplification of CBG-C from pCBG-C-FKBP with a linker sequence (15 amino acid Gly-Ser repeats) using the following primers: forward primer pCBG-C-βTrCP: AAGCTTGCCACCATGAAAGGCTACGTGAACAATGTGGAGGCCAC; reverse primer pCBG-C-βTrCP: TGCTAGCTCGAGCCACCGCTGGACCCGGATCCACCGCTAGAGCC. The PCR product was subcloned into a topoisomerase plasmid (pZeroBlunt PCR4Blunt) for sequencing, and then digested with EcoRI and BamHI in EcoRI buffer with BSA. pβ-TrCP (kind gift from Peter Howley, Harvard Medical School) was digested with BamHI and NotI in BamHI buffer. Finally pcDNA3.1-V5/His A (Invitrogen) was digested with EcoRI and NotI in EcoRI buffer with BSA. The three fragments were ligated using T4 Rapid Ligase (Invitrogen).
pIκBα-CBR-N was created by amplifying CBR-N from pFRB-CBR-N using the following primers. pIκBα-CBR-N forward: GGATCCGGCGGTGGCTCATCTGGCGGAGGT; pIκBα-CBR-N reverse: GCGGCCGCTTAGCCGTCGTCGTCGATGGCCTCCTTG. The PCR product was subcloned into a topoisomerase plasmid (pZeroBlunt PCR4Blunt) for sequencing, digested with NotI and BamHI, and ligated into pIkBα-FLuc (Gross and Piwnica-Worms, 2005) which had been similarly digested.
pβ-cat-CBG-N was similarly created from pFRB-CBG-N using the following primers. pβ-cat-CBG-N forward: GGATCCGGCGGTGGCTCATCTGGCGGAGGT; pβ-cat-CBG-N reverse: GCGGCCGCTTAGCCGTCGTCGTCGATGGCCTCCTTG. The PCR product again was subcloned into a topoisomerase plasmid (pZeroBlunt PCR4Blunt) for sequencing, digested with ApaI and BamHI and ligated into pβ-cat-FLuc (Naik and Piwnica-Worms, 2007) which had been similarly digested.
pκB5-IκBα-FLuc, the transcriptionally-coupled IκBα-firefly luciferase fusion reporter, has been described (Moss et al., 2008). Plasmids expressing the IκBα S32A and S36A mutants, singly or together, were prepared by Quikchange mutagenesis (Stratagene, San Diego, CA, USA) following the protocol provided by the manufacturer. pNFκB-Luc was purchased from Stratagene.
Luciferase PCA imaging in cellulo
HEK293T cells (1 × 104 per well of a 96-well plate) were transfected with a total of 155 ng of DNA (75 ng of each PCA fusion construct plus 5 ng Renilla luciferase [pRlucN1, Biosignal Packard] for normalization of transfection efficiency and plating uniformity) using 3 μl Fugene6 (Roche Applied Sciences) for each 1 μg DNA in batch transfections. After 48 hours in Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco/BRL, Bethesda, MD) with 10% ΔFBS and 1% glutamine (Sigma-Aldrich), 100 nM rapamycin or vehicle was added to wells and incubated in media for 6 hours. The cells were then washed with PBS and imaged in colorless MEBSS [Modified Earle’s balanced salt solution: 144 mM NaCl, 5.4 mM KCl, 0.8 mM MgSO4, 0.8 mM NaH2PO4, 1.2 mM CaCl2, glucose 5.6 mM, HEPES 4 mM, pH 7.4] containing 1% heat-inactivated fetal bonive serum (ΔFBS) (HyClone) and 150 μg/ml D-luciferin (Biosynth). Live cells were imaged with a cooled charge-coupled device (CCD) camera in a light-tight box (Caliper Lifesciences, IVIS 100) at 37°C using open, 540AF20 band pass, and 590 long pass or 650A long pass filters in rapid succession to image all light, green light and red light, respectively (f-stop, 1; FOV, 20 cm; binning, 4; exposure time, varies from 10 sec to 5 minutes depending on transfection efficiency and total photon output). For RLuc imaging, D-luciferin-containing solution was replaced with colorless MEBSS supplemented with 1% ΔFBS and 1 μg/ml coelenterazine (Biotium Inc.). Acquisition parameters were: f/stop, 1; FOV, 15 cm; binning, 4; exposure time, 10 sec; <510 filter. Data were analyzed using LivingImage (Caliper) and Igor (WaveMetric) analysis software packages and presented as photon flux (photons/sec) without or with normalization to initial (t = 0) values (fold-initial).
Detailed methods for determination of drug titration curves, reversibility of PCA systems, EGFR protein conformation sensing, D-luciferin concentration-dependence, and spectral deconvolution in cellulo as well as bioluminescence imaging of PCA systems in mice in vivo are described in Supplemental Materials.
Imaging a two-color PCA protein interaction switch in cellulo
HEK293T cells (1 × 104 per well of a 96-well plate) were batch transfected with pIκBα-CBR-N, pβ-cat-CBG-N, pCBG-C-β-TrCP in ratios of 1:9:10, respectively, at a total DNA of 155 ng (7.5 ng, 67.5 ng, and 75 ng, respectively) using 3 μl Fugene6 for each 1 μg DNA. Empty vector (pTriEx3Neo) was used as a control to replace either pIκBα-CBR-N or pβ-cat-CBG-N in the aforementioned proportions when isolating one protein interaction. After 48 hours, cells were washed with PBS, MEBSS containing 10%ΔFBS, 1% glutamine and 150 μg/ml D-luciferin was added, and cells allowed to equilibrate at 37° for 15 minutes within the IVIS 100 imaging instrument. Imaging sequences were performed immediately upon addition of concentrated ligand (TNFα, final concentration 20 ng/ml) and/or drug (GSK3β inhibitor SB-216763 (Frame and Cohen, 2001), final concentration 50 μM) or vehicle (DMSO, 1 μl/ml) in pre-warmed MEBSS imaging buffer. Live cells were imaged at 37°C using open, 540AF20 band pass, and 650A long pass filters in rapid succession to image all light, green light and red light, respectively (f-stop, 1; FOV, 20 cm; binning, 4; exposure time, 1 minute for each filter taken in succession for 35 minutes). Data were analyzed using LivingImage and Igor analysis software packages. Images were then spectrally unmixed using the Biolumunmix v.1.0 software plug-in on ImageJ (Gammon et al., 2006) and data normalized to time t = 0 (fold-initial), and untreated cells (fold-untreated).
Imaging NFκB-dependent transcription and IκBα processing in cellulo
For NFκB-dependent transcriptional analysis, HEK293T cells were transfected with pNFκB-Luc (200 ng/well) in a black 24-well plate using Fugene6 reagent, and 48 hours after transfection, media was replaced with DMEM imaging media. A pre-treatment image was obtained after a 15 min pre-equilibration period in the IVIS imaging system, following which cells were treated with TNFα (final concentration 20 ng/ml) and/or drug (GSK3β inhibitor SB-216763, final concentration 50 μM) or vehicle control (DMSO). After 2 hours of incubation in a humidified 5% CO2 incubator, a post-treatment image was acquired.
For analysis of IκBα processing, QuikChange Site-Directed Mutagenesis (Stratagene) was utilized to mutate IκBα at Ser-32, Ser-36, or both to Ala in the pκB5-IκBα-FLuc plasmid (Moss et al., 2008). Fugene 6 (Roche) was used to transiently transfect HepG2 cells with wild-type and mutant reporters. After 48 hrs, media was replaced with fresh media containing D-luciferin, cells were stimulated with TNFα (20ng/mL) or vehicle, and imaging performed in an IVIS 100 under 5% CO2 as above. Photon flux data were analyzed and presented as fold-initial, fold-untreated using LivingImage and Igor analysis software packages.
Western blot analysis
For analysis of luciferase PCA fusions, HEK293T cells were batch transfected with 20 μg total of appropriate mixtures of plasmids expressing luciferase PCA fusions and pRlucN1 in 10 cm dishes so as to be confluent within 48 hours. Aliquots of the batch transfections were also plated into a 96-well plate to validate expression and inducibility. 48 hours post-transfection, the cells in the 96-well plate were imaged with or without addition of rapamycin to measure fold-inducibility (luciferase PCA fusions) and confirm equal transfection (RLuc) as above, while the cells in the 10 cm dishes were washed with 10 ml of cold PBS, then lysed for 15 minutes using 1 ml of reporter lysis buffer (Promega) and scraped. Lysates were spun at 14,000 rpm and the supernatant removed and frozen at −80°C. Thawed samples (50 μg of protein by Biorad Protein Assay per well) were separated by 12% SDS-PAGE and transferred to PVDF membranes. Membranes were blocked in 5% milk/TBS-T and probed overnight at 4°C with a 1:500 dilution of a primary rabbit anti-click beetle luciferase antiserum (Supplemental Material). Membranes were washed 3 × 10 min and incubated with 1:1000 dilution of goat anti-rabbit secondary antibody conjugated to horseradish peroxidase (Amersham Bioscience). In related Western blot analysis, specimens were similarly probed for all luciferase fragments using rabbit polyclonal antibody reactive to FRB (H-266; Santa Cruz Biotechnology) at 1:100 dilution and goat polyclonal antibody to FKBP (N-19; Santa Cruz Biotechnology) at 1:100 dilution, and then incubated with anti-rabbit secondary antibody conjugated to horseradish peroxidase (HRP) (GE Healthcare) for FRB blotting at 1:1000 dilution and anti-goat secondary antibody conjugated to HRP (Jackson ImmunoResearch Laboratories) at 1:2,000 dilution. Control blots confirmed the absence of specific bands in mock transfected cells. A non-specific band at ~55 kD confirmed equal loading of lanes. Images were resolved with luminol reagent (Amersham) in an IVIS 100 instrument and analyzed using LivingImage and Igor software.
For comparison of endogenous and PCA fusion protein levels, HEK293T cells were transfected in 6-well dishes with various combinations of pIκBα-CBR-N, pβ-cat-CBG-N, pCBG-C-β-TrCP for 48 hours, followed by treatment with SB216763 (50μM) and TNFα (20 ng/mL) for 30 mins. Western blot analysis was performed as above following 10% SDS-PAGE. Antibodies to β-cat, IκBα and β-TrCP (Santa Cruz Biotechnology, sc-7199, sc-203 and sc-15354, respectively) were used at 1:1000 dilutions for β-cat and IκBα, and 1:200 for β-TrCP. All secondary antibodies were used at 1:1000 dilutions for visualization of proteins. Blots were then stripped and reprobed using an antibody to actin (Sigma-Aldrich) to validate equal loading. Untransfected and cross-transfected samples as well as electrophoretic mobility confirmed the identify of endogenous proteins (data not shown).
Highlights.
New luciferase fragments enable reversible, multi-color PCA in live cells and animals in vivo.
Simultaneous dual-color quantification of three distinct interacting proteins is possible.
The strategy identified GSK3β as a novel candidate kinase regulating IκBα processing.
Supplementary Material
Acknowledgments
The authors thank Dr. A. Pichler-Wallace for performing hydrodynamic injections, Dr. S. Gammon for assistance with spectral unmixing of multi-colored luciferases, and Scott Harpstrite for figure preparation. The study was funded by National Institutes of Health grant P50 CA94056. This work also was supported by the Medical Scientist Training Program (VV), the Cancer Biology Pathway training program through the Alvin J. Siteman Cancer Center at Barnes-Jewish Hospital and Washington University School of Medicine (SN, M-HP), a National Science Foundation Graduate Award (BM), and an American Cancer Society Post-doctoral Fellowship Award (RSD).
Abbreviations
- β-cat
β-catenin
- β-TrCP
β-transducin repeat containing protein
- CBG
click beetle green luciferase
- CBR
click beetle red luciferase
- CBG-N
FRB-click beetle green luciferase N-terminus
- CBG-C
click beetle green luciferase C-terminus-FKBP
- CBR-N
FRB-click beetle red luciferase N-terminus
- CBR-C
click beetle red luciferase C-terminus-FKBP
- ΔCBG-N
mutant S2035I FRB-click beetle green luciferase N-terminus
- ΔCBR-N
mutant S2035I FRB-click beetle red luciferase N-terminus
- EGFR
epithelial growth factor receptor
- EGFR-CBG-N
EGFR-click beetle green luciferase N-terminus
- EGFR-CBG-C
EGFR-click beetle green luciferase C-terminus
- FACS
fluorescence-activated cell sorting
- FKBP
FK506-binding protein-12
- FLuc
firefly luciferase
- FLuc-N
FRB-firefly luciferase N-terminus
- FLuc-C
firefly luciferase C-terminus-FKBP
- FRB
rapamycin binding fragment of mTOR
- GSK3β
glycogen synthase kinase 3β
- IκBα
inhibitor of κB
- IKK
inhibitor of κB kinase
- MAPK
mitogen activated protein kinase
- mTOR
mammalian target of rapamycin
- NFκB
nuclear factor κB
- PCA
protein fragment complementation assay
- TNFα
tumor necrosis factor α
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. Beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierer BE, Mattila PS, Standaert RF, Herzenberg LA, Burakoff SJ, Crabtree G, Schreiber SL. Two distinct signal transmission pathways in T lymphocytes are inhibited by complexes formed between an immunophilin and either FK506 or rapamycin. Proc Natl Acad Sci U S A. 1990;87:9231–9235. doi: 10.1073/pnas.87.23.9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zheng XF, Brown EJ, Schreiber SL. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci U S A. 1995;92:4947–4951. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci U S A. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E, Franks NP, Brick P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure. 1996;4:287–298. doi: 10.1016/s0969-2126(96)00033-0. [DOI] [PubMed] [Google Scholar]
- Deshaies R. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- Frame S, Cohen P. GSK3 takes centre stage more than 20 years after its discovery. Biochem J. 2001:1–16. doi: 10.1042/0264-6021:3590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S, Spiegelman V, Kumar K. The many faces of beta-TrCP E3 ubiquitin ligases: reflections in the magic mirror of cancer. Oncogene. 2004;23:2028–2036. doi: 10.1038/sj.onc.1207389. [DOI] [PubMed] [Google Scholar]
- Galarneau A, Primeau M, Trudeau LE, Michnick S. β-Lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein-protein interactions. Nat Biotechnol. 2002;20:619–622. doi: 10.1038/nbt0602-619. [DOI] [PubMed] [Google Scholar]
- Gammon ST, Leevy WM, Gross S, Gokel GW, Piwnica-Worms D. Spectral unmixing of multicolored bioluminescence emitted from heterogeneous biological sources. Anal Chem. 2006;78:1520–1527. doi: 10.1021/ac051999h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(Suppl):S81–96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- Gross S, Piwnica-Worms D. Real-time imaging of ligand-induced IKK activation in intact cells and in living mice. Nat Methods. 2005;2:607–614. doi: 10.1038/nmeth779. [DOI] [PubMed] [Google Scholar]
- Hart M, Concordet JP, Lassot I, Albert I, del los Santos R, Durand H, Perret C, Rubinfeld B, Margottin F, Benarous R, Polakis P. The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activity in the cell. Curr Biol. 1999;9:207–210. doi: 10.1016/s0960-9822(99)80091-8. [DOI] [PubMed] [Google Scholar]
- Hida N, Awals M, Takeuchi M, Ueno N, Tashiro M, Takagi C, Singh T, Hayashi M, Ohmiya Y, Ozawa T. High-sensitivity real-time imaging of dual protein-protein interactions in living subjects using multi-color luciferases. PLoS ONE. 2009;4:e5868. doi: 10.1371/journal.pone.0005868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CD, Kerppola TK. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat Biotechnol. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol Cell. 2006;21:737–748. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Inoue T, Heo W, Grimley J, Wandless T, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson N, Varshavsky A. Split ubiquitin as a sensor of protein interactions in vivo. Proc Natl Acad Sci USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaihara A, Kawai Y, Sato M, Ozawa T, Umezawa Y. Locating a protein-protein interaction in living cells via split Renilla luciferase complementation. Anal Chem. 2003;75:4176–4181. doi: 10.1021/ac0300800. [DOI] [PubMed] [Google Scholar]
- Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- Kesarwala A, Samrakandi M, Piwnica-Worms D. Proteasome inhibition blocks ligand-induced dynamic processing and internalization of epidermal growth factor receptor via altered receptor ubiquitination and phosphorylation. Cancer Res. 2009;69:976–983. doi: 10.1158/0008-5472.CAN-08-2938. [DOI] [PubMed] [Google Scholar]
- Kim S, Kanno A, Ozawa T, Tao H, Umezawa Y. Nongenomic activity of ligands in the association of androgen receptor with SRC. ACS Chem Biol. 2007;2:484–492. doi: 10.1021/cb7000439. [DOI] [PubMed] [Google Scholar]
- Kim SB, Ozawa T, Watanabe S, Umezawa Y. High-throughput sensing and noninvasive imaging of protein nuclear transport by using reconstitution of split Renilla luciferase. Proc Natl Acad Sci U S A. 2004;101:11542–11547. doi: 10.1073/pnas.0401722101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Hatakeyama S, Shirane M, Matsumoto M, Ishida N, Hattori K, Nakamichi I, Kikuchi A, Nakayama K. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of beta-catenin. Embo J. 1999;18:2401–2410. doi: 10.1093/emboj/18.9.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H. Quality control against misfolded proteins in the cytosol: a network for cell survival. J Biochem. 2009;146:609–616. doi: 10.1093/jb/mvp139. [DOI] [PubMed] [Google Scholar]
- Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- Luker GD, Sharma V, Piwnica-Worms D. Visualizing protein-protein interactions in living animals. Methods. 2003;29:110–122. doi: 10.1016/s1046-2023(02)00285-2. [DOI] [PubMed] [Google Scholar]
- Luker K, Piwnica-Worms D. Optimizing luciferase protein fragment complementation for bioluminescent imaging of protein-protein interactions in live cells and animals. Methods Enzymol. 2004;385:349–360. doi: 10.1016/S0076-6879(04)85019-5. [DOI] [PubMed] [Google Scholar]
- Luker KE, Smith MC, Luker GD, Gammon ST, Piwnica-Worms H, Piwnica-Worms D. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc Natl Acad Sci USA. 2004;101:12288–12293. doi: 10.1073/pnas.0404041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michnick S, Ear P, Manderson E, Remy I, Stefan E. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat Rev Drug Discov. 2007;6:569–582. doi: 10.1038/nrd2311. [DOI] [PubMed] [Google Scholar]
- Moss BL, Gross S, Gammon ST, Vinjamoori A, Piwnica-Worms D. Identification of a ligand-induced transient refractory period in nuclear factor-kappaB signaling. J Biol Chem. 2008;283:8687–8698. doi: 10.1074/jbc.M706831200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Piwnica-Worms D. Real-time imaging of beta-catenin dynamics in cells and living mice. Proc Natl Acad Sci USA. 2007;104:17465–17470. doi: 10.1073/pnas.0704465104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T, Umezawa Y. Detection of protein-protein interactions in vivo based on protein splicing. Curr Opin Chem Biol. 2001;5:578–583. doi: 10.1016/s1367-5931(00)00244-1. [DOI] [PubMed] [Google Scholar]
- Paulmurugan R, Gambhir S. Monitoring protein-protein interactions using split synthetic Renilla luciferase protein-fragment-assisted complementation. Anal Chem. 2003;75:1584–1589. doi: 10.1021/ac020731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulmurugan R, Gambhir SS. Firefly luciferase enzyme fragment complementation for imaging in cells and living animals. Anal Chem. 2005;77:1295–1302. doi: 10.1021/ac0484777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter J, Stains C, Jester B, Ghosh I. A general and rapid cell-free approach for the interrogation of protein-protein, protein-DNA, and protein-RNA interactions and their antagonists utilizing split-protein reporters. J Am Chem Soc. 2008;130:6488–6497. doi: 10.1021/ja7114579. [DOI] [PubMed] [Google Scholar]
- Remy I, Michnick S. Clonal selection and in vivo quantitation of protein interactions with protein-fragment complementation assays. Proc Natl Acad Sci USA. 1999;96:5394–5399. doi: 10.1073/pnas.96.10.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy I, Michnick SW. Mapping biochemical networks with protein-fragment complementation assays. Methods Mol Biol. 2004;261:411–426. doi: 10.1385/1-59259-762-9:411. [DOI] [PubMed] [Google Scholar]
- Remy I, Michnick SW. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat Methods. 2006;3:977–979. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- Remy I, Wilson I, Michnick S. Erythropoietin receptor activation by a ligand-induced conformation change. Science. 1999;283:990–993. doi: 10.1126/science.283.5404.990. [DOI] [PubMed] [Google Scholar]
- Sali A, Glaeser R, Earnest T, Baumeister W. From words to literature in structural proteomics. Nature. 2003;422:216–225. doi: 10.1038/nature01513. [DOI] [PubMed] [Google Scholar]
- Siegfried E, Chou TB, Perrimon N. Wingless signaling acts through zeste-white 3, the Drosophila homolog of glycogen synthase kinase-3, to regulate engrailed and establish cell fate. Cell. 1992;71:1167–1179. doi: 10.1016/s0092-8674(05)80065-0. [DOI] [PubMed] [Google Scholar]
- Stagljar I, Korostensky C, Johnsson N, te Heesen S. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc Natl Acad Sci USA. 1998;95:5187–5192. doi: 10.1073/pnas.95.9.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan E, Aquin S, Berger N, Landry C, Nyfeler B, Bouvier M, Michnick S. Quantification of dynamic protein complexes using Renilla luciferase fragment complementation applied to protein kinase A activities in vivo. Proc Natl Acad Sci USA. 2007;104:16916–16921. doi: 10.1073/pnas.0704257104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toby G, Golemis E. Using the yeast interaction trap and other two-hybrid-based approaches to study protein-protein interactions. Methods. 2001;24:201–217. doi: 10.1006/meth.2001.1182. [DOI] [PubMed] [Google Scholar]
- Villalobos V, Naik S, Piwnica-Worms D. Current state of imaging protein-protein interactions in vivo with genetically encoded reporters. Annu Rev Biomed Eng. 2007;9:321–349. doi: 10.1146/annurev.bioeng.9.060906.152044. [DOI] [PubMed] [Google Scholar]
- von Degenfeld G, Wehrman T, Hammer M, Blau H. A universal technology for monitoring G-protein-coupled receptor activation in vitro and noninvasively in live animals. FASEB J. 2007;21:3819–3826. doi: 10.1096/fj.07-9597com. [DOI] [PubMed] [Google Scholar]
- Wehrman T, Kleaveland B, Her JH, Balint RF, Blau HM. Protein-protein interactions monitored in mammalian cells via complementation of beta-lactamase enzyme fragments. Proc Natl Acad Sci U S A. 2002;99:3469–3474. doi: 10.1073/pnas.062043699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T, Hastings JW. Bioluminescence. Annu Rev Cell Dev Biol. 1998;14:197–230. doi: 10.1146/annurev.cellbio.14.1.197. [DOI] [PubMed] [Google Scholar]
- Yang KS, Ilagan MX, Piwnica-Worms D, Pike LJ. Luciferase fragment complementation imaging of conformational changes in the EGF receptor. J Biol Chem. 2009;284:7474–7482. doi: 10.1074/jbc.M808041200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.