Abstract
Mutations in RAS proteins occur widely in human cancer. Prompted by confirmation of KRAS mutation as a predictive biomarker of response to EGFR targeted therapies, limited clinical testing for RAS pathway mutations has recently been adopted. We performed a multiplatform genomic analysis to characterize, in a non-biased manner, the biologic, biochemical and prognostic significance of Ras pathway alterations in colorectal tumors and other solid tumor malignancies. Mutations in coding exon 4 of KRAS were found to occur commonly and to predict for a more favorable clinical outcome in patients with colorectal cancer. Exon 4 KRAS mutations, all of which were identified at amino acid residues K117 and A146, were associated with lower levels of GTP-bound RAS in isogenic models. These same mutations were also often accompanied by conversion to homozygosity and increased gene copy number, in both human tumors and tumor cell lines. Models harboring exon 4 KRAS mutations exhibited MEK-dependence and resistance to EGFR-targeted agents. Our findings suggest that RAS mutation is not a binary variable in tumors, and that the diversity in mutant alleles and variability in gene copy number may also contribute to the heterogeneity of clinical outcomes observed in cancer patients. These results also provide a rationale for broader KRAS testing beyond the most common hotspot alleles in exons 2 and 3.
Introduction
Constitutive MAPK activation is frequent in human cancer and is often the result of activating mutations in RAS 1-2. Mutationally activated forms of RAS were first identified in the Harvey and Kirsten sarcoma viruses, in which they were determined to be oncogenic 3-5. Shortly thereafter, somatic RAS mutations were detected in human tumors 4-6. The most common of these mutations, occurring at the G12, G13 and Q61 positions, result in impaired intrinsic and GAP-mediated GTP hydrolysis, leading to elevated levels of cellular RAS-GTP 7.
Despite evidence that oncogenic RAS plays a central role in mediating transformation in a diverse set of human tumors, only recently has limited KRAS mutational testing entered clinical practice. Testing of lung and colorectal tumors for KRAS mutations was prompted by the demonstration that KRAS mutational status is a predictive marker of response to EGFR targeted therapies such as erlotinib, cetuximab, and panitumumab 8-13. Clinical testing, however, has been restricted to the identification of mutations involving only a small number of the most commonly mutated alleles 14-15.
Recent technological advances have made a more comprehensive assessment of RAS gene alterations feasible but widespread adoption of broader testing beyond the most commonly mutated alleles at codons 12 and 13 has been limited by a lack of knowledge about the frequency and biological significance of non-exon 2 KRAS mutations 16-17. We therefore employed a multiplatform approach to define the incidence, biologic and prognostic significance of RAS mutations beyond the well-characterized hotspots in KRAS coding exon 2.
Materials and Methods
Mutation detection
Clinical data was collected on patients under an Institutional Review Board-approved protocol or waiver of authorization. Genomic DNA was obtained by using the DNeasy Tissue Kit (Qiagen, Valencia, CA). Mutations were detected using the iPLEX assay (Sequenom, Inc., San Diego, CA), which is based on a single-base primer extension assay 18. Briefly, multiplexed PCR and extension primers are designed for a panel of known mutations. After PCR and extension reactions, the resulting extension products are analyzed using a MALDI-TOF mass spectrometer. For mutation detection by the Sanger method, PCR primer sequences were used for exon amplification as previously reported 19. All primer sequences are available upon request.
MS-based genetic fingerprinting assay
Colorectal cancer cell lines and tumors were checked for mislabeling, contamination, and misidentification using a multiplexed PCR/MS-based genetic fingerprinting assay developed specifically for this purpose. Briefly, forty-two highly polymorphic SNPs, covering all chromosomes, were selected and a 4-well, multiplexed assay was designed. The assays were run on the Sequenom platform as described in the supplemental methods.
Array CGH
For CGH studies, labeled tumor DNA was co-hybridized to Agilent 244K aCGH microarrays with a pool of reference normal. Raw copy number estimates were normalized 20, segmented with Circular Binary Segmentation 21, and analyzed with RAE 22, all as previously described. The status of genomic gain was determined for segments spanning the KRAS locus as those with A0 > 0.9 and A1 > 0.01 per the multi-component model in RAE 22. Regions of significant alteration were excluded as either known or presumed germline copy-number polymorphisms if they overlapped previously identified variants 23. Segmented copy number data were visualized in the Integrative Genomics Viewer and all genome coordinates were standardized to NCBI build 36.1 (hg18) of the reference human genome.
Site-directed mutagenesis and RAS-GTP measurement
KRAS mutations were engineered into pcDNA3.1+2XMycKRAS4B using the QuickChange XLII (Stratagene, La Jolla, CA) as per the manufacturer’s instructions. All constructs were verified by Sanger sequencing. The level of GTP bound, active RAS was measured using the recombinant Ras binding domain (RBD) of RAF (Millipore, Temecula, CA). Briefly, 0.5 mg of lysate was immuno-precipitated using beads containing recombinant RAS binding domain (RBD). After washing, the beads were mixed with sample buffer and separated using SDS-PAGE. The membrane was probed with pan-RAS antibody to detect the levels of GTP bound, active RAS. Total RAS levels were detected using whole cell lysates.
Animal Studies
Four to six week old nu/nu athymic BALB/c mice were maintained in pressurized ventilated cages. All studies were performed in compliance with IACUC guidelines. Tumors were established by injecting 1 × 107 cells suspended 1:1 (vol) with reconstituted basement membrane (Matrigel). Tumor volumes were calculated using the formula π/6 × larger diameter × (smaller diameter)2. PD0325901 was formulated in 0.5% hydroxypropyl methylcellulose + 0.2% Tween-80 and administered by oral gavage.
RESULTS
Identification of exon 4 KRAS mutations by mass spectrometric genotyping
Clinical data demonstrating that KRAS mutations predict for resistance to EGFR targeted agents has led to the adoption of limited KRAS testing in lung and colorectal cancer patients. Testing beyond the most commonly mutated exon 2 alleles, however, has not been adopted over uncertainty regarding the frequency and clinical significance of lower frequency alleles. To determine the prevalence of non-hotspot KRAS mutations, we genotyped 1,183 human tumor samples which included 415 colorectal tumors and 70 colorectal cell lines for alterations in all three RAS proteins and BRAF. This panel included colorectal tumors from all clinical states including adenomas (n = 39), primary invasive colorectal adenocarcinomas (n = 322), and lung and liver metastases (n = 54).
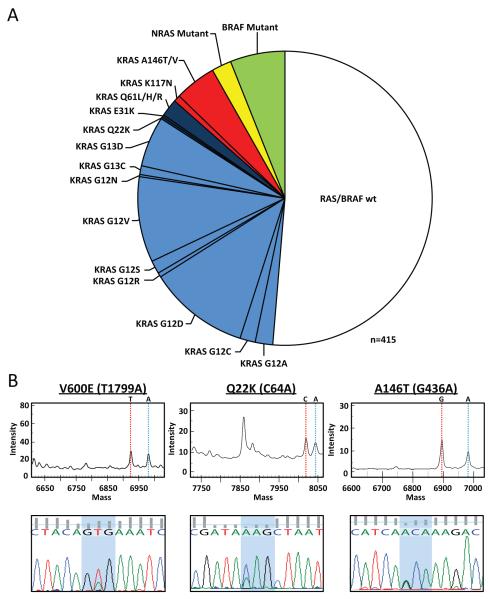
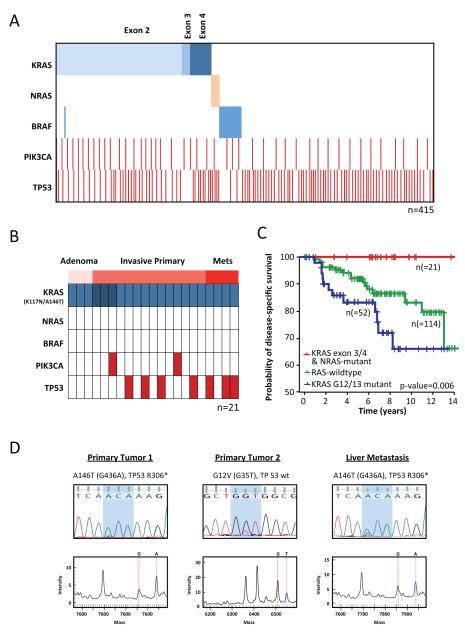
To facilitate the genomic characterization of large sample sets, we developed a MALDI-TOF mass spectrometry-based assay to screen for not only the most common hotspot alterations in KRAS, NRAS and BRAF but also less commonly reported somatic mutations in these genes 16-17. To validate the sensitivity, specificity, and coverage of the assay, we also sequenced all coding exons of all three RAS isoforms in the 415 colorectal cancers. As shown in Figure 1A, 49% of the tumors harbored a mutation in KRAS, NRAS or BRAF. Consistent with prior reports, the majority of these alterations (33%) were at amino acids 12 or 13 of KRAS. Nevertheless, we also detected mutations in exons 3 and 4 of KRAS and in exons 2 and 3 of NRAS in approximately 10% of tumors. Specifically, 5.5% of tumors harbored exon 4 KRAS mutations (19 A146T, 1 A146V, and 3 K117N). Representative mass spectrometry and Sanger sequencing traces for three of these mutations (V600E BRAF, Q22K KRAS and A146T KRAS) are shown in Figure 1B. Only two mutations (K117N identified in 3 cases and E31K in a single case) were detected exclusively by Sanger sequencing. The exon 4 (K117 and A146) mutations were confirmed somatic by directly sequencing the corresponding normal tissue DNA (See Fig. S1). Nine tumors also harbored mutations in NRAS (three in exon 2 and six in exon 3). All NRAS and exon 3 and 4 KRAS mutations were found in a mutually exclusive pattern with mutations in KRAS exon 2 and BRAF (Fig. 1 and 2). The KRAS, NRAS and BRAF mutations were not mutually exclusive with mutations in PIK3CA and TP53 (Fig. 2A).
Figure. 1. Prevalence of KRAS, NRAS and BRAF mutations in patients with colorectal cancer.
A. 415 colorectal tumors were screened for mutations in RAS and BRAF. Exon 2 KRAS mutations are shown in light blue, exon 3 KRAS mutations in dark blue, exon 4 KRAS mutations (K117 and A146) in red, NRAS mutations in yellow and BRAF mutations in green. B. Representative mass spectrometry and Sanger sequencing traces are shown for tumors harboring a V600E BRAF, Q22K KRAS, and A146T KRAS mutation.
Figure. 2. Concordance of KRAS, BRAF, PIK3CA and TP53 mutations.
A. Exon 4 KRAS mutations were non-overlapping in distribution with mutations in KRAS exons 2 and 3 and BRAF. B. A146 KRAS mutations were identified in samples derived from all clinical states including adenomas, invasive primary colorectal adenocarcinomas, and liver metastases. C. Kaplan-Meier plot of disease-specific survival of 186 patients with stages 1-3 colorectal cancer as a function of KRAS/NRAS mutational status (p-value as indicated, log-rank test). No patients with stage 1-3 colorectal cancer whose tumor expressed an exon 3 or 4 KRAS or NRAS mutation died of colorectal cancer. D. Sanger and mass spectrometry traces of three tumors resected from a 75 year old woman who presented with synchronous primaries in the rectosigmoid (primary 1), cecum (primary 2), and four liver metastases. She is without evidence of disease 20 months following surgical resection of all disease sites. Primary 1 and the liver metastasis harbored A146T KRAS and R306* TP53 mutations. Primary 2 was G12V KRAS mutant and TP53 wild-type.
Exon 4 KRAS mutations were identified not only in primary invasive colorectal tumors, but also adenomas, suggesting that these alterations occur early within the natural history of the disease (Fig. 2B and Table S1). Notably, the clinical outcome of patients with non-G12/G13 KRAS mutations and NRAS mutations was more favourable than that of patients with mutations in KRAS at the G12 and G13 positions (Fig. 2C, p value = 0.006). In our series, not a single patient with Stage 1-3 colorectal cancer whose tumor expressed an exon 3 or 4 KRAS mutation or an NRAS mutation died of colorectal cancer (median clinical follow-up of 6.5 years). Furthermore, all four patients who underwent curative-intent liver resection and whose liver metastases harbored A146T KRAS mutation were disease-free following liver resection (followup intervals of 1.7, 12.5, 13 and 19.5 years, see Table S1). This latter observation, if confirmed in larger datasets, would suggest that exon 4 KRAS mutation might identify a population of patients more amenable to aggressive surgical treatment of low volume metastatic disease. Consistent with some but not all prior studies, a trend towards improved disease-specific survival was observed in a pairwise comparison of the KRAS wild-type cohort versus the cohort of patients with G12/G13 KRAS mutations but this was not statistically significant (p-value = 0.07, Fig. S2) 24-26. Notably, patients whose tumors harbored G12/G13 KRAS mutations did exhibit a worse disease-specific survival versus those wild-type for KRAS when the non-G12/G13 KRAS mutants and NRAS mutants were included in the wild-type cohort (p-value = 0.02, Fig. S2).
Cells harbouring exon 4 KRAS mutations exhibit elevated RAS-GTP expression and KRAS-dependence
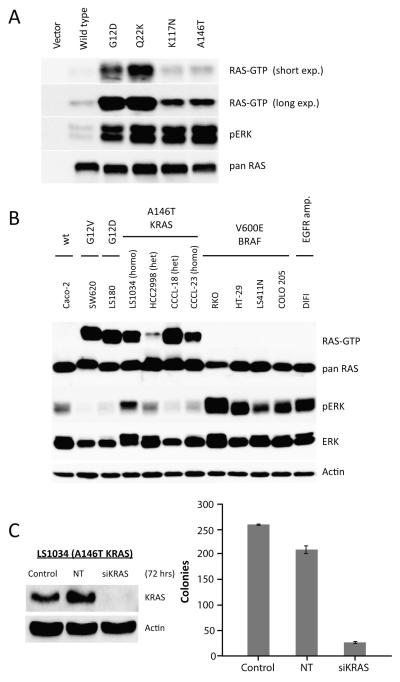
The mutually exclusive distribution of the A146T/V and K117N KRAS mutations with those in BRAF and exons 2 and 3 of KRAS suggests that these alterations confer overlapping downstream effects. To biologically characterize the A146T and K117N mutations, we transiently expressed them as myc-tagged constructs in HEK-293FT cells. Expression of K117N and A146T KRAS mutants in HEK-293FT cells resulted in elevated RAS-GTP and phosphorylated ERK expression as compared to wild-type RAS (Fig. 3A). In contrast, the E31K KRAS allele exhibited RAS-GTP levels similar to the wild-type construct (data not shown). The level of RAS-GTP expression induced by the K117N and A146T mutants was, however, lower than that observed in cells transfected with the G12D and Q22K KRAS mutants.
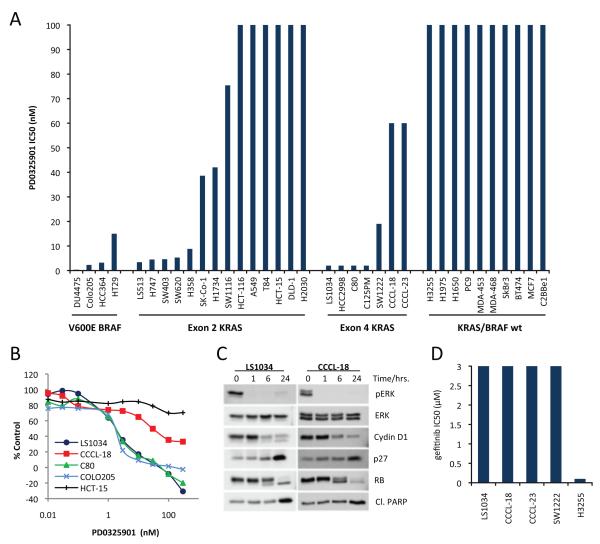
Figure. 3. Cell lines harboring an A146T KRAS mutation demonstrate elevated RAS-GTP expression and KRAS-dependence.
A. HEK-293FT cells were transfected with KRAS mutants and GTP-bound RAS was measured by immuno-precipitating active RAS with recombinant Ras binding domain (RBD) of RAF. B. Colorectal cancer cell lines expressing A146T KRAS demonstrate elevated RAS-GTP expression as compared to cell lines with V600E BRAF mutation or those wild-type for both KRAS and BRAF. C. Knockdown of KRAS expression by siRNA in LS1034 cells (homozygous A146T KRAS mutant) inhibited colony formation.
To further define the biological importance of the K117N and A146T/V KRAS mutations, we used our Sequenom assay to screen 351 cancer cell lines of which 70 were derived from colorectal cancers for RAS pathway mutations (Table S2 and Fig. S3). To exclude the possibility of redundancy among the colorectal cancer cell lines due to mislabelling or cross contamination, DNA from each cell line was characterized for 42 highly polymorphic SNPs using a mass spectrometry-based assay generated specially for this purpose (see Supplemental Methods for a detailed description of the assay methods and validation). Seven unique colorectal cancer cell lines were identified which harbored exon 4 KRAS mutations (4 A146T, 2 A146V and 1 K117N). Matched tumor tissue was available for the two cell lines (CCCL-18 and CCCL-23) 27 and in both cases, we were able to confirm the presence of A146T KRAS alterations in the primary tumors from which the cell lines were derived (Fig. S1C). None of the 281 non-colorectal cancer cell lines were found to harbor exon 4 KRAS mutations.
Cell lines expressing A146T KRAS exhibited elevated RAS-GTP expression compared to those with V600E BRAF mutation or EGFR amplification (Fig. 3B). To characterize the KRAS-dependence of cells expressing an A146T KRAS mutation, we used siRNA to knock down KRAS expression in LS1034 (A146T KRAS) cells. Transfection of KRAS but not non-targeting control siRNA effectively downregulated KRAS expression (greater than 90%) and led to inhibition of colony formation (Fig. 3C).
Whereas expression of A146T KRAS in the HEK-293FT model was associated with lower RAS-GTP expression than G12D KRAS, three of four A146T KRAS cell lines expressed levels of RAS-GTP comparable to that expressed in models harboring G12D and G12V KRAS mutations (Fig. 3B). We hypothesized that the variable RAS-GTP expression in the A146T KRAS mutant cell lines could be the result of increased gene dosage due to KRAS gene amplification or conversion to homozygosity. In fact, we found two of the four A146T KRAS cell lines (LS1034 and CCCL-23) were homozygous for the mutant allele.
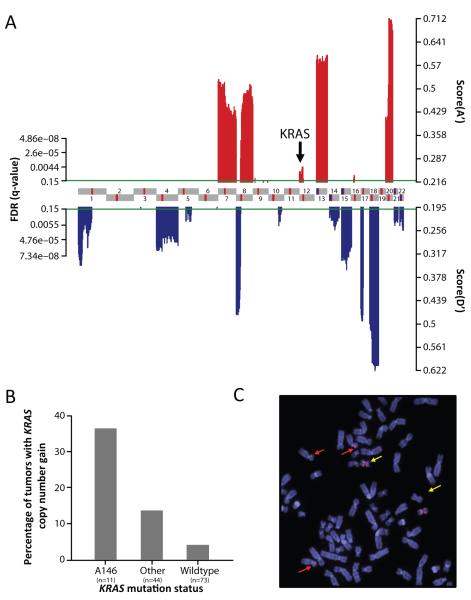
To explore the hypothesis that focal KRAS amplification may be common in tumors harboring less potent KRAS alleles, we performed DNA copy number profiling on 128 colorectal tumors and three of the A146T mutant cell lines using the Agilent 244K aCGH platform. We identified a profile of statistically significant copy-number alterations using the RAE framework 22, with results consistent with previous genome-wide characterization efforts (Fig. 4A). A detailed listing of the focal areas of copy number gain and loss and genes of interest located within these regions is included in Tables S3 and S4 28-29. In our analysis, focal KRAS amplification was uncommon (2.3%, all KRAS mutant samples) and was not identified as a statistically significant region of focal copy number gain (Fig 4A and Table S3). We did, however, detect broader copy number gains at the KRAS locus in 24% of tumors. Although focal KRAS amplification was rare in the overall dataset, the percentage of samples with KRAS copy number gain was significantly higher among tumors harboring A146T KRAS mutations versus those with exon 2 or 3 KRAS mutations or those wild-type for KRAS (36 versus 14 and 4% respectively, p-value = 0.014, Fig. 4B). An increase in KRAS copy number was also observed in two of the three A146T KRAS mutant cell lines. As shown by FISH for the LS1034 cell line, the mechanism responsible for increased KRAS copy number was complex. LS1034 exhibits a hypertriploid karyotype with three normal copies of chromosome 12 and two copies of an isochromosome for 12p. In summary, these data suggest that the lower potency exon 4 KRAS alleles are more frequently associated with increased KRAS gene dosage.
Figure. 4. Colorectal tumors and cell lines harboring A146 KRAS mutations demonstrate evidence of copy number gain at the KRAS gene locus.
A. Statistically significant genomic aberrations in 128 colorectal adenocarcinomas or adenomas (red and blue for amplifications and deletions respectively; FDR ≤ 15%). Profile is shown for the 22 autosomes in genomic coordinates (centromeres in red). B. Copy number gain at the KRAS locus was more common in tumors expressing A146T mutation versus those with exon 2 KRAS mutation or KRAS wild-type tumors (p-value = 0.014, one-tailed Fisher exact test). C. FISH analysis of the LS1034 (KRAS A146T) cell line demonstrating a hyper-triploid karyotype with three copies of chromosome 12 (red arrow) and two copies of an isochromosome for 12p (yellow arrows).
MEK-dependence and EGFR inhibitor resistance of colorectal cancers with exon 4 KRAS mutation
One strategy for treating RAS-mutant tumors is to inhibit the signaling cascades downstream of RAS that mediate RAS-dependent transformation. We previously reported that cells harboring BRAF mutations are selectively sensitive to MEK inhibition whereas tumors harboring G12/13 KRAS and Q61 NRAS mutants are variably dependent on MEK/MAPK 19,30. To determine the MEK/ERK-dependence of cell lines expressing exon 4 KRAS mutations, we used PD0325901, a selective allosteric inhibitor of MEK1/2. As shown in Figure 5, treatment of exon 4 KRAS-expressing cells with PD0325901 resulted in inhibition of MAPK signaling as assessed by downregulation in the expression of phosphorylated ERK1/2 (Fig. 5C). All seven exon 4 KRAS mutant colorectal cancer cell lines were MEK-dependent for proliferation (IC50 < 100nM, Fig. 5A, B). In BRAF-mutant tumors, MEK/ERK signaling is required for both D-cyclin expression and assembly of the cyclin D/cdk4 complex 30. Similarly, treatment of LS1034 and CCCL-18 (both A146T KRAS) cells with PD0325901 caused a marked decline in D-cyclin protein levels, induction of p27, hypo-phosphorylation of RB and a profound G1 cell cycle arrest (Fig. 5C). G1 arrest was accompanied by apoptosis in the LS1034 cell lines, but not in the other three A146T KRAS mutant models suggesting that additional genetic or epigenetic alterations exist in these tumors that likely diminish KRAS-dependence (data not shown). One candidate would be PIK3CA which is concurrently mutated in both CCCL-23 and CCCL18, both of which demonstrate a purely cytostatic response to MEK inhibition. Consistent with prior data showing that exon 2 KRAS mutation confers resistance to EGFR-directed therapies, cell lines harboring A146T/V KRAS mutations were also resistant to the selective EGFR inhibitor gefitinib (Fig. 5D).
Figure. 5. MEK-dependence of A146T KRAS mutant cell lines.
A. IC50 values for the selective MEK1/2 inhibitor PD0325901 for thirty-five colorectal, lung and breast cell lines including all seven models harboring exon 4 KRAS mutation (4 A146T, 2 A146V and 1 K117N). B. Representative IC50 curves for the LS1034 (A146T KRAS), C80 (A146V KRAS), CCCL-18 (A146T KRAS, PIK3CA E542K), HCT-15 (KRAS G13D, PIK3CA E545K) and COLO-205 (V600E BRAF) models. C. Treatment of A146T KRAS expressing cell lines with PD0325901 was associated with a decrease in pERK and cyclin D1 expression, induction of p27 and hypo-phosphorylation of RB. (D). Cell lines expressing exon 4 KRAS mutations were resistant to the EGFR inhibitor gefitinib. The gefitinib-sensitive H3255 (L858R EGFR) model is included for comparison.
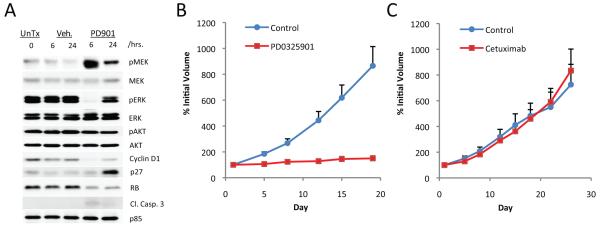
To explore the potential clinical utility of MEK inhibition in tumors driven by A146T KRAS mutation, mice with established A146T KRAS xenografts were treated with PD0325901. Treatment of mice bearing established LS1034 (A146T KRAS) xenografts with a single 25 mg/kg dose of PD0325901 resulted in >95% downregulation of phosphorylated ERK expression at 6 hours (Fig. 6A). MAPK pathway inhibition was associated with downregulation of cyclin D1 and a durable increase in p27 expression and hypo-phosphorylation of RB. Furthermore, chronic treatment of established LS1034 xenografts with non-toxic doses of PD0325901 was associated with complete growth suppression (Fig. 6B). In contrast to the marked sensitivity of the LS1034 model to the MEK inhibitor, this model was resistant to the EGFR targeted antibody cetuximab (Fig. 6C). These data suggest that exon 4 KRAS mutations may predict for sensitivity to MEK inhibition and resistance to EGFR targeted inhibitors.
Figure. 6. The growth of A146T KRAS mutant xenografts was MEK-dependent.
A. Treatment of mice bearing established LS1034 xenografts with a single oral dose of PD0325901 (25 mg/kg) resulted in downregulation of pERK and cyclin D1 expression at 6 hours and upregulation of p27 at 24 hours. B. Mice with established LS1034 (A146T KRAS) xenografts were treated with PD0325901 (25 mg/kg 5x/week × 3 weeks) or vehicle only as control. (C). Mice were treated with 10 mg/kg of cetuximab or vehicle only as control twice weekly for three weeks.
DISCUSSION
Several decades have passed since oncogenic RAS was first identified as the transforming factor in the Harvey and Kirsten strains of the Mouse Sarcoma Virus 1,3-5. Since these discoveries, all three RAS family genes (KRAS, NRAS and HRAS) have been shown to be somatically mutated in human cancer, most commonly as a result of single point mutations at codons 12, 13 and 61.
Despite overwhelming evidence that oncogenic RAS plays a central role in mediating transformation in human tumors, only recently has limited testing for somatic RAS mutations entered routine clinical practice. Widespread adoption of mutational profiling in the clinic has been delayed for several reasons. First, prior to recent advances in sequencing technology, RAS mutational testing was expensive and time-intensive. Second, until recently there was no definitive evidence that routine testing for RAS mutations would meaningfully impact clinical practice. This changed with the identification of KRAS mutations as a predictor of resistance to EGFR kinase inhibitors in patients with lung adenocarcinoma 8. Similar data soon followed in patients with colorectal cancer, where mutations in exon 2 of KRAS were associated with a lack of clinical benefit with panitumumab and cetuximab 9-13. On the basis of these data, routine testing of patients with lung and colorectal cancers has become increasingly common, and some clinical practice guidelines and regulatory agencies have proposed the restriction of anti-EGFR therapy to patients whose tumors lack G12 or G13 KRAS mutations.
In the vast majority of studies to date, tumors have been genotyped only for KRAS mutations at the most commonly altered G12 and G13 positions 31-34. The frequency, predictive, and prognostic value of other RAS mutations has therefore remained poorly defined 16-17. To facilitate the identification of low frequency RAS pathway mutations, we developed a multiplexed MALDI-TOF based genotyping assay using the Sequenom platform. Consistent with prior studies we observed that approximately one third of colorectal tumors harbored mutations at the G12 and G13 codons. Notably, an additional 10%, that would have been characterized as KRAS wild-type in clinical practice, harbored mutations in exons 3 or 4 of KRAS or in NRAS. These latter mutations were mutually exclusive with those at G12 and G13 suggesting overlapping roles in tumorigenesis.
Our dataset suggests that the underrepresentation of these mutations in the literature and their low reported frequency in the COSMIC database (0.002% of the KRAS mutations reported in the large intestine) is the result of detection bias 35. To explore this possibility further, we used our MALDI-TOF assay to characterize the frequency of exon 4 KRAS mutations in several additional lineages. In an analysis of 698 non-colorectal cancer tumors and cell lines, we identified only two additional samples with A146 mutations (one ovarian and one endometrial cancer). A146 mutations in KRAS were also not identified in two recent analyses comprising 449 non-small cell lung cancers in which the entire coding region of the gene was sequenced 36-37. The basis for the higher relative frequency of exon 4 KRAS mutations in colorectal cancer versus other cancers such as lung cancer is unknown, but may be the result of differences in the underlying mutagenic insults responsible for cancer initiation at these sites. We also sequenced exon 4 of both NRAS and HRAS, but detected no mutations in these exons in our colorectal tumors and cell lines. NRAS A146T mutation has, however, been reported in the leukemic cell lines NALM6 and ML-2 16 and germline HRAS mutations at the K117 and A146 codons have been reported in a small number of patients with Costello’s Syndrome 38.
The RAS family proteins function as small GTPases that cycle between an inactive GDP-bound and an active GTP-bound state. The slow intrinsic GTPase activity of RAS is enhanced by several orders of magnitude by GTPase activating proteins (RAS GAPs), which include p120 GAP and NF1, which facilitate GTP hydrolysis by stabilizing an intermediate high-energy transitional state 7. The most common site of RAS mutation located at position 12 results in substitution of glycine for a residue with a side chain. Crystal structure modeling predicts that this substitution is associated with steric interference with GAP-mediated GTP hydrolysis. As glycine is the only amino acid lacking a side chain, a diversity of mutations at this position confer similar phenotypic effects. Mutations at the codon 61 position also impair RAS GTPase activity, but in this case by disrupting a hydrogen bond between the glutamine residue at position 61 of RAS and Arg789 of GAPp120 39-40.
Although the mutual exclusivity of the exons 2, 3 and 4 mutations in KRAS suggest significant functional overlap, the cohort of patients with non-exon 2 mutations in KRAS exhibited a better prognosis than patients whose tumors expressed G12 or G13 KRAS mutations. The most common site of KRAS mutation in exon 4 in our series was at amino acid A146. This site is within an evolutionarily conserved region which in structural modeling is predicted to interact with the guanine base of GDP 41. In contrast to mutations at codons 12 and 13, mutations at codon 146 do not impair RAS GTPase activity 42. Rather, the transforming potential of the A146 HRAS mutations has been attributed to an increase in guanine nucleotide exchange 42. As discussed above, mutations of A146 and of the biologically conserved K117 positions of HRAS have been reported in Costello’s syndrome. The most common mutant allele in Costello’s syndrome is G12S and notably, its transforming effects are lower than that of the G12V mutation, which is among the most common mutant alleles in human cancer 43. Based upon this observation it has been speculated that only low activation alleles of RAS may be compatible with viability when found in the germline.
Given the favorable prognosis of colorectal cancer patients with exon 4 KRAS mutations and the observation that the K117 and A146 mutations are found in Costello’s syndrome, we hypothesized that these mutations may be less potent than mutations at codons 12 and 13. Although we found lower RAS-GTP expression in an isogenic model of K117 and A146T KRAS, in some A146T KRAS expressing cancer cell lines, we observed levels of RAS-GTP comparable to that of cell lines harboring the G12D/V mutations. Our data suggest that whereas A146T KRAS mutation may confer lower intrinsic RAS activity, this may be augmented in part by frequent conversion to homozygosity and low-level copy number gain of the KRAS gene locus.
Our analysis suggests that a broader assessment of RAS mutations beyond the most common mutations in exon 2 is warranted and would lead to the identification of a mutation predicted to confer EGFR inhibitor resistance in close to 50% of patients with colorectal cancer 12,19. While such testing would decrease the use of toxic and expensive agents in this population unlikely to derive benefit, it would also further limit the available treatment options in a disease in which few currently exist. This and the inability to date to identify a clinically effective inhibitor of RAS have contributed to the reluctance of many clinicians to advocate broader RAS mutation testing. It should be noted, however, that the prospective identification of RAS mutations could also have the secondary benefit of accelerating the clinical development of novel therapies in this class of patients by facilitating the identification of those most likely to benefit. Promising therapeutic approaches include targets that function as synthetic lethals in RAS mutant tumors and inhibitors of downstream effectors such as MEK 30,44-47. Our data demonstrating complete growth inhibition in A146T KRAS expressing xenografts with the selective MEK inhibitor PD0325901 support the clinical feasibility of this latter approach.
In summary, our data support a more comprehensive assessment of RAS mutational status beyond the most frequently mutated alleles at positions 12, 13 and 61. The ability to use a multiplexed platform makes such an approach feasible even in the case of low frequency alleles. Our data also support the hypothesis that different RAS alleles have overlapping but not identical biologic activities, and may thus confer differential prognostic effects. These differences may impact the choice of therapy in individual cases and may be exploited to therapeutic advantage.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Igor Dolgalev, Sabrena Thomas, and Olga Aminova from the Geoffrey Beene Translational Oncology Core and Agnes Viale of the Genomics Core, and Elisa De Stanchina for technical assistance. We also thank Walter Bodmer and Rachael Hancox (Cancer Research UK), Diego Arango (Hospital Universitario Vall d’Hebron, Barcelona, Spain) and Cliff Stanners and Mary Luisa DeMarte (McGill University, Montreal, Canada) for cell lines. This study was supported by grants from the National Institutes of Health, the Kimmel Foundation, and the Abrams Foundation. The MSKCC Sequenom facility is supported by the Anbinder Fund.
REFERENCES
- 1.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–65. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 2.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 3.Chang EH, Furth ME, Scolnick EM, Lowy DR. Tumorigenic transformation of mammalian cells induced by a normal human gene homologous to the oncogene of Harvey murine sarcoma virus. Nature. 1982;297:479–83. doi: 10.1038/297479a0. [DOI] [PubMed] [Google Scholar]
- 4.Shimizu K, et al. Three human transforming genes are related to the viral ras oncogenes. Proc Natl Acad Sci U S A. 1983;80:2112–6. doi: 10.1073/pnas.80.8.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Der CJ, Krontiris TG, Cooper GM. Traxnsforming genes of human bladder and lung carcinoma cell lines are homologous to the ras genes of Harvey and Kirsten sarcoma viruses. Proc Natl Acad Sci U S A. 1982;79:3637–40. doi: 10.1073/pnas.79.11.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang EH, Gonda MA, Ellis RW, Scolnick EM, Lowy DR. Human genome contains four genes homologous to transforming genes of Harvey and Kirsten murine sarcoma viruses. Proc Natl Acad Sci U S A. 1982;79:4848–52. doi: 10.1073/pnas.79.16.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boguski MS, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–54. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 8.Pao W, et al. KRAS Mutations and Primary Resistance of Lung Adenocarcinomas to Gefitinib or Erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lievre A, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–5. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 10.Di Fiore F, et al. Clinical relevance of KRAS mutation detection in metastatic colorectal cancer treated by Cetuximab plus chemotherapy. Br J Cancer. 2007;96:1166–9. doi: 10.1038/sj.bjc.6603685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amado RG, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–34. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 12.Di Nicolantonio F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–12. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 13.Van Cutsem E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–17. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 14.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 15.Brose MS, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- 16.Edkins S, et al. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol Ther. 2006;5:928–32. doi: 10.4161/cbt.5.8.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tyner JW, et al. High-throughput sequencing screen reveals novel, transforming RAS mutations in myeloid leukemia patients. Blood. 2009;113:1749–55. doi: 10.1182/blood-2008-04-152157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas RK, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–51. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 19.Pratilas CA, et al. Genetic predictors of MEK dependence in non-small cell lung cancer. Cancer Res. 2008;68:9375–83. doi: 10.1158/0008-5472.CAN-08-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–63. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 22.Taylor BS, et al. Functional copy-number alterations in cancer. PLoS ONE. 2008;3:e3179. doi: 10.1371/journal.pone.0003179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCarroll SA, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–74. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 24.Ogino S, et al. A cohort study of cyclin D1 expression and prognosis in 602 colon cancer cases. Clin Cancer Res. 2009;15:4431–8. doi: 10.1158/1078-0432.CCR-08-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richman SD, et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol. 2009;27:5931–7. doi: 10.1200/JCO.2009.22.4295. [DOI] [PubMed] [Google Scholar]
- 26.Roth AD, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28:466–74. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 27.Gerdes H, Elahi A, Chen Q, Jhanwar S. Characterization of newly established colorectal cancer cell lines:correlation between cytogenetic abnormalities and allelic deletions associated with multistep tumorigenesis. Journal of Genetics. 2000;79:113–123. [Google Scholar]
- 28.Martin ES, et al. Common and distinct genomic events in sporadic colorectal cancer and diverse cancer types. Cancer Res. 2007;67:10736–43. doi: 10.1158/0008-5472.CAN-07-2742. [DOI] [PubMed] [Google Scholar]
- 29.Tsafrir D, et al. Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer Res. 2006;66:2129–37. doi: 10.1158/0008-5472.CAN-05-2569. [DOI] [PubMed] [Google Scholar]
- 30.Solit DB, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–62. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neumann J, Zeindl-Eberhart E, Kirchner T, Jung A. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol Res Pract. 2009 doi: 10.1016/j.prp.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 32.Andreyev HJ, et al. Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer. 2001;85:692–6. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst. 1998;90:675–84. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 34.Barault L, et al. Mutations in the RAS-MAPK, PI(3)K (phosphatidylinositol-3-OH kinase) signaling network correlate with poor survival in a population-based series of colon cancers. Int J Cancer. 2008;122:2255–9. doi: 10.1002/ijc.23388. [DOI] [PubMed] [Google Scholar]
- 35.Forbes SA, et al. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr Protoc Hum Genet. 2008 doi: 10.1002/0471142905.hg1011s57. Chapter 10, Unit 10 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marks JL, et al. Novel MEK1 mutation identified by mutational analysis of epidermal growth factor receptor signaling pathway genes in lung adenocarcinoma. Cancer Res. 2008;68:5524–8. doi: 10.1158/0008-5472.CAN-08-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zampino G, et al. Diversity, parental germline origin, and phenotypic spectrum of de novo HRAS missense changes in Costello syndrome. Hum Mutat. 2007;28:265–72. doi: 10.1002/humu.20431. [DOI] [PubMed] [Google Scholar]
- 39.Krengel U, et al. Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules. Cell. 1990;62:539–48. doi: 10.1016/0092-8674(90)90018-a. [DOI] [PubMed] [Google Scholar]
- 40.Scheffzek K, et al. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–8. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 41.Milburn MV, et al. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–45. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 42.Feig LA, Cooper GM. Relationship among guanine nucleotide exchange, GTP hydrolysis, and transforming potential of mutated ras proteins. Mol Cell Biol. 1988;8:2472–8. doi: 10.1128/mcb.8.6.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seeburg PH, Colby WW, Capon DJ, Goeddel DV, Levinson AD. Biological properties of human c-Ha-ras1 genes mutated at codon 12. Nature. 1984;312:71–5. doi: 10.1038/312071a0. [DOI] [PubMed] [Google Scholar]
- 44.Engelman JA, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–6. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo J, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–48. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scholl C, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–34. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 47.Singh A, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.