Abstract
The regulated activation of NF-κB by antigen receptor signaling is required for normal B and T lymphocyte activation during the adaptive immune response. Dysregulated NF-κB activation is associated with several types of lymphoma, including Diffuse Large B Cell Lymphoma (DLBCL). During normal antigen receptor signaling, the multidomain scaffold protein CARD11 undergoes a transition from a closed, inactive state to an open, active conformation that recruits several signaling proteins into a complex, leading to IKK kinase activation. This transition is regulated by the CARD11 Inhibitory Domain (ID), which participates in intramolecular interactions that prevent cofactor binding to CARD11 prior to signaling, but which is neutralized after receptor engagement by phosphorylation. Several oncogenic CARD11 mutations have been identified in DLBCL that enhance activity and that are mostly found in the Coiled-coil domain. However, the mechanisms by which these mutations cause CARD11 hyperactivity and spontaneous NF-κB activation are poorly understood. In this report, we provide several lines of evidence that oncogenic mutations F123I and L225LI induce CARD11 hyperactivity by disrupting autoinhibition by the CARD11 ID. These mutations disrupt ID-mediated intramolecular interactions, ID-dependent inhibition, and bypass the requirement for ID phosphorylation during T cell receptor signaling. Intriguingly, these mutations selectively enhance the apparent affinity of CARD11 for Bcl10, but not for other signaling proteins that are recruited to CARD11 in an ID-dependent manner during normal antigen-receptor signaling. Our results establish a mechanism that explains how DLBCL-associated mutations in CARD11 can initiate spontaneous, receptor-independent activation of NF-κB.
The activation of the NF-κB transcription factor by antigen receptor signaling is critical for the proliferation and activation of B and T lymphocytes in the adaptive immune response (1). NF-κB regulates many genes responsible for lymphocyte function, including cytokines, cell surface receptors, and pro-proliferative and anti-apoptotic genes. Mice and humans with mutations that prevent antigen-induced activation of NF-κB fail to mount productive immune responses (2).
Both T cell receptor (TCR) and B cell receptor (BCR) signaling pathways activate NF-κB through the IKK complex, which is composed of two kinase subunits, IKKα and IKKβ, and a regulatory subunit, IKKγ. The IKK complex phosphorylates IκB proteins that bind and stably retain NF-κB in an inactive state in the cytoplasm. Once phosphorylated, IκB is ubiquitinated and degraded by the 26S proteasome, allowing NF-κB to stably translocate to the nucleus to regulate target genes.
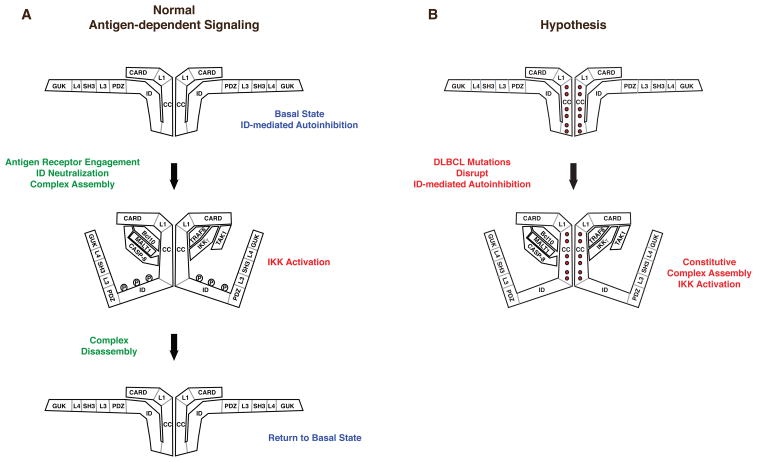
The TCR and BCR signaling pathways share many common components that function to activate the IKK complex in a signal-inducible manner. A key scaffold molecule that is required in both pathways is CARD11 (CARMA1, BIMP3), a multi-domain signaling scaffold composed of CARD, Coiled-coil, PDZ, SH3, and GUK domains (3–5). CARD11 functions in these pathways to translate upstream signaling from the BCR and TCR complexes into the dynamic assembly of a protein complex, the components of which function together to induce IKK activity (Figure 1A). CARD11 scaffold activity is inhibited in the absence of receptor engagement by an Inhibitory Domain (ID) that interacts with the CARD and Coiled-coil domains to prevent cofactor recruitment in the basal state (6). Antigen recognition results in the inducible phosphorylation of the ID mediated in part by PKCβ in B cells and PKCθ in T cells (7, 8). This phosphorylation neutralizes the inhibitory effect of the ID and causes a conformational change in CARD11 from an autoinhibited inactive scaffold to an active scaffold that allows cofactor recruitment and subsequent signaling to occur (Figure 1A). In this manner, the ID controls the inducible association of CARD11 with Bcl10, TAK1, TRAF6, Caspase-8, and IKKγ during signaling (6) (Figure 1A). Each of these protein cofactors requires the CARD or Coiled-coil domains, or both, for high affinity association with CARD11, the same domains targeted by the ID in the inactive state (6). The association of these proteins with CARD11 during signaling is highly dynamic; complex disassembly occurs shortly after assembly to return CARD11 to the basal inactive state (6). The precise roles of CARD11-associated factors in IKK kinase activation are incompletely defined, but there is evidence that the process involves the K63-linked ubiquitination of IKKγ by MALT1 (9) or TRAF6 (10), the ubiquitination of Bcl10 and its subsequent binding to IKKγ (11), the phosphorylation of IKKβ by TAK1 (10, 12), the TRAF6-mediated ubiquitination of MALT1 (13), and the proteolytic action of Caspase-8 on an unknown substrate (14).
Figure 1.
Conformational changes in CARD11 during signaling. (A) Prior to TCR or BCR engagement, CARD11 rests in an inactive, “closed” conformation promoted by the interaction of the ID with the CARD and Coiled-coil domains. Receptor engagement results in ID phosphorylation, which causes the ID to disengage from the CARD and Coiled-coil domains, converting CARD11 to an active, ”open” conformation, and allowing the depicted signaling proteins to associate with CARD11, leading to IKK activation. The signaling complex is rapidly disassembled to return CARD11 to the basal state. (B) Hypothesized action of oncogenic CARD11 mutations to spontaneously convert CARD11 into the “open” active conformation. The mutations are predicted to disrupt ID-mediated autoinhibition, thus allowing CARD11 to associate with signaling cofactors independently of receptor engagement and ID phosphorylation.
Antigen receptor signaling to NF-κB is dysregulated in several types of lymphoma, including DLBCL, the most common type of non-Hodgkin’s lymphoma. DLBCL has been categorized into three different subtypes based upon profiles of gene expression, the activated B cell-like (ABC) subtype, the germinal center B cell-like (GCB) subtype, and primary mediastinal B cell lymphoma (15). A signature feature of the ABC subtype is constitutive, antigen-independent activation of NF-κB, which is required for the dysregulated proliferation of the lymphoma. Inhibition of NF-κB activity in ABC DLBCL cell lines leads to apoptotic death in culture. An RNAi screen designed to identify molecules required for this dysregulated signaling and growth independently identified CARD11, Bcl10, and MALT1 (16). In addition, oncogenic CARD11 mutations have recently been reported in cell lines and patient samples of ABC DLBCL (17, 18). These mutations enhance the signaling activity of CARD11, and RNAi of CARD11 inhibits the growth of the cells harboring the mutations. Most of the reported oncogenic mutations in CARD11 are found in the Coiled-coil domain of CARD11, but the precise mechanisms by which these mutations confer hyperactive signaling have not been elucidated.
Based on our previous characterization of the role of the ID in regulating CARD11 activity during normal antigen receptor signaling (6), we hypothesized that the oncogenic CARD11 mutations found in DLBCL might initiate hyperactive CARD11 signaling activity by disrupting the association of the ID with the Coiled-coil domain of CARD11 that normally keeps CARD11 inactive in the basal state (Figure 1B). As a result, oncogenic CARD11 mutants would spontaneously convert into an active signaling scaffold in a manner that is independent of antigen receptor engagement and ID phosphorylation, and recruit the group of signaling proteins that are normally regulated in an ID-dependent manner, leading to IKK activation. In this report, we test several predictions of this hypothesis and provide mechanistic insight into how oncogenic CARD11 mutations found in DLBCL initiate unregulated CARD11-dependent signaling.
EXPERIMENTAL PROCEDURES
Expression Constructs
CARD11 variants F123I, L225LI, ΔID-F123I, ΔID-L225LI, S564A, F123I S564A, and L225LI S564A were constructed in pcDNA3 by Quickchange mutagenesis (Stratagene) using as substrates pcCARD11 and pcΔID (6). FLAG-tagged variants of F123I and L225LI contain a FLAG epitope at the C-terminus of murine CARD11. Although these constructs were made in the context of murine CARD11, for simplicity we have preserved the numbering scheme of Lenz et al. (17); the murine residues that correspond to F123 and L225 are actually F130 and L232 (19). The ID-GST, GST, FLAG-Bcl10, FLAG-TAK1, FLAG-IKKγ, FLAG-Caspase-8 C360S, and FLAG-TRAF6 expression constructs used in Figures 3, 6, and 7 have been described (6). The HA-MALT1 expression construct was kindly provided by Peter Lucas. In the untagged Bcl10 construct, the C-terminal FLAG epitope from FLAG-Bcl10 was replaced with the dipeptide LD. The FLAG-Bcl10 and FLAG-Bcl10 K31R, K63R expression constructs used in Figure 9 are FLAG-tagged at the N-terminus. The FLAG-Bcl10 K31R, K63R vector was constructed by Quickchange mutagenesis (Stratagene).
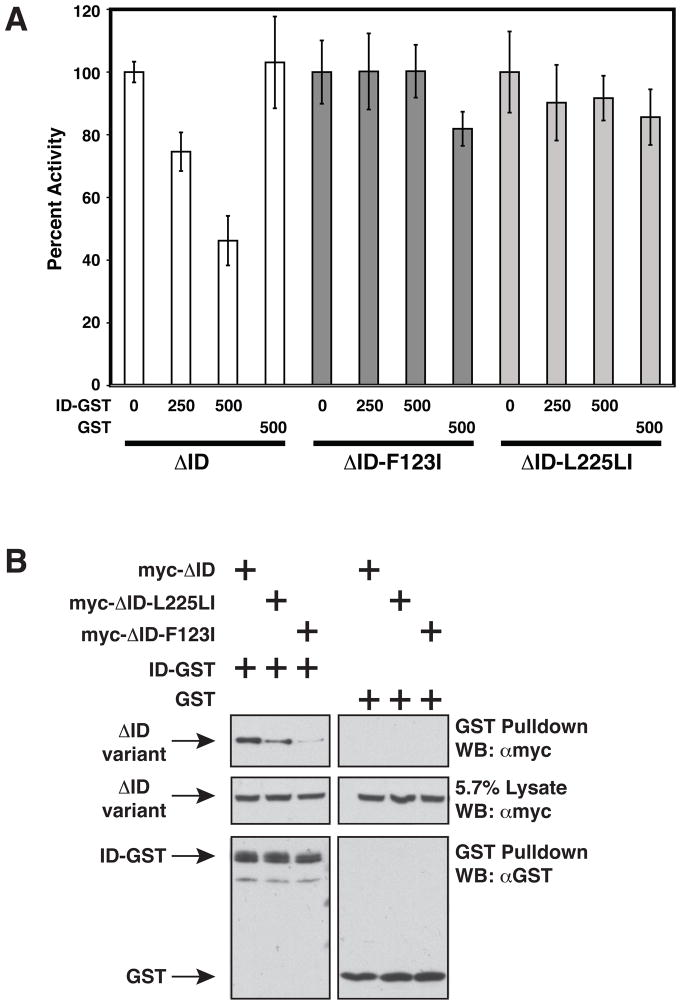
Figure 3.
F123I and L225LI oncogenic mutations confer resistance to ID inhibition and impair ID binding in trans. (A) Jurkat T cells were transfected with 200ng pCSK-LacZ and 1800ng Igκ2-IFN-LUC in the presence of expression vectors for ΔID (100 ng), ΔID-F123I (200 ng), or ΔID-L225LI (150 ng) and the indicated amounts (in ng) of expression vectors for GST (pEBG) or ID-GST (pEBB-HA-ID-GST). Fold reporter activation was normalized on a percent basis to that observed with each CARD11 variant in the absence of ID-GST or GST. (B) HEK293T cells were transfected with the following amounts in ng of expression vectors for ΔID (165–660 ng), ΔID-F123I (235–940 ng), ΔID-L225LI (125–500 ng), ID-GST(380–420 ng), or GST(200 ng), and glutathione-sepharose pulldowns were performed as described in Experimental Procedures and analyzed by western analysis with the indicated primary antibodies. WB, western blot; α, anti.
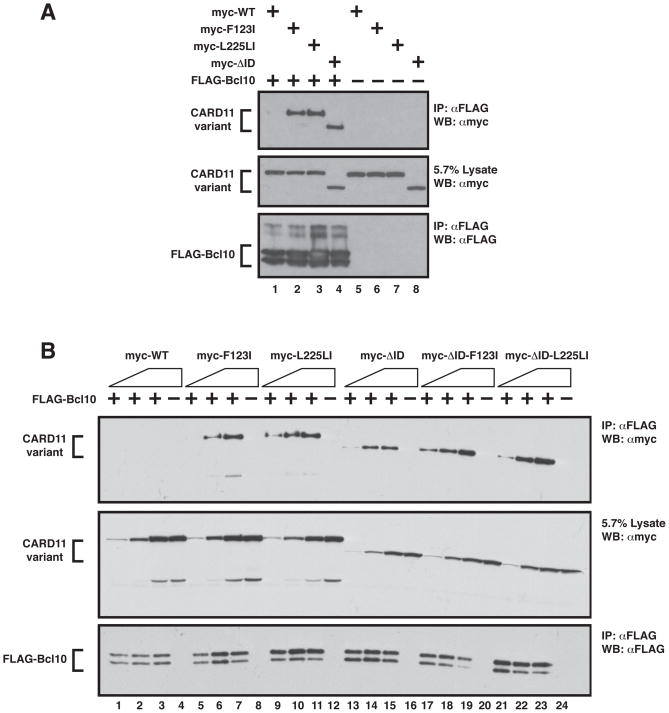
Figure 6.
F123I and L225LI mutations enhance the apparent affinity of CARD11 for Bcl10. (A) HEK293T cells were transfected with expression vectors for wild-type CARD11 (120 ng), F123I (120 ng), L225LI (120 ng), ΔID (200 ng), and FLAG-Bcl10 (100 ng), as indicated and anti-FLAG immunoprecipitations were performed as described in Experimental Procedures and analyzed by western analysis with the indicated primary antibodies. (B) Anti-FLAG immunoprecipitations were performed as in (A) using expression vectors for wild-type CARD11 (10–100 ng), F123I (15–150 ng), L225LI (10–100 ng), ΔID (10–100 ng), ΔID-F123I (14–140 ng), ΔID-L225LI (7–70 ng), and FLAG-Bcl10 (40–150 ng), as indicated. IP, Immunoprecipitate; WB, western blot; α, anti
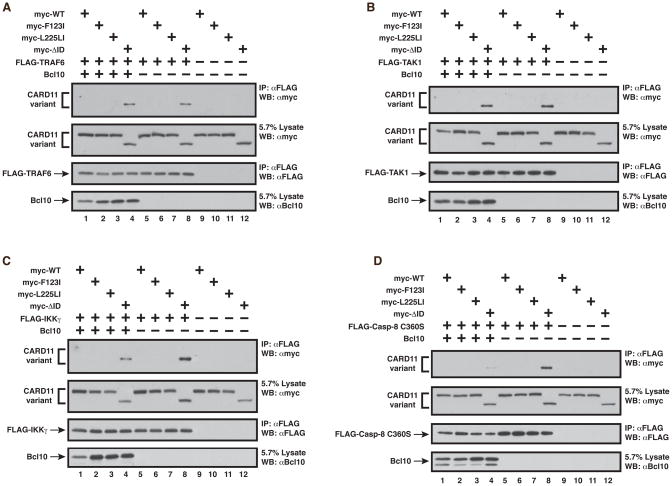
Figure 7.
The ID-mediated prevention of binding of TRAF6, TAK1, IKKγ, and Caspase-8 is intact in the F123I and L225LI oncogenic mutants. Anti-FLAG immunoprecipitations were performed as described in Experimental Procedures using the following expression vectors for each panel, as indicated: (A) wild-type CARD11 (200–400 ng), F123I (300–400 ng), L225LI (400 ng), ΔID (400 ng), FLAG-TRAF6 (350–450 ng), and Bcl10 (150–600 ng); (B) wild-type CARD11 (50–600 ng), F123I (280–600 ng), L225LI (300–600 ng), ΔID (500–850 ng), FLAG-TAK1 (100–200 ng), and Bcl10 (150–800 ng); (C) wild-type CARD11 (200–800 ng), F123I (350–800 ng), L225LI (400–800 ng), ΔID (400–800 ng), FLAG-IKKγ (1 ug), and Bcl10 (120–800 ng); (D) wild-type CARD11 (200–400 ng), F123I (350–400 ng), L225LI (400 ng), ΔID (400 ng), FLAG-Caspase-8 C360S (1 ug), and Bcl10 (150–600 ng). IP, Immunoprecipitate; WB, western blot; α, anti.
Figure 9.
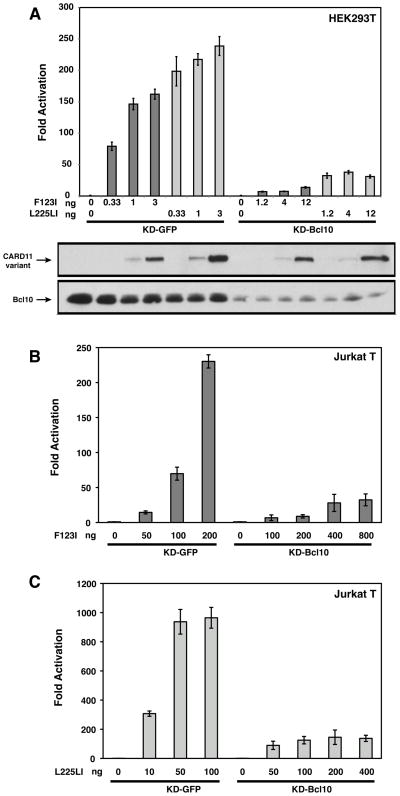
F123I and L225LI oncogenic mutants require Bcl10 for signaling to NF-κB. (A) HEK293T cells stably expressing either a hairpin that targets human Bcl10 (KD-Bcl10), or a control hairpin that targets GFP (KD-GFP), were transfected with 6 ng of pCSK-LacZ and 20 ng of Igκ2-IFN-LUC in the presence of the indicated amounts (in ng) of expression vectors for the indicated myc-tagged CARD11 variants. The lower panels display western blots of corresponding lysates probed with anti-myc and anti-Bcl10 primary antibodies to indicate relative expression levels. β-galactosidase activity, driven by pCSK-LacZ, was used to normalize luciferase activity and to calculate equivalent amounts of transfected cell-lysate for western analysis. (B, C) Jurkat T cells stably expressing either a hairpin that targets human Bcl10 (KD-Bcl10), or a control hairpin that targets GFP (KD-GFP), were transfected with 200 ng of pCSK-LacZ and 1500 ng of Igκ2-IFN-LUC in the presence of the indicated amounts (in ng) of expression vectors for myc-tagged F123I (B) or L225LI (C) CARD11 variants.
Reporter assays
HEK293T and Jurkat T cell lines were maintained and reporter assays were performed using Igκ2-IFN-LUC and Csk-LacZ as described (6). To determine relative specific activity of CARD11 variants, expression vectors were titrated into transient transfection reporter assays in HEK293T cells over the range of concentrations in which there was a linear relationship between fold Igκ2-IFN-LUC reporter activation and protein concentration. Fold reporter activation was determined as described (19) and protein concentration was determined by densitometric analysis of western blots of lysates performed under subsaturating exposure conditions using ImageJ. Relative specific activity was determined by taking the fold reporter activation elicited by a CARD11 variant at the same protein concentration at which the wild-type CARD11 elicited two-fold activation and dividing by two. The sihCARD11-2 Jurkat T cell line, and KD-GFP and KD-Bcl10 HEK293T and Jurkat T cell lines have been described (6).
Immunoprecipitations and Glutathione-sepharose pull downs
Immunoprecipitations were done as described (6) except as follows: For Figure 5, cells were lysed in 440ul lysis buffer, 8.5% was saved for analysis of the input, and the rest was used for the immunoprecipitation. For Figures 6A and 8, the cells were lysed in 500ul lysis buffer and precleared 2×30min with 7ul Protein G Sepharose 4 Fast Flow (GE Healthcare) at 4°C with rotation. From the precleared lysate, 5.7% was saved for the input and 350ul was incubated with either 1ug anti-FLAG (Sigma F7425; for Figure 6A) or anti-HA (Santa Cruz sc-805-G; for Figure 8) for 1.5 hours at 4°C with rotation followed by incubation with 7ul bed volume Protein G Sepharose previously blocked with 1% insulin (Sigma I9278). The resulting immunocomplex was washed and boiled as described before being resolved on a 10% SDS-PAGE gel (6). For the experiments in Figures 6B and 7A-D, the immunoprecipitations were completed as described above except that the resulting immunocomplexes were eluted by two successive incubations in 30ul of elution buffer containing FLAG peptide (Sigma F3290; 100ug/ml) for 30 minutes with rotation at room temperature. The two eluates were pooled and 30ul was used for subsequent western analysis. Gluthathione-sepharose pull downs were done as described above except that after the preclearing steps, 350ul of lysate was incubated with 10ul bed volume glutathione sepharose (GE Healthcare) for 2 hours at 4°C with rotation. Samples were washed and analyzed as described (6). Western blotting was completed using the following antibodies: anti-GST (Santa Cruz, sc-459), anti-myc (Santa Cruz, sc-40), anti-HA (Santa Cruz; sc7392), anti-Bcl10 (Santa Cruz, sc-5273), and anti-FLAG (M2; Eastman Kodak IB13026).
Figure 5.
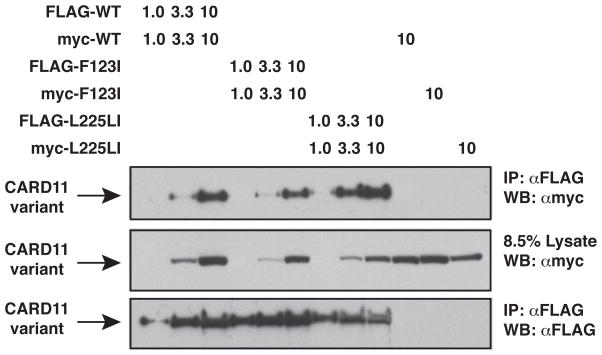
Effect of F123I and L225LI oncogenic mutations on CARD11:CARD11 association. HEK293T cells were transfected with the indicated amounts (in ng) of the indicated FLAG- and myc-tagged CARD11 variants and anti-FLAG immunoprecipitations were performed and analyzed by western blot using the indicated primary antibodies. IP, Immunoprecipitate; WB, western blot; α, anti
Figure 8.
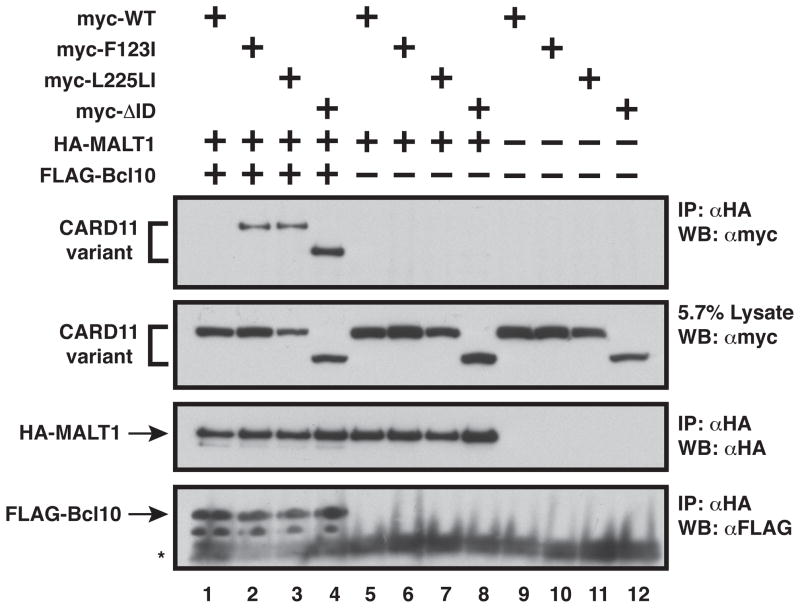
Bcl10 bound to F123I and L225LI mutants is competent to recruit MALT1 to CARD11. Anti-HA immunoprecipitations were performed as described in Experimental Procedures and analyzed by western blot with the indicated primary antibodies using the following expression vectors as indicated: wild-type CARD11 (400–600 ng), F123I (600 ng), L225LI (600 ng), ΔID (900 ng), HA-MALT1 (500–800 ng), and FLAG-Bcl10 (300–500 ng). The asterisk indicates Protein G. IP, Immunoprecipitate; WB, western blot; α, anti,
RESULTS
To test our hypothesis, we chose to characterize two of the most active CARD11 oncogenic mutants, F123I and L225LI (17). The F123I mutation is located in the L1 region of CARD11 that we have previously defined between the CARD and Coiled-coil domains (6). The L225LI mutation lies within the Coiled-coil domain.
To begin to address whether oncogenic CARD11 mutations interfere with ID function, we quantitatively determined the specific signaling activity of wild-type and mutant CARD11 variants in the absence and presence of the ID. If an oncogenic CARD11 mutation confers hyperactivity by interfering with ID function, then the effect of the mutation should be comparable to that of deleting the ID. Furthermore, the mutation should have a minimal effect in the absence of the ID.
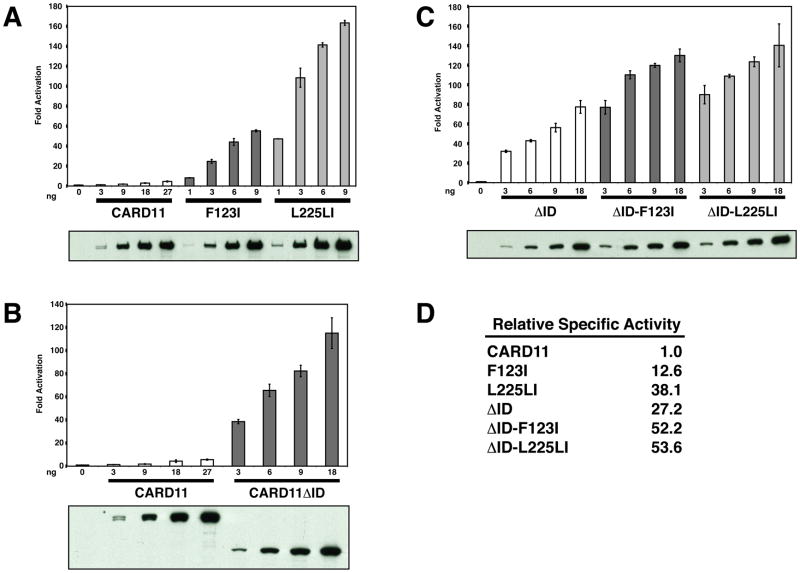
We determined the relative specific activity of wild-type CARD11 and oncogenic mutants F123I and L225LI by titrating the expression of these proteins in an NF-κB activation assay in HEK293T cells that included the NF-κB reporter, Igκ2-IFN-LUC, and the CSK-LacZ normalization control vector. While HEK293T cells do not express CARD11 endogenously, they do express all of the factors that function downstream of CARD11 and that are required for CARD11-mediated NF-κB activation. Each expression vector was titrated under subsaturating conditions, and the signaling activity and levels of protein expression were quantitatively determined as described in Experimental Procedures. As shown in Figure 2A and D, the F123I mutation increased the specific activity of CARD11 by 12.6 fold, while the L225LI mutation increased activity by 38.1 fold. These effects are comparable to that achieved by the deletion of the ID, which conferred a 27.2 fold increase in specific activity (Figure 2B and D). The F123I and L225LI mutations had a much smaller effect in the absence of the ID. The ΔID-F123I variant displayed only a 1.9 fold enhanced activity, as compared to the ΔID, while ΔID-L225LI was only 2.0 fold more active than the ΔID (Figure 2C and D). Since the oncogenic mutations had a much larger effect in the presence of the ID than in the absence of the ID, the results are consistent with the hypothesis that these mutations interfere with the autoinhibitory action of the ID.
Figure 2.
Effect of F123I and L225LI oncogenic mutations on CARD11 specific signaling activity. (A–C) HEK293T cells were transfected with 6 ng of pCSK-LacZ and 20 ng of Igκ2-IFN-LUC in the presence of the indicated amounts (in ng) of expression vectors for the indicated myc-tagged CARD11 variants. The panel below each titration displays western blots of corresponding lysates probed with anti-myc primary antibody to indicate the relative expression level of each variant. β-galactosidase activity, driven by pCSK-LacZ, was used to normalize luciferase activity and to calculate equivalent amounts of transfected cell-lysate for western analysis. (D) The relative specific activity of each variant is shown, normalized to that of wild-type CARD11, determined as described in Experimental Procedures.
We have previously demonstrated that the ID can inhibit the ΔID when expressed in trans by associating in a CARD- and Coiled-coil-dependent manner (6). We tested whether the F123I and L225LI oncogenic mutations, in the context of the ΔID, would confer resistance to the ID inhibition in trans, as predicted by our hypothesis. We titrated the expression of an ID-GST fusion protein in presence of either ΔID, ΔID-F123I, or ΔID-L225LI in Jurkat T cells, and determined that while the ΔID could readily be inhibited by the ID in trans in a dose-dependent manner, the ΔID-F123I and ΔID-L225LI variants were not inhibited by the ID in trans (Figure 3A).
To address the effects of these mutations on the association of the ID with the ΔID, we co-expressed the ID-GST with either ΔID, ΔID-F123I, or ΔID-L225LI in HEK293T cells and determined the effect of the mutations on the ability of the ID-GST to bind the ΔID in a GST-pulldown assay. Both F123I and L225LI mutations reduced the association of the ΔID with the ID (Figure 3B), consistent with the hypothesis that these oncogenic mutations disrupt the intramolecular interaction between the ID and the Coiled-coil domain.
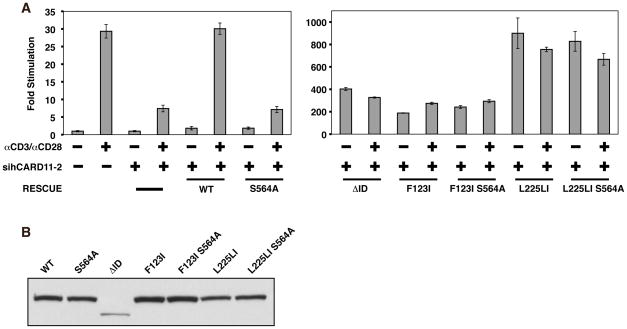
During antigen receptor signaling, the autoinhibitory effect of the ID on CARD11 scaffold activity is neutralized by the inducible phosphorylation of ID residues including S564, S577, and S657 (4, 7, 8). This phosphorylation is thought to cause the ID to disengage from the CARD and Coiled-coil domains, allowing the recruitment of signaling proteins to CARD11 into a complex that activates the IKK complex. The mutation of either S564, S577, or S657 to alanine prevents their phosphorylation and renders CARD11 activity uninducible by upstream signaling (7, 8). A prediction of our hypothesis that oncogenic CARD11 mutations disrupt ID-mediated inhibition is that these oncogenic mutations should bypass the effect of serine-to-alanine mutations that render the ID resistant to phosphorylation-mediated neutralization. To test this prediction, we assayed the activity of wild-type and mutant CARD11 variants in an RNAi-rescue assay using Jurkat T cells in which endogenous CARD11 is knocked down by the stable expression of an shRNA (sihCARD11-2) that targets the human CARD11 mRNA. As shown in Figure 4A, the stable knockdown of CARD11 reduced the activation of NF-κB by anti-CD3/anti-CD28 treatment by ~75%, as compared to a control Jurkat T cell line, as previously described (6). This effect could be rescued by the expression of wild-type murine CARD11, which is resistant to knockdown by this hairpin, but not by the S564A mutant of murine CARD11, which cannot support ID neutralization. As predicted by our hypothesis, the effect of the S564A mutation was completely bypassed by both F123I and L225LI oncogenic mutations. When expressed in the sihCARD11-2 line, the F123I S564A double mutant displayed an enhanced activity that was similar to that observed with the ΔID and the F123I single mutant, and this activity was not further enhanced by anti-CD3/anti-CD28 crosslinking (Figure 4A). The L225LI S564A double mutant also displayed an activity indistinguishable from the L225LI mutant and was also not further enhanced by TCR engagement (Figure 4A). Importantly, the S564A mutation did not affect protein expression levels in the context of wild-type, F123I, or L225LI variants (Figure 4B). These results indicate that both F123I and L225LI mutations obviate the need for the TCR-induced PKCθ-mediated ID phosphorylation and ID neutralization that is required for activation of wild-type CARD11 during signaling. The data support the hypothesis that the oncogenic mutations disrupt the inhibitory intramolecular interaction mediated by the ID.
Figure 4.
F123I and L225LI oncogenic mutations bypass the need for ID neutralization in T cells. (A) Wild-type Jurkat T cells, or Jurkat T cells stably expressing the sihCARD11-2 hairpin that targets human CARD11, were transfected with 200ng pCSK-LacZ, 1800 ng Igκ2-IFN-LUC, and expression vectors for wild-type murine CARD11 (150 ng), S564A (150 ng), ΔID (150 ng), F123I (150ng), F123I S564A (150 ng), L225LI (75 ng), or L225LI S564A (75 ng). Cells were stimulated with anti-CD3/anti-CD28 crosslinking for 4.5 hours as indicated. (B) HEK293T cells were transfected with 200ng pCSK-LacZ, 1800 ng Igκ2-IFN-LUC, and expression vectors for wild-type murine CARD11 (150 ng), S564A (150 ng), ΔID (150 ng), F123I (150ng), F123I S564A (150 ng), L225LI (75 ng), or L225LI S564A (75 ng). Lysates were probed by western blot using anti-myc primary to indicate the relative expression level of each variant. β-galactosidase activity, driven by pCSK-LacZ, was used to calculate equivalent amounts of transfected cell-lysate for western analysis. α, anti
Since it has been suggested that oncogenic mutations result in the aggregation of CARD11 (17) we tested the effect of the F123I and L225LI mutations on the ability of FLAG-CARD11 to associate with myc-CARD11 in an immunoprecipitation assay in HEK293T cells under subsaturating conditions (Figure 5). The F123I mutation had no apparent effect on the extent of CARD11 oligomerization, while the L225LI mutation displayed a modest, but reproducible three-fold increase in apparent oligomerization. Since one mutation affected oligomerization while the other did not, the data suggest that the enhancement of CARD11:CARD11 association is not an obligate part of the mechanism by which oncogenic mutations in CARD11 increase activity. However, it remains possible that the increased apparent oligomerization of the L225LI variant may contribute to the enhanced activity of that variant.
We have previously demonstrated that the ID controls the recruitment of Bcl10, TAK1, TRAF6, IKKγ, and Caspase-8 to CARD11 (6). The hyperactive ΔID variant can constitutively associate with these proteins and behaves as a CARD11 in which the ID has been neutralized by upstream signaling. An important prediction of the hypothesis that the oncogenic mutations interfere with ID function is that these mutations should lead to an enhanced ability of CARD11 to recruit signaling proteins whose association is regulated by the ID. Using an immunoprecipitation assay in HEK293T cells, we first tested whether the F123I or L225LI mutations increased the apparent affinity of CARD11 for Bcl10. We expressed FLAG-Bcl10 with myc-tagged wild-type CARD11, ΔID, the F123I mutant, or the L225LI mutant and assayed the extent to which each CARD11 variant coimmunoprecipitated with Bcl10 in an anti-FLAG IP. As shown in Figure 6A, both the F123I and L225LI mutants displayed an enhanced ability to associate with Bcl10, indistinguishable from that of the ΔID (Figure 6A cf. lanes 1–4).
We then tested whether the F123I or L225LI mutations enhance affinity for Bcl10 in the absence of the ID. Both the ΔID-F123I and ΔID-L225LI variants did display a slightly higher apparent affinity for Bcl10 in the coimmunoprecipitation assay, as compared to the ΔID (Figure 6B, cf. lanes 13–24), indicating that each mutation does modestly enhance Bcl10 association in an ID-independent manner. However, the effect of these mutations was much larger in the presence of the ID (Figure 6B, cf. lanes 1–12), than in the absence of the ID (Figure 6B, cf. lanes 13–24). These data support the hypothesis that the main effect of the mutations is to disrupt ID-mediated intramolecular interactions to expose a surface on CARD11 for Bcl10 binding. The minimal effects in the absence of the ID indicate that the mutations do not dramatically increase the affinity of CARD11 for Bcl10 at the CARD11:Bcl10 protein-protein interface.
We next tested whether the F123I or L225LI mutants displayed enhanced apparent affinities for other protein cofactors that we have previously shown to associate with CARD11 in an ID-regulated manner. Surprisingly, and in contrast to the predictions of our hypothesis, neither F123I nor L225LI mutants displayed enhanced associations with any of the other FLAG-tagged cofactors tested, including TRAF6 (Figure 7A cf. lanes 5–8), TAK1 (Figure 7B cf. lanes 5–8), IKKγ (Figure 7C cf. lanes 5–8), and Caspase-8 (Figure 7D cf. lanes 5–8). These results indicate that the oncogenic mutations F123I and L225LI selectively disrupt ID function. While the mutations disrupt the ability of the ID to prevent Bcl10 association with CARD11, they do not appear to affect the inhibitory action of the ID on the association of other signaling proteins.
To test whether Bcl10 binding to the F123I or L225LI mutants could influence the recruitment of other cofactors, we also coexpressed FLAG-tagged TRAF6, IKKγ, Caspase-8, or TAK1, with the F123I and L225LI mutants in the presence of overexpressed untagged Bcl10. Even in the presence of high concentrations of Bcl10, none of these cofactors were recruited to the F123I or L225LI mutants at concentrations at which they clearly associated with the ΔID (Figures 7A, 7B, 7C, 7D cf. lanes 1–4). The results confirm that ID-mediated inhibition of CARD11 binding to TAK1, TRAF6, Caspase-8, and IKKγ is intact in the F123I and L225LI oncogenic mutants, and that these mutants have a selective, enhanced ability to recruit Bcl10.
Since MALT1 is an obligate signaling cofactor that directly associates with Bcl10 (20–22), we tested whether Bcl10 could mediate the recruitment of MALT1 to the F123I and L225LI mutants. We coexpressed HA-tagged MALT1 with myc-tagged wild-type CARD11, the ΔID, or the F123I or L225LI mutants in the absence and presence of FLAG-tagged Bcl10 and conducted an anti-HA coimmunoprecipitation assay (Figure 8). MALT1 did not associate with any of the CARD11 variants in the absence of overexpressed Bcl10 (Figure 8, cf. lanes 5–8). In the presence of overexpressed Bcl10, MALT1 did not associate with wild-type CARD11, but did associate with the F123I and L225LI mutants to an extent that was comparable to that observed with the ΔID (Figure 8, cf. lanes 1–4). Thus, the Bcl10 that is selectively recruited to the F123I and L225LI mutants is competent to co-recruit MALT1.
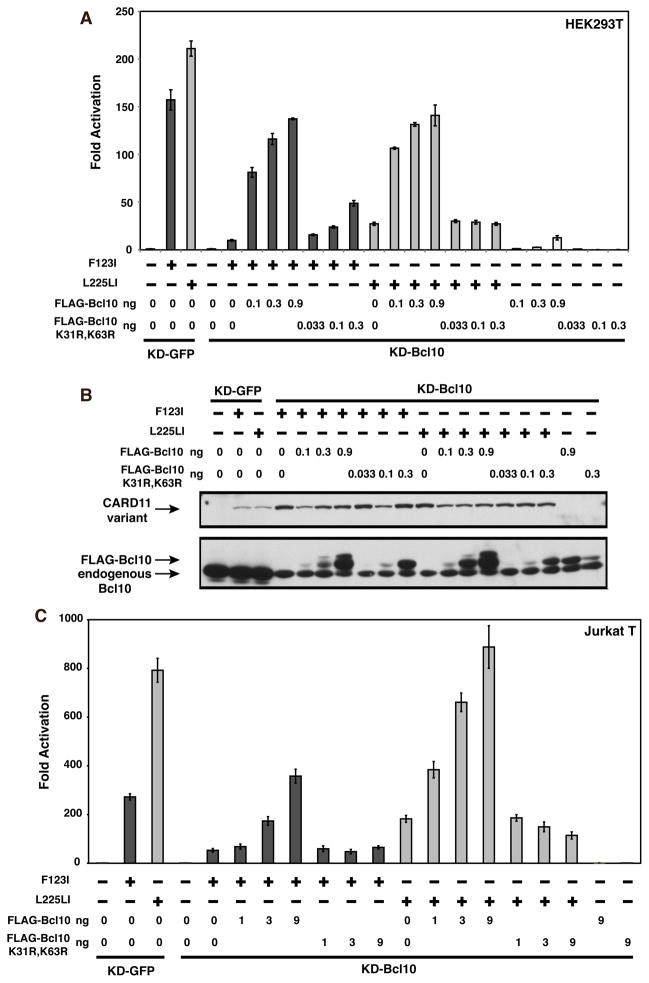
T cell receptor signaling to the IKK complex has been shown to require the ubiquitination of Bcl10 on lysines 31 and 63, which facilitates the transient association of Bcl10 with IKKγ during IKK activation (11). To test whether the F123I and L225LI CARD11 variants also depend on this modification of Bcl10, we first assessed the NF-κB-inducing activity of these mutants in an HEK293T cell line, KD-Bcl10, which stably expresses an shRNA that targets the Bcl10 mRNA and results in ~90% Bcl10 knockdown (6). We compared activities in the KD-Bcl10 line to that observed in the KD-GFP HEK293T cell line, which stably expresses an shRNA that targets GFP. As expected, Bcl10 deficiency impaired the ability of both F123I and L225LI mutants to activate the Igκ2-IFN-LUC reporter, indicating that both variants require Bcl10 for signaling activity (Figure 9A). Assessment of F123I and L225LI activities in KD-GFP and KD-Bcl10 Jurkat T cell lines revealed that Bcl10 was also required for the full activities of these oncogenic mutants in Jurkat T cells (Figures 9B and 9C). We next compared wild-type Bcl10 with the K31R, K63R double mutant for the potential to rescue signaling in the KD-Bcl10 HEK293T and Jurkat T cell lines. The Bcl10 K31R, K63R mutant has previously been shown to be functionally deficient in TCR signaling (11). At comparable levels of expression, the K31R, K63R double mutant was less able than wild-type Bcl10 to rescue signaling downstream of either the F123I or L225LI CARD11 variants in either HEK293T cells (Figure 10A and B) or Jurkat T cells (Figure 10C). These data suggest that both of these oncogenic mutants depend upon the same ubiquitination of Bcl10 that occurs during physiological TCR signaling.
Figure 10.
Bcl10 residues K31 and K63 are required for maximal signaling by F123I and L225LI oncogenic mutants. (A) KD-GFP or KD-Bcl10 HEK293T cell lines were transfected as in Figure 9A with 2–8 ng of myc-F123I or 1–8 ng of myc-L225LI, and the indicated amounts of expression vectors for either FLAG-Bcl10 or the FLAG-Bcl10 K31R, K63R mutant. (B) The corresponding lysates from (A) were analyzed by western blot and probed with anti-myc or anti-Bcl10 primary antibodies to indicate relative expression level. β-galactosidase activity, driven by pCSK-LacZ, was used to normalize luciferase activity and to calculate equivalent amounts of transfected cell-lysate for western analysis. (C) KD-GFP or KD-Bcl10 Jurkat T cell lines were transfected as in Figure 9B with 200 ng (for KD-GFP) or 800 ng (for KD-Bcl10) of myc-F123I or 50 ng (for KD-GFP) or 200 ng (for KD-Bcl10) of myc-L225LI, and the indicated amount (in ng) of either FLAG-Bcl10 or FLAG-Bcl10 K31R, K63R.
DISCUSSION
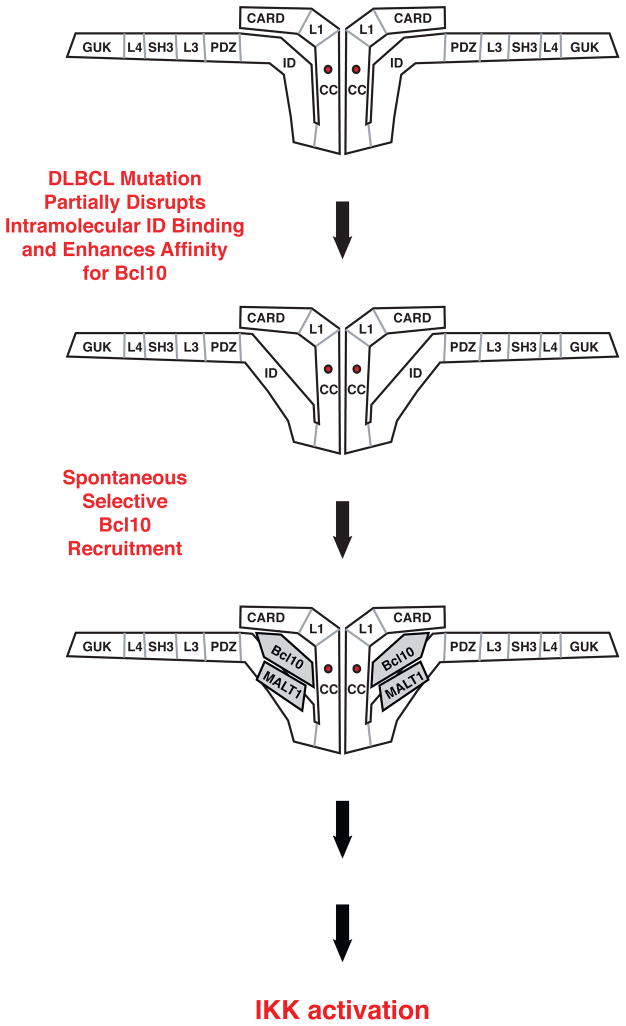
The presence of CARD11 mutations that confer hyperactive signaling accounts for the dysregulated activation of NF-κB in a number of DLBCL cases, but the mechanism by which these mutations initiate aberrant signaling have been largely undefined. We present five pieces of evidence to support the hypothesis that oncogenic CARD11 mutations induce hyperactivity by disrupting the autoinhibition mediated by the ID that normally keeps CARD11 inactive in the absence of antigen receptor engagement (Figure 11). First, the F123I and L225LI mutations increase specific activity to an extent comparable to that achieved by deletion of the ID, and have a much greater effect in the presence of the ID than in the absence. Second, the F123I and L225LI mutations impede the binding of the ID to the ΔID in trans. Third, in the context of the ΔID, these mutations confer resistance to the inhibitory effect of the ID on NF-κB activation. Fourth, in T cells, the mutations bypass the requirement for the phosphorylation of serine 564 that normally serves to neutralize ID inhibitory activity during signaling. Fifth, the mutations allow the recruitment of Bcl10 to CARD11, which is normally prevented by the ID prior to antigen receptor engagement. Our results provide a satisfying mechanistic explanation for how the emergence of the F123I and L225LI mutations in human DLBCL tumors can lead to spontaneous, receptor-independent activation of NF-κB, and the subsequent induction of NF-κB-dependent targets that promote tumor growth and survival.
Figure 11.
Model of the mechanism by which oncogenic mutations initiate dysregulated signaling to NF-κB in Diffuse Large B Cell Lymphoma.
Surprisingly, the F123I and L225LI mutations selectively affect Bcl10 binding and do not promote the association of other cofactors whose association with CARD11 is regulated by the ID, including TAK1, TRAF6, IKKγ, or Caspase-8. In the context of the F123I and L225LI mutants, most intramolecular interactions involving the ID must therefore be intact, and the mutations likely disrupt only those ID-mediated interactions that prevent spontaneous Bcl10 binding. Intriguingly, since the hyperactive F123I and L225LI mutants only appear capable of spontaneously recruiting Bcl10, the data suggest that the enhanced selective recruitment of Bcl10 is sufficient to initiate signaling to the IKK complex and NF-κB activation (Figure 11). Consistent with this, we observed a clear correlation among the CARD11 variants analyzed between their relative abilities to associate with Bcl10 (Figure 6B) and their specific activities in the NF-κB reporter assay (Figure 2). The F123I and L225LI mutations each enhanced reporter activity and Bcl10 association to an extent similar to that resulting from ID deletion, and in the context of the ΔID, each mutation only had a mild enhancing effect on NF-κB activation and Bcl10 binding.
How does spontaneous, selective Bcl10 recruitment to CARD11 oncogenic mutants initiate signaling? Our data indicate that Bcl10 can recruit MALT1 to each F123I and L225LI mutant, but not TAK1, TRAF6, Caspase-8, or IKKγ. This suggests that the Bcl10 or Bcl10/MALT complex that is recruited to the oncogenic variant is somehow “activated” in a manner that depends upon its association with CARD11 and that once activated, may dissociate from CARD11 and act upon other signaling proteins in a subsequent step that leads to IKK activation. In this scenario, the actions of the other signaling proteins in the pathway occur off of CARD11, with the IKK complex likely activated when unbound to CARD11. Alternatively, it is possible that Bcl10 co-recruits a protein to CARD11 that we have not examined, or that is unknown, but that may bridge an interaction between Bcl10/MALT1 and the examined cofactors in a CARD11-nucleated complex. We have so far not been able to demonstrate stable association between either F123I or L225LI mutant and endogenous Bcl10 during reporter activation (data not shown), consistent with the notion that the induced Bcl10:mutant CARD11 association is transient during oncogenic signaling. A similar transient association occurs between wild-type CARD11 and Bcl10 during physiological antigen receptor signaling (Figure 1A), as receptor engagement triggers first the assembly of a multiprotein complex on CARD11 followed by its rapid disassembly (6).
Since both F123I and L225LI mutants optimally depend on the K31 and K63 residues of Bcl10, it is highly likely that IKK activation by the oncogenic mutants involves the ubiquitination of these Bcl10 residues and the subsequent transient association of ubiquitinated Bcl10 with IKKγ, as has been demonstrated to occur during TCR signaling. The E3 ligase responsible for Bcl10 ubiquitination has not been identified, but since Bcl10 ubiquitination requires CARD11 (11), it is likely that Bcl10 recruitment to CARD11 facilitates the action of this putative ligase on Bcl10. Agents that target the Bcl10:CARD11 association, or Bcl10 ubiquitination may emerge as attractive therapeutics for the treatment of DLBCL cases that display dysregulated NF-κB activation as a result of oncogenic mutations that disrupt the autoinhibitory action of the CARD11 ID.
Acknowledgments
We thank T. Schaffer and Z. Wang for technical assistance; P. Lucas for the HA-MALT1 construct; W. Chan, R. Jattani, and d. Mackie for critical reading of the manuscript; and M. Meffert and S. Desiderio for helpful discussions and advice.
Abbreviations
- DLBCL
Diffuse Large B Cell Lymphoma
- ID
inhibitory domain
- TCR
T cell receptor
- BCR
B cell receptor
- ABC
activated B cell-like
- GCB
germinal center B cell-like
Footnotes
This work was supported by RO1AI078980 and PO1AI072677 from the NIH, RSG-06-172-01-LIB from the American Cancer Society, and funds from the Johns Hopkins University Institute for Cell Engineering. R.L. is supported by Ruth L. Kirschstein National Research Service Award F31AG031689. S.M.L. is supported by a Dr. Richard and Mavis Fowler and The Foundation for Advanced Research in the Medical Sciences, Inc. (FARMS) Fellowship. J.L.P is a recipient of a Kimmel Scholar Award from the Sidney Kimmel Foundation for Cancer Research and a Rita Allen Foundation Scholar.
References
- 1.Schulze-Luehrmann J, Ghosh S. Antigen-receptor signaling to nuclear factor kappa B. Immunity. 2006;25:701–715. doi: 10.1016/j.immuni.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 2.Gerondakis S, Siebenlist U. Roles of the NF-kappaB pathway in lymphocyte development and function. Cold Spring Harb Perspect Biol. 2:a000182. doi: 10.1101/cshperspect.a000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blonska M, Lin X. CARMA1-mediated NF-kappaB and JNK activation in lymphocytes. Immunol Rev. 2009;228:199–211. doi: 10.1111/j.1600-065X.2008.00749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thome M, Weil R. Post-translational modifications regulate distinct functions of CARMA1 and BCL10. Trends Immunol. 2007;28:281–288. doi: 10.1016/j.it.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Rawlings DJ, Sommer K, Moreno-Garcia ME. The CARMA1 signalosome links the signalling machinery of adaptive and innate immunity in lymphocytes. Nat Rev Immunol. 2006;6:799–812. doi: 10.1038/nri1944. [DOI] [PubMed] [Google Scholar]
- 6.McCully RR, Pomerantz JL. The protein kinase C-responsive inhibitory domain of CARD11 functions in NF-kappaB activation to regulate the association of multiple signaling cofactors that differentially depend on Bcl10 and MALT1 for association. Molecular and cellular biology. 2008;28:5668–5686. doi: 10.1128/MCB.00418-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsumoto R, Wang D, Blonska M, Li H, Kobayashi M, Pappu B, Chen Y, Lin X. Phosphorylation of CARMA1 Plays a Critical Role in T Cell Receptor-Mediated NF-kappaB Activation. Immunity. 2005;23:575–585. doi: 10.1016/j.immuni.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Sommer K, Guo B, Pomerantz JL, Bandaranayake AD, Moreno-Garcia ME, Ovechkina YL, Rawlings DJ. Phosphorylation of the CARMA1 Linker Controls NF-kappaB Activation. Immunity. 2005;23:561–574. doi: 10.1016/j.immuni.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 9.Zhou H, Wertz I, O’Rourke K, Ultsch M, Seshagiri S, Eby M, Xiao W, Dixit VM. Bcl10 activates the NF-kappaB pathway through ubiquitination of NEMO. Nature. 2004;427:167–171. doi: 10.1038/nature02273. [DOI] [PubMed] [Google Scholar]
- 10.Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol Cell. 2004;14:289–301. doi: 10.1016/s1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- 11.Wu CJ, Ashwell JD. NEMO recognition of ubiquitinated Bcl10 is required for T cell receptor-mediated NF-kappaB activation. Proc Natl Acad Sci U S A. 2008;105:3023–3028. doi: 10.1073/pnas.0712313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shambharkar PB, Blonska M, Pappu BP, Li H, You Y, Sakurai H, Darnay BG, Hara H, Penninger J, Lin X. Phosphorylation and ubiquitination of the IkappaB kinase complex by two distinct signaling pathways. Embo J. 2007;26:1794–1805. doi: 10.1038/sj.emboj.7601622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oeckinghaus A, Wegener E, Welteke V, Ferch U, Arslan SC, Ruland J, Scheidereit C, Krappmann D. Malt1 ubiquitination triggers NF-kappaB signaling upon T-cell activation. Embo J. 2007;26:4634–4645. doi: 10.1038/sj.emboj.7601897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su H, Bidere N, Zheng L, Cubre A, Sakai K, Dale J, Salmena L, Hakem R, Straus S, Lenardo M. Requirement for caspase-8 in NF-kappaB activation by antigen receptor. Science. 2005;307:1465–1468. doi: 10.1126/science.1104765. [DOI] [PubMed] [Google Scholar]
- 15.Rosenwald A, Staudt LM. Gene expression profiling of diffuse large B-cell lymphoma. Leuk Lymphoma. 2003;44(Suppl 3):S41–47. doi: 10.1080/10428190310001623775. [DOI] [PubMed] [Google Scholar]
- 16.Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, Staudt LM. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 17.Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, Dave SS, Zhao H, Xu W, Rosenwald A, Ott G, Muller-Hermelink HK, Gascoyne RD, Connors JM, Rimsza LM, Campo E, Jaffe ES, Delabie J, Smeland EB, Fisher RI, Chan WC, Staudt LM. Oncogenic CARD11 Mutations in Human Diffuse Large B Cell Lymphoma. Science. 2008 doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 18.Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, Bertoni F, Ponzoni M, Scandurra M, Califano A, Bhagat G, Chadburn A, Dalla-Favera R, Pasqualucci L. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pomerantz JL, Denny EM, Baltimore D. CARD11 mediates factor-specific activation of NF-kappaB by the T cell receptor complex. Embo J. 2002;21:5184–5194. doi: 10.1093/emboj/cdf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lucas PC, Yonezumi M, Inohara N, McAllister-Lucas LM, Abazeed ME, Chen FF, Yamaoka S, Seto M, Nunez G. Bcl10 and MALT1, independent targets of chromosomal translocation in malt lymphoma, cooperate in a novel NF-kappa B signaling pathway. J Biol Chem. 2001;276:19012–19019. doi: 10.1074/jbc.M009984200. [DOI] [PubMed] [Google Scholar]
- 21.Ruland J, Duncan GS, Wakeham A, Mak TW. Differential requirement for Malt1 in T and B cell antigen receptor signaling. Immunity. 2003;19:749–758. doi: 10.1016/s1074-7613(03)00293-0. [DOI] [PubMed] [Google Scholar]
- 22.Che T, You Y, Wang D, Tanner MJ, Dixit VM, Lin X. MALT1/paracaspase is a signaling component downstream of CARMA1 and mediates T cell receptor-induced NF-kappaB activation. J Biol Chem. 2004;279:15870–15876. doi: 10.1074/jbc.M310599200. [DOI] [PubMed] [Google Scholar]