Abstract
The extracellular matrix (ECM) plays a key role in cell–cell communication and signaling, and the signals it propagates are important for tissue remodeling and survival. However, signals from disease-altered ECM may lead to anoikis—apoptotic cell death triggered by loss of ECM contacts. Previously, we found that an altered fibronectin matrix triggers anoikis in human primary ligament cells via a pathway that requires p53 transcriptional downregulation. Here we show that this p53 reduction is suppressed by transfecting cells with Mdm2 antisense oligonucleotides or small interfering RNA. Similar results were found in cells treated to prevent p53 and Mdm2 interactions. When p53 was overexpressed in cells lacking Mdm2 and p53, p53 levels were unaffected by anoikis conditions. However, cells cotransfected with p53 and wild type Mdm2, but not a mutant Mdm2, exhibited decreased p53 levels in response to anoikis conditions. Thus, cells under anoikis conditions undergo p53 degradation that is mediated by Mdm2.
Keywords: Extracellular matrix, Fibronectin, Mdm2, p53, Proteasome, Periodontitis
Introduction
The extracellular matrix (ECM)1 provides structural integrity to tissues and organs. The ECM is important for cell adhesion and migration, serves as a conduit for cell-to-cell communication, and promotes cell survival by providing a structural matrix and reservoir for growth factors. However, changes in the ECM due to infection, inflammation, or metastasis may disrupt the homeostasis of the extracellular environment. Under such conditions, ECM proteins undergo proteolytic cleavage or alternative splicing, resulting in fragmented or altered forms [1–5]. These alterations lead to aberrant signaling in surrounding cells and catalyze further degradation of the matrix, exacerbating the disease. For example, disease-associated fibronectin fragments can cause anoikis and further tissue catabolism by inducing the expression of matrix metalloproteinases, nitric oxide, and proinflammatory cytokines [6–14]. The mechanisms by which ECM fragments elicit these adverse cellular effects may involve alterations in signaling processes and receptor regulation [13, 15–18], such as altered regulation and signaling via p53 and the c-Jun NH2-terminal stress-related kinases [13, 16, 17, 19].
p53 is a tumor suppressor gene that plays a critical role in safeguarding the integrity of the genome [20]. It exerts tumor suppressor effects through various mechanisms, including cell-cycle arrest, apoptosis, and cellular senescence [21]. Given its pivotal role in coordinating cellular events in response to stress signals, its expression and activation are tightly controlled within the cell by ubiquitination, acetylation, phosphorylation, and other processes. Often, p53 regulatory networks are perturbed by stress signals, such as DNA damage, oncogene activation, hypoxia, and nitric oxide production, resulting in ubiquitination of p53 [22]. An important regulator of p53 is Mdm2 [23]. p53 and Mdm2 form a feedback loop in which p53 positively regulates Mdm2 by activating Mdm2 transcription and Mdm2 negatively regulates p53 by promoting its ubiquitination and degradation. Proteins destined for degradation in the proteasome are tagged with ubiquitin [24, 25]. Depending on the degree of ubiquitination, a protein may be activated or targeted for degradation in the proteasome [26]. Mdm2 functions as a ubiquitin ligase for p53. After ubiquitination by Mdm2, p53 is rapidly degraded by the proteasome.
Previously, we showed that an altered fibronectin matrix triggers anoikis in cells, and this process is mediated by decreases in p53 and c-Myc at the transcriptional and protein levels [2, 11, 15]. In these studies, we used the disease-associated fibronectin fragments that are found in chronic inflammatory fluids and are known to be important in the pathogenesis of chronic inflammatory diseases including arthritis and periodontal disease [1, 2, 5–7, 14, 15]. The disease-associated fibronectin fragment (AFn) or a control fibronectin fragment (cAFn) was expressed as recombinant protein and were purified and used in our studies. Thus, using these altered fibronectin matrices, we have extensively characterized this novel anoikis mechanism with respect to the involvement of p53, c-myc, focal adhesion kinase (FAK), JNK-1, and JNK-2 in our earlier publications [11–13]. We showed that signals emanating from an altered fibronectin matrix resulted in decreases in p53 and c-Myc levels that were propagated by decreased FAK phosphorylation and upregulated JNK phosphorylation. Our experiments showed that FAK is physically and spatially linked to JNK and p53, and p53 relocalizes from the nucleus to the cell membrane to mediate this interaction. Further, p53 participates in a feedback mechanism with JNK to regulate this cross-talk and p53 is oppositely regulated by JNK1 and JNK2. Nonetheless, the reductions in p53 promoter activity and mRNA levels did not explain the significant loss of p53 protein in cells exposed to the anoikis-inducing altered fibronectin matrix. In a recent study, we showed that the loss of p53 in cells exposed to anoikis conditions is in part due to enhanced ubiquitination of p53 [27]. In this study, we investigated the involvement of Mdm2, the other key regulator of p53, in the context of anoikis conditions.
Materials and methods
Fibroblast cell culture
Human primary ligament fibroblasts were isolated and cultured as described [12]. Their use in these studies was approved by the University of Michigan Health Sciences Institutional Review Board. p53-null fibroblasts were a gift from Dr. Gerard P. Zambetti (St. Jude Children's Research Hospital, Memphis, TN) [28]. p53/Mdm2-null fibroblasts were a gift from Dr. Gigi Lozano (University of Texas M.D. Anderson Cancer Center, Houston, TX) [29]. The primary fibroblasts were cultured in α-minimum essential medium (Invitrogen) and p53-null and p53/Mdm2-null cells were cultured in high-glucose Dulbecco's modified Eagle's medium (Invitrogen); both media contained 10% fetal calf serum (Hyclone) and penicillin and streptomycin.
Plasmids/DNA constructs
Human wildtype p53 expression plasmid pC53-SN3 and human Mdm2 pCMV were from Dr. Bert Vogelstein (Johns Hopkins University School of Medicine, Baltimore, MD) [30]. Mutant human Mdm2 pCHDMΔ222-437 expression plasmid was provided by Dr. Arnold J. Levine (Princeton University, Princeton, NJ) [31].
Recombinant fibronectin proteins
For these studies, we used two previously described recombinant fibronectin fragments [10] that consisted of the alternatively spliced V region and contained either an intact (cAFn) or a mutated, nonfunctional high-affinity heparin-binding domain (AFn). The control fragment (cAFn), the mutated anoikis-inducing fragment (AFn), and intact fibronectin were used at a concentration of 0.1 mM.
Western blot analysis
For Western blot analysis, the cells were lysed in ice-cold RIPA buffer (Sigma) containing protease inhibitors (Sigma). Protein concentration was determined with the BCA protein assay kit (Pierce, Rockford, IL). Equal amounts of protein were loaded into each well and resolved by SDS-PAGE with 4–20% gels (Novex, Invitrogen) and electroblotted onto polyvinylidene membranes (Immobilon-P, Millipore, Billerica, MA) by semidry transfer blot (Biorad) according to the manufacturer's instructions. The membranes were incubated with 5% nonfat dry milk in 25 mM Tris, 150 mM NaCl, 0.05% Tween-20, pH 7.4 (TBST) for 1 h at room temperature and then with primary and horseradish peroxidase-conjugated secondary antibodies in blocking buffer at room temperature for 2 h or at 4°C overnight, washed with TBST and developed with the West-Pico ECL kit (Pierce). The primary antibodies were horseradish peroxidase conjugated mouse anti-human p53 (DO-1 HRP), and goat anti-human actin (I-19) (Santa Cruz Biotechnology, Santa Cruz, CA) and rabbit anti-human Mdm2 (AF1244, R & D Systems, Minneapolis, MN).
Transient transfection
Cells were plated in six-well tissue-culture plates at 60–80% confluency. One day later, the cells were transfected with 0.5 μg, 1 μg, or 100 ng of p53 or 400 ng of plasmids expressing the cDNAs for wildtype or mutant human Mdm2 using Lipofectamine 2000 (Invitrogen). Thirty-six hours after transfection, the cells were washed with phosphate-buffered saline (PBS), treated with the recombinant fibronectin fragments for 7 h, and processed for Western blot experiments.
Antisense experiments
Primary ligament fibroblasts were transfected with antisense Mdm2 oligonucleotides or Mdm2 small interfering RNA (siRNA). One day before transfection, the cells were plated at 60% confluency in six-well dishes. The Mdm2 oligonucleotides synthesized by Invitrogen have been described previously [32]: antisense, 5′-UGACACCTGTTCTCACUCAC-3′; control, 5′-UGTCACCCTTTTTCATUCAC-3′. These oligonucleotides were phosphorothioated at all positions to minimize intracellular degradation and increase stability [33]. The oligonucleotides were mixed with Oligofectamine (Invitrogen), incubated for 20 min, and added to the cells in serum-free medium. After 4 h, serum was added, and the cells were cultured for 36 h and then treated with the fibronectin fragments in serum-free medium. The siRNAs were from Dharmacon, NM_002392 (human Mdm2). Stealth RNAi Negative Control (Invitrogen) was used for control transfections. siRNA transfections were done with Lipofectamine 2000. Thirty-six hours after transfection, the cells were treated with fibronectin fragments in serum-free medium.
ELISA assay
Cells were pretreated with Nutlin and then treated with the fibronectin fragments or control media. Thereafter, all the cells were lysed and equal amounts of cell lysate protein were processed to quantitate apoptosis using the kit, Cell Death Detection ELISAPLUS (Roche) as recommended by the manufacturer.
Other reagents
Nutlin, Mdm2 inhibitor (trans-4-iodo, 4′-boranyl-chalcone) and lactacystin were from Calbiochem. All other chemicals were from Sigma unless mentioned otherwise.
Results
Mdm2 is required for p53 degradation mediated by anoikis
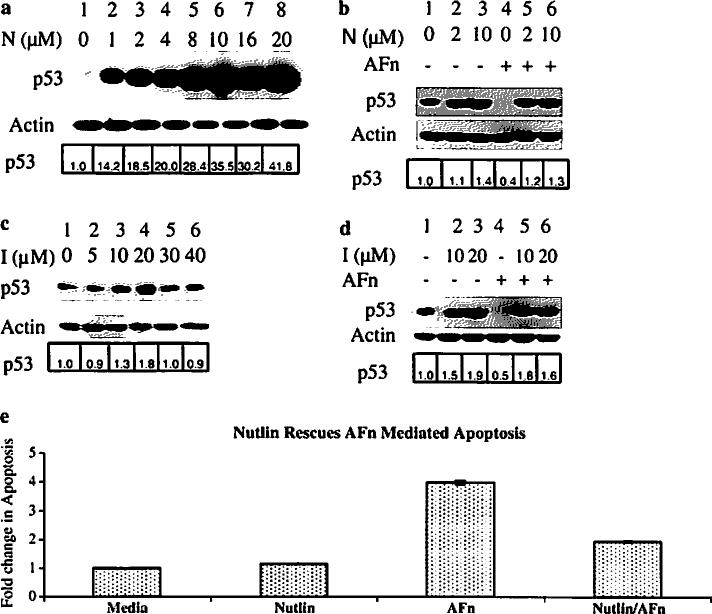
Mdm2, an important regulator of p53 [23], has a p53 binding pocket that is essential for its ubiquitin ligase activity [34]. To investigate the role of Mdm2 in p53 degradation in this mechanism, we used Nutlin to prevent Mdm2 from binding to p53 [35]. Treatment with increasing concentrations of this small molecule increased p53 levels in primary ligament fibroblasts (Fig. 1a, lanes 2–8) compared to the untreated cells (Fig. 1a, lane 1). p53 stability was also enhanced in cells pretreated with different doses of Nutlin despite treatment under anoikis conditions for 7 h (Fig. 1b, lanes 5 and 6).
Fig. 1.
Inhibiting the binding of Mdm2 to p53 prevents p53 degradation under anoikis conditions. Human primary ligament fibroblasts were treated with various concentrations of Nutlin (N) or (c), a different Mdm2 inhibitor (I) for 7 h and lysed, and the p53 level was determined by Western blotting with an anti-p53 antibody. b Based on the results from the dose–response experiments in (a) and (c), cells were pretreated with chosen concentrations of Nutlin or (d), Mdm2 inhibitor or control medium for 2 h and then treated with the fibronectin fragments in presence of the drugs for 7 h. Cell lysates were prepared and analyzed by immunoblotting with antibodies to p53 and Mdm2. Actin served as a loading control. e Human primary ligament fibroblasts were treated with 10 μM Nutlin for 2 h and then treated with the fibronectin fragments or control medium in presence of the drugs for 7 h. Cell lysates were prepared and analyzed for apoptosis using the Cell Death Detection ELISAPLUS (Roche). The relative fold change in p53 expression was analyzed by densitometry and expressed with respect to p53 in lane 1 for all the figures
To further test the importance of Mdm2 in this mechanism, we used trans-4-iodo, 4′-boranyl-chalcone, an Mdm2 inhibitor that also prevents Mdm2 from interacting with p53 [36]. This inhibitor induced concentration-dependent increases in p53 levels up to 20 μM; higher concentrations were toxic (Fig. 1c). We next subjected cells to treatment with this Mdm2 inhibitor at two different doses and found that cells could be rescued from p53 degradation mediated by anoikis (Fig. 1d, lanes 5 and 6) than the untreated cells (Fig. 1d, lane 4). These findings show that p53 degradation in response to anoikis requires a physical interaction between p53 and Mdm2.
To test whether inhibiting the interaction between Mdm2 with p53 could prevent anoikis in these cells, we pretreated the cells with Nutlin and then subjected them to the anoikis condition and thereafter quantified cell death using an ELISA assay. As shown in Fig. 1e, Nutlin pretreatment greatly reduced anoikis levels, indicating the importance of Mdm2 and p53 interactions in this mechanism.
Inhibition of Mdm2 expression prevents p53 degradation induced by anoikis
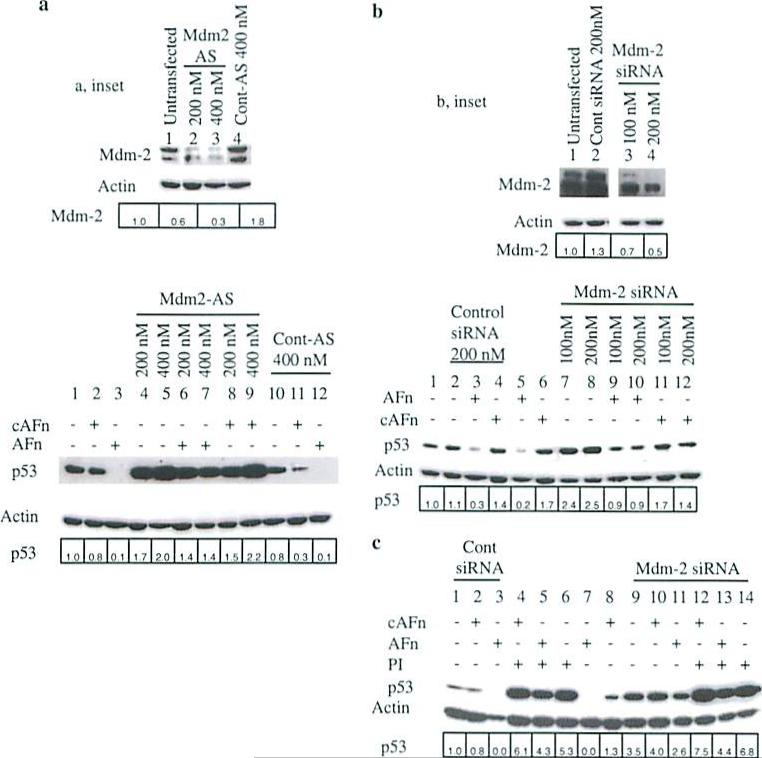
To further assess the requirement for Mdm2 in p53 degradation induced under anoikis, we transfected ligament fibroblasts with antisense oligonucleotides or siRNA to inhibit Mdm2 expression. p53 levels increased in a dose-dependent fashion in cells transfected with Mdm2 antisense oligonucleotide (Fig. 2a, bottom, lanes 1, 4, and 5) but were unchanged in cells transfected with control oligonucleotide (Fig. 2a, bottom, lane 10). Importantly, cells transfected with the antisense oligonucleotide were resistant to p53 degradation triggered by anoikis (Fig. 2a, bottom, lanes 3, 6, and 7). Similarly, Mdm2 siRNA also caused a dose-dependent increase in p53 protein levels (Fig. 2b, bottom, lanes 1, 7, and 8) and reduced p53 degradation under anoikis conditions (Fig. 2b, bottom, lanes 5, 9, and 10).
Fig. 2.
Inhibition of Mdm2 expression with either an antisense oligonucleotide or siRNA blocks p53 degradation under anoikis conditions. a Inhibition of Mdm2 expression with an antisense oligonucleutides (Mdm2-AS) or (b), Mdm2 siRNA blocked the degradation of p53 by an altered fibronectin matrix. Human primary ligament fibroblasts were transfected with either Mdm2 antisense or control oligonucleotides or Mdm2 siRNA or control siRNA for 36 h, treated with recombinant fibronectin molecules for 7 h, and lysed, and the cell lysates were analyzed for Mdm2 and p53 expression. c Synergistic effects of the proteasome inhibitor and Mdm2 siRNA on altered fibronectin matrix-mediated degradation of p53 was examined. Cells were transfected with either 200 nM Mdm2 or control siRNA as in (b), pretreated wilh lactacystin (PI) or control medium, and treated with the recombinant fibronectin fragments. Cells were lysed, and p53 and Mdm2 levels were determined by immunoblotting with anti-p53 and anti-Mdm2 antibodies. Actin served as a loading control. a, b inset Cells upon transfection with Mdm2 AS oligonucleotide or Mdm2 siRNA showed decreased levels of Mdm2 protein expression. The relative fold change in p53 or Mdm2 expression was analyzed by densitometry and expressed with respect to p53 or Mdm2 in lane 1 for all the figures
To rule out residual Mdm2 activity and ensure complete stabilization of endogenous p53 in primary ligament fibroblasts transfected with Mdm2 siRNA, we pretreated the cells with lactacystin, a proteasome inhibitor. Lactacystin treatment resulted in higher p53 levels in untransfected cells (Fig. 2c, lanes 4–6)and cells transfected with Mdm2 siRNA (Fig. 2c, lanes 12–14) than in non-pretreated cells transfected with control siRNA (Fig. 2c, lanes 1–3) or Mdm2 siRNA (Fig. 2c, lanes 9–11). Under anoikis conditions, lactacystin, and Mdm2 siRNA synergistically increased p53 stability in these cells, further confirming the importance of Mdm2 and proteasomal pathways in anoikis mediated p53 degradation.
Anoikis downregulates exogenous p53 Via an Mdm2-dependent pathway in p53-null cells
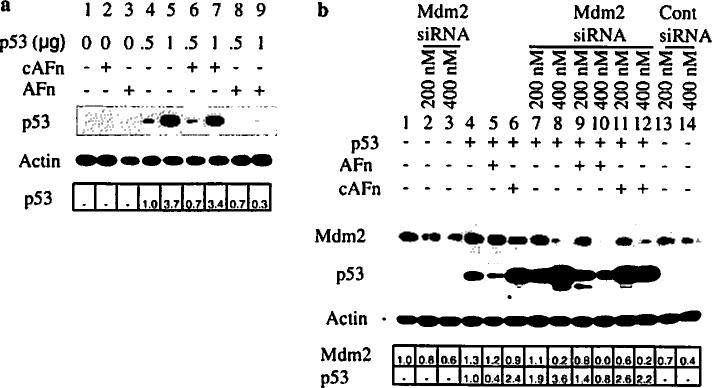
To further explore the Mdm2 dependency in p53 degradation induced by anoikis, we overexpressed p53 in p53-null fibroblasts that have normal Mdm2 expression. As expected, p53 expression increased in a dose-dependent fashion with increasing DNA concentrations (Fig. 3a, lanes 4 and 5). Anoikis conditions efficiently degraded the exogenously expressed p53 (Fig. 3a, lanes 8 and 9).
Fig. 3.
Mdm2 mediates the degradation of exogenous p53 triggered by anoikis. a p53-null fibroblasts were transfected with different concentrations of wildtype p53, treated with fibronectin fragments, and then the level of p53 was analyzed by Western blotting with anti-p53 antibody. b p53-null fibroblasts were cotransfected with wildtype p53 and Mdm2 siRNA or control siRNA for 36 h, treated with recombinant fibronectin fragments for 7 h, and lysed. Lysates were examined for p53 and Mdm2 expression by immunoblotting with anti-p53 and anti-Mdm2 antibodies. Cells upon transfection with Mdm2 siRNA showed decreased levels of Mdm2 protein expression. The relative fold change in p53 or Mdm2 expression was analyzed by densitometry and expressed with respect to p53 in lane 4 in Fig. 3a and b and lane 1 for Mdm2 in Fig. 3b
Next, we cotransfected the p53-null cells with Mdm2 siRNA and wild type p53 and then induced anoikis. Transfection with Mdm2 siRNA decreased Mdm2 protein levels in a dose-dependent fashion (Fig. 3b, lanes 1–3 and 7–12). Mdm2 siRNA and wild type p53 cotransfection resulted in a dose-dependent increase in p53 (Fig. 3b, lanes 7 and 8 vs. lane 4). Treatment of p53-transfected cells under anoikis conditions reduced exogenous p53 levels (Fig. 3b, lanes 4 and 5). In cotransfected cells, however, p53 was resistant to degradation induced by anoikis (Fig. 3b, lanes 9 and 10 vs. lane 5).
p53 degradation induced by anoikis is reduced in Mdm2-null cells and cells expressing mutant nonfunctional Mdm2
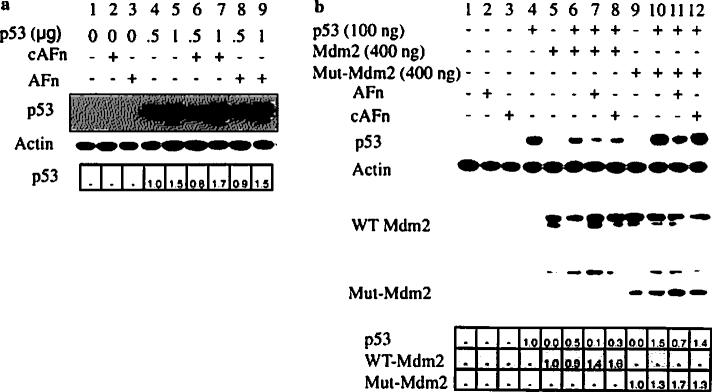
To confirm the requirement for Mdm2 in p53 degradation, we used a fibroblast cell line deficient in both p53 and Mdm2. Transfection with p53 cDNA caused a dose-dependent increase in p53 expression (Fig. 4a, lanes 4 and 5). However, anoikis conditions did not reduce p53 levels (Fig. 4a, lanes 8 and 9), which is consistent with a requirement for Mdm2 in this mechanism.
Fig. 4.
Mdm2 null and mutant conditions resist p53 degradation triggered by anoikis. a p53/Mdm2-null cells were transfected with wildtype p53, treated with recombinant fibronectin fragments, lysed, and analyzed for p53 expression by Western blotting. b p53/Mdm2-null cells were transfected with wildtype p53 or cotransfected with wildtype p53 and wildtype Mdm2 (WT Mdm2) or wildtype p53 and mutant Mdm2 (Mut-Mdm2). Thirty-six hours after transfection, the cells were treated with recombinant fibronectin fragments or control medium, and lysed. p53 and Mdm2 protein expressions were determined by Western blot. Cells upon transfection with wild type Mdm2 or mutant Mdm2 cDNA showed decreased levels of Mdm2 protein expression. The relative fold change in p53 or Mdm2 expression was analyzed by densitometry and expressed with respect to p53 in lane 4 in Fig. 4a and b and lane 5 for wild type Mdm2 and lane 9 for mutant Mdm2 in Fig. 4b
Finally, we transfected p53/Mdm2-null cells with wildtype p53, wildtype Mdm2 and a mutant Mdm2 (human Mdm2 Δ222–437) that binds p53 but cannot target it for proteasomal degradation [31, 37], or a combination of these. As expected, coexpression of Mdm2 and p53 resulted in lower levels of p53 protein than overexpression of p53 alone (Fig. 4b, lanes 4 and 6), and anoikis conditions caused a further reduction in p53 levels (Fig. 4b, lane 7). However, in cells cotransfected with p53 and mutant Mdm2, anoikis conditions did not reduce p53 levels (Fig. 4b, lane 11), which was similar to that in cells transfected with p53 alone (Fig. 4b, lane 4), Control conditions (cAFn) did not affect p53 protein levels. Thus, degradation of p53 triggered by anoikis conditions is mediated through an Mdm2-dependent pathway.
Discussion
This study shows that primary fibroblasts under anoikis conditions exhibit Mdm2-mediated degradation of p53. Mdm2 is known to negatively regulate p53 by mediating its ubiquitination and subsequent degradation in the proteasome [37, 38]. Our previous observation that an anoikis-inducing fibronectin matrix signals the degradation of p53 by ubiquitination [27] led us to inquire about the role of p53 regulatory molecules, i.e., Mdm2, in the downregulation of p53 under anoikis conditions. p53 and Mdm2 form a complex in many regulatory pathways, and several studies have focused on disrupting this complex formation using peptide or chemical inhibitors. Disruption of the p53–Mdm2 complex would lead to the stabilization of p53 and thereby apoptosis of cancerous cells. Indeed, several natural products and synthetic small molecules have been identified [34, 39, 40] for this purpose. In our experiments, we pretreated cells with Nutlin or an Mdm2 inhibitor, both of which block the p53 binding pocket of Mdm2 and prevent it from interacting with p53. As shown by Western blot experiments, each of these agents prevented p53 degradation under anoikis conditions. Furthermore, prevention of Mdm2–p53 interaction using Nutlin greatly reduced apoptosis in fibroblasts as determined by the ELISA assay. We then transfected cells with Mdm2 antisense oligonucleotides or siRNA. As expected, inhibiting Mdm2 expression resulted in accumulation of p53 in both untreated cells and cells under anoikis conditions. Although Mdm2 siRNA stabilized the p53 protein in treated cells, the p53 levels were lower than in cells treated with a proteasome inhibitor (Fig. 2c, lanes 4–6 vs. lanes 9–11). Proteasome inhibitor treatment of cells transfected with Mdm2 siRNA resulted in even greater stabilization of p53.
There are at least two explanations for this observation. First, although siRNAs can inhibit the expression of their target proteins, they can be “leaky” in the sense that some fraction of the target protein gets expressed. For example, in this case a low level of Mdm2 was being expressed, which would ubiquitinate and degrade a small fraction of p53. Second, ubiquitin ligases other than Mdm2, such as COP1 and Pirh2, both of which interact with p53 [41–43], might contribute to the downregulation of p53. A ubiquitin-independent proteasomal p53 degradation has also been discovered [44] and it would be interesting to evaluate the contribution of these pathways in our system.
To further validate the requirement for Mdm2 in p53 downregulation induced by anoikis, we used p53-null and p53/Mdm2-null mouse embryonic fibroblast cells. Transfecting p53-null cells with wildtype p53 gave a robust expression of the p53 protein, which was degraded when the cells were treated under anoikis conditions. When p53-null cells were cotransfected with Mdm2 siRNA and p53, the p53 level increased markedly, even in cells under anoikis conditions. When wildtype p53 was expressed in p53/Mdm2-null cells, treatment with the anoikis promoting fibronectin fragment had minimal effects on p53 levels, further supporting the need for Mdm2 in mediating the degradation of p53 in this process. However, when these cells were cotransfected with functional Mdm2, p53 was degraded by the anoikis promoting fibronectin fragment. Cotransfecting these cells with a mutant Mdm2, which can bind to p53 but cannot target it for degradation, resulted in higher p53 levels than that in cells transfected with wildtype Mdm2, even in the presence of the anoikis promoting fibronectin fragment.
Our result showing that anoikis mediated p53 degradation is Mdm2 and ubiquitin dependent is in agreement with previous findings that Mdm2 downregulates p53 by ubiquitination through the proteasome [45-47]. Interestingly, our western blot data in Fig. 4b showed that the mutant Mdm2, which can bind to p53 but cannot target it for degradation, gave the expected mutant Mdm2 band at the 37 KD position but also some high molecular weight bands. These high molecular weight bands can be attributed to other proteins, which can also associate with Mdm2, like the ribosomal protein S7, the tumor suppressor RASSF1A, Cu14A, insulin-like growth factor 1 receptor and cyclin G1 [48–52] and thereby form a complex with mutant Mdm2 that is detected with the anti-Mdm2 antibody. Recently, other than Mdm2, a wide variety of E3 ligases like COP1, Pirh2, and ARF-BP1 have been shown to regulate and maintain the level of p53 [41, 42, 53]. However, it is uncertain how these E3 ligases are specifically regulated and differentially activated in stressed and unstressed cells. Moreover, there are molecules like NAD(P)H quinone oxidoreductase 1 (NQO1) which regulate p53 degradation in the proteasome by a mechanism that is independent of Mdm2 and ubiquitin [54]. In future studies it may be promising to investigate whether there is potential cross-talk between these other molecules and Mdm2 towards the degradation of p53 in primary fibroblasts undergoing anoikis.
In summary, anoikis triggers the degradation of p53 by a mechanism that requires the association of p53 with Mdm2 leading to its ubiquitination and subsequent degradation by the proteasome. Since, inhibiting either the interaction of Mdm2 with p53 or the expression of Mdm2 resulted in decreased degradation of p53, these molecular pathways might represent potential therapeutic targets for inflammatory diseases like arthritis and periodontal disease that cause alterations in the ECM.
Acknowledgments
We thank Dr. Gerard P. Zambetti (St. Jude Children's Research Hospital, Memphis, TN) for p53-null fibroblasts, Dr. Gigi Lozano (University of Texas M.D. Anderson Cancer Center, Houston, TX) for p53/Mdm2-null fibroblasts, Dr. Bert Vogelstein (Johns Hopkins University School of Medicine, Baltimore, MD) for human wildtype p53 and human Mdm2 pCMV expression plasmids, Dr. Arnold J. Levine (Princeton University, Princeton, NJ) for human Mdm2 pCHDMΔ222-437 expression plasmid, Dr. Paul W. Johnson for mentoring and the fibronectin fragments and Stephen Ordway for editorial assistance. This study was support by an NIH grant ROI DE013725 (to Y. L. K.).
Footnotes
The abbreviations used are: ECM, extracellular matrix; cAFn and AFn, alternatively spliced V region (V+) and containing intact (H+) or a mutated, nonfunctional (H–) high-affinity heparin binding domain; TBST, 25 mM Tris, 150 mM NaCl, 0.05% Tween-20.
References
- 1.Homandberg GA, Wen C, Hui F. Cartilage damaging activities of fibronectin fragments derived from cartilage and synovial fluid. Osteoarthr Cartil. 1998;6:231–244. doi: 10.1053/joca.1998.0116. [DOI] [PubMed] [Google Scholar]
- 2.Huynh QN, Wang S, Tafolla E, Gansky SA, Kapila S, Armitage GC, Kapila YL. Specific fibronectin fragments as markers of periodontal disease status. J Periodontol. 2002;73:1101–1110. doi: 10.1902/jop.2002.73.10.1101. [DOI] [PubMed] [Google Scholar]
- 3.Talonpoika J, Heino J, Larjava H, Hakkinen L, Paunio K. Gingival crevicular fluid fibronectin degradation in periodontal health and disease. Scand J Dent Res. 1989;97:415–421. doi: 10.1111/j.1600-0722.1989.tb01455.x. [DOI] [PubMed] [Google Scholar]
- 4.Talonpoika J, Paunio K, Soderling E. Molecular forms and concentration of fibronectin and fibrin in human gingival crevicular fluid before and after periodontal treatment. Scand J Dent Res. 1993;101:375–381. doi: 10.1111/j.1600-0722.1993.tb01135.x. [DOI] [PubMed] [Google Scholar]
- 5.Xie DL, Meyers R, Homandberg GA. Fibronectin fragments in osteoarthritic synovial fluid. J Rheumatol. 1992;19:1448–1452. [PubMed] [Google Scholar]
- 6.Homandberg GA, Meyers R, Williams JM. Intraarticular injection of fibronectin fragments causes severe depletion of cartilage proteoglycans in vivo. J Rheumatol. 1993;20:1378–1382. [PubMed] [Google Scholar]
- 7.Homandberg GA, Meyers R, Xie DL. Fibronectin fragments cause chondrolysis of bovine articular cartilage slices in culture. J Biol Chem. 1992;267:3597–3604. [PubMed] [Google Scholar]
- 8.Kapila YL, Kapila S, Johnson PW. Fibronectin and fibronectin fragments modulate the expression of proteinases and proteinase inhibitors in human periodontal ligament cells. Matrix Biol. 1996;15:251–261. doi: 10.1016/s0945-053x(96)90116-x. [DOI] [PubMed] [Google Scholar]
- 9.Kapila YL, Lancero H, Johnson PW. The response of periodontal ligament cells to fibronectin. J Periodontol. 1998;69:1008–1019. doi: 10.1902/jop.1998.69.9.1008. [DOI] [PubMed] [Google Scholar]
- 10.Kapila YL, Niu J, Johnson PW. The high affinity heparin-binding domain and the V region of fibronectin mediate invasion of human oral squamous cell carcinoma cells in vitro. J Biol Chem. 1997;272:18932–18938. doi: 10.1074/jbc.272.30.18932. [DOI] [PubMed] [Google Scholar]
- 11.Kapila YL, Wang S, Dazin P, Tafolla E, Mass MJ. The heparin-binding domain and V region of fibronectin regulate apoptosis by suppression of p53 and c-myc in human primary cells. J Biol Chem. 2002;277:8482–8491. doi: 10.1074/jbc.M108932200. [DOI] [PubMed] [Google Scholar]
- 12.Kapila YL, Wang S, Johnson PW. Mutations in the heparin binding domain of fibronectin in cooperation with the V region induce decreases in pp125(FAK) levels plus proteoglycan-mediated apoptosis via caspases. J Biol Chem. 1999;274:30906–30913. doi: 10.1074/jbc.274.43.30906. [DOI] [PubMed] [Google Scholar]
- 13.Tafolla E, Wang S, Wong D, Leong J, Kapila YL. JNK1 and JNK2 oppositely regulate p53 in signaling linked to apoptosis triggered by an altered fibronectin matrix: JNK links FAK and p53. J Biol Chem. 2005;280:19992–19999. doi: 10.1074/jbc.M500331200. [DOI] [PubMed] [Google Scholar]
- 14.Xie D, Homandberg GA. Fibronectin fragments bind to and penetrate cartilage tissue resulting in proteinase expression and cartilage damage. Biochim Biophys Acta. 1993;1182:189–196. doi: 10.1016/0925-4439(93)90140-v. [DOI] [PubMed] [Google Scholar]
- 15.Dai R, Iwama A, Wang S, Kapila YL. Disease-associated fibronectin matrix fragments trigger anoikis of human primary ligament cells: p53 and c-myc are suppressed. Apoptosis. 2005;10:503–512. doi: 10.1007/s10495-005-1880-5. [DOI] [PubMed] [Google Scholar]
- 16.Forsyth CB, Pulai J, Loeser RF. Fibronectin fragments and blocking antibodies to alpha2beta1 and alpha5beta1 integrin stimulate mitogen-activated protein kinase signaling and increase collagenase 3 (matrix metalloproteinase 13) production by human articular chondrocytes. Arthritis Rheum. 2002;46:2368–2376. doi: 10.1002/art.10502. [DOI] [PubMed] [Google Scholar]
- 17.Gemba T, Valbracht J, Alsalameh S, Lotz M. Focal adhesion kinase and mitogen-activated protein kinases are involved in chondrocyte activation by the 29-kDa amino-terminal fibronectin fragment. J Biol Chem. 2002;277:907–911. doi: 10.1074/jbc.M109690200. [DOI] [PubMed] [Google Scholar]
- 18.Homandberg GA, Costa V, Wen C. Fibronectin fragments active in chondrocytic chondrolysis can be chemically cross-linked to the alpha5 integrin receptor subunit. Osteoarthr Cartil. 2002;10:938–949. doi: 10.1053/joca.2002.0854. [DOI] [PubMed] [Google Scholar]
- 19.Yasuda T, Kakinuma T, Julovi SM, Yoshida M, Hiramitsu T, Akiyoshi M, Nakamura T. COOH-terminal heparin-binding fibronectin fragment induces nitric oxide production in rheumatoid cartilage through CD44. Rheumatology. 2004;43:1116–1120. doi: 10.1093/rheumatology/keh274. [DOI] [PubMed] [Google Scholar]
- 20.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y, Li CC, Weissman AM. Regulating the p53 system through ubiquitination. Oncogene. 2004;23:2096–2106. doi: 10.1038/sj.onc.1207411. [DOI] [PubMed] [Google Scholar]
- 23.Michael D, Oren M. The p53-Mdm2 module and the ubiquitin system. Semin Cancer Biol. 2003;13:49–58. doi: 10.1016/s1044-579x(02)00099-8. [DOI] [PubMed] [Google Scholar]
- 24.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 25.Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 26.Mani A, Gelmann EP. The ubiquitin-proteasome pathway and its role in cancer. J Clin Oncol. 2005;23:4776–1789. doi: 10.1200/JCO.2005.05.081. [DOI] [PubMed] [Google Scholar]
- 27.Ghosh A, Joo NE, Chen TC, Kapila YL. Proapoptotic fibronectin fragment induces the degradation of ubiquitinated p53 via proteasomes in periodontal ligament cells. J Periodontal Res. 2010 doi: 10.1111/j.1600-0765.2009.01261.x. doi:10.1111/j.1600-0765.2009.01261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frazier MW, He X, Wang J, Gu Z, Cleveland JL, Zambetti GP. Activation of c-myc gene expression by tumor-derived p53 mutants requires a discrete C-terminal domain. Mol Cell Biol. 1998;18:3735–3743. doi: 10.1128/mcb.18.7.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonnell TJ, Montes de Oca Luna R, Cho S, Amelse LL, Chavez-Reyes A, Lozano G. Loss of one but not two mdm2 null alleles alters the tumour spectrum in p53 null mice. J Pathol. 1999;188:322–328. doi: 10.1002/(SICI)1096-9896(199907)188:3<322::AID-PATH372>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 30.Baker SJ, Markowitz S, Fearon ER, Willson JK, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 31.Chen J, Wu X, Lin J, Levine AJ. mdm-2 inhibits the G1 arrest and apoptosis functions of the p53 tumor suppressor protein. Mol Cell Biol. 1996;16:2445–2452. doi: 10.1128/mcb.16.5.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z, Li M, Wang H, Agrawal S, Zhang R. Antisense therapy targeting MDM2 oncogene in prostate cancer: Effects on prolifertion, apoptosis, multiple gene expression, and chemotherapy. Proc Natl Acad Sci USA. 2003;100:11636–11611. doi: 10.1073/pnas.1934692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akhtar S, Kole R, Juliano RL. Stability of antisense DNA oligodeoxynucleotide analogs in cellular extracts and sera. Life Sci. 1991;49:1793–1801. doi: 10.1016/0024-3205(91)90480-y. [DOI] [PubMed] [Google Scholar]
- 34.Klein C, Vassilev LT. Targeting the p53-MDM2 interaction to treat cancer. Br J Cancer. 2004;91:1415–1419. doi: 10.1038/sj.bjc.6602164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Karmmlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 36.Kumar SK, Hager E, Pettit C, Gurulingappa H, Davidson NE, Khan SR. Design, synthesis, and evaluation of novel boronic-chalcone derivatives as antitumor agents. J Med Chem. 2003;46:2813–2815. doi: 10.1021/jm030213+. [DOI] [PubMed] [Google Scholar]
- 37.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 38.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 39.Duncan SJ, Gruschow S, Williams DH, McNicholas C, Purewal R, Hajek M, Gerlitz M, Martin S, Wrigley SK, Moore M. Isolation and structure elucidation of Chlorofusin, a novel p53–MDM2 antagonist from a Fusarium sp. J Am Chem Soc. 2001;123:554–560. doi: 10.1021/ja002940p. [DOI] [PubMed] [Google Scholar]
- 40.Zhao J, Wang M, Chen J, Luo A, Wang X, Wu M, Yin D, Liu Z. The initial evaluation of non-peptidic small-molecule HDM2 inhibitors based on p53-HDM2 complex structure. Cancer Lett. 2002;183:69–77. doi: 10.1016/s0304-3835(02)00084-8. [DOI] [PubMed] [Google Scholar]
- 41.Dornan D, Wertz I, Shimizu H, Amott D, Frantz GD, Dowd P, O'Rourke K, Koeppen H, Dixit VM. The ubiquitin ligase COPI is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 42.Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, Parant JM, Lozano G, Hakem R, Benchimol S. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 43.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 44.Asher G, Tsvetkov P, Kahana C, Shaul Y. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 2005;19:316–321. doi: 10.1101/gad.319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 46.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 47.Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 48.Girnita L, Girnita A, Larsson O. Mdm2-dependent ubiquitination and degradation of the insulin-like growth factor 1 receptor. Proc Natl Acad Sci USA. 2003;100:8247–8252. doi: 10.1073/pnas.1431613100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kimura SH, Nojima H. Cyclin G1 associates with MDM2 and regulates accumulation and degradation of p53 protein. Genes Cells. 2002;7:869–880. doi: 10.1046/j.1365-2443.2002.00564.x. [DOI] [PubMed] [Google Scholar]
- 50.Nag A, Bagchi S, Raychaudhuri P. Cul4A physically associates with MDM2 and participates in the proteolysis of p53. Cancer Res. 2004;64:8152–8155. doi: 10.1158/0008-5472.CAN-04-2598. [DOI] [PubMed] [Google Scholar]
- 51.Song MS, Song SJ, Kim SY, Oh HJ, Lim DS. The tumour suppressor RASSFIA promotes MDM2 self-ubiquitination by disrupting the MDM2-DAXX-HAUSP complex. EMBO J. 2008;27:1863–1874. doi: 10.1038/emboj.2008.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, Prives C. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell. 2009;35:316–326. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 54.Asher G, Lotem J, Sachs L, Kahana C, Shaul Y. Mdm-2 and ubiquitin-independent p53 proteasomal degradation regulated by NQO1. Proc Natl Acad Sci USA. 2002;99:13125–13130. doi: 10.1073/pnas.202480499. [DOI] [PMC free article] [PubMed] [Google Scholar]