Abstract
Autism spectrum disorders (ASDs) represent a group of childhood neurodevelopmental and neuropsychiatric disorders characterized by deficits in verbal communication, impairment of social interaction, and restricted and repetitive patterns of interests and behaviour. To identify common genetic risk factors underlying ASDs, here we present the results of genome-wide association studies on a cohort of 780 families (3,101 subjects) with affected children, and a second cohort of 1,204 affected subjects and 6,491 control subjects, all of whom were of European ancestry. Six single nucleotide polymorphisms between cadherin 10 (CDH10) and cadherin 9 (CDH9)—two genes encoding neuronal cell-adhesion molecules—revealed strong association signals, with the most significant SNP being rs4307059 (P = 3.4 × 10−8, odds ratio = 1.19). These signals were replicated in two independent cohorts, with combined P values ranging from 7.4 × 10−8 to 2.1 × 10−10. Our results implicate neuronal cell-adhesion molecules in the pathogenesis of ASDs, and represent, to our knowledge, the first demonstration of genome-wide significant association of common variants with susceptibility to ASDs.
ASDs encompass a range of clinically defined conditions, including autism and pervasive developmental disorder not otherwise specified, which are more common and severe, as well as Asperger's syndrome, which appears less frequently and is milder1. ASDs are about four times more common in boys than girls, and at present around 1 in 150 children in the United States have a diagnosis of an ASD2. Several sources of evidence suggest that strong genetic components are involved in susceptibility to ASDs: there are much higher concordance rates of ASDs in monozygotic twins (92%) than dizygotic twins (10%)3, and recent estimate of the sibling recurrence risk ratio (λs) is 22 for autism4. Despite being highly heritable, ASDs show heterogeneous clinical symptoms and genetic architecture, which have hindered the identification of common genetic susceptibility factors5. Although previous linkage studies, candidate gene association studies and cytogenetic studies have implicated several chromosomal regions for the presence of autism susceptibility loci6-9, they have failed to consistently identify and replicate common genetic variants that increase the risk of ASDs.
Besides well-known genetic conditions reported in ASDs, recent studies have identified a growing number of distinct and individually rare genetic causes, suggesting that the genetic architecture of ASDs may have a significant contribution from heterogeneous rare variants. For example, rare de novo copy number variants have been implicated in 7% of families with ASDs, but only in 1% of control families10. In addition, 16p11.2 microdeletions and microduplications have been found in approximately 1% of autism cases11,12. Several hundred rare structural variations have also been catalogued in families with ASDs13. Although these reported variants indicate a role for rare genomic variation in a proportion of families, no common variants have been previously associated with ASDs in genome-wide studies. The latter is consistent with reports from previous genome-wide association studies of other neuropsychiatric disorders, including bipolar disorder14,15, schizophrenia16 and attention deficit/hyperactivity disorder16, all of which have failed to identify common susceptibility loci with genome-wide significance, when individual data sets with small sample sizes were analysed. However, recent meta-analysis reported evidence for common variants in both schizophrenia17 and bipolar disorder18, suggesting that the search for common genetic variation that confer susceptibility to ASDs may benefit from the combined analysis of several studies.
Genome-wide association studies on ASDs
To identify common genetic risk factors for ASDs, we carried out a genome-wide association study on 943 ASDs families (4,444 subjects) from the Autism Genetic Resource Exchange (AGRE cohort, Table 1)19. The subjects with ASDs in the AGRE cohort were diagnosed using both the Autism Diagnostic Interview-Revised (ADI-R)20 and Autism Diagnostic Observation Schedule (ADOS)21 diagnostic tools, which are the gold standard diagnostic tools for individuals with ASDs. All subjects were genotyped using the Illumina HumanHap550 BeadChip with over 550,000 single nucleotide polymorphism (SNP) markers. We applied stringent quality control criteria (Supplementary Methods), including call rates, Mendelian inconsistencies and genetically inferred ancestry, to identify a set of 3,101 subjects of European ancestry in 780 AGRE families for association tests. We performed analysis with the Pedigree Disequilibrium Test (PDT)22 for autosomes, and with X-APL23 for the X chromosome, using genotypes from 486,864 markers. The complete sets of SNP genotype data and signal intensity data were released to the academic research community in April 2008 (http://www.agre.org).
Table 1.
Description of the four data sets used in the study
| Data set | Purpose | Study design | Measurements | Subjects |
Analysis |
||||
|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Families | Total | Genotyped markers | Imputed markers | ||||
| AGRE | Discovery | Family-based | 550K Illumina | 1,299 | 780 | 3,101 | PDT, X-APL | ||
| ACC | Discovery | Case-control | 550K Illumina | 1,204 | 6,491 | 7,695 | PLINK | SNPTEST | |
| CAP* | Replication | Family-based | 1M Illumina | 504 | 447 | 1,390 | PDT | ||
| CART | Replication | Case-control | 300K Illumina | 108 | 540 | 648 | PLINK | SNPTEST | |
The CAP data set has been fully described in ref. 51.
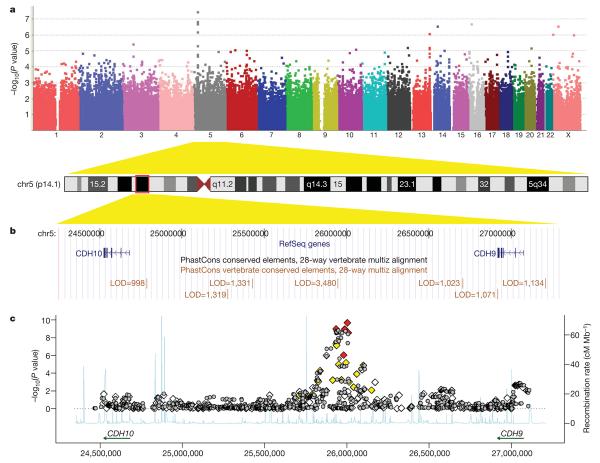
We did not observe genome-wide significant association (P < 5 × 10−8) to ASDs in the AGRE cohort, but we proposed that meaningful associations were contained within the lowest P values. To boost power for identifying these associations, we examined an Autism Case-Control cohort (ACC cohort, Table 1), comprising 1,453 subjects with ASDs from several US sites, and 7,070 control subjects without ASDs from the Children's Hospital of Philadelphia, who were also genotyped on the same platform. The subjects with ASDs in this cohort were diagnosed using the ADI and ADOS tools. After conducting thorough quality control measures on the geno-types, association analyses were conducted on 1,204 subjects with ASDs and 6,491 control subjects of inferred European ancestry. We did not detect genome-wide significant association (P < 5 × 10−8) to ASDs in the ACC cohort either. Therefore, we subsequently performed a combined analysis of these two independent data sets using recommended meta-analysis approaches24. From examining autosomes and the X chromosome, one SNP located on 5p14.1 reached genome-wide significance (rs4307059, P = 3.4 × 10−8), and five further SNPs at the same locus had Pvalues below 1 × 10−4 (Table 2 and Fig. 1a). Several other loci contain SNPs with suggestive association signals (Table 3), such as 13q33.3 (near MYO16 (myosin XVI)), 14q21.1 (between FBXO33 (F-box protein 33) and LRFN5 (leucine rich repeat and fibronectin type III domain containing 5)) and Xp22.32 (between PRKX (protein kinase, X-linked) and NLGN4X (neuroligin 4, X-linked)). We also analysed ten markers on the Y chromosome in the ACC cohort, with the most significant SNP being rs2032597 (P = 1.1 × 10−4) located within USP9Y (ubiquitin specific protease 9, Y-linked) (Supplementary Table 1). Furthermore, we have analysed 15 markers in pseudoautosomal regions of sex chromosomes in the two discovery cohorts, but no markers showed evidence of association (Supplementary Table 2).
Table 2.
The most significantly associated SNPs (P<1×10−4 in the discovery phase) between CDH10 and CDH9 on 5p14.1
| SNP | Position* | Minor/ major allele* |
Discovery cohorts |
Replication cohorts |
P value (combined) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF† (AGRE) |
P value (AGRE) |
Z score‡ (AGRE) |
Case MAF (ACC) |
Control MAF (ACC) |
P value (ACC) |
Odds ratio§ (ACC) |
P value (discovery cohorts) |
P value (CAP) | P value (CART) | ||||
| rs4307059 | 26003460 | C/T | 0.38 | 1.1×10−5 | 4.40 | 0.35 | 0.39 | 2.2×10−4 | 1.19 | 3.4×10−8 | 1.2×10−2 | 1.6×10−2 | 2.1×10−10 |
| rs7704909 | 25934678 | C/T | 0.39 | 1.6×10−5 | 4.31 | 0.36 | 0.40 | 6.2×10−4 | 1.17 | 1.4×10−7 | 9.1×10−3 | 4.0×10−2 | 9.9×10−10 |
| rs12518194 | 25987318 | G/A | 0.39 | 1.3×10−5 | 4.36 | 0.36 | 0.39 | 1.0×10−3 | 1.16 | 2.0×10−7 | 9.3×10−3 | 1.8×10−2 | 1.1×10−9 |
| rs4327572 | 26008578 | T/C | 0.39 | 2.2×10−5 | 4.24 | 0.36 | 0.39 | 2.0×10−3 | 1.15 | 6.2×10−7 | 7.3×10−3 | 1.5×10−2 | 2.7×10−9 |
| rs1896731 | 25934777 | C/T | 0.34 | 1.7×10−3 | −3.14 | 0.38 | 0.34 | 1.7×10−3 | 0.87 | 1.7×10−5 | 7.7×10−5 | 9.9×10−1 | 4.8×10−8 |
| rs10038113 | 25938099 | C/T | 0.40 | 1.4×10−3 | −3.19 | 0.43 | 0.39 | 2.4×10−3 | 0.87 | 2.1×10−5 | 2.8×10−5 | 4.5×10−1 | 7.4×10−8 |
The chromosome coordinates and allele designation are on the basis of the forward strand of the NCBI 36 genome assembly.
The minor allele frequencies (MAF) are calculated on the basis of AGRE parents of European ancestry.
Positive Z score indicates overtransmission of the major allele.
The odds ratio is calculated with respect to the major allele.
Figure 1. Genome-wide association results for the 5p14.1 region.
a, A Manhattan plot showing the −log10(P values) of SNPs from the combined association analysis of the AGRE and ACC cohorts. b, The 5p14.1 genomic region is displayed in UCSC Genome Browser, with conserved genomic elements in the PhastCons track. c, Both genotyped (diamonds) and imputed (grey circles) SNPs are plotted with their combined P values in all four cohorts. Genotyped SNPs were coloured on the basis of their correlation with rs4307059 (red: r2 ≥ 0.5; yellow: 0.2 ≤ r2 < 0.5; white: r2 < 0.2). Estimated recombination rates from HapMap data are plotted to reflect the local linkage disequilibrium structure.
Table 3.
Genotyped and imputed markers (other than those on 5p14.1) with suggestive association signals (P<1×10−5) in the two discovery cohorts
| Chr | SNP | Position | Locus | Gene | Type | MACH Rsq* |
Minor / major allele |
MAF (AGRE) |
Z score† (AGRE) |
Case MAF (ACC) |
Control MAF (ACC) |
Odds ratio† (ACC) |
P value (combined) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | rs3755827 | 62335411 | 3p14.2 | Near FEZF2 | Genotyped | C/T | 0.13 | 3.37 | 0.14 | 0.11 | 1.27 | 3.5×10−6 | |
| 6 | rs9395885 | 53853436 | 6p12.1 | LRRC1 | Imputed | 0.99 | T/C | 0.08 | 2.27 | 0.10 | 0.07 | 1.43 | 5.8×10−6 |
| 6 | rs9349688 | 53870051 | 6p12.1 | LRRC1 | Genotyped | G/A | 0.09 | 2.10 | 0.10 | 0.07 | 1.43 | 8.1×10−6 | |
| 6 | rs9384952 | 116066757 | 6q22.1 | Between HS3ST5 and FRK |
Genotyped | C/T | 0.41 | 2.33 | 0.42 | 0.37 | 1.20 | 9.4×10−6 | |
| 10 | rs9651325 | 68649859 | 10q21.3 | CTNNA3 | Imputed | 0.97 | A/G | 0.11 | −3.24 | 0.09 | 0.12 | 0.80 | 9.2×10−6 |
| 10 | rs807023 | 102760072 | 10q24.31 | Near LZTS2 | Genotyped | A/C | 0.14 | 1.12 | 0.15 | 0.10 | 1.48 | 7.8×10−6 | |
| 12 | rs10774538 | 118888180 | 12q24.23 | Near CCDC64 | Genotyped | T/C | 0.13 | −2.51 | 0.12 | 0.15 | 0.77 | 6.2×10−6 | |
| 13 | rs9521337 | 108823637 | 13q33.3 | Near MYO16 | Imputed | 0.98 | T/G | 0.13 | 2.41 | 0.14 | 0.11 | 1.34 | 6.8×10−6 |
| 13 | rs4771632 | 108846743 | 13q33.3 | Near MYO16 | Imputed | 0.99 | A/G | 0.13 | 2.32 | 0.14 | 0.11 | 1.32 | 9.5×10−6 |
| 13 | rs7996916 | 108855628 | 13q33.3 | Near MYO16 | Imputed | 0.99 | A/C | 0.12 | 3.07 | 0.15 | 0.12 | 1.27 | 5.5×10−6 |
| 13 | rs1328250 | 108856632 | 13q33.3 | Near MYO16 | Imputed | 1.00 | T/C | 0.12 | 3.07 | 0.14 | 0.12 | 1.27 | 5.9×10−6 |
| 13 | rs4771633 | 108858788 | 13q33.3 | Near MYO16 | Imputed | 1.00 | G/A | 0.12 | 2.98 | 0.14 | 0.12 | 1.26 | 9.2×10−6 |
| 13 | rs9521354 | 108865125 | 13q33.3 | Near MYO16 | Genotyped | C/A | 0.13 | 3.38 | 0.14 | 0.12 | 1.25 | 5.8×10−6 | |
| 13 | rs9521355 | 108865183 | 13q33.3 | Near MYO16 | Genotyped | T/C | 0.13 | 3.20 | 0.14 | 0.12 | 1.25 | 7.1×10−6 | |
| 13 | rs1328244 | 108881899 | 13q33.3 | Near MYO16 | Genotyped | C/T | 0.08 | 3.92 | 0.10 | 0.08 | 1.31 | 8.2×10−7 | |
| 14 | rs12897470 | 39895590 | 14q21.1 | Between FBXO33 and LRFN5 |
Imputed | 0.99 | A/G | 0.50 | 3.39 | 0.51 | 0.47 | 1.17 | 2.6×10−6 |
| 14 | rs7147817 | 39901754 | 14q21.1 | Between FBXO33 and LRFN5 |
Genotyped | G/A | 0.54 | 3.38 | 0.54 | 0.50 | 1.20 | 2.7×10−7 | |
| 14 | rs17783432 | 76141161 | 14q24.3 | Between ESRRB and VASH1 |
Genotyped | T/G | 0.15 | 1.97 | 0.15 | 0.11 | 1.36 | 9.4×10−6 | |
| 16 | rs7206043 | 8412954 | 16p13.2 | Between A2BP1 and C16orf68 |
Imputed | 0.98 | G/A | 0.41 | −3.03 | 0.39 | 0.43 | 0.84 | 4.8×10−6 |
| 16 | rs9932538 | 19116070 | 16p12.3 | SYT17 | Genotyped | G/A | 0.16 | 1.97 | 0.17 | 0.12 | 1.47 | 1.9×10−7 | |
| 20 | rs6131030 | 44241393 | 20q13.12 | CDH22 | Genotyped | A/G | 0.42 | 3.24 | 0.41 | 0.37 | 1.17 | 6.5×10−6 | |
| X | rs11798405 | 4940801 | Xp22.32 | Between PRKX and NLGN4X |
Genotyped | G/A | 0.09 | 2.71 | 0.10 | 0.06 | 1.66 | 9.0×10−7 | |
| X | rs5972577 | 32390211 | Xp21.1 | DMD | Genotyped | A/G | 0.33 | 3.90 | 0.35 | 0.30 | 1.26 | 2.7×10−7 | |
| X | rs6646569 | 119125802 | Xq24 | Near RHOXF1 | Genotyped | T/C | 0.13 | 2.49 | 0.14 | 0.09 | 1.56 | 9.7×10−7 |
MACH Rsq estimates the squared correlation between imputed and true genotypes, and a value less than 0.3 flags poorly imputed SNPs.
The Z score and odds ratio are calculated with respect to the major allele.
To identify other variants that associate with ASDs but were not captured by the SNP genotyping array, we analysed the discovery cohorts using whole-genome imputed genotypes on autosomes generated by the MACH software (Supplementary Methods). The most significant association signals were still those in the 5p14.1 region. However, several other genomic loci, such as 10q21.3 (within CTNNA3 (catenin, alpha 3)) and 16p13.2 (between A2BP1 (ataxin 2-binding protein 1) and C16orf68 (chromosome 16 open reading frame 68)), contain imputed SNPs with suggestive association signals (Table 3). Follow-up studies with larger sample sizes are required to determine whether these represent genuine ASD susceptibility loci.
Replication of the association signals
To replicate our genome-wide significant association signals at the 5p14.1 locus, we examined the association statistics for these markers in a third independently generated and analysed cohort, including 1,390 subjects from 447 autism families genotyped with ~1 million markers on the Illumina HumanHap1M BeadChip (CAP cohort, Table 1). The association signals for all the aforementioned SNPs were replicated in this cohort with the same direction of association, with Pvalues ranging from 0.01 to 2.8 × 10−5 (Table 2). To seek further evidence of replication, we examined association statistics from a fourth independent cohort of 108 ASD cases and 540 genetically matched control subjects, genotyped on the Illumina HumanCNV370 BeadChip, an array with over 300,000 SNP markers (CART cohort, Table 1). Because rs7704909 and rs10038113 were not present in this array platform, we analysed association on imputed genotypes. Most of the SNPs were replicated (P < 0.05) in the CART cohort with the same direction of association (Table 2). Combined analysis on all four data sets indicates that all six SNPs are associated with ASDs, with Pvalues ranging from 7.4 × 10−8 to 2.1 × 10−10 (Table 2 and Supplementary Table 3). Taken together, several sources of converging evidence firmly established that common genetic variants on 5p14.1 confer susceptibility to ASDs.
Genomic features of the 5p14.1 region
Closer examination of the 5p14.1 region indicated that all genotyped and imputed SNPs with P values below 1 × 10−7 reside within the same ~100 kilobase (kb) linkage disequilibrium block, suggesting that these SNPs are tagging the same variants (Supplementary Figs 1 and 2). The linkage disequilibrium block is located within a 2.2-megabase (Mb) intergenic region between CDH10 (cadherin 10) and CDH9 (cadherin 9) (Fig. 1b, c). Both CDH10 and CDH9 encode type II classical cadherins from the cadherin superfamily, which represent transmembrane proteins that mediate calcium-dependent cell-cell adhesion. To search for other types of variants, including copy number variations (CNVs), in the intergenic region, we used the PennCNV software25 on the signal intensity data and identified five CNV loci (Supplementary Fig. 3). All of these CNVs are present in control subjects in our study, and three of the five CNVs are also reported in the Database for Genomic Variants that annotates healthy individuals (Supplementary Fig. 4), suggesting that CNVs in the region are unlikely to be causal variants for ASDs.
We next focused on the ~100 kb linkage disequilibrium block containing the most significant SNPs, and determined whether other transcripts or functional elements are located in the block. By examining the UCSC Genome Browser annotations26, we did not identify predicted genes, predicted transcription start sites, spliced human expressed sequence tag (EST) sequences, known microRNA genes or predicted microRNA targets that overlap with the linkage disequilibrium block (Supplementary Fig. 5). However, we note that the linkage disequilibrium block contains several highly conserved genomic elements, including a 849-base pair (bp) element that ranks as the top 0.026% most-conserved elements in the entire human genome (log odds (LOD) score 5 3,480 by PhastCons27, Fig. 1b). Consistent with previous reports that large stable gene deserts typically contain regulatory elements for genes involved in development or transcription28, we hypothesized that these tagging SNPs were capturing the association of functional variants that regulate the expression and action of either CDH10 or CDH9.
Expression of CDH10 and CDH9 in brain
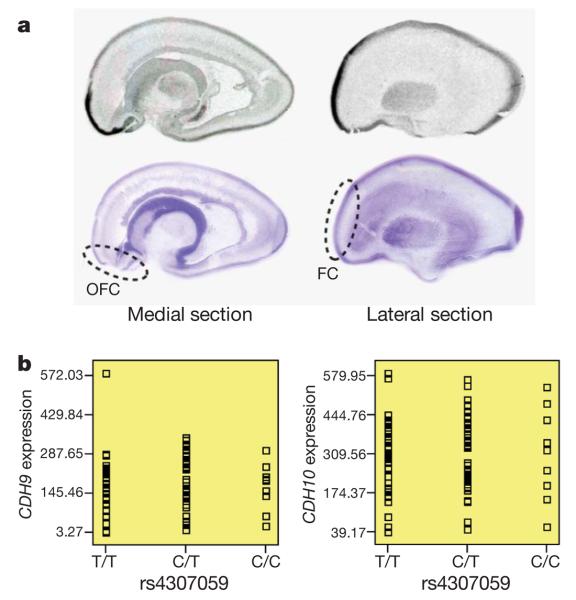
Because CDH10 and CDH9 are expressed at low levels in non-neural tissues (Supplementary Figs 6 and 7), we evaluated their messenger RNA distribution in human fetal brain by in situ hybridization. Multiple sagittally sectioned human fetal brains, each between 19 and 20 weeks gestation, were hybridized with riboprobes against CDH10 or CDH9. Results for CDH9, showing uniformly low levels of expression at the time points evaluated, were largely uninformative. In contrast, a marked pattern of enrichment for CDH10 was observed in the frontal cortex (Fig. 2a)—a region known to be important in ASDs. The expression pattern was similar to that for CNTNAP2 (contactin-associated protein-like 2)29, a molecule now well-established to be involved in the ASDs1. These results are consistent with previous work showing high levels of CDH10 in the human fetal brain30 and a prominent enrichment of Cdh10 mRNA in the anterior cortical plate of the developing mouse brain31.
Figure 2. Examination of brain expression for CDH10 and CDH9.
a, The in situ hybridization of CDH10 in human fetal brain sectioned in the sagittal plane. Medial and lateral sections from a representative sample are shown above corresponding cresyl-violet-stained marker slides. Orbitofrontal cortex (OFC) and frontal cortex (FC) are highlighted, with marked expression enrichment. b, The SNP genotypes of rs4307059 are not associated with CDH9 or CDH10 transcript levels in 93 cortical brain tissues.
To examine whether the SNP genotypes associate with gene expression for CDH10 and CDH9, we next examined the SNPExpress database32 that profiles gene expression in 93 human cortical brain tissues from genotyped subjects. However, none of the SNPs in Table 2 was associated with expression levels for either CDH9 (P = 0.92 for rs4307059) or CDH10 (P = 0.86 for rs4307059) (Fig. 2b). Although the small sample size may not have sufficient power to detect subtle effect sizes, it is also possible that the causal variants regulate gene expression only in the developing brain, or that the causal variants target an unidentified functional element, similar to the variants reported in the intergenic region on 8q24, which have been implicated in various cancers33,34.
Pathway analysis of cell-adhesion genes
Recent genetic studies have identified several neuronal cell-adhesion genes, including NRXN1 (neurexin 1)35,36, CNTNAP2 (refs 37-39) and PCDH10 (protocadherin 10)40, as potentially disrupted in rare ASD cases. Cadherins represent a large group of transmembrane proteins that are involved in cell adhesion and the generation of synaptic complexity in the developing brain41. In light of the information described earlier, we note that several other cadherin genes were also tagged by the top 1,000 most significant SNPs of the combined discovery cohorts (Supplementary Table 4). In addition, SNPs surrounding several prominent ASD candidate loci1, including CACNA1C (L type voltage-gated calcium channel), CNTNAP2, GRIK2 (glutamate receptor, ionotropic, kainate 2), NRXN1 and NLGN4X, also show suggestive evidence of association (Supplementary Table 5). These sources of evidence indicate a potential role for cell-adhesion molecules in the pathogenesis of ASDs.
To examine if cell-adhesion molecules, as a gene family, associate with ASDs, we applied two pathway-based association approaches on the genotype data (Supplementary Methods). First, we assign each SNP to the overlapping or the closest gene, summarize the significance of each gene using the Simes-adjusted P value42 from its SNPs, and then test whether the distribution of P values differ between a group of genes and all other genes using a nonparametric rank sum test. Using the combined P values from the two discovery cohorts, we found that a group of 25 related cadherin genes show more significant association with ASDs than all other genes (P = 0.02), whereas a stronger enrichment signal (P = 0.004) was obtained when the 25 cadherin genes were combined with eight neurexin family genes (NRXN1 to NRXN3, CNTNAP1 to CNTNAP5). Second, we analysed the ACC cohort using a formal pathway-association method for case-control data sets43. This method examines whether statistics for a group of genes have modest yet consistent deviation from what is expected by chance, through shuffling case/control labels many times, each time recalculating P values for all SNPs. We confirmed that the set of cadherin genes is associated with ASDs (permutation P = 0.02), whereas the combined cadherin/neurexin genes show more significant association (permutation P = 0.002). Therefore, our pathway analysis suggests that neuronal cell-adhesion molecules may be collectively associated with ASDs.
Discussion
Besides recent genetic findings supporting the role of neuronal cell-adhesion molecules in the pathogenesis of autism, an increasing number of functional neuroimaging studies have suggested the presence of cortical underconnectivity in subjects with ASDs44,45. Furthermore, neuroanatomy studies have implicated abnormal brain development of the frontal lobes in autism46,47. The genetic findings, when coupled with anatomical and functional imaging studies, convergently indicate that ASDs may result from structural and functional disconnection of brain regions that are involved in higher-order associations48-50, suggesting that ASDs may represent a neuronal disconnection syndrome.
In the current study, we have completed a genetic analysis in a large number of ASD cases and families, with a combined sample set of more than 10,000 subjects of European ancestry. We have identified and replicated common genetic variants on 5p14.1 that are associated with susceptibility to ASDs. Besides the potential roles of the nearby CDH10 and CDH9 genes, pathway-based association analysis lend further support to neuronal cell-adhesion molecules in conferring susceptibility to ASDs, suggesting that specific genetic variants in this gene class may be involved in shaping the physical structure and functional connectivity of the brain, that leads to the clinical manifestations of ASDs. Apart from highlighting the genetic complexity of ASDs and the need for large sample sizes in unveiling their genetic causes, our study represents a successful application of the genome-wide association approach in identifying common susceptibility alleles, as part of a larger effort to interrogate the complex genetic architecture of ASDs. Because the genetic aetiologies of ASDs may be linked to the neurobiological components that build and modify connectivity of the brain, by comprehensively identifying the relevant genes, genomic variants and genetic pathways, more focused analysis on gene expression, as well as structural and functional imaging, can be performed on subjects carrying specific genetic defects. Together with studies addressing epigenetic modifications and comprehensive analysis of environmental risk factors, these pieces of information can be better integrated to improve our understanding of the molecular basis of ASDs, and foster the development of early preventive and corrective strategies.
METHODS SUMMARY
All genome-wide SNP genotyping for the discovery cohorts was performed using the Illumina HumanHap550 BeadChip at the Center for Applied Genomics at the Children's Hospital of Philadelphia. For family-based cohorts, the association tests for markers in autosomes and pseudoautosomal region of sex chromosomes were performed by PDT, whereas tests for markers in the X chromosome were performed by X-APL. For case-control cohorts, the association tests were performed by PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/). Pathway association analysis was performed by GenGen (http://www.openbioinformatics.org/gengen/), using the genotype data. The whole-genome genotype imputation was performed by MACH (http://www.sph.umich.edu/csg/abecasis/MaCH/) on the autosomal markers, on the basis of phased haplotypes (release 22) for the HapMap CEU population (http://ftp.hapmap.org/phasing/2007-08_rel22/). We removed all markers with MACH Rsq measure of less than 0.3, and zeroed out imputed genotypes with a posterior probability of less than 0.9. The case-control association tests for imputed genotypes were performed by SNPTEST (http://www.stats.ox.ac.uk/~marchini/software/gwas/snptest.html), which can handle genotype imputation uncertainty. CNV calls were generated by PennCNV (http://www. openbioinformatics.org/penncnv/) on genotyping signal intensity data. For CNV validation by multiplex ligation-dependent probe amplification (MLPA), we used the Universal Probe Library system from Roche, and all reactions were performed in triplicate with an ABI Prism 7900HT Sequence Detection System (Applied Biosystems). For CNV validation by quantitative PCR (qPCR), TaqMan probes were custom-designed using Primer Express 3.0 (Applied Biosystems). For in situ hybridization, multiple sagittally sectioned human fetal brains were obtained from the Developmental Brain and Tissue Bank at the University of Maryland. Riboprobes against CDH9 or CDH10 were used for hybridization. The SNPExpress database and software (http://people. genome.duke.edu/~dg48/SNPExpress/) were used to examine the genotype-expression relationships.
Supplementary Material
Acknowledgements
We gratefully thank all the children with ASDs and their families at the participating study sites who were enrolled in this study and all the control subjects who donated blood samples to Children's Hospital of Philadelphia (CHOP) for genetic research purposes. We also acknowledge the resources provided by the AGRE Consortium (D. H. Geschwind, M. Bucan, W. T. Brown, J. D. Buxbaum, R. M. Cantor, J. N. Constantino, T. C. Gilliam, C. M. Lajonchere, D. H. Ledbetter, C. Lese-Martin, J. Miller, S. F. Nelson, G. D. Schellenberg, C. A. Samango-Sprouse, S. Spence, M. State, R. E. Tanzi) and the participating families. AGRE is a program of Autism Speaks and is at present supported, in part, by grant 1U24MH081810 from the National Institute of Mental Health to C. M. Lajonchere (PI), and formerly by grant MH64547 to D. H. Geschwind (PI). We thank the technical staff at the Center for Applied Genomics at CHOP for producing the genotypes used for analyses, and the nursing, medical assistant and medical staff for their help with recruitment of patient and control subjects for the study. We thank R. Liu and I. Lindquist for helping with CNV validation. We thank D. J. Hedges, H. N. Cukier, J. L. McCauley, G. W. Beecham, H. H. Wright, R. K. Abramson, E. R. Martin and J. P. Hussman for their comments, advice and statistical support, and the laboratory core and the autism clinical personnel at the Miami Institute for Human Genomics and the autism clinical staff at the Vanderbilt Center for Human Genetics Research. A subset of the CAP participants was ascertained while M.A.P.-V. was a faculty member at Duke University. We thank the National Institutes of Health (NIH)-funded Developmental Brain and Tissue Bank at University of Maryland for access to the fetal brain tissues used in these studies (National Institute of Child Health and Human Development Contract no. NO1-HD-4-3368 and NO1-HD-4-3383). All genotyping of the AGRE and ACC cohort was supported by an Institutional Development Award to the Center for Applied Genomics (H.H.) at the Children's Hospital of Philadelphia. The study was supported in part by a Research Award from the Margaret Q. Landenberger Foundation (H.H.), a Research Development Award from the Cotswold Foundation (H.H. and S.F.A.G), UL1-RR024134-03 (H.H.), an Alavi-Dabiri fellowship from Mental Retardation and Developmental Disability Research Center at CHOP (K.W.), the Beatrice and Stanley A. Seaver Foundation (J.D.B.), the Department of Veterans Affairs (G.D.S.), NIH grants HD055782-01 (J.Munson, A.E., O.K., G.D. and G.D.S.), MH0666730 (J.D.B.), MH061009 and NS049261 (J.S.S.), HD055751 (E.H.C.), MH69359, M01-RR00064 and the Utah Autism Foundation (H.C., J.Miller and W.M.M.), MH64547, MH081754 (D.H.G.), HD055784 (D.H.G. and M.S.), NS26630, NS36768, MH080647 and a gift from the Hussman Foundation (M.A.P.-V.), the Autism Genome Project Consortium (B.S.A., J.P., C.W.B., D.H.G., T.H.W., W.M.M., H.C., J.I.N., J.S.S., E.H.C., J.Munson, A.E., O.K., J.D.B., B.D. and G.D.S.) funded by Autism Speaks, the Medical Research Council (UK) and the Health Research Board (Ireland). We also acknowledge the partial support to CAP cohort from the Autism Genome Project.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nature Rev. Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Autism and Developmental Disabilities Monitoring Network. 2007 〈 http://www.cdc.gov/mmwr/pdf/ss/ss5601.pdf 〉 .
- 3.Bailey A, et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol. Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- 4.Lauritsen MB, Pedersen CB, Mortensen PB. Effects of familial risk factors and place of birth on the risk of autism: a nationwide register-based study. J. Child Psychol. Psychiatry. 2005;46:963–971. doi: 10.1111/j.1469-7610.2004.00391.x. [DOI] [PubMed] [Google Scholar]
- 5.Sykes NH, Lamb JA. Autism: the quest for the genes. Expert Rev. Mol. Med. 2007;9:1–15. doi: 10.1017/S1462399407000452. [DOI] [PubMed] [Google Scholar]
- 6.Gupta AR, State MW. Recent advances in the genetics of autism. Biol. Psychiatry. 2007;61:429–437. doi: 10.1016/j.biopsych.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 7.Freitag CM. The genetics of autistic disorders and its clinical relevance: a review of the literature. Mol. Psychiatry. 2007;12:2–22. doi: 10.1038/sj.mp.4001896. [DOI] [PubMed] [Google Scholar]
- 8.Veenstra-VanderWeele J, Cook EH., Jr. Molecular genetics of autism spectrum disorder. Mol. Psychiatry. 2004;9:819–832. doi: 10.1038/sj.mp.4001505. [DOI] [PubMed] [Google Scholar]
- 9.Vorstman JAS, et al. Identification of novel autism candidate regions through analysis of reported cytogenetic abnormalities associated with autism. Mol. Psychiatry. 2006;11:18–28. doi: 10.1038/sj.mp.4001781. [DOI] [PubMed] [Google Scholar]
- 10.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss LA, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 12.Kumar RA, et al. Recurrent 16p11.2 microdeletions in autism. Hum. Mol. Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 13.Marshall CR, et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sklar P, et al. Whole-genome association study of bipolar disorder. Mol. Psychiatry. 2008;13:558–569. doi: 10.1038/sj.mp.4002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alkelai A, et al. The XVth world congress of psychiatric genetics, October 7–11, 2007: rapporteur summaries of oral presentations. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2008;174B:233–277. doi: 10.1002/ajmg.b.30711. [DOI] [PubMed] [Google Scholar]
- 17.O'Donovan MC, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nature Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 18.Ferreira MA, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nature Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geschwind DH, et al. The autism genetic resource exchange: a resource for the study of autism and related neuropsychiatric conditions. Am. J. Hum. Genet. 2001;69:463–466. doi: 10.1086/321292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lord C, Rutter M, Le Couteur A. Autism diagnostic interview-revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J. Autism Dev. Disord. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- 21.Lord C, et al. The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J. Autism Dev. Disord. 2000;30:205–223. [PubMed] [Google Scholar]
- 22.Martin ER, Monks SA, Warren LL, Kaplan NL. A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am. J. Hum. Genet. 2000;67:146–154. doi: 10.1086/302957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung RH, Morris RW, Zhang L, Li YJ, Martin ER. X-APL: an improved family-based test of association in the presence of linkage for the X chromosome. Am. J. Hum. Genet. 2007;80:59–68. doi: 10.1086/510630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Bakker PI, et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn RM, et al. The UCSC genome browser database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ovcharenko I, et al. Evolution and functional classification of vertebrate gene deserts. Genome Res. 2005;15:137–145. doi: 10.1101/gr.3015505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abrahams BS, et al. Genome-wide analyses of human perisylvian cerebral cortical patterning. Proc. Natl Acad. Sci. USA. 2007;104:17849–17854. doi: 10.1073/pnas.0706128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kools P, Vanhalst K, Van den Eynde E, van Roy F. The human cadherin-10 gene: complete coding sequence, predominant expression in the brain, and mapping on chromosome 5p13–14. FEBS Lett. 1999;452:328–334. doi: 10.1016/s0014-5793(99)00672-9. [DOI] [PubMed] [Google Scholar]
- 31.Visel A, Thaller C, Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–D556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinzen EL, et al. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 2008;6:e1000001. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Witte JS. Multiple prostate cancer risk variants on 8q24. Nature Genet. 2007;39:579–580. doi: 10.1038/ng0507-579. [DOI] [PubMed] [Google Scholar]
- 34.Ghoussaini M, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J. Natl. Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim HG, et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am. J. Hum. Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szatmari P, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nature Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arking DE, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am. J. Hum. Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alarcon M, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am. J. Hum. Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bakkaloglu B, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am. J. Hum. Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morrow EM, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Redies C. Cadherins in the central nervous system. Prog. Neurobiol. 2000;61:611–648. doi: 10.1016/s0301-0082(99)00070-2. [DOI] [PubMed] [Google Scholar]
- 42.Simes RJ. An improved Bonferroni procedure for multiple tests of significance. Biometrika. 1986;73:751–754. [Google Scholar]
- 43.Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am. J. Hum. Genet. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Just MA, Cherkassky VL, Keller TA, Minshew NJ. Cortical activation and synchronization during sentence comprehension in high-functioning autism: evidence of underconnectivity. Brain. 2004;127:1811–1821. doi: 10.1093/brain/awh199. [DOI] [PubMed] [Google Scholar]
- 45.Koshino H, et al. fMRI investigation of working memory for faces in autism: visual coding and underconnectivity with frontal areas. Cereb. Cortex. 2008;18:289–300. doi: 10.1093/cercor/bhm054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amaral DG, Schumann CM, Nordahl CW. Neuroanatomy of autism. Trends Neurosci. 2008;31:137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 47.Courchesne E, Pierce K. Why the frontal cortex in autism might be talking only to itself: local over-connectivity but long-distance disconnection. Curr. Opin. Neurobiol. 2005;15:225–230. doi: 10.1016/j.conb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 48.Hughes JR. Autism: the first firm finding = underconnectivity? Epilepsy Behav. 2007;11:20–24. doi: 10.1016/j.yebeh.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 49.Frith C. Is autism a disconnection disorder? Lancet Neurol. 2004;3:577. doi: 10.1016/S1474-4422(04)00875-0. [DOI] [PubMed] [Google Scholar]
- 50.Geschwind DH, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Curr. Opin. Neurobiol. 2007;17:103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 51.Ma DQ, et al. A genome-wide association study of autism reveals a common novel risk locus at 5p14.1. Ann. Hum. Genet. doi: 10.1111/j.1469-1809.2009.00523.x. doi:10.1111/j.1469-1809.2008.00523.x (in the press) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.