Abstract
Objective
A meta-analysis of genome-wide linkage studies allows us to summarize the extensive information available from family-based studies, as the field moves into genome-wide association studies.
Methods
Here we apply the genome scan meta-analysis (GSMA) method, a rank-based, model-free approach, to combine results across eight independent genome-wide linkages performed on celiac disease (CD), including 554 families with over 1,500 affected individuals. We also investigate the agreement between signals we identified from this meta-analysis of linkage studies and those identified from genome-wide association analysis using a hypergeometric distribution.
Results
Not surprisingly, the most significant result was obtained in the HLA region. Outside the HLA region, suggestive evidence for linkage was obtained at the telomeric region of chromosome 10 (10q26.12-qter; p = 0.00366), and on chromosome 8 (8q22.2-q24.21; p = 0.00491). Testing signals of association and linkage within bins showed no significant evidence for co-localization of results.
Conclusion
This meta-analysis allowed us to pool the results from available genome-wide linkage studies and to identify novel regions potentially harboring predisposing genetic variation contributing to CD. This study also shows that linkage and association studies may identify different types of disease-predisposing variants.
Key Words: Celiac disease, Meta-analysis, Genome-wide linkage analysis, Genome-wide association analysis, Dermatitis herpetiformis
Introduction
Celiac disease (CD) is an inflammatory disorder of the small intestine with a complex etiology, triggered by dietary gluten. The disease prevalence is estimated to be up to 1% in Western populations. In addition to the classical gastrointestinal form, a variety of other clinical manifestations of the disease have been described, including atypical and asymptomatic forms [1]. The most common extra-intestinal manifestation is dermatitis herpetiformis (DH), a blistering skin condition, currently regarded as a variant of CD, affecting one-quarter of the patients with gluten intolerance. Asymptomatic forms are characterized by the presence of histological changes, identified through screening programs on apparently healthy subjects.
The only genetic factor indisputably involved in CD is the human leukocyte antigen (HLA) locus, where a clear association has been found with HLA-DQ variants [2]. However, the HLA association alone is insufficient to explain the hereditary nature of the disease [3], indicating that additional, non-HLA genes must also be involved in disease susceptibility.
Several independent genome-wide scans in multiplex families have identified genomic regions that may harbor susceptibility genes for CD, but few studies have shown significant evidence for linkage outside the HLA region, and there is little convincing replication of linked regions across studies. This is typical of linkage studies for complex diseases, and is likely due to the low power of studies to detect genes of relatively small effect and to the high degree of genetic heterogeneity among families. Promising loci include a region on 5q31-33 (CELIAC2) which was identified in several linkage studies [4,5,6] and in a meta-analysis and a pooled genotype analysis of data from the European Genetics Cluster on Coeliac Disease [7]. A locus on chromosome 19p13.1 (CELIAC 4), which was identified in a Dutch linkage study [8], harbors a functional candidate gene implicated in intestinal barrier integrity (MYO9B, MIM: 602129) for which an intronic SNP shows significant association with CD [9].
More recently, one genome-wide association (GWA) study has been published [10], with follow-up of top regions confirming eight novel, non-HLA regions associated with CD [11]. These studies, and other previous studies focusing on candidate genes, have identified evidence of association with common variants, generally with only minor effect on disease risk.
A meta-analysis of genome-wide linkage studies, to summarize the available information in these extensive family-based studies, is timely as the field moves into genome-wide association studies. We use the genome scan meta-analysis (GSMA) method [12] to combine results across linkage studies, and identify regions which may harbor genetic determinants for CD. We also investigate the agreement between signals of genome-wide linkage and association analysis to test whether CD-associated SNPs are over-represented in linked regions.
Material and Methods
GSMA
The genome scan meta-analysis (GSMA) method [12] was used to combine linkage results from CD genome screens. The GSMA method is designed to deal with dissimilarity in study design, marker panels, analysis methods, and linkage statistics used in different genome scans. The GSMA uses only genome-wide linkage results, with no original genotype or family data required. To apply the GSMA method the genome needs to be fragmented into bins of approximately equal length. Using a bin width of approximately 30 cM, 118 bins provided the most even coverage across autosomes based on the Marshfield linkage maps (http://research.marshfieldclinic.org/genetics/home/index.asp) [13], with chromosome 1 having 10 bins, and chromosomes 21 and 22 having 2 bins each [14].
For each scan, the maximum value of the linkage statistic obtained in each bin was identified. The bins were then ranked, from the lowest (rank = 1) to the highest (rank = 118) value of the linkage statistic across the genome. Bin ranks were then summed across studies and the summed rank (SR) was compared to its probability distribution under the null hypothesis of no linkage (i.e. assuming ranks were randomly assigned to a bin). The significance of the SR in each bin was assessed with the GSMA software which obtains an empirical p value for the observed SR in each bin, using Monte Carlo simulations permuting the bin location of the ranks within each study (http://www.kcl.ac.uk/schools/medicine/depts/memoge/research/epidemiology/gsma/index.html) [15]. To control for multiple testing of bins, we used a Bonferroni correction. For 30 cM width bins, a p value of 0.05/118 = 0.00042 was required for genome-wide evidence of linkage, or a p value of 1/118 = 0.00847 for suggestive evidence of linkage over the autosomes, corresponding to the SR p values expected to arise by chance once in 20 GSMA studies, and once in a single GSMA study, respectively.
Meta-analysis of CD was performed both unweighted (assuming equal contribution from each study) and weighted by study size. The weighting factor for each study was defined as the square root of (# affecteds – # families), scaled to a mean of 1. Analyses under different weighting functions produced similar results, and are not reported. The primary statistical analysis used 30 cM bin widths; chromosomes showing suggestive evidence for linkage (under weighted or unweighted analysis) were analysed further using bins of 10 and 20 cM width in order to improve the localization of linkage signals detected.
CD Linkage Studies
In total, 13 published genome scans of CD families were identified through a review of the literature and through PubMed search. Only whole genome-wide linkage scan analyses were considered, with partial scans and follow-up studies in candidate regions excluded. One study [16] was an update of a previous study, with a larger family collection, and the earlier study was therefore omitted [17]. We also excluded a study of a single four-generation Dutch pedigree (17 affecteds) with disease transmission following a Mendelian pattern [18], and two studies performed in extended, consanguineous families from a Finnish genetic isolate [19] and a Bedouin population in the Negev Desert [20]. These studies were not included as the loci involved in these families may not play a major role in CD as a multifactorial disorder represented in the other studies. Finally, we also excluded a small, low-powered study for which only best peaks were available from the published paper [21].
Authors of identified studies were contacted by e-mail and invited to contribute linkage results required for the meta-analysis. All groups agreed to participate in the meta-analysis, providing multipoint non-parametric linkage statistics across the genome, with results of one study being extracted from published graphs [22].
The eight independent CD studies analysed consisted of 554 families and over 1,500 affected individuals (table 1). All studies were of European ancestry and comprised sibships with at least two affected children or families with more distant affected relative pairs. X chromosome results were available for only four studies and were therefore not analysed.
Table 1.
Summary of genome-wide linkage studies included in the GSMA (ordered by year of publication)
| First author | Year | Population | No. families | No. affecteds | GSMA weight | Linkage statistica | Diagnostic criteria | Diseaseb |
|---|---|---|---|---|---|---|---|---|
| Garner | 2006 | US | 160 | 562 | 1.95 | NPL | biopsy; antibodies; DH (3%)c | symptomatic; asymptomatic |
| Rioux | 2004 | Finnish | 54 | 168d | 1.04 | NPL | biopsy; antibodies; DH (13%)c | symptomatic; asymptomatic |
| Van Belzen | 2003 | Dutch | 67 | 142d | 0.84 | MLS | biopsy | symptomatic; asymptomatic |
| Popat | 2002 | UK, Sweden, other | 24 | 79d | 0.72 | NPL | ESPGAN criteria (biopsy) | symptomatic |
| Liue | 2002 | Finnish | 60 | 159 | 0.97 | NPL | biopsy; DH (22%)c | symptomatic; asymptomatic |
| Naluaie | 2001 | Scandinavian | 70 | 156d | 0.90 | NPL | ESPGAN criteria (biopsy) | symptomatic; asymptomatic |
| Kinge | 2000 | UK | 16 | 50 | 0.57 | NPL | biopsy; antibodies | symptomatic; asymptomatic |
| Grecoe | 1998 | I | 103 | 213d | 1.02 | MLS | biopsy; antibodies | symptomatic; asymptomatic |
MLS = Maximum likelihood statistic; NPL = non parametric linkage score
Studies included patients with symptomatic CD, or asymptomatic CD, diagnosed through screening but without the presence of well-defined CD symptoms
DH = dermatitis herpetiformis; percentage of DH cases among affecteds are indicated
Number of affecteds derived from information on number of affected sib pairs/trios etc or from the family trees of affected families
Study included in the previous meta/mega analysis by Babron et al. [7].
Five studies included only patients diagnosed with classic CD, while three studies [5, 16, 22] also included patients with dermatitis herpetiformis (DH). DH and CD share a common immunogenetic background but may be heterogeneous gluten-sensitive diseases, and we therefore performed a separate analysis of the five CD-only.
Data Extraction
Linkage results obtained from investigators consisted of computer files from analysis programs showing linkage statistics (NPL score, p value, etc.) at each marker genotyped or at specific genetic locations. For results with marker data [4, 5, 23], we first mapped markers to locations on the Marshfield linkage map and then determined the bin location of each marker using the previously defined bin boundaries. For results indexed by genetic location [6, 8, 16, 24], we determined the locations of the first and last markers genotyped on each chromosome and then rescaled the genetic locations of the linkage statistics accordingly, before dividing the chromosome into the required number of bins. This correction overcomes the problem of bin shifting when the first or last marker genotyped on a chromosome is located several cM from the telomere, and ensures that bins from each study cover the same genetic region, regardless of data format. Data was extracted from genome-wide linkage graphs [22], using Engauge Digitizer (http://digitizer.sourceforge.net/) and chromosomes divided into the required number of equal width bins [14].
After markers and linkage statistic locations for each study had been localized onto the Marshfield map, we then identified the maximum linkage statistic per bin. R-scripts facilitating these data extraction steps are available from the authors.
Correlation between Linkage and Association Findings
In order to compare results from the linkage studies with the recent CD GWA study [10], we extracted SNPs showing association with a p value of <10–4 from their online supplementary table 3 (www.karger.com/doi/10.1159/000228920), and mapped these onto the 30 cM linkage bins using the Rutgers combined linkage-physical map for Build 36 (http://compgen.rutgers.edu/mapomat/). We tested for significant co-occurrence of nominally significant linkage findings with these association findings using the hypergeometric distribution. Specifically, we tested whether the associated SNPs were more likely to be located in bins identified in the GSMA than expected by chance, using different p value thresholds in the weighted and unweighted 30 cM GSMA.
Results
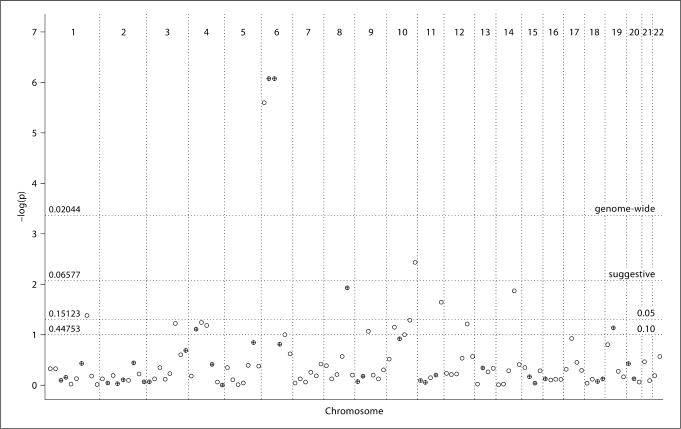
Meta-analysis of eight genome-wide linkage studies in CD was performed both unweighted and weighted. Results based on 30 cM bin widths are summarized in figure 1 and table 2, which lists all nominally significant bins (p value <0.05), highlighting those with suggestive and genome-wide evidence for linkage.
Fig. 1.
Linkage results across the genome for eight CD studies for the weighted analysis, showing −log10 (p value) against chromosomal location, with a single point plotted for each 30 cM bin. Thresholds for nominal, suggestive, and genome-wide significance are shown. Crossed points represent bins where SNPs with p < 10−4 were identified in the genome-wide association study [10]. p values from the hypergeometric distribution for specific linkage thresholds are indicated on the left.
Table 2.
Summary of all nominally significant bins (p < 0.05) in the unweighted and weighted GSMA analyses using 30 cM bin widths
| Chr | Bin | cM (Marshfield) | Mb (build 36) | All studies (8), p value |
CD studies (5), p value |
GWAa |
|||
|---|---|---|---|---|---|---|---|---|---|
| unweighted | weighted | unweighted | weighted | Kb | SNP | ||||
| 1 | 1_8 | 203–232 | 183–208 | – | 0.0416 | – | – | 190,803 | rs2816316 |
| 4 | 4_4 | 91–121 | 81–115 | 0.0411 | – | – | – | ||
| 6 | 6_1 | 0–32 | 0–16 | <0.0001 | <0.0001 | 0.0026 | 0.0012 | ||
| 6_2 | 32–64 | 16–42 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |||
| 6_3 | 64–97 | 42–89 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 52,573 | rs6936059 | |
| 8 | 8_5 | 112–140 | 100–129 | 0.0063 | 0.0118 | 0.0484 | – | 103,207 | rs648119 |
| 9 | 9_3 | 56–84 | 37–85 | 0.0481 | – | – | – | ||
| 10 | 10_5 | 115–144 | 94–123 | 0.0360 | – | 0.0298 | 0.0369 | ||
| 10_6 | 144–173 | 123–135 | 0.0027 | 0.0037 | 0.0153 | 0.0227 | |||
| 11 | 11_5 | 118–148 | 123–134 | 0.0150 | 0.0229 | 0.0407 | – | ||
| 14 | 14_4 | 83–111 | 74–93 | 0.0208 | 0.0137 | – | – | ||
| 19 | 19_2 | 26–53 | 8–36 | 0.0121 | – | 0.0037 | 0.0031 | 34,672 | rs1036229 |
Bins with suggestive (p < 0.00847; in bold) and genome-wide (p < 0.00042; in bold and underlined) evidence for linkage are highlighted.
SNPs with p < 10−4 reported in supplementary table 3 from the GWA by van Heel et al. [10].
Genome-wide evidence for linkage was obtained in the HLA region (bin 6_2, i.e. the second bin on chromosome 6). A strong signal was also seen in the flanking bins (6_1 and 6_3), due to a carry-over effect from elevated multipoint linkage scores in an extended region of chromosome 6.
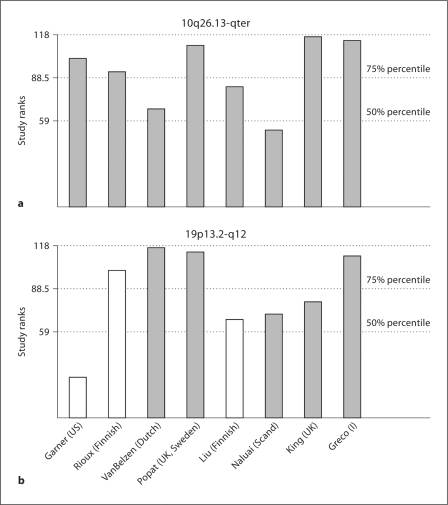
Outside the HLA region, the strongest evidence for linkage occurred in bin 10_6, with suggestive evidence for linkage in both weighted and unweighted analyses (p = 0.00366, p = 0.00271, respectively). This telomeric region on chromosome 10 corresponds to the genetic interval 144.3–173.1 cM in the Marshfield map, and to the physical interval 123.3–135.1 Mb in Build 36 (cytogenetic band 10q26.12-qter). The contribution of individual studies to this bin suggests that most linkage evidence arises in the studies with broad European-ancestry studies, with weaker evidence from the Finnish, Scandinavian, and Dutch studies (fig. 2). A further region on chromosome 8 showed suggestive evidence for linkage in the unweighted analysis (p = 0.00635), but only nominal significance in the weighted analysis. This region (bin 8_5; 8q22.2-q24.21; 101.5–139.5 Mb), includes rs648119 (at 103.2 Mb) which was associated with CD in the GWA study with p = 4 × 10–5[10]. In the analysis of five studies based on classic CD patients, we observed only nominal significance in bins 10_6 and 8_5, but suggestive significance for linkage on chromosome 19 (bin 19_2, 19p13.2-q12). This bin contains SNP rs1036229 (at 34.7 Mb), which achieved a p value of 2.5 × 10–5 in the GWA, and MYO9B (at 17.2 Mb).
Fig. 2.
Contribution of each study at the identified regions on 10q26.13-qter (a) and 19p13.2-q12 (b). The study-specific ranks at bin 10_6 and bin 19_2, respectively, for the 30 cM bin definition are shown, with 50th and 75th percentile of the study ranks. White bars represent studies that included also DH cases.
We re-analyzed chromosomes showing suggestive evidence for linkage using two narrower bin widths of 20 cM (giving 173 bins) and 10 cM (giving 349 bins) to assess whether narrower bins provide finer localization of linkage findings. Results of the weighted GSMA are shown in the supplementary material for chromosome 8 and 10 (using 8 studies) and chromosome 19 (for the 5 CD-only studies). Similar results were obtained in the unweighted analysis. For chromosome 8, suggestive evidence for linkage (p = 0.00491) was obtained with the 20 cM bin at 99.2–118.6 Mb. No improved localization for chromosome 10 linkage finding was obtained. On chromosome 19, analysis of 10 cM bins showed that the strongest evidence for linkage occurred at 13.4–17.9 Mb, an interval which includes the MYO9B gene.
Testing signals of association and linkage within bins showed no significant evidence for co-localization of results (see fig. 1 for bin location of association signals with p < 10–4 and for the p values from the hypergeometric distribution), except for the genome-wide threshold that identified in the GSMA analysis the HLA region (p = 0.02044). For example, 8 out of 117 bins showed nominal significance in the weighted GSMA (p < 0.05), and only 3 of these were among the 33 bins containing significant association signals (p = 0.15123).
Discussion
CD is an interesting model disorder for complex diseases, as the main environmental factor (gluten) and genetic factor (HLA) have been identified. Several genome-wide linkage scans and candidate gene studies have been carried out to identify other genetic factors underlying CD, with no clear definition of further genetic contributions to CD. In this study, we performed meta-analysis of eight genome-wide linkage studies in CD, to summarize the evidence for linkage in these extensive family studies.
Highly significant evidence for linkage was obtained in the HLA region and flanking regions on chromosome 6. We also obtained suggestive evidence for linkage to the telomeric region of chromosome 10q. This locus provided only marginal evidence in some studies and would have not be identified by ad hoc assessment of individual scan results, showing the strength of the GSMA in identifying regions with modest but consistently elevated linkage scores across studies. Further, looking at the specific contributions of individual genome-wide linkage studies to the 10q26.12-qter region, it is apparent that most linkage evidence derives from studies with broad European ancestry, including the Italian study. An additional region on chromosome 8 showed suggestive significance in some analyses, and includes SNP rs648119 which was identified in the GWA [10], although not confirmed in the subsequent follow-up [11].
The chromosome 5 region highlighted in the previous meta-analysis [3] did not achieve even nominal significance in this study. This inconsistency is not surprising given current study doubled both the number of studies and the number of affected individuals included. Application of different weighting functions did not affect results, but other explanations include the different test statistics (NPL and MLS) included in the current study (the previous analysis had used Kong and Cox's Zlr statistic for all studies) [25], or a false positive finding in the initial meta-analysis. Analysis of the subset of four studies from the original meta-analysis recaptured the 5q linkage evidence, implying that heterogeneity between studies, rather than differences in statistical methodology may explain the discrepancy. Notably, the 5q results may be correlated with family structure. When we stratified the studies by affected sibling pairs and more extended pedigrees, we obtained suggestive significance in the same 5q region when analyzing the four affected sib pairs studies [4,5,6, 8]. Indeed, in the previous meta-analysis, 3 of the 4 studies included only affected sib pairs.
In general, we observed no concordance between regions showing significant linkage identified in this GSMA and the occurrences of the significant SNPs in genome-wide association analysis study [10], except for the HLA locus. The number of ‘linked’ bins with significant association findings was not significant under either GSMA analysis method (weighted, unweighted) or the GSMA thresholds considered (suggestive significance, nominal significance, and p < 0.10). Association studies with current SNPs platforms are aimed at identifying common variants involved in disease etiology, whereas linkage approaches could be used efficiently to identify multiple rare variants (even variation that is unique at the individual level) of moderate effect, which are not possible to detect using traditional association methods. On the other hand, common variants identified by van Heel et al. [10] would have very low power to be detected in a linkage study.
We sub-divided studies by phenotype, under the hypothesis that more phenotypically homogenous studies may have higher power to detect linkage. Excluding three studies that included DH cases gave an additional region with suggestive evidence for linkage at 19p13.2-q12 (8–36 Mb), which was further localised to 14.9–17.9 Mb under analysis with 10 cM bins. This region contains the MYO9B gene located at 17.2 Mb (which was not highlighted in the GWA study). MYO9B is thought to have a role in epithelial barrier functions, and association with CD has been replicated, but does not arise in all populations [9]. Association with inflammatory bowel disease, systemic lupus erythematosus, rheumatoid arthritis, and type 1 diabetes have also been reported, making this gene a potential shared risk factor across autoimmune disorders.
These results, in conjunction with data emerging from dense SNP typing of specific regions or future genome-wide association studies will help guide efforts to identify the actual predisposing genetic variation contributing to this complex genetic disease. In particular, results from a meta-analysis of linkage studies can be used in a GWA study to enhance power by weighting association evidence using linkage results. This approach had been shown to have considerable powerful, if the linkage study is informative [26].
Acknowledgements
This research was funded by the UK MRC (G0400960 to CML), and the NIH (DK50678 to SLN). The Dutch linkage study was supported by the Coeliac Disease Consortium, an innovative cluster approved by the Netherlands Genomics Initiative and partially funded by the Dutch government (grant BSIK03009). We thank all authors of the original genome wide linkage studies for their contribution to this study.
References
- 1.Fasano A, Catassi C. Current approaches to diagnosis and treatment of celiac disease: An evolving spectrum. Gastroenterology. 2001;120:636–651. doi: 10.1053/gast.2001.22123. [DOI] [PubMed] [Google Scholar]
- 2.Spurkland A, Sollid LM, Polanco I, Vartdal F, Thorsby E. Hla-dr and -dq genotypes of celiac disease patients serologically typed to be non-dr3 or non-dr5/7. Hum Immunol. 1992;35:188–192. doi: 10.1016/0198-8859(92)90104-u. [DOI] [PubMed] [Google Scholar]
- 3.Bevan S, Popat S, Braegger CP, Busch A, O'Donoghue D, Falth-Magnusson K, Ferguson A, Godkin A, Hogberg L, Holmes G, Hosie KB, Howdle PD, Jenkins H, Jewell D, Johnston S, Kennedy NP, Kerr G, Kumar P, Logan RF, Love AH, Marsh M, Mulder CJ, Sjoberg K, Stenhammer L, Walker-Smith J, Marossy AM, Houlston RS. Contribution of the mhc region to the familial risk of coeliac disease. J Med Genet. 1999;36:687–690. [PMC free article] [PubMed] [Google Scholar]
- 4.Greco L, Corazza G, Babron MC, Clot F, Fulchignoni-Lataud MC, Percopo S, Zavattari P, Bouguerra F, Dib C, Tosi R, Troncone R, Ventura A, Mantavoni W, Magazzu G, Gatti R, Lazzari R, Giunta A, Perri F, Iacono G, Cardi E, de Virgiliis S, Cataldo F, De Angelis G, Musumeci S, Clerget-Darpoux F, et al. Genome search in celiac disease. Am J Hum Genet. 1998;62:669–675. doi: 10.1086/301754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Juo SH, Holopainen P, Terwilliger J, Tong X, Grunn A, Brito M, Green P, Mustalahti K, Maki M, Gilliam TC, Partanen J. Genomewide linkage analysis of celiac disease in Finnish families. Am J Hum Genet. 2002;70:51–59. doi: 10.1086/338453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naluai AT, Nilsson S, Gudjonsdottir AH, Louka AS, Ascher H, Ek J, Hallberg B, Samuelsson L, Kristiansson B, Martinsson T, Nerman O, Sollid LM, Wahlstrom J. Genome-wide linkage analysis of scandinavian affected sib-pairs supports presence of susceptibility loci for celiac disease on chromosomes 5 and 11. Eur J Hum Genet. 2001;9:938–944. doi: 10.1038/sj.ejhg.5200752. [DOI] [PubMed] [Google Scholar]
- 7.Babron MC, Nilsson S, Adamovic S, Naluai AT, Wahlstrom J, Ascher H, Ciclitira PJ, Sollid LM, Partanen J, Greco L, Clerget-Darpoux F. Meta and pooled analysis of European coeliac disease data. Eur J Hum Genet. 2003;11:828–834. doi: 10.1038/sj.ejhg.5201051. [DOI] [PubMed] [Google Scholar]
- 8.Van Belzen MJ, Meijer JW, Sandkuijl LA, Bardoel AF, Mulder CJ, Pearson PL, Houwen RH, Wijmenga C. A major non-hla locus in celiac disease maps to chromosome 19. Gastroenterology. 2003;125:1032–1041. doi: 10.1016/s0016-5085(03)01205-8. [DOI] [PubMed] [Google Scholar]
- 9.Monsuur AJ, de Bakker PI, Alizadeh BZ, Zhernakova A, Bevova MR, Strengman E, Franke L, van't Slot R, van Belzen MJ, Lavrijsen IC, Diosdado B, Daly MJ, Mulder CJ, Mearin ML, Meijer JW, Meijer GA, van Oort E, Wapenaar MC, Koeleman BP, Wijmenga C. Myosin ixb variant increases the risk of celiac disease and points toward a primary intestinal barrier defect. Nat Genet. 2005;37:1341–1344. doi: 10.1038/ng1680. [DOI] [PubMed] [Google Scholar]
- 10.van Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M, Wapenaar MC, Barnardo MC, Bethel G, Holmes GK, Feighery C, Jewell D, Kelleher D, Kumar P, Travis S, Walters JR, Sanders DS, Howdle P, Swift J, Playford RJ, McLaren WM, Mearin ML, Mulder CJ, McManus R, McGinnis R, Cardon LR, Deloukas P, Wijmenga C. A genome-wide association study for celiac disease identifies risk variants in the region harboring il2 and il21. Nat Genet. 2007;39:827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunt KA, Zhernakova A, Turner G, Heap GA, Franke L, Bruinenberg M, Romanos J, Dinesen LC, Ryan AW, Panesar D, Gwilliam R, Takeuchi F, McLaren WM, Holmes GK, Howdle PD, Walters JR, Sanders DS, Playford RJ, Trynka G, Mulder CJ, Mearin ML, Verbeek WH, Trimble V, Stevens FM, O'Morain C, Kennedy NP, Kelleher D, Pennington DJ, Strachan DP, McArdle WL, Mein CA, Wapenaar MC, Deloukas P, McGinnis R, McManus R, Wijmenga C, van Heel DA. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wise LH, Lanchbury JS, Lewis CM. Meta-analysis of genome searches. Ann Hum Genet. 1999;63:263–272. doi: 10.1046/j.1469-1809.1999.6330263.x. [DOI] [PubMed] [Google Scholar]
- 13.Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: Individual and sex-specific variation in recombination. Am J Hum Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forabosco P, Ng MY, Bouzigon E, Fisher SA, Levinson DF, Lewis CM. Data acquisition for meta-analysis of genome-wide linkage studies using the genome search meta-analysis method. Hum Hered. 2007;64:74–81. doi: 10.1159/000101425. [DOI] [PubMed] [Google Scholar]
- 15.Pardi F, Levinson DF, Lewis CM. Gsma: Software implementation of the genome search meta-analysis method. Bioinformatics. 2005;21:4430–4431. doi: 10.1093/bioinformatics/bti725. [DOI] [PubMed] [Google Scholar]
- 16.Garner CP, Ding YC, Steele L, Book L, Leiferman K, Zone JJ, Neuhausen SL. Genome-wide linkage analysis of 160 North American families with celiac disease. Genes Immun. 2007;8:108–114. doi: 10.1038/sj.gene.6364361. [DOI] [PubMed] [Google Scholar]
- 17.Neuhausen SL, Feolo M, Camp NJ, Farnham J, Book L, Zone JJ. Genome-wide linkage analysis for celiac disease in North American families. Am J Med Genet. 2002;111:1–9. doi: 10.1002/ajmg.10527. [DOI] [PubMed] [Google Scholar]
- 18.van Belzen MJ, Vrolijk MM, Meijer JW, Crusius JB, Pearson PL, Sandkuijl LA, Houwen RH, Wijmenga C. A genomewide screen in a four-generation Dutch family with celiac disease: Evidence for linkage to chromosomes 6 and 9. Am J Gastroenterol. 2004;99:466–471. doi: 10.1111/j.1572-0241.2004.04072.x. [DOI] [PubMed] [Google Scholar]
- 19.Woolley N, Holopainen P, Ollikainen V, Mustalahti K, Maki M, Kere J, Partanen J. A new locus for coeliac disease mapped to chromosome 15 in a population isolate. Hum Genet. 2002;111:40–45. doi: 10.1007/s00439-002-0745-z. [DOI] [PubMed] [Google Scholar]
- 20.Ding YC, Weizman Z, Yerushalmi B, Elbedour K, Garner CP, Neuhausen SL: An autosomal genome-wide screen for celiac disease in Bedouin families. Genes Immun 2007. [DOI] [PubMed]
- 21.Zhong F, McCombs CC, Olson JM, Elston RC, Stevens FM, McCarthy CF, Michalski JP. An autosomal screen for genes that predispose to celiac disease in the western counties of Ireland. Nat Genet. 1996;14:329–333. doi: 10.1038/ng1196-329. [DOI] [PubMed] [Google Scholar]
- 22.Rioux JD, Karinen H, Kocher K, McMahon SG, Karkkainen P, Janatuinen E, Heikkinen M, Julkunen R, Pihlajamaki J, Naukkarinen A, Kosma VM, Daly MJ, Lander ES, Laakso M. Genomewide search and association studies in a Finnish celiac disease population: Identification of a novel locus and replication of the hla and ctla4 loci. Am J Med Genet A. 2004;130:345–350. doi: 10.1002/ajmg.a.30072. [DOI] [PubMed] [Google Scholar]
- 23.King AL, Yiannakou JY, Brett PM, Curtis D, Morris MA, Dearlove AM, Rhodes M, Rosen-Bronson S, Mathew C, Ellis HJ, Ciclitira PJ. A genome-wide family-based linkage study of coeliac disease. Ann Hum Genet. 2000;64:479–490. doi: 10.1046/j.1469-1809.2000.6460479.x. [DOI] [PubMed] [Google Scholar]
- 24.Popat S, Bevan S, Braegger CP, Busch A, O'Donoghue D, Falth-Magnusson K, Godkin A, Hogberg L, Holmes G, Hosie KB, Howdle PD, Jenkins H, Jewell D, Johnston S, Kennedy NP, Kumar P, Logan RF, Love AH, Marsh MN, Mulder CJ, Sjoberg K, Stenhammar L, Walker-Smith J, Houlston RS. Genome screening of coeliac disease. J Med Genet. 2002;39:328–331. doi: 10.1136/jmg.39.5.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong A, Cox NJ. Allele-sharing models: Lod scores and accurate linkage tests. Am J Hum Genet. 1997;61:1179–1188. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roeder K, Bacanu SA, Wasserman L, Devlin B. Using linkage genome scans to improve power of association in genome scans. Am J Hum Genet. 2006;78:243–252. doi: 10.1086/500026. [DOI] [PMC free article] [PubMed] [Google Scholar]