Abstract
A variety of mutations in the gene encoding DJ-1 protein are causal of autosomal recessive early-onset parkinsonism. Recently, a novel pathogenic homozygous DJ-1 missense mutation resulting in the L10P amino acid substitution was reported. In a separate study, a novel homozygous mutation resulting in the deletion of DJ-1 residue P158 was also reported to be causative of disease. The specific effects of the novel L10P and P158DEL mutations on protein function have not been studied. Herein, L10P and P158DEL DJ-1 proteins were assessed for protein stability, dimerization, solubility, subcellular localization, and protective function in comparison to WT and the L166P DJ-1 pathogenic variant. It was discovered that in comparison to WT protein, L10P, L166P, and P158DEL DJ-1 variants exhibited dramatically reduced protein stabilities. Degradation of each of the respective pathogenic mutants appeared to be mediated in-part by the proteasome. Interestingly, unlike L166P DJ-1, the L10P and P158DEL DJ-1 variants retained the ability to dimerize with WT DJ-1 protein; however, neither of these mutants was able to form homodimers. Additionally, the L10P, L166P, and P158DEL DJ-1 variants exhibited altered profiles on size-exclusion chromatography and demonstrated reduced solubilities in comparison to WT protein, and the latter aberration could be exacerbated in the presence of MG-132. Further, cells stably expressing L10P DJ-1 were more vulnerable to treatments with proteasome inhibitors, suggesting that L10P DJ-1 may be toxic to cells under conditions of proteasome stress. Taken together, these findings suggest that diverse aberrant mechanisms, including alterations in protein stability and protein folding, are associated with the pathogenicity of the L10P and P158DEL DJ-1 variants.
Keywords: dimerization, DJ-1, mutation, Parkinson disease, proteasome, solubility, stability
INTRODUCTION
Several genes, designated PARK1-PARK13, that can cause Parkinson disease (PD) have been indentified (Lesage and Brice 2009; Gasser. 2009). A variety of mutations in PARK7/DJ-1, including large genetic deletions, splice-site alteration and missense mutations, are also known to be causal of autosomal recessive early-onset parkinsonism (Kahle et al. 2009). Recently, two novel recessive mutations in the PARK7/DJ-1 gene have been reported which result in early-onset parkinsonian phenotypes in the affected patients (Guo et al. 2008; Macedo et al. 2009). A homozygous missense mutation resulting in the DJ-1 amino acid substitution L10P was identified in a consanguineous Chinese family (Guo et al. 2008). The affected patients presented with disease symptoms at 19 years of age which is the earliest age of onset reported for any PD case specifically linked to DJ-1 mutations. Following this report, another DJ-1 mutation carrier was identified in a genetic study conducted on early-onset PD patients from the Netherlands (Macedo et al. 2009). The affected individual harbored a small homozygous deletion in DJ-1 exon 7 which resulted in the deletion of the highly conserved DJ-1 residue, Pro 158 (P158DEL) (Macedo et al. 2009).
Although the first causative PARK7/DJ-1 mutations were reported nearly a decade ago (Bonifati et al. 2003) and DJ-1 has been implicated in many biological pathways (Kahle et al. 2009 and the references therein; Fitzgerald and Plun-Favreau 2008), the specific role of DJ-1 in disease pathogenesis is still unclear. DJ-1 is a relatively small 189 amino acids protein that is evolutionarily conserved across many species (Nagakubo et al. 1997; Bonifati et al. 2003; Bandyopadhyay and Cookson 2004). It is ubiquitously expressed in most tissues and is present in cell nuclei and cytoplasm (Nagakubo et al. 1997; Bonifati et al. 2003; Baulac et al. 2004). Crystallization studies reveal that DJ-1 is composed of eight α-helices and eleven β-strands that are arranged into a helix-strand-helix sandwich (Huai et al. 2003; Anderson and Daggett. 2008). These structural features are common to members of the ThiJ/Pfp superfamily of proteins (Wilson et al. 2003; Honbou et al. 2003; Huai et al. 2003). Further, it is known that DJ-1 tightly associates into a homodimer and that the dimer interface is composed of α-helices 1, 7, and 8 and β-strands 3 and 4 (Huai et al. 2003; Anderson and Daggett 2008; Wilson et al. 2003; Honbou et al. 2003). DJ-1 dimer formation may be required for the protein to function properly, though this hypothesis has never been proven. It is well known that the pathogenic DJ-1 mutant L166P fails to dimerize and that this deficit is likely caused by structural perturbations of the dimer interface (Wilson et al. 2003; Miller et al. 2003; Macedo et al. 2003; Olzmann et al. 2004; Blackinton et al. 2005; Gorner et al. 2007; Anderson and Daggett 2008). Such conformational abnormalities target L166P DJ-1 protein for rapid degradation by the proteasome, and the associated reductions in mutant protein abundance may be causative of disease phenotypes (Miller et al. 2003; Lockhart et al. 2004; Gorner et al. 2007). Studies to assess for the effects of pathogenic DJ-1 mutations on protein function will give insights into the cellular mechanisms that may be implicated in the etiology of PD.
In the current study, the biochemical properties of the novel pathogenic DJ-1 mutants, L10P and P158DEL are explored. The novel mutants are assessed for protein stability, dimerization, solubility, subcellular localization, and protective function in comparison to wild-type (WT) and L166P DJ-1. These analyses reveal that L10P, L166P and P158DEL DJ-1 mutants are dramatically destabilized in comparison to WT protein. Further, L10P and P158DEL mutants retain the ability to dimerize with WT protein but are unable to form homodimers. Additionally, both mutants demonstrate altered protein solubilities and aberrations in native protein size. Taken together, the studies herein suggest that L10P, L166P, and P158DEL DJ-1 mutations may be causative of disease by mechanisms involving altered protein stability and protein folding.
MATERIALS AND METHODS
Antibodies
DJ-5 is a mouse monoclonal antibody that is specific for human DJ-1 protein (Meulener et al. 2005). Affinity purified mouse anti-actin (clone C4) monoclonal antibody reacts with all forms of vertebrate actin (Millipore, Billerica, MA). 691 is a rabbit polyclonal antibody raised against recombinant human DJ-1 protein but that reacts with DJ-1 from various species (Meulener et al. 2005). The affinity purified polyclonal antibody, HA.11 (Covance, Emeryville, CA), reacts with proteins with the amino acid sequence, YPYCVPVYA. Vimentin (C-20) is an affinity purified polyclonal antibody that reacts with vimentin (Santa Cruz Biotechnology, Inc, Santa Cruz, CA). Anti-histone H3, CT, pan (Millipore, Temecula, CA) is a polyclonal antibody that recognizes histone H3 protein. Affinity purified glyceraldehyde-3-phosphate dehydrogeanase (GAPDH) (clone 6C5) monoclonal antibody reacts with GAPDH from various species (Advanced Immunochemical, Long Beach, CA).
Cloning of Human DJ-1 Constructs
Human full-length wild-type (WT) DJ-1 cDNA was cloned into the pZeoSV2 (Invitrogen, Carlsbad, CA) mammalian expression vector at the HindIII and Xho I restriction sites. Using the WT DJ-1 construct, the QuickChange® Site Directed Mutagenesis Kit (Stratagene, La Jolla, CA) was used in order to generate L10P, L166P, and P158DEL mutant forms of DJ-1 in the pZeoSV2 vector. The sequences of the oligonucleotides used for mutagenesis are listed in Table 1. The respective DJ-1 plasmid sequences were verified by DNA sequencing as a service offered by the DNA Sequencing Facility of the University of Pennsylvania.
Table 1.
| Human DJ-1 Variant |
Forward Oligonucleotide sequence |
Reverse Oligonucleotide Sequence |
|---|---|---|
| L10P | 5’-GCT CTG GTC ATC CCG GCT AAA GGA GCA GAG-3’ |
5’-CTC TGC TCC TTT AGC CGG GAT GAC CAG AGC- 3’ |
| L166P | 5’-TTC GAG TTT GCG CCT GCA ATT GTT GAA-3’ |
5’-TTC AAC AAT TGC AGG CGC AAA CTC GAA-3’ |
| P158DEL | 5’-CTT ACA AGC CGG GGG GGG ACC AGC TTC GAG-3’ |
5’-CTC GAA GCT GGT CCC CCC CCG GCT TGT AAG-3’ |
| HA-tagged WT | 5’-AAG CTT GCC ACC ATG TAC CCA TAC GAT GTT CCA GAT TAC GCT ATG GCT TCC AAA AGA GCT CTG GTC ATC- 3’ |
5’-CTC GAG CTA GTC TTT AAG AAC AAG-3’ |
A human N-terminal HA-tagged WT DJ-1 construct was generated by PCR, using the human full-length WT pZeoSV2 construct as a template. The sequences for the oligonucleotides are listed in Table 1. The tagged insert was cloned into the pCR 2.1 TOPO vector (Invitrogen) and subsequently cloned into the pZeoSV2 vector at the Hind III and Xho I restriction sites. Using the N-terminal HA-tagged WT DJ-1 construct, the QuickChange® Site Directed Mutagenesis Kit (Stratagene) was used in order to generate L10P, L166P, and P158DEL mutant N-terminal HA-tagged DJ-1 constructs. The sequences of the respective plasmids were verified by DNA sequencing as described above.
Cell Culture
Chinese Hamster Ovary (CHO) cells were cultured in Dulbecco-modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), 100 U/mL penicillin and 100 µg/mL streptomycin (Invitrogen). Cells were incubated at 37°C and 95% air/5% CO2 atmosphere.
Western Blot Analysis
Protein samples were resolved by SDS-PAGE followed by electrophoretic transfer onto nitrocellulose membranes. Membranes were blocked in Tris buffered saline (TBS) with 5% dry milk, and incubated overnight with primary antibodies diluted in TBS/ 5% dry milk. Each incubation was followed by goat anti-mouse conjugated horseradish peroxidase (HRP) (Amersham Biosciences, Piscataway, NJ) or goat anti-rabbit HRP (Santa Cruz Biotechnology, Inc, Santa Cruz, CA), and immunoreactivity was detected using chemiluminescent reagent (NEN, Boston, MA) followed by exposure onto X-ray film.
Pulse-chase Protein Turnover Analysis
CHO cells were cultured in 100 mm dishes. Cells were transfected with pZeoSV2 full-length WT, L10P, L166P, or P158DEL human DJ-1 constructs using Lipofectamine Reagent (Invitrogen), following the manufacturer’s protocol. At 24 hours post transfection, cells were split into 35 mm dishes and cultured in complete DMEM for an additional 24 hours. At 48 hours post transfection, cells were methionine-deprived for 15 minutes by incubation in pre-warmed methionine-free DMEM (Invitrogen)/10% dialyzed FBS (Hyclone) before adding 100 µCi [35S]-methionine (Perkin-Elmer, Waltham, MA) per ml of methionine free DMEM/10% dialyzed FBS for 30 min. Chase experiments were conducted in triplicates with normal DMEM/FBS with or without 20 µM MG-132 for 0, 1, 3, 6, and 10 hours. Cells were then rinsed with PBS and harvested in cytoskeleton (CSK) buffer (100 mM NaCl, 50 mM Tris, pH 7.5, 2 mM EDTA, 1 % Triton X-100) containing 1 % SDS and boiled at 100 °C for 5 minutes. CSK buffer was added to the lysates in order to bring the final concentration of SDS to 0.25%. Lysates were frozen on dry ice and kept frozen at −20 °C until the last time point was harvested. The radiolabelled protein extracts were pre-cleared with a rabbit serum pre-incubated with protein A-agarose (Santa Cruz Biotechnology, Inc.) for 3 hours at 4°C and radiolabelled extracts were then immunoprecipitated overnight at 4°C with antibody DJ5 pre-incubated with protein A/G PLUS-agarose (Santa Cruz Biotechnology, Inc). The antibody-protein complexes were washed 3 times with 10 volumes of CSK buffer, resuspended in 2 volumes of 2X SDS sample buffer and boiled at 100 °C for 5 minutes. The beads were removed by centrifugation and the samples were loaded onto 13 % polyacrylamide gels. Following electrophoresis, gels were fixed with 50% methanol/5% glycerol, dried and exposed to a PhosphorImager plate and the signal was quantified using ImageQuant software (Molecular Dynamics, Inc., Sunnyvale, CA).
Steady-State Protein Analysis
CHO cells were cultured in 35 mm dishes. Cells were transfected in triplicates with pZeoSV2 full-length WT, L10P, L166P, or P158DEL human DJ-1 constructs using Lipofectamine 2000 Transfection Reagent (Invitrogen), following the manufacturer’s protocol. At 48 hours post transfection, cells were rinsed with PBS and harvested in 2% SDS/50 mM Tris, pH 7.5 with protease inhibitors. Total cell lysates were boiled at 100 °C for 5 minutes, quantified using the bicinchoninic acid (BCA) protein assay (Thermo Scientific, Rockford, IL), and analyzed by western blot analysis with the monoclonal antibodies DJ5 and actin.
Size Exclusion Chromatography
Gel filtration chromatography was performed as previous described (Ramsey and Giasson. 2008). CHO cells transiently expressing human full-length WT, L10P, L166P, or P158DEL DJ-1 were cultured in 100 mm dishes, grown to confluency, and harvested. To harvest cells, the cells were rinsed and scraped in PBS. After recovery by centrifugation, cells were lysed in PBS/0.1% Triton and the cell debris was sedimented at 13,000 × g for 5 min. The extracts were filtered through a 0.22 µm filter and loaded onto the column. Fractions were analyzed by immunoblotting with anti-human DJ-1 specific antibody DJ5 or the polyclonal DJ-1 antibody, 691. Experiments were repeated at least 3 times and similar results were obtained.
Co-Immunoprecipitation and Heterodimer or Homodimer Analysis
CHO cells were cultured in 100 mm plates. For heterodimer analysis of WT DJ-1 with mutant DJ-1, cells were transfected with N-terminal HA-tagged full-length WT human DJ-1 construct (HA-WThDJ/pZeoSV2) or co-transfected with HA-WThDJ/pZeoSV2 and pZeoSV2 full-length untagged WT, L10P, L166P, or P158DEL human DJ-1 constructs. For homodimer analysis, cells were co-transfected with the pZeoSV2 plasmids expressing HA-tagged and untagged WT, L10P, L166P, or P158DEL human DJ-1. After 24 hours, cells were rinsed and scraped in PBS and lysed by vortexing into 500 uL of ice-cold 1X CSK buffer with protease inhibitors. Cell debris was sedimented at 13,000 × g for 1 minute and supernatants were removed to fresh tubes. 50 uL of each supernatant was saved as the “Start” fraction. The remainders of the supernatants were incubated for 3 hours at 4 °C with anti-HA tag antibody, HA.11 (Covance) preabsorbed to protein A-agarose beads (Santa Cruz Biotechonogy, Inc.). The beads were sedimented, repeatedly rinsed with ice-cold 1X CSK buffer, resuspended in 2X SDS sample buffer, heated to 100 °C and saved as the “IP” fractions. The supernatants remaining after immunoprecipitation were saved as the “Unbound” fractions. Sample buffer was added to the “Start” and “Unbound” fractions and then they were heated to 100 °C for 5 minutes. The Start, IP, and Unbound fractions were resolved by SDS-PAGE and immunoblotted with DJ5 antibody.
Crosslinking Analysis with Disuccinimidyl suberate (DSS)
CHO cells were cultured in 100 mm plates. Cells were transfected with pZeoSV2 full-length untagged WT, L10P, P158DEL, or L166P human DJ-1 constructs. At 48 hours post transfection, cells were lysed in PBS/0.1% Triton. Cell debris were removed at 13, 000 × g. Disuccinimidyl suberate (DSS, Thermo Scientific, Rockford, IL) was added at various concentrations (1–5 mM) and proteins were cross-linked for 2 hrs at 4 °C. The reactions were quenched by adding 50 mM Tris, pH 7.5 for 30 min. SDS-sample buffer was added and samples were heated to 100 °C for 5 min and analyzed by western blot with DJ-5 antibody.
Biochemical Fractionation and Cell Compartment Fractionation
CHO cells were cultured in 100 mm dishes and transfected with pZeoSV2 full-length WT, L10P, L166P, or P158DEL DJ-1 constructs using Lipofectamine transfection reagent. At 24 hours post transfection, the cells were cultured for an additional 18 hours in fresh DMEM with or without the addition of either 20 µM MG-132 or 1 µM epoxomicin. For biochemical fractionation, cells were scraped into PBS, sedimented, and lysed into buffers of increasing solubilization strengths using previously described methods (Ramsey and Giasson 2008). The respective biochemical fractions were analyzed by western blot with the monoclonal antibodies DJ5 and anti-actin.
For cell compartment fractionation, cells were scraped into ice-cold PBS and fractionated into cellular compartments using the Qproteome Cell Compartment Kit (Qiagen, Valencia, CA) following the manufacture’s protocol. Cell lysates were quantified using the BCA assay and analyzed by western blot with the monoclonal antibodies DJ5 and GAPDH and the polyclonal antibodies anti-histone H3 and vimentin (C-20).
Double-immunofluorescence Analyses
CHO cells were cultured in 35 mm dishes and transfected with pZeoSV2 full-length WT, L10P, L166P, or P158DEL DJ-1 constructs using Lipofectamine transfection reagent. At 48 hours post transfection, the cells were cultured for an additional 12 hours in fresh DMEM with or without the addition of 20 µM MG-132. Cells were rinsed in PBS and fixed by incubation in ice-cold acetic-methanol (1 part acetic acid to 20 parts methanol) at −20 °C for at least 30 minutes. Cells were rehydrated with water, rinsed with PBS, and blocked in PBS/1% FBS/1% skin fish gelatin (Sigma, Saint Louis, MO)/1% milk/0.1% Triton for 30 minutes. Primary antibodies were diluted into blocking solution and cells were labeled for 1–2 h at room temperature. Following PBS washed, coverslips were incubated with anti-mouse IgG antibody secondary conjugated to Alexa 488 and anti-rabbit IgG antibody conjugated to Alexa 594. Nuclei were counterstained with Hoechst trihydrochloride trihydrate 33342 (Invitrogen), and coverslips were mounted using Fluoromount-G (Southern Biotech, Birmingham, AL). Images were captured using an Olympus BX51 fluorescence microscope mounted with a DP71 digital camera (Olympus, Center Valley, PA). For quantification of cells with DJ-1 inclusions, images were captured with a 20x objective and all cells in the field were counted.
Generation of Stable Cells Lines Expressing WT and L10P DJ-1 and Cell Viability Studies
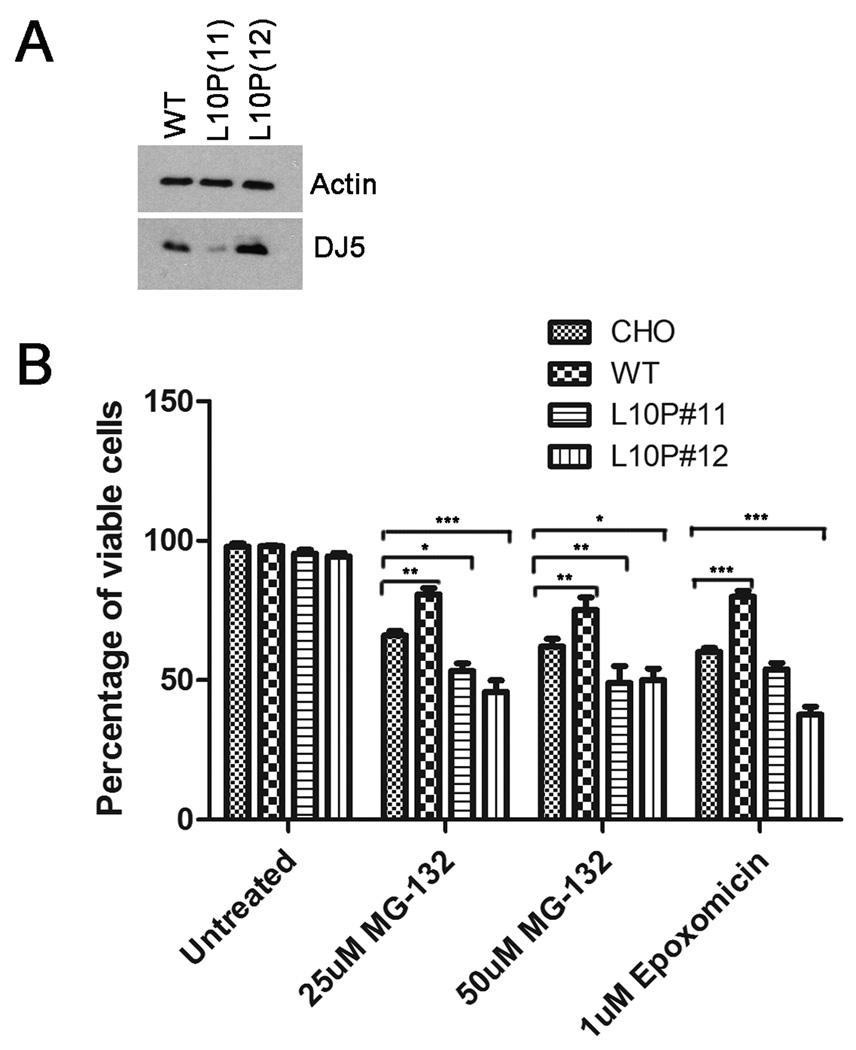
Human full-length WT or L10P pZeoSV2 DJ-1 constructs were used to transfect CHO cells using Lipofectamine reagent (Invitrogen) following the manufacterer’s protocol. Stably expressing clones were isolated and selected with Zeocin (Invitrogen) at 250 µg/mL and screened by western blotting analysis for the expression of DJ-1 using antibody DJ5. Stably expressing clones expressing WT or L10P human DJ-1 were used in the cell viability studies (See Figure 5A).
Figure 5. L10P DJ-1 is toxic during conditions of proteasome stress.
CHO cells stably expressing either WT or L10P DJ-1 were generated as described in “Materials and Methods”. A) Cells were harvested and total protein lysates were extracted and assessed by western blot analysis for DJ-1 with antibody DJ5. Blots were also probed with an actin antibody to assess equal protein loading. B) CHO cells and stable cell clones of CHO cells expressing WT or L10P (clone 11 and clone 12) human DJ-1 were cultured for 21 hours with DMEM (“untreated”) or DMEM containing the indicated concentrations of MG-132 or epoxomicin. Trypan blue exclusion assay was used to assess viability. Results were plotted as the percentage of viable cells over time of drug treatment. The error bars show standard deviation (n=6). Two-way ANOVA determined the results to be statistically significant, p<.0001. Bonferonni’s post hoc test were performed to determine the degrees of statistical significance between groups, ***p<.001, **p<.01, *p<.05.
To assess cell viability, native CHO or CHO cell lines expressing WT or L10P DJ-1 were cultured separately into 48 well plates. Each cell type was treated for 21 hours in sextuplicates with DMEM/FBS containing either 25 µM or 50 µM MG-132 or 1 µM epoxomicin. After treatment, the media from each well was collected into separate 1.5 mL microfuge tubes. The cells remaining in the wells were trypsinized and harvested in corresponding microfuge tubes. Cells were pelleted and resuspended in fresh DMEM/FBS, and then 3-fold volume of Trypan blue solution (Sigma) was added. Live and dead cells were counted manually using a hemacytometer and an Olympus CKX41 microscope. The percentage of live cells relative to the total number in each well was calculated.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 5.0 (GraphPad Software Inc, La Jolla, CA).
RESULTS
Pathogenic DJ-1 mutants demonstrate reduced protein stabilities and are partially regulated by the proteasome
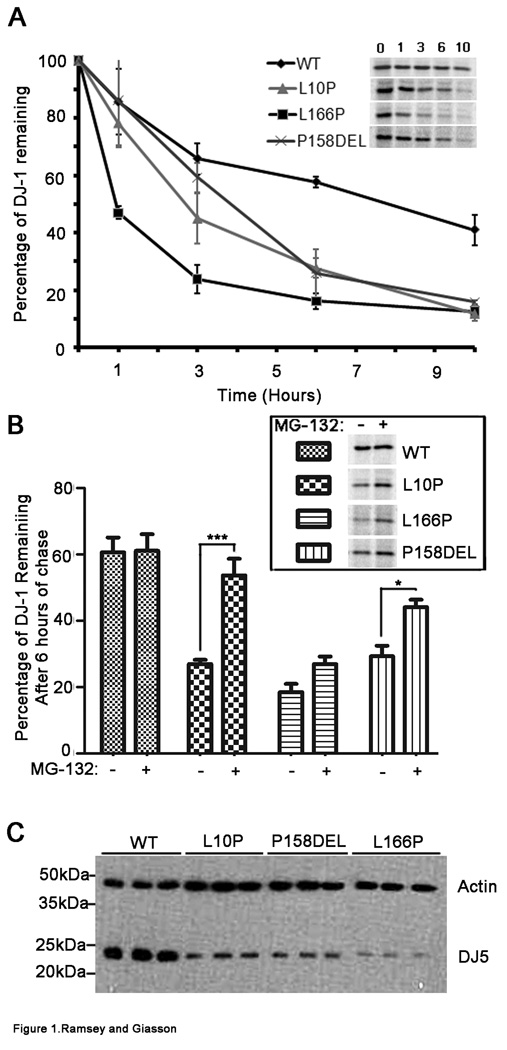
It has been previously shown that L166P mutant DJ-1 demonstrates dramatically reduced protein stability in comparison to WT protein (Miller et al. 2003; Moore et al. 2003; Macedo et al. 2003; Gorner et al. 2004; Olzmann et al. 2004; Blackinton et al. 2005; Gorner et al. 2007) and that this aberration is due in part to rapid degradation of mutant protein by the proteasome (Miller et al. 2003; Moore et al. 2003; Gorner et al. 2004; Olzmann et al. 2004; Takahashi-Niki et al. 2004; Gorner et al. 2007). To determine the effects of the recently reported L10P and P158DEL DJ-1 mutations on protein stability, protein turn-over pulse-chase experiments were performed with 35 S-methionine. The turn-over rates of WT, L10P, L166P, and P158DEL DJ-1 protein were compared. Following these analyses it was determined that each of the DJ-1 mutants analyzed showed significantly reduced stabilities in comparison to WT protein, albeit to different degrees (Figure 1A). Bonferroni’s post hoc analyses indicated that the differences between the turnover of WT and L166P DJ-1 were statistically significant at 1, 3, 6, and 10 hours of chase (p<.001, p<.001, p<.001, p<.01 respectively). The differences between the turnover of WT and L10P DJ-1 were statistically significant at 3, 6, and 10 hours of chase (p<.05, p<.01, p<.01 respectively), indicating that L10P DJ-1 is more stable than L166P DJ-1. The differences between the turnover of WT and P158DEL DJ-1 were significant at 6 and 10 hours of chase (p<.001 and p<.01 respectively), indicating that although P158DEL DJ-1 is less stable than WT protein, the P158DEL DJ-1 variant is more stable than the other DJ-1 mutants analyzed (Moore et al. 2003; Olzmann et al. 2004). To determine the effects of the proteasome complex on the turn-over rates of WT, L10P, L166P, and P158DEL DJ-1 protein variants, protein pulse-chase experiments were performed in the presence of 20 µM MG-132. The percentage of DJ-1 protein remaining with the addition of MG-132 after 6 hours of chase was calculated and compared to that without the addition of MG-132 for each DJ-1 variant. The 6 hour chase time point was chosen for analyses because as shown in Figure 1A, in comparison to WT protein, all of the DJ-1 mutant proteins analyzed showed significant degradation at 6 hours of chase. It was determined that the turn-over rate for WT DJ-1 was not significantly affected by proteasome inhibition (Figure 1B). Conversely, treatment of cells with MG-132 had different effects on the stabilities of all of the DJ-1 mutant proteins analyzed. The effect was most striking for the L10P DJ-1 mutant. Following treatment with MG-132, L10P DJ-1 showed a ∼2-fold increase in stability whereas both P158DEL and L166P DJ-1 showed ∼1.5 fold increases in stability. Interestingly, proteasome inhibition slowed the decay of L10P, L166P, P158DEL DJ-1 mutant proteins, but it did not completely block the degradation of these mutants. This observation has been previously reported for the L166P DJ-1 mutant (Miller et al. 2003; Olzmann et al. 2004; Gorner et al. 2007), suggesting that in addition to proteasome-mediated degradation, turn-over of L10P, L166P, and P158DEL DJ-1 mutant proteins can also be regulated by proteasome-independent mechanisms.
Figure 1. Pathogenic DJ-1 mutants demonstrate dramatically reduced protein stabilities that are partially due to proteasome degradation.
A) CHO cells were transfected with full-length WT, L10P, L166P, or P158DEL human DJ-1 pZeoSV2 “(+)” constructs. At 48 hours post transfection, cells were labeled with 35S-methionine for 30 mins and chased for 0, 1, 3, 6, or 10 hours. The graph represents the protein turnover results for the respective DJ-1 variants. The results are plotted as percentage of protein remaining over time standardized to the 0 hrs time point. The error bars show standard deviation (n=6). The inset shows representative pulse-chase experiments. The results were analyzed by two-way ANOVA and were determined to be statistically significant, p=.0015. B) At 48 hours post transfection, cells were labeled with 35S-methionine for 30 mins and chased for 6 hours in the presence or absence of 20 µM MG-132. The graph depicts the average percentage of DJ-1 remaining after 6 hours of chase in the absence and presence of MG-132. The error bars indicate standard deviation (n=5). Two-way ANOVA was performed and the results were stastically significant, p=.0061. The data was further analyzed by Bonferroni’s post hoc test. ***p<.001, *p<.05. The inset image represents the DJ-1 signal remaining after 6 hours of chase for each of the DJ-1 variants in the absence and presence of MG-132. C) At 48 hours post transfection, total cell lysates were extracted. Equal amounts of each sample were loaded onto polyacrylamide gels and analyzed by western blot for DJ-1 with the DJ5 antibody. Blots were also probed simultaneously with an actin antibody to assess equal protein loading. The image shows the results from experiments performed in triplicate. The mobility of molecular mass markers is indicated on the left.
Since it has been shown that enhanced turnover of L166P DJ-1 acts to diminish the levels of L166P DJ-1 protein when assessed at steady-state in comparison to WT DJ-1 protein (Miller et al. 2003; Moore et al. 2003; Macedo et al. 2003; Gorner et al. 2004; Olzmann et al. 2004; Blackinton et al. 2005; Gorner et al. 2007), it was of interest to determine the steady-state levels of WT, L10P, L166P, and P158DEL DJ-1 protein variants. The steady-state levels of L166P DJ-1 were greatly reduced in comparison to WT DJ-1 protein (Figure 1C). Similarly, L10P and P158DEL DJ-1 protein levels were also less abundant than WT DJ-1. This suggests that increased turnover of mutant L10P, L166P, and P158DEL DJ-1 variants results in attenuated levels of the respective exogenously added proteins in comparison to WT DJ-1 protein.
The L10P and P158DEL DJ-1 mutations impair dimerization and alter native DJ-1 protein folding
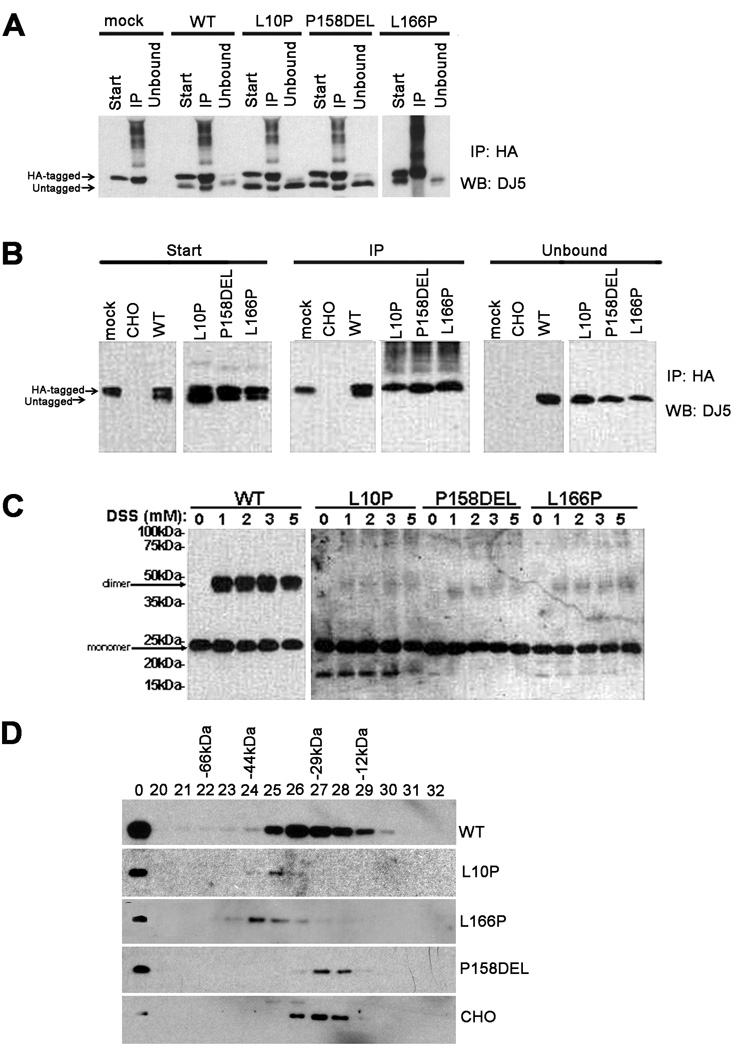
Previous studies reveal that in comparison to WT DJ-1 protein, L166P mutant DJ-1 displays aberrant folding (Anderson and Daggett 2008;Wilson et al. 2003; Olzmann et al. 2004; Gorner et al. 2007) that impairs the dimerization of this mutant and that is associated with a dramatic instability of this mutant protein (Miller et al. 2003; Macedo et al. 2003; Blackinton et al. 2005; Gorner et al. 2007; Anderson and Daggett 2008; Moore et al. 2003; Olzmann et al. 2004). Thus, it is possible that L10P and P158DEL DJ-1 mutants are destabilized due to disrupted dimerization and/or improper protein folding. To assess the ability of L10P and P158DEL DJ-1 variants to form dimers, co-transfection and co-immunoprecipitation experiments were conducted. Experiments with WT and L166P DJ-1 were also conducted for comparison. CHO cells were transiently co-transfected with human HA-tagged full-length WT DJ-1 and human untagged full-length WT, L10P, L166P, or P158DEL DJ-1. Cells extracts were immunoprecipitated with an anti-HA antibody followed by immunoblot with antibody DJ5. These analyses revealed that the untagged WT, L10P, and P158DEL DJ-1 variants were all co-immunoprecipitated with HA-tagged WT DJ-1 protein (Figure 2A). However, L166P DJ-1 was not co-immunoprecipitated with HA-tagged WT DJ-1. This indicates that L10P and P158DEL DJ-1 variants retain the ability to dimerize with WT DJ-1 protein.
Figure 2. The L10P and P158DEL DJ-1 mutations impair dimer formation and alter native protein folding patterns.
A) CHO cells were co-transfected with HA-tagged full-length WT DJ-1 and pZeoSV2 (+) mock vector (“mock”) or with untagged full-length WT, L10P, P158DEL, or L166P human DJ-1 pZeoSV2 (+) constructs. At 24 hours post transfection, cells were harvested and soluble protein lysates were extracted (“Start”). The cell lysates were then immunoprecipitated using anti-HA polyclonal antibody (“IP”). Equal amounts of the start, IP, and unbound supernatant (Unbound) fractions were resolved by SDS-PAGE and analyzed by western blot (“WB”) with the monoclonal DJ-1 antibody, DJ5. DJ5 antibody does not cross-react with hamster DJ-1 and thus endogenous DJ-1 is not visible in the immunoprecipitates. B) HA-tagged and untagged WT, L10P, P158DEL, or L166P human DJ-1 pZeoSV2 constructs were co-transfected in CHO cells to assess for homodimerization. Cells were harvested, extracted, and analyzed as described above. “CHO” represents the results from untransfected CHO cells. “Mock” represents the results from CHO cells co-transfected with HA-tagged full-length WT DJ-1 and pZeoSV2 (+) mock vector. Because L10P, L166P, and P158DEL are expressed at much lower levels than WT DJ-1, for the western blot analysis of these DJ-1 mutants, the immunoblot were exposed to film for 10-times longer than that for WT DJ-1. C) CHO cells were transfected with untagged full-length WT, L10P, P158DEL, or L166P human DJ-1 constructs. At 48 hours post transfection, cells were lysed in PBS/0.1% Triton. Cell debris were removed at 13, 000 × g. DSS was added at the concentrations indicated and proteins were cross-linked for 2 hrs at 4 °C. The reactions were quenched by adding 50 mM Tris, pH 7.5 for 30 min. SDS-sample buffer was added and samples were heated to 100 °C for 5 min and analyzed by Western blot with DJ-5. Because L10P, L166P, and P158DEL are expressed at much lower levels than WT DJ-1, for the western blot analysis of these DJ-1 mutants, 3-times more protein extract was loaded on the SDS-polyacrylamide gel and the immunoblot were exposed to film for 10-times longer than that for WT DJ-1. D) CHO cells were transiently transfected with full-length WT, L10P, L166P, or P158DEL human DJ-1 pZeoSV2 (+) constructs. Soluble extracts from native CHO cells or CHO cells expressing the respective exogenous human DJ-1 proteins were loaded on a precalibrated Superose 6 column as described in the “Materials and Methods” section. The total cell lysates ("total") and fractions collected from the size-exclusion column were analyzed by western blot analysis with DJ5 antibody to detect human DJ-1 or antibody 691 to detect endogenous DJ-1. Fractions 20–32 are shown. The elution profile of known molecular mass standards [BSA (66 kDa), ovalbumin (44 kDa), carbonic anhydrase (29kDa) and cytochrome C (12kDa)] are indicated above.
Since L10P and P158DEL DJ-1 mutations cause PD with an autosomal-recessive pattern of inheritance (Guo et al. 2008; Macedo et al. 2009), it was also of interest to determine the effects of these mutations on DJ-1 homodimer formation. To address this question, co-transfection and co-immunoprecipitation experiments were conducted to compare the self-association of human HA-tagged full-length WT, L10P, L166P, or P158DEL DJ-1 with the respective untagged human full-length WT, L10P, L166P, or P158DEL DJ-1 proteins. While untagged WT DJ-1 was co-immunoprecipitated with HA-tagged WT DJ-1 protein, none of the untagged DJ-1 mutants analyzed were co-immunoprecipitated with their respective HA-tagged forms (Figure 2B). This reveals that similar to L166P DJ-1, L10P and P158DEL DJ-1 mutations disrupt homodimer formation.
To confirm these findings, homodimer formation of WT, L10P, L166P, and P158DEL DJ-1 were assessed by in vitro chemical crosslinking experiments using the DSS crosslinking agent. Consistent with the co-immunoprecipitation results, crosslinking analyses revealed that WT DJ-1 formed homodimers while L10P, L166P, or P158DEL DJ-1 was deficient in the ability to form homodimers under the same experimental conditions (Figure 2C).
To further analyze the consequences of DJ-1 mutations on protein structure, size exclusion chromatography experiments were conducted. Soluble lysates were fractionated from CHO cells expressing WT, L10P, P158DEL, or L166P human DJ-1. The assay was standardized by elution of purified proteins of known molecular masses. Fractions were resolved by SDS-PAGE and analyzed by immunoblot with the anti-human specific DJ-1 antibody DJ5. The peak elution fraction for WT human DJ-1 was fraction 26 (Figure 2D), consistent with dimer formation. It has been previously reported that L166P DJ-1 forms high molecular weight complexes which increase its apparent molecular weight when analyzed by gel filtration analyses (Macedo et al. 2003; Olzmann et al. 2004). In the studies herein, L166P DJ-1 eluted in peak fraction 24, suggesting that it also forms high-molecular weight complexes in CHO cells. Interestingly, L166P DJ-1 that would elute in fractions that would be consistent with monomeric forms of DJ-1 was not detected in these analyses. Surprisingly, the peak elution for L10P DJ-1 was fraction 25, indicating that the L10P mutant adopts an apparent larger conformation than WT DJ-1 protein, even though it cannot form a homodimer. Conversely, P158DEL DJ-1 primarily eluted into fractions 27 and 28, suggesting that it is natively smaller in size than WT DJ-1 protein, which may be consistent with elongated monomeric forms. Endogenous hamster DJ-1 protein, which is known to be smaller in size in comparison to WT human DJ-1 (Siva et al. 2004), was primarily eluted into fraction 27.
L10P, L166P and P158DEL DJ-1 mutants show reduced protein solubilities
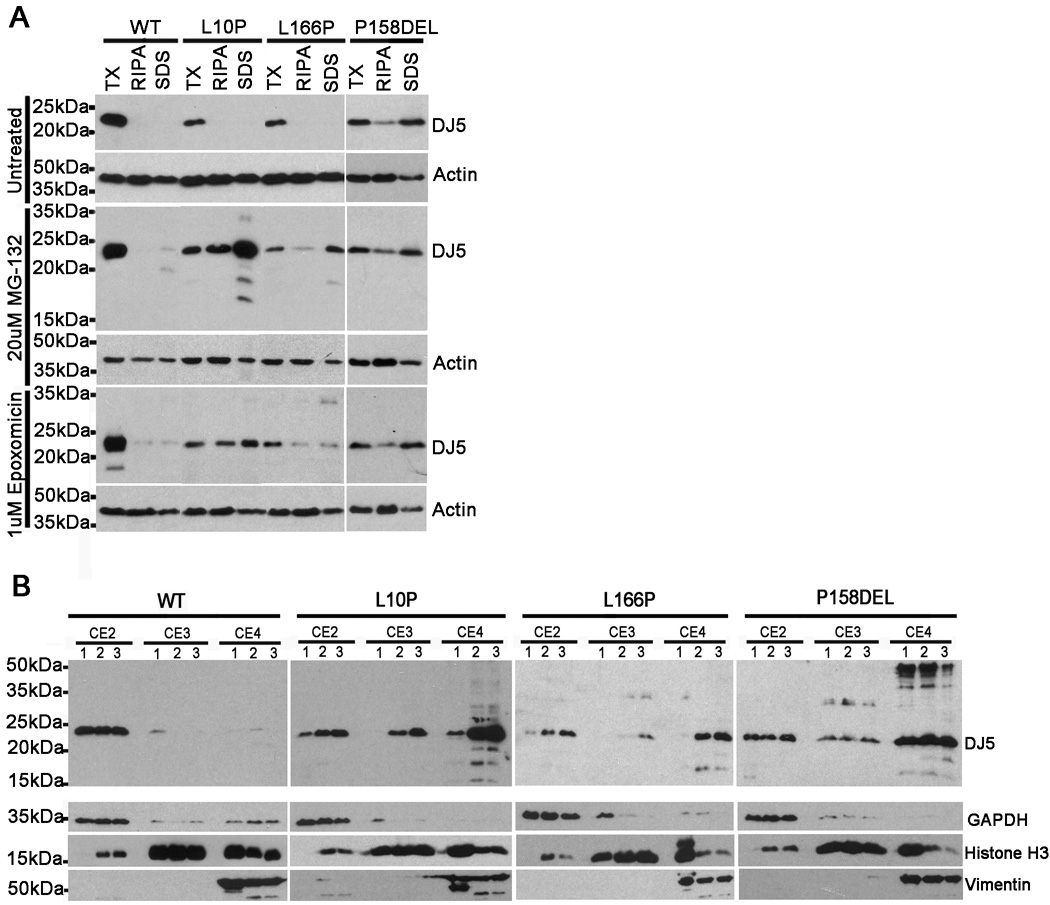
Protein misfolding and aggregation is a prominent feature of many neurodegenerative disorders (Soto and Estrada 2008) and reduced solubility of DJ-1 is correlated with human disease (Rizzu et al. 2004; Neumann et al. 2004; Meulener et al. 2005; Moore et al. 2005; Kumaran et al. 2007). L166P DJ-1 is known to be misfolded (Olzmann et al. 2004; Gorner et al. 2007; Anderson and Daggett 2008) and this may result in reduced solubility of the mutant in comparison to WT DJ-1 (Shinbo et al. 2006). Gel filtration analyses revealed that the L10P and P158DEL DJ-1 variants may adopt abnormal folding patterns (Figure 2D); however, it is unknown whether protein solubility is affected by these mutations. To assess for changes in solubility, CHO cells expressing full-length WT, L10P, L166P, or P158DEL human DJ-1 were harvested and cell lysates were extracted into buffers of increasing solubilization strengths. The lysates were then analyzed by western blot with antibody DJ5. WT, L10P, L166P, and P158DEL DJ-1 variants were all predominantly extracted in the soluble PBS/0.1% Triton (TX) (Figure 3A). However, a significant fraction of P158DEL DJ-1 protein was extracted in the RIPA and SDS fractions.
Figure 3. L10P, L166P and P158DEL DJ-1 mutants show reduced protein solubilities.
CHO cells were transfected with full-length WT, L10P, L166P, or P158DEL human DJ-1 pZeoSV2 (+) constructs. At 24 hours post transfection, cells were cultured for an additional 18 hours in fresh DMEM with or without the addition of either 20 µM MG-132 or 1 µM epoxomicin. A) Cells were harvested and sequentially extracted into PBS/0.1% Triton (TX) buffer, RIPA buffer, and 2% SDS/17mM Tris (SDS) buffer. Equal amounts (10ug) of each sample were loaded onto SDS-polyacrylamide gels and analyzed by western blot for DJ-1 distribution with the DJ5 antibody. Blots were also probed with an actin antibody to assess equal protein loading. The mobility of molecular mass markers is indicated on the left. B) Cells were harvested and extracted using the Qproteome Cell Compartment Kit (Qiagen), following the manufacturer’s instructions. Equal amounts (10ug) of each sample were loaded onto SDS-polyacrylamide gels and analyzed by western blot with antibody DJ5 to assess the distribution of human DJ-1, GAPDH monoclonal antibody as a cytoplasmic marker, histone H3 polyclonal antibody as a nuclear marker, and vimentin polyclonal antibody as a cytoskeletal marker. “CE2” represents the cytoplasmic fractions, “CE3” represents the nuclear fractions and “CE4” represents the cytoskeletal/insoluble fractions. “1” lanes were loaded with extracts from cells treated with fresh DMEM only. “2” lanes were loaded with extracts from cells treated with fresh DMEM + epoxomicin. “3” lanes were loaded with extracts from cells treated with fresh DMEM +MG-132.
Proteasome inhibitors have been shown to stabilize insoluble forms of L166P DJ-1 protein when assessed in cell culture studies (Moore et al. 2003; Olzmann et al. 2007); however, it is not known whether L10P and P158DEL DJ-1 protein mutants display similar biochemical properties. To analyze the effects of proteasome inhibitors on DJ-1 solubility, CHO cells expressing WT, L10P, L166P, or P158DEL human DJ-1 were treated for 18 hours with 20µM MG-132 or 1µM epoxomicin followed by harvesting and extraction into buffers of increasing solubilization strengths. The extracts were then assessed by immunoblot with antibody DJ5. Treatments with either MG-132 or epoxomicin resulted in only trace amounts of WT DJ-1 detected in the RIPA or SDS-soluble fractions (Figure 3A). Both MG-132 and epoxomicin dramatically promoted the accumulation of RIPA and SDS-soluble forms of L10P DJ-1, though the effects were more provocative with MG-132. Similarly, MG-132 and epoxomicin resulted in the buildup of Triton-insoluble pools of L166P DJ-1. Interestingly, however, neither MG-132 nor epoxomicin treatments affected the amount of Triton-insoluble P158DEL DJ-1 protein.
To expand on these biochemical fractionation results and to assess for possible changes in cellular localization, protein fractionation experiments were performed using the Qproteome Cell Compartment Kit. CHO cells expressing WT, L10P, L166P, or P158DEL human DJ-1 were compared when untreated or challenged with 20 µM MG-132 or 1 µM epoxomicin for 18 hours. Cytoplasmic (CE2), nuclear (CE3), and cytoskeletal/insoluble (CE4) protein fractions were generated. Cell extracts were analyzed by western blot with the antibodies to human DJ-1 (DJ-5), GAPDH, histone-H3, and vimentin. These analyses revealed that under normal conditions, WT DJ-1 protein primarily localized to cytoplasmic biochemical fractions and to a much lesser extent into nuclear fractions (Figure 3B). WT DJ-1 protein was absent from cytoskeletal/insoluble protein fractions. As expected, MG-132 and epoxomicin treatments had little effect on WT DJ-1 protein. Under normal conditions, L10P DJ-1 was predominantly detected in both cytoplasmic and cytoskeletal/insoluble protein fractions. Following treatments with MG-132 or epoxomicin, L10P DJ-1 accumulated in the nucleus and cytoskeletal/insoluble fractions, suggesting that inhibition of the proteasome may act to stabilize insoluble forms of the L10P DJ-1. Under normal conditions, L166P DJ-1 was predominantly observed in the cytoplasmic fraction; however, following treatments with MG-132 or epoxomicin, L166P DJ-1 dramatically accumulated in cytoplasmic and cytoskeletal/insoluble protein fractions, and to a much lesser extent in nuclear fractions. In the absence of proteasome inhibitors, the P158DEL DJ-1 variant was extracted in all three fractions and treatments with either MG-132 or epoxomicin had no significant effect on the biochemical extractability of P158DEL DJ −1 protein as detected by this method (Figure 3B). This finding further implies that this mutant exhibits reduced solubility even in the absence of induced proteasome challenge.
L10P, L166P, and P158DEL DJ-1 form intracellular inclusions following proteasome inhibition
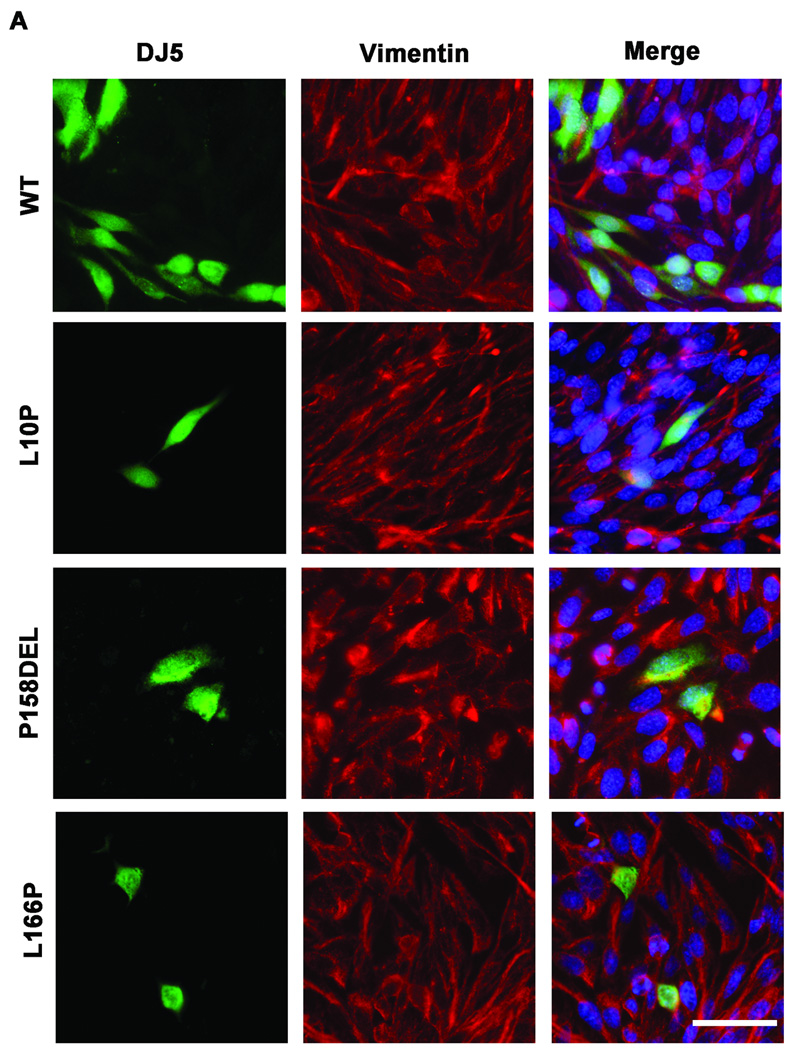
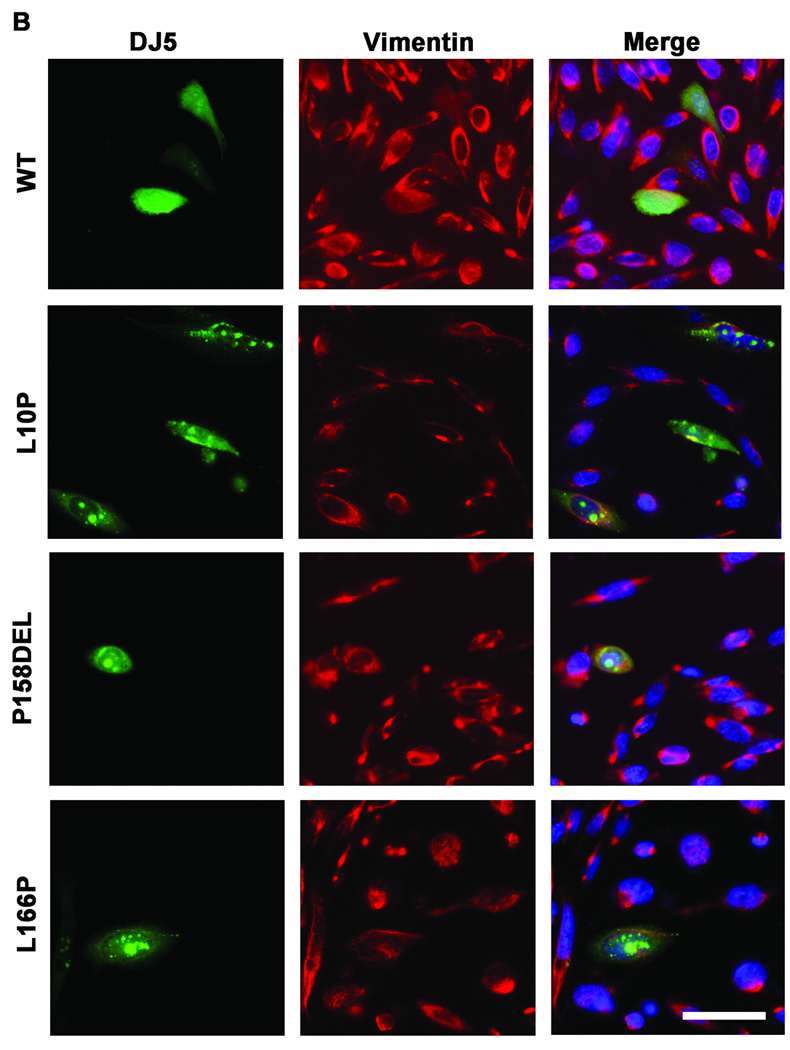
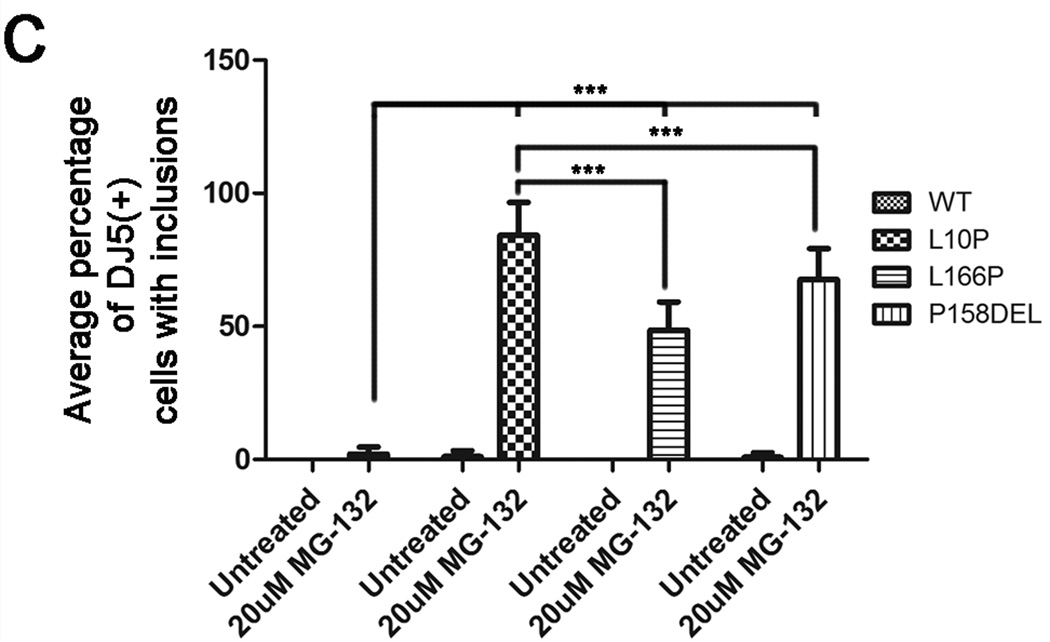
Olzmann et al showed that MG-132 induced the recruitment of L166P DJ-1 into aggresomes (Olzmann et al. 2007). To determine whether a similar effect would be observed for the L10P and P158DEL DJ-1 mutants, co-immunofluorescence studies were conducted comparing untreated and treated CHO cells expressing WT, L10P, L166P, or P158DEL human DJ-1 challenged for 12 hours with 20 µM MG-132. Cells were co-labeled with DJ5 and vimentin antibodies to assess for DJ-1 staining and inclusion formation, respectively. Under normal culturing conditions, WT and mutant DJ-1 proteins exhibited diffuse staining (Figure 4A). However, nuclear and cytoplasmic DJ-1 positive inclusions were detected in a small percentage of L10P and P158DEL DJ-1 expressing cells (Figure 4A and C and data not shown). MG-132 treatment induced the formation of intranuclear and cytoplasmic DJ-1 positive protein inclusions in cells expressing L10P, L166P, and P158DEL DJ-1 but had little effect on cells expressing WT DJ-1 (Figure 4B and C). Interestingly, co-immunofluorescence analyses with DJ5 and vimentin antibodies did not reveal any accumulation or redistribution of vimentin surrounding L10P or P158DEL DJ-1 protein inclusions, although vimentin was co-localized with these inclusions on rare occasions, suggesting that the observed DJ-1 positive inclusions are not classical aggresomes. MG-132 caused inclusion formation most provocatively in cells expressing L10P DJ-1 since ∼84±13% of these cells had inclusions. P158DEL DJ-1 formed inclusions in 68±11% of cells, while L166P DJ-1 formed inclusions in 49±10% of cells.
Figure 4. L10P, L166P, and P158DEL DJ-1 form intracellular inclusions following proteasome inhibition.
CHO cells were transfected with full-length WT, L10P, L166P, or P158DEL human DJ-1 pZeoSV2 (+) constructs. At 48 hours post transfection, cells were cultured for an additional 12 hours in fresh DMEM with or without the addition of 20µM MG-132. Cells were stained with the mouse monoclonal anti-DJ-1 antibody DJ5 (green), a rabbit polyclonal anti-vimentin (red) and Hoechst 33342 (blue) and visualized by microscopy. Representative images are shown for cells that were not treated with MG-132 in A) and cells that were treated with MG-132 in B). Bar = 50µm (C) The proportion of cells depicting DJ-1 inclusions was quantified as the percentage of the total cells expressing full-length WT, L10P, L166P, or P158DEL human DJ-1 with or without MG-132 challenge. Three fields from three independent experiments with at least 20 cells per field were counted. The graph represents the percentage of cells with inclusions averaged between replicate images. The error bars show standard deviation. Two-way ANOVA determined the results to be statistically significant, p<.0001. Bonferroni’s post hoc tests were also performed to determine the level of statistical significance between groups, ***p<.001.
L10P DJ-1 may be toxic to cells under proteasome stress
DJ-1 protects against proteasome impairments (Yokota et al. 2003; Ramsey and Giasson 2008); however, over-expression of pathogenic L166P mutant DJ-1 is toxic under these conditions (Yokota et al. 2003). Protein misfolding and aggregation is associated with many neurodegenerative disorders and may even be causal of neurodegeneration (Gregersen et al. 2006; Soto and Estrada 2008). Since both L10P and P158DEL DJ-1 mutant proteins exhibited reduced solubility and formed inclusions following proteasome inhibition (Figure 3 and 4), it is plausible to hypothesize that L10P and P158DEL DJ-1 proteins may have impaired protective function and may even be toxic when expressed under conditions of proteasome stress. To test this hypothesis, we tried to generate stable cell lines that express these DJ-1 mutations using methods described in “Material and Methods” that have been used successfully for other DJ-1 mutants (Ramsey and Giasson 2008). Two CHO stable cell lines expressing different levels of L10P mutant human DJ-1 (clone #11 and clone #12) were established (Figure 5A). A cell line expressing human WT DJ-1 with levels equivalent to the highest L10P DJ-1 cell line (clone #12) was also generated. No CHO cell line stably expressing detectable amount of P158DEL DJ-1 could be established.
Native CHO cells or CHO cells stably expressing human WT or L10P mutant DJ-1 were challenged for 21 hours with 25 µM MG-132, 50 µM MG-132, or 1 µM epoxomicin. Following these treatments, cells were assessed for cell viability using the Trypan blue exclusion assay. In comparison to the effects of 25 µM MG-132, 50 µM MG-132, or 1 µM epoxomicin on native CHO cells, WT DJ-1 was significantly protective against all treatments with the inhibitors (Figure 5B). Conversely, neither of the L10P DJ-1 clones tested demonstrated protection against any of the proteasome inhibitor treatment. Further, both of the tested L10P DJ-1 clones demonstrated significantly reduced viabilities in comparison to native CHO cells following 25 µM and 50 µM MG-132 treatments. Interestingly, L10P DJ-1 clone #12, which expressed higher levels of mutant DJ-1 protein than clone #11, also exhibited significantly reduced viability in comparison to native CHO cells following epoxomicin exposure, suggesting that the level of L10P mutant DJ-1 protein expression may affect cell toxicity during proteasome stress.
DISCUSSSION
A short nucleotide deletion resulting in P158DEL DJ-1 and a missense mutation resulting in the L10P DJ-1 amino acid substitution are novel recently reported PARK7/DJ-1 mutations causative of autosomal recessive early-onset parkinsonism (Guo et al. 2008; Macedo et al. 2009). In the current study, the effects of these mutations on DJ-1 protein stability, dimerization, folding, solubility, localization, and cell viability were assessed in cultured cells. Additionally, the previously extensively characterized pathogenic L166P DJ-1 mutant was also analyzed in parallel. In comparison to WT DJ-1 protein, the L10P, L166P, and P158DEL DJ-1 mutants all exhibited reduced protein stabilities when assessed by pulse-chase protein turn-over analyses; however, the extent of the effects of the respective mutations on protein stability was variable. The L10P and P158DEL mutants were significantly less unstable than L166P. To begin to explain these findings, and because it was previously reported that the instability of L166P DJ-1 was at least partially mediated by the proteasome (Miller et al. 2003; Moore et al. 2003; Gorner et al. 2004; Gorner et al. 2007), pulse-chase analyses were conducted in the presence of the reversible proteasome inhibitor, MG-132. As previously shown by others (Gorner et al. 2004; Gorner et al. 2007), MG-132 only partially stabilized L166P DJ-1 protein in the studies herein. Similarly, MG-132 failed to completely block degradation of L10P and P158DEL DJ-1 mutants, though turn-over was dramatically slowed for both variants. Noteworthily, in comparison to the other mutants, MG-132 most effectively slowed the degradation of L10P DJ-1, suggesting that this mutation renders DJ-1 to be more susceptible to degradation by the proteasome. Based on the findings herein, is it also likely that proteasome-independent mechanisms can act to degrade L10P, L166P, and P158DEL DJ-1 mutant proteins. It is worth mentioning that autophagy inhibitors did not impede the degradation of any of the DJ-1 protein variants analyzed in these studies (data not shown). This observation has also been reported by other groups (Olzmann et al. 2004; Blackinton et al. 2005). Though a recent paper by Giaime and colleagues supports a role for caspase-6 mediated cleavage of DJ-1, it is not clear what additional proteases may act to regulate DJ-1 protein (Giaime et al. 2010). The current findings suggest that reduced DJ-1 protein stability resulting from the L10P and P158DEL mutations may at least be partially responsible for DJ-1 loss-of-function.
It has been previously shown that the L166P mutation causes DJ-1 to unfold and it is believed that this disconformity effectively disrupts dimer formation, which may be associated with rapid degradation (Miller et al. 2003; Moore et al. 2003; Olzmann et al. 2004; Gorner et al. 2007; Anderson and Daggett 2008). The L166P mutation is located in α-helix 7 that is part of the structure involved in dimer interface (Wilson et al. 2003; Honbou et al. 2003; Huai et al. 2003;Bandyopadhyay and Cookson 2004). Interestingly, it was determined that unlike L166P DJ-1, L10P and P158DEL DJ-1 mutants were able to dimerize with WT DJ-1 protein, but they were unable to self-associate into homodimers. These findings indicate that both the L10P and P158DEL DJ-1 mutations alter protein structure in a manner that prevents self-interaction, but both protein variants maintain sufficient protein folding to interact with WT DJ-1. The L10P mutation is located in the first β-strand of DJ-1, which is not directly involved in the dimer interface, but this type of mutation should result in significant disrupting of this β-strand (Wilson et al. 2003; Honbou et al. 2003; Huai et al. 2003). Residue P158 is also outside of the DJ-1 dimer interface; however it located right before the beginning of DJ-1 α-helix 7 which is required for dimer formation (Wilson et al. 2003; Honbou et al. 2003; Huai et al. 2003). Interestingly, P158 is a highly conserved residue in the DJ-1 protein family (Bandyopadhyay and Cookson 2004).
It is still unknown whether the ability for DJ-1 to form dimers correlates directly to protein stability. It is possible that either improper protein folding and/or dimerization impairments may be causal of the decreased stability observed for pathogenic L10P and P158DEL DJ-1 mutants. Data from size exclusion chromatography analyses support that both L10P and P158DEL DJ-1 variants adopt altered structural conformations in comparison to WT DJ-1 protein. Surprisingly, the L10P DJ-1 mutant exhibited an apparent elution profile that is consistent with a slightly larger conformation than WT protein, despite the finding that it cannot homodimize. This type of finding has previously been reported for L166P DJ-1, and has been explained by L166P DJ-1 forming higher-order protein complexes (Macedo et al. 2003; Olzmann et al. 2004). Consistent with these studies notion, we showed that although L166P DJ-1 cannot dimerize, it eluted on gel filtration as a species with an apparent molecular mass greater than WT DJ-1. In addition, monomeric forms of L166P or L10P DJ-1 of the expected ∼21 kDa size were not detected by size exclusion chromatography suggesting that monomeric L166P and L10P DJ-1 may be very unstable. In comparison, the elution of WT DJ-1 was primarily consistent with dimer formation, although the broad elution profile would also suggest equilibrium with a monomeric pool. Conversely, P158DEL DJ-1 exhibited an elution profile indicating that it is natively smaller than WT protein, suggesting that it may be predominantly monomeric. Nevertheless, it is clear that the L10P, L166P and P158DEL DJ-1 proteins migrate differently with respect to each other suggesting that the respective mutations have distinct effects on protein conformation and/or the degree of protein-protein interactions.
Without proper regulation, misfolded proteins can accumulate and aggregate with each other, and this can eventually lead to impaired cellular processes (Olanow and McNaught 2006; McNaught and Olanow 2006). Since our data indicate that the L10P, L166P, and P158DEL mutations can alter protein folding, it was of interest to determine the effects of these mutations on protein solubility. The respective solubilities of WT, L10P, L166P, and P158DEL DJ-1 proteins were assessed by two different biochemical fractionation methods. When expressed in CHO cells under normal culturing conditions, P158DEL DJ-1 and to a lesser extent L10P DJ-1 demonstrated reduced solubilities. Interestingly, when biochemical fractionation experiments were performed following treatments with MG-132 or epoxomicin, L10P and L166P DJ-1 proteins demonstrated dramatic accumulations of insoluble proteins while WT DJ-1 was unaffected by these treatments. This indicated that L10P and L166P DJ-1 protein variants, which are inclined to misfolding and being degraded, accumulate as aggregates when degradation is impaired. Surprisingly, while P158 DEL formed insoluble aggregates under native conditions, proteasome inhibition did not enhance the accumulation of insoluble species. These findings suggest that P158DEL DJ-1 can spontaneously accumulate as misfolded aggregates that are largely regulated by proteasome-independent mechanisms.
Aggregation of DJ-1 mutant proteins also was studied at the microscopic level. Under normal conditions, only a small percent of cells expressing L10P and 158DEL DJ-1 depicted DJ-1 inclusions. Such a finding has not been reported for other pathogenic DJ-1 mutants, suggesting that these mutants may be more prone to aggregation. MG-132 treatments induced the formation of L10P, L166P, and P158DEL DJ-1 intranuclear and intracytoplasmic protein inclusions that were not detected in cells expressing WT DJ-1 protein under the same conditions. Most of the observed inclusions did not co-localize with common aggresome markers, suggesting that they likely represent deposits of aggregated DJ-1. The formation of these inclusions did not correlate with formation of insoluble DJ-1 observed biochemically. For example, biochemical analysis demonstrated that P158 DEL DJ-1 forms an intrinsic insoluble pool of protein and this is not significantly altered by MG132 treatment. It is possible that MG132 treatment induced this pool of P158DEL DJ-1 to coalesce into inclusions. Remarkably, inclusions were most prominent in cells expressing L10P DJ-1 following MG132 challenge. Biochemical analysis demonstrated that MG132 resulted in a dramatic accumulation of insoluble L10P DJ-1 and it is possible that such a pool is required for inclusion formation. Similarly, MG132 treatment resulted in the accumulation of insoluble L166P DJ-1 and inclusion formation. Olzmann and colleagues showed that L166P DJ-1 formed perinuclear aggresomes in cultured cells following treatments with MG-132; however it was determined that this phenomenon only occurred in the presence of the E3 ubiquitin-protein ligase, parkin (Olzmann et al. 2007). However since co-immunoprecipitation analyses did not reveal any interactions between parkin and L10P or P158DEL DJ-1 variants in the studies herein (data not shown), it is likely that L10P and P158DEL DJ-1 inclusion formation is regulated by alternative mechanisms.
In addition to characterizing the biochemical properties of L10P and P158DEL DJ-1 mutants, it was of interest to determine how these mutations affect protein function. To address this question, native CHO cells or CHO cells stably expressing WT or L10P DJ-1 were assessed for cell viability following proteasome insults. Interestingly, although WT DJ-1 was protective against MG-132 and epoxomicin treatments, L10P DJ-1 was unable to rescue CHO cells under the same conditions. Further, cells stably expressing L10P DJ-1 were more vulnerable to MG-132 and epoxomicin insults than native CHO cells, suggesting that L10P DJ-1 is toxic under conditions of proteasome stress. Yokota and colleagues have previously shown that expression of L166P DJ-1 is harmful to cells that are treated with proteasome inhibitors (Yokota et al. 2003). However, it has not yet been explored how pathogenic DJ-1 mutants mechanistically induce toxicity during proteasome insult. In the present study, L10P DJ-1 demonstrated decreased solubility and provocative inclusion formation in response to inhibition of the proteasome. Thus it is possible that L10P DJ-1 might be toxic in relation to its propensity to associate into inclusions. Of course it is of interest to determine whether similar effects on cell viability would be observed in CHO cells stably expressing P158DEL or L166P DJ-1; however, we were unsuccessful in our attempts to generate stably expressing CHO cell clones for the L166P and P158DEL DJ-1 mutants.
In conclusion, these studies demonstrate that the pathogenic L10P and P158DEL mutants result in the destabilization of DJ-1 and that proteasome degradation is at least partially involved in this process. Additionally, the L10P and P158DEL mutations impair homodimer formation, indicating that that the observed decreases in protein stability may be associated with these observed changes in quaternary structure, but local perturbations in protein folding may also be important. Alterations in protein structure are likely responsible for the observed reductions in protein solubility and formation of intracellular protein inclusions. These aberrant properties of L10P and P158DEL DJ-1 mutants may collectively contribute to DJ-1 loss-of-function by depleting the pool of functional protein. Additionally, since L10P DJ-1 can induce cell death as a consequence of proteasome impairments, it is possible that under conditions of proteolytic stress, the expression of L10P DJ-1 may itself also have adverse affects. Future studies to assess for specific effects of pathogenic DJ-1 mutations on biological pathways in vivo and studies in patients would give insights into the physiological role for DJ-1 protein especially as it relates to the etiology of PD.
ACKNOWLEDGEMENTS
This work was funded by grants from the National Institute on Aging (AG09215) and the National Institute of Neurological Disorders and Stroke (NS053488). C.P.R. is supported by a pre-doctoral NRSA fellowship (F31GM082026) from the National Institute on General Medicine Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- Anderson PC, Daggett V. Molecular basis for the structural instability of human DJ-1 induced by the L166P mutation associated with Parkinson's disease. Biochemistry. 2008;47:9380–9393. doi: 10.1021/bi800677k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Cookson MR. Evolutionary and functional relationships within the DJ1 superfamily. BMC Evol Biol. 2004;4:6. doi: 10.1186/1471-2148-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulac S, LaVoie MJ, Strahle J, Schlossmacher MG, Xia W. Dimerization of Parkinson's disease-causing DJ-1 and formation of high molecular weight complexes in human brain. Mol Cell Neurosci. 2004;27:236–246. doi: 10.1016/j.mcn.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Blackinton J, Ahmad R, Miller DW, van der Brug MP, Canet-Aviles RM, Hague SM, Kaleem M, Cookson MR. Effects of DJ-1 mutations and polymorphisms on protein stability and subcellular localization. Brain Res Mol Brain Res. 2005;134:76–83. doi: 10.1016/j.molbrainres.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M, van Dongen JW, Vanacore N, van Swieten JC, Brice A, Meco G, van Duijn CM, Oostra BA, Heutink P. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- Fitzgerald JC, Plun-Favreau H. Emerging pathways in genetic Parkinson's disease: autosomal-recessive genes in Parkinson's disease--a common pathway? FEBS J. 2008;275:5758–5766. doi: 10.1111/j.1742-4658.2008.06708.x. [DOI] [PubMed] [Google Scholar]
- Gasser T. Mendelian forms of Parkinson's disease. Biochim Biophys Acta. 2009;1792:587–596. doi: 10.1016/j.bbadis.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Giaime E, Sunyach C, Druon C, Scarzello S, Robert G, Grosso S, Auberger P, Goldberg MS, Shen J, Heutink P, Pouyssegur J, Pages G, Checler F, Alves dC. Loss of function of DJ-1 triggered by Parkinson's disease-associated mutation is due to proteolytic resistance to caspase-6. Cell Death Differ. 2010;17:158–169. doi: 10.1038/cdd.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorner K, Holtorf E, Odoy S, Nuscher B, Yamamoto A, Regula JT, Beyer K, Haass C, Kahle PJ. Differential Effects of Parkinson's Disease-associated Mutations on Stability and Folding of DJ-1. J Biol Chem. 2004;279:6943–6951. doi: 10.1074/jbc.M309204200. [DOI] [PubMed] [Google Scholar]
- Gorner K, Holtorf E, Waak J, Pham TT, Vogt-Weisenhorn DM, Wurst W, Haass C, Kahle PJ. Structural determinants of the C-terminal helix-kink-helix motif essential for protein stability and survival promoting activity of DJ-1. J Biol Chem. 2007;282:13680–13691. doi: 10.1074/jbc.M609821200. [DOI] [PubMed] [Google Scholar]
- Gregersen N, Bross P, Vang S, Christensen JH. Protein misfolding and human disease. Annu Rev Genomics Hum Genet. 2006;7:103–124. doi: 10.1146/annurev.genom.7.080505.115737. [DOI] [PubMed] [Google Scholar]
- Guo JF, Xiao B, Liao B, Zhang XW, Nie LL, Zhang YH, Shen L, Jiang H, Xia K, Pan Q, Yan XX, Tang BS. Mutation analysis of Parkin, PINK1, DJ-1 and ATP13A2 genes in Chinese patients with autosomal recessive early-onset Parkinsonism. Mov Disord. 2008;23:2074–2079. doi: 10.1002/mds.22156. [DOI] [PubMed] [Google Scholar]
- Honbou K, Suzuki NN, Horiuchi M, Taira T, Niki T, Ariga H, Inagaki F. Crystallization and preliminary crystallographic analysis of DJ-1, a protein associated with male fertility and parkinsonism. Acta Crystallogr D Biol Crystallogr. 2003;59:1502–1503. doi: 10.1107/s090744490301271x. [DOI] [PubMed] [Google Scholar]
- Huai Q, Sun Y, Wang H, Chin LS, Li L, Robinson H, Ke H. Crystal structure of DJ-1/RS and implication on familial Parkinson's disease. FEBS Lett. 2003;549:171–175. doi: 10.1016/s0014-5793(03)00764-6. [DOI] [PubMed] [Google Scholar]
- Kahle PJ, Waak J, Gasser T. DJ-1 and prevention of oxidative stress in Parkinson disease and other age-related disorders. Free Radic Biol Med. 2009;47:1354–1361. doi: 10.1016/j.freeradbiomed.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Kumaran R, Kingsbury A, Coulter I, Lashley T, Williams D, de Silva R, Mann D, Revesz T, Lees A, Bandopadhyay R. DJ-1 (PARK7) is associated with 3R and 4R tau neuronal and glial inclusions in neurodegenerative disorders. Neurobiol Dis. 2007;28:122–132. doi: 10.1016/j.nbd.2007.07.012. [DOI] [PubMed] [Google Scholar]
- Lesage S, Brice A. Parkinson's disease: from monogenic forms to genetic susceptibility factors. Hum Mol Genet. 2009;18:R48–R59. doi: 10.1093/hmg/ddp012. [DOI] [PubMed] [Google Scholar]
- Lockhart PJ, Lincoln S, Hulihan M, Kachergus J, Wilkes K, Bisceglio G, Mash DC, Farrer MJ. DJ-1 mutations are a rare cause of recessively inherited early onset parkinsonism mediated by loss of protein function. J Med Genet. 2004;41 doi: 10.1136/jmg.2003.011106. e22-e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macedo MG, Anar B, Bronner IF, Cannella M, Squitieri F, Bonifati V, Hoogeveen A, Heutink P, Rizzu P. The DJ-1L166P mutant protein associated with early onset Parkinson's disease is unstable and forms higher-order protein complexes. Hum Mol Genet. 2003;12:2807–2816. doi: 10.1093/hmg/ddg304. [DOI] [PubMed] [Google Scholar]
- Macedo MG, Verbaan D, Fang Y, van Rooden SM, Visser M, Anar B, Uras A, Groen JL, Rizzu P, van Hilten JJ, Heutink P. Genotypic and phenotypic characteristics of Dutch patients with early onset Parkinson's disease. Mov Disord. 2009;24:196–203. doi: 10.1002/mds.22287. [DOI] [PubMed] [Google Scholar]
- McNaught KS, Olanow CW. Protein aggregation in the pathogenesis of familial and sporadic Parkinson's disease. Neurobiol Aging. 2006;27:530–545. doi: 10.1016/j.neurobiolaging.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Meulener MC, Graves CL, Sampathu DM, Armstrong-Gold CE, Bonini NM, Giasson BI. DJ-1 is present in a large molecular complex in human brain tissue and interacts with alpha-synuclein. J Neurochem. 2005;93:1524–1532. doi: 10.1111/j.1471-4159.2005.03145.x. [DOI] [PubMed] [Google Scholar]
- Miller DW, Ahmad R, Hague S, Baptista MJ, Canet-Aviles R, McLendon C, Carter DM, Zhu PP, Stadler J, Chandran J, Klinefelter GR, Blackstone C, Cookson MR. L166P mutant DJ-1, causative for recessive Parkinson's disease, is degraded through the ubiquitin-proteasome system. J Biol Chem. 2003;278:36588–36595. doi: 10.1074/jbc.M304272200. [DOI] [PubMed] [Google Scholar]
- Moore DJ, Zhang L, Troncoso J, Lee MK, Hattori N, Mizuno Y, Dawson TM, Dawson VL. Association of DJ-1 and parkin mediated by pathogenic DJ-1 mutations and oxidative stress. Human Molecular Genetics. 2005;14:71–84. doi: 10.1093/hmg/ddi007. [DOI] [PubMed] [Google Scholar]
- Moore DJ, Zhang L, Dawson TM, Dawson VL. A missense mutation (L166P) in DJ-1, linked to familial Parkinson's disease, confers reduced protein stability and impairs homo-oligomerization. J Neurochem. 2003;87:1558–1567. doi: 10.1111/j.1471-4159.2003.02265.x. [DOI] [PubMed] [Google Scholar]
- Nagakubo D, Taira T, Kitaura H, Ikeda M, Tamai K, Iguchi-Ariga SM, Ariga H. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem Biophys Res Commun. 1997;231:509–513. doi: 10.1006/bbrc.1997.6132. [DOI] [PubMed] [Google Scholar]
- Neumann M, Muller V, Gorner K, Kretzschmar HA, Haass C, Kahle PJ. Pathological properties of the Parkinson's disease-associated protein DJ-1 in alpha-synucleinopathies and tauopathies: relevance for multiple system atrophy and Pick’s disease. Acta Neuropathol (Berl) 2004;107:489–496. doi: 10.1007/s00401-004-0834-2. [DOI] [PubMed] [Google Scholar]
- Olanow CW, McNaught KS. Ubiquitin-proteasome system and Parkinson's disease. Mov Disord. 2006;21:1806–1823. doi: 10.1002/mds.21013. [DOI] [PubMed] [Google Scholar]
- Olzmann JA, Brown K, Wilkinson KD, Rees HD, Huai Q, Ke H, Levey AI, Li L, Chin LS. Familial Parkinson's disease-associated L166P mutation disrupts DJ-1 protein folding and function. J Biol Chem. 2004;279:8506–8515. doi: 10.1074/jbc.M311017200. [DOI] [PubMed] [Google Scholar]
- Olzmann JA, Li L, Chudaev MV, Chen J, Perez FA, Palmiter RD, Chin LS. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J Cell Biol. 2007;178:1025–1038. doi: 10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey CP, Giasson BI. The E163K DJ-1 mutant shows specific antioxidant deficiency. Brain Res. 2008;1279:1–11. doi: 10.1016/j.brainres.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzu P, Hinkle DA, Zhukareva V, Bonifati V, Severijnen LA, Martinez D, Ravid R, Kamphorst W, Eberwine JH, Lee VM, Trojanowski JQ, Heutink P. DJ-1 colocalizes with tau inclusions: a link between parkinsonism and dementia. Ann Neurol. 2004;55:113–118. doi: 10.1002/ana.10782. [DOI] [PubMed] [Google Scholar]
- Shinbo Y, Niki T, Taira T, Ooe H, Takahashi-Niki K, Maita C, Seino C, Iguchi-Ariga SM, Ariga H. Proper SUMO-1 conjugation is essential to DJ-1 to exert its full activities. Cell Death Differ. 2006;13:96–108. doi: 10.1038/sj.cdd.4401704. [DOI] [PubMed] [Google Scholar]
- Siva AB, Sundareswaran VR, Yeung CH, Cooper TG, Shivaji S. Hamster contraception associated protein 1 (CAP1) Mol Reprod Dev. 2004;68:373–383. doi: 10.1002/mrd.20085. [DOI] [PubMed] [Google Scholar]
- Soto C, Estrada LD. Protein misfolding and neurodegeneration. Arch Neurol. 2008;65:184–189. doi: 10.1001/archneurol.2007.56. [DOI] [PubMed] [Google Scholar]
- Takahashi-Niki K, Niki T, Taira T, Iguchi-Ariga SM, Ariga H. Reduced anti-oxidative stress activities of DJ-1 mutants found in Parkinson's disease patients. Biochem Biophys Res Commun. 2004;320:389–397. doi: 10.1016/j.bbrc.2004.05.187. [DOI] [PubMed] [Google Scholar]
- Wilson MA, Collins JL, Hod Y, Ringe D, Petsko GA. The 1.1-A resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson's disease. Proc Natl Acad Sci U S A. 2003;100:9256–9261. doi: 10.1073/pnas.1133288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T, Sugawara K, Ito K, Takahashi R, Ariga H, Mizusawa H. Down regulation of DJ-1 enhances cell death by oxidative stress, ER stress, and proteasome inhibition. Biochem Biophys Res Commun. 2003;312:1342–1348. doi: 10.1016/j.bbrc.2003.11.056. [DOI] [PubMed] [Google Scholar]