Abstract
Tn916 and similar elements are very common in clinical enterococcal isolates, and are responsible for transmission of a variety of resistance determinants. It is commonly assumed that clinical strains carrying Tn916 have a single copy, although the actual number of copies in clinical isolates has never been systematically studied. We report a clinical isolate of Enterococcus faecium in which three distinct and excision-proficient copies of Tn916-like elements are present in the genome. All of the elements contain tet(M) genes, at least one of which confers resistance to tetracycline and minocycline. Two elements (Tn6085a, Tn6085b) are indistinguishable, containing an inserted 2758 bp Group II intron at the start of open reading frame Tn916ORF_06. The third (Tn6084) also contains the intron, but also has an ISEfa11 integrated upstream of tet(M). All three copies are able to excise from plasmid vectors when cloned in E. coli, and at least two of the elements can transfer to an E. faecium recipient strain. These data indicate that nearly identical Tn916-like elements encoding Tet(M)-mediated tetracycline/minocycline resistance can coexist in clinical E. faecium isolates.
Conjugative transposons form a subgroup of a larger class of mobile elements known as Integrating Conjugative Elements (ICEs)(Burrus et al., 2002). The prototype conjugative transposon is Tn916, originally described from Enterococcus faecalis DS16(Franke and Clewell, 1980). Tn916, 18 kb in length, is most commonly chromosomally-encoded. It transposes using a non-replicative lambda-like integration-excision mechanism and has the capacity to transfer to a wide range of hosts using its own conjugation genes(Clewell et al., 1995). Transfer to a recipient strain is generally accompanied by integration into the recipient chromosome, although integration into recipient plasmids has also been demonstrated (Scott et al., 1994).
Study of conjugative transfer of Tn916 in the laboratory has demonstrated multiple integrations into the recipient genome(Rice et al., 1992). The mechanism for these multiple integrations is not clear, nor is it clear whether multiple integrations occur in vivo with any frequency. Although the copy number of conjugative transposons found in specific strains has not been systematically studied in any species, most strains within which Tn916 (as well as most other conjugative elements) has been identified appear to have only a single copy of the transposon. One potential exception was reported by Spigaglia and colleagues(Spigaglia et al., 2006), who noted hybridization of two different sized tet(M)-int HincII fragments in 8 of 19 Clostridium difficile isolates examined. The authors concluded that these eight isolates each contained two different copies of Tn916-like elements, although they did not demonstrate that any of the transposons was either complete or functional.
We report an Enterococcus faecium strain that contains three variants of Tn916. All of these variants contain tet(M) genes in the typical location, along with functional excision mechanisms.
MATERIALS AND METHODS
Bacterial strains and plasmids
Bacterial strains and plasmids used in these studies are listed and described in Table 1.
Table 1.
Strains, plasmids and primers used in these studies
| Strain | Resistance (phenotype) | Origin (reference) |
|---|---|---|
| E. faecium C68 | Apr, Vmr, Tcr/Mcr | Clinical isolate |
| E. faecium TX1330 | Kanr, Rifr, Tcs | Provided by Barbara E. Murray (Nallapareddy et al., 2006); Used as a recipient in mating experiments with C68 |
|
E faecium CV621 to CV632 |
Rifr,Fusr, Tcr | transconjugants from mating of C68 with TX1330, selected for Tcr transfer |
| E. coli DH10B | ΔM15 ΔlacX74 deoR recA1 ara Δ139 Δ(ara leu)7697 galU galK λ- rpsL endA1 nupG |
Transformation-competent E. coli (Invitrogen, Carlsbad, Calif.) |
| E. coli EPI300 | F− mcrA Δ(mrr-hsdRMS- mcrBC) Φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ− rpsL nupG trfA dhfr |
Strain used for maintaining copy control of pCC1 plasmids (Epicentre Biotechnologies, Madison, Wis.) |
| Plasmid | ||
| pACYC184 | Cmr, Tcr | Cloning vector |
| PCR-XL-TOPO | Kmr |
E. coli vector for cloning PCR products directly (Invitrogen) |
| pIndigoBAC-5 | Cmr |
E. coli vector for cloning (Epicentre Biotechnologies, Madison, Wis.) |
| CopyControl pCC1(Blunt) |
Cmr |
E. coli F-factor replicon (maintains one copy per cell) with an inducible high-copy oriV origin of replication (Epicentre Biotechnologies, Madison, Wis.) |
| pCWR1216 | Kmr, Tcr | 21 kb Tn6085a-containing BgIII fragment from C68 cloned into BamHI-digested pIndigoBAC cloning vector |
| pCWR1217 | Kmr, Tcr | 23 kb Tn6084-containing BgIII fragment from C68 cloned into BamHI-digested pIndigoBAC cloning vector |
| pCWR1262 | Cmr | 3 kb HincII fragment from C68 chromosome containing the int916xis916 genes and termini from Tn6085b and junction sequence cloned into EcoRV-digested pACYC184 |
| pCWR1782 | Cmr, Tcr | 20 kb PCR amplification product containing intact Tn6085b from the C68 chromosome cloned into CopyControl pCC1 vector |
| Primer name | Primer Sequence | Purpose of Primer |
| Tn916-1 | 5’- GTTTTGACCTTGATAA AGTGTGATAAGTCC-3’ |
Used to detect circularization of Tn916 construct (Rice and Carias, 1994) |
| Tn916-17890 | 5’- CACTTCTGACAGCTAA GACATGAG-3’ |
Used to detect circularization of Tn916 construct (Rice and Carias, 1994) |
| Tn916INT_A | 5’- CGCAAAACAGAACGC TCAAAAGACC-3’ |
Tn916 integrase amplification |
| Tn916INT_B | 5’- GTGTCAAACCACCAAA CTCTGAAATACG-3’ |
Tn916 integrase amplification |
| p1262_C68 | 5'- AGATGATCGAAATCAT AATGGGATGGTAAGT- 3' |
derived from E. faecium DO to use with Tn916-1 |
| 1266_B | 5’- TCTTTTAGATACAGCT TTGAGTC-3’ |
used to amplify Tn6085b |
| 1262_C | 5’- TATTCTTCCCTCATAG CTTCC-3’ |
used to amplify Tn6085b |
| Tn916_7 | 5’- GCATAAAAATCTAGTT ATCCGC-3’ |
used to amplify Tn916 then HincII digest PCR product |
| Tn916_4449_R | 5’- GATACCATTTCCCATA GTCG-3’ |
used to amplify Tn916 then HincII digest PCR product |
| Tn916_4228 | 5’- GGCTTGAATGATGAAT ATGAG-3’ |
used to amplify Tn916 then HincII digest PCR product |
| Tn916_10083_R | 5’- GCATAACATCTTCCGC AG-3’ |
used to amplify Tn916 then HincII digest PCR product |
| Tn916_9961 | 5’- GCTCATAAGCCTATGG TAG-3’ |
used to amplify Tn916 then HincII digest PCR product |
| Tn916_14956_R | 5’- GATGTACTTCATGGCG ACG-3’ |
used to amplify Tn916 then HincII digest PCR product |
| Tn916_14865 | 5’- TGGTAGTGCTATTTAC GCTG-3’ |
used to amplify Tn916 then HincII digest PCR product |
| Tn916_17978_R | 5’- GTAAAGCCTTATTCTA TGTGC-3’ |
used to amplify Tn916 then HincII digest PCR product |
| ACJQ_149_7990 | 5’- TTA CAA ATA TGC TCT TAC GTG CT -3’ |
used with 6773_B to amplify Tn6084, distinguishing it from Tn6085 |
| 6773_B | 5’- AAT GAG CTT TAT TTG CGC AGA TG -3’ |
Within ISEfa11 used with ACJQ_149 to amplify Tn6084, distinguishing it from Tn6085 |
PCR amplification experiments
Primers used in PCR amplifications experiments are listed and described in Table 1. PCR amplification protocols were performed as previously described (Rice and Carias, 1994). Extension times were prolonged to 12 to 20 minutes when using GeneAmp XL PCR kit (Applied Biosystems) for amplification of the entire Tn6085b. Smaller amplification of segments of the cloned elements were column purified prior to digestion with HincII followed by separation on 1% agarose gels
Cloning of chromosomal fragments and sequencing
Genomic DNA was isolated from E. faecium C68 and digested with restriction enzymes as previously described (Rice and Carias, 1994). Digested fragments were then separated on 0.7% agarose gels and transferred to nylon membranes. Membranes were hybridized with digoxigenin-labelled probes according to the specifications of the manufacturer (Roche Diagnostics). After stringent washes hybridized fragments were detected using an anti-digoxigenin antibody and a chemiluminescent substrate. In subsequent gels, appropriate sized fragments were removed and cloned into an appropriate E. coli cloning vector. Colonies containing plasmids with appropriate inserts were identified by colony hybridization as described elsewhere (Dahl et al., 2000). Sequence of selected inserts was accomplished using services of Cogenics, Houston TX.
Excision experiments
All PCR primers used in these experiments are listed in Table 1. Circularization of Tn6084,Tn6085a and Tn6085b was assayed from E coli clones as previously described (Rice and Carias, 1994).
Conjugation experiments
Transferability of tetracycline resistance was tested by performing matings between E. faecium C68 and E. faecium TX1330 (fusr, rifr). Matings were performed overnight on BHI agar plates by cross-streak technique as previously described (Rice and Carias, 1998). Transconjugants were selected on BHI agar plates containing tetracycline (15 µg/ml) fusidic acid (25 µg/ml) and rifampin (100 µg/ml). Transconjugant colonies were streaked to single colonies and retested for resistance to tetracycline. The presence of Tn6084 and Tn6085A or Tn6085b in the transconjugants was confirmed by PCR using primers designed to amplify the HincII fragment containing the tet(M) determinant and additionally with primers designed to amplify from upstream of tet(M) to within the ISEfa11.
RESULTS
Identification of three copies of Tn916-like elements in E. faecium C68
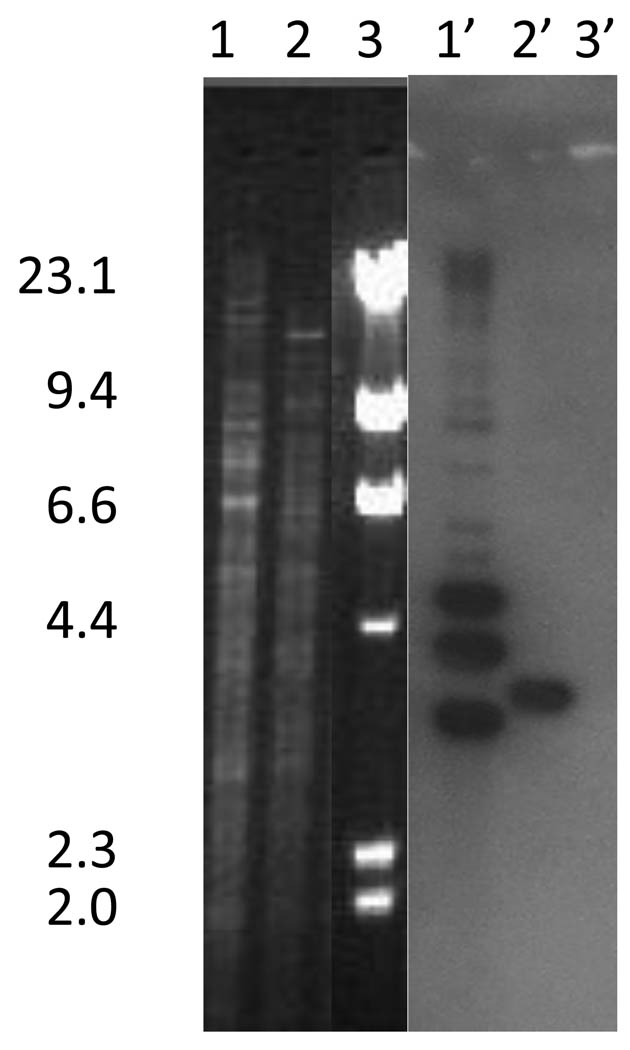
We have previously identified the Tn916-like VanB-type glycopeptide resistance transposon Tn5382 in E. faecium C68 (Carias et al., 1998). Resistance to tetracycline and minocycline in C68 suggested the presence of a tet(M) gene, which was confirmed by PCR amplification (data not shown). In an effort to determine whether the tet(M) gene was incorporated into a Tn916-like element, and if so whether Tn916 and Tn5382 were in close proximity within the C68 genome, we performed BstZI digestions of C68 genomic DNA and hybridized with internal fragments of tet(M) and the integrase (int) genes from Tn916 and Tn5382.(data not shown) To our surprise, three different bands were identified that hybridized with the Tn916 int internal fragment, suggesting the possibility that there were three copies of a Tn916-like element (in addition to Tn5382, whose int gene does not have sufficient homology with Tn916 int to hybridize) present in the genome of C68. HincII digestions of genomic DNA from C68 and E. faecium D344R (with a single copy of Tn916) hybridized with an internal fragment of int916, confirming three copies, are shown in Figure 1.
Figure 1.
Hybridizations of HincII-digested genomic DNA from enterococcal strains using an internal fragment of int916 as a probe. Lane 1: E. faecium C68 digested with HincII; Lane 2: E. faecium D344R (with a single copy of Tn916) digested with HincII; Lane 3: λ HindIII size standards – sizes in kb to the left of the figure; Lanes 1’–3’: Southern hybridization of the DNA pictured at the left using the internal fragment of int916.
Two of the resident Tn916-like elements were cloned directly into E. coli DH10B by ligating int-hybridizing BglII restriction fragments to the pIndigoBAC cloning vector. HincII digestion of these inserts suggested that they contained all of the elements of Tn916 (data not shown), although with some variations discussed below. In order to clone the final Tn916 copy in E. coli, the above int916 probe was used to identify the HincII fragment containing this end and its junction with the C68 genome (distinguishable by size from HincII fragments derived from the other copies). This 3kb HincII fragment was cloned into E. coli by ligation to cloning vector pACYC184 and the sequence of the junction region was determined. This junction sequence was used to query the publicly available E. faecium DO database and identify the target sequence within which the transposon inserted, since a Tn916-like transposon is not present at this location in E. faecium DO (Genbank NZ_ACIY00000000). This sequence was used to create an additional primer that would amplify the opposite junction when used with a primer directed outward from the nt 1 end of Tn916. Sequence analysis confirmed that this amplification product included the transposon-C68 junction. Finally, in order to confirm that this insertion represented a functional Tn916-like element, PCR primers were synthesized based on the flanking sequences and the entire transposon with flanking sequences was PCR amplified and cloned into E. coli by ligating it to cloning vector pCC1.
Sequence analysis of the three transposon variants
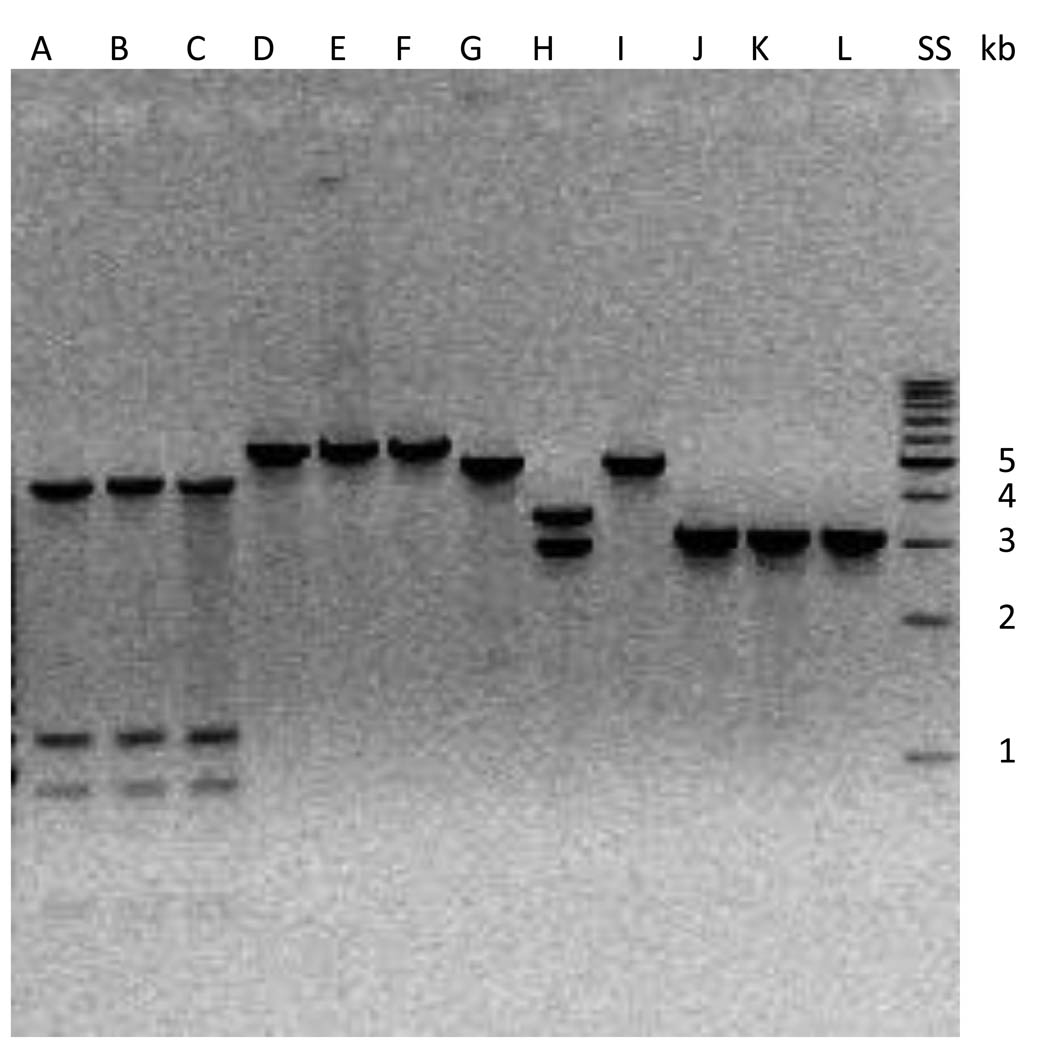
The fact that the three cloned transposons represented three separate insertions was confirmed by demonstrating distinct junction sequences for each (Figure 2). HincII digests of amplification products spanning 4 segments of each cloned transposon revealed identical sized internal restriction fragments for two of the elements, with differences observed in one of the elements (Tn6084) for restriction digest of one amplification product (Figure 3). Sequence analysis revealed that this difference was due to insertion of ISEfa11 (Genbank accession number FJ866609) 56 nucleotides upstream of tet(M) (data not shown). While it is not clear whether this insertion exerted an effect on tet(M) transcription, it should be noted that previous work has suggested that the presence of sequences further upstream of the structural tet(M) genes in Tn916-like elements enhances transcription (Hill et al., 1988). The newly available genome assembly for C68 [provided through support by the National Institute of Allergy and Infectious Diseases (NIAID), Genome Sequencing Centers] allowed us to determine the sequences of the remainder of the elements. All three of the transposons contained a 2.7 kb putative group II intron inserted at Tn916 nucleotide 3913 (Genbank accession NC_006372). This insertion occurred at the beginning of open reading frame Tn916_06 (formerly orf19), whose function remains unknown. One (Tn6084) had the above-described insertion of ISEfa11. The other two transposons were identical and therefore were given the designations Tn6085a and Tn6085b. Comparison of the remainder of the sequence with the sequence of Tn916 reveal only 7 nucleotide changes in the non-tet(M) region of the transposons. The tet(M) gene was found to be identical to the tet(M) reported in two Tn916 like elements found in human isolates of E. faecalis (18854-s-1; 20028-s-1)(Agerso et al., 2006), which differs from the Tn916 tet(M) sequence by 47 nucleotides. The sequences of the Tn6084, Tn6085a and Tn6085b and their flanking sequences can be found in Genbank under the accession numbers HM243622, HM243621 and HM43623.
Figure 2.
Termini and junction sequences of each of the three E. faecium C68 transposon insertions. Transposon nucleotides are in upper case. Junction regions (underlined) and genomic nucleotides are in lower case.
Figure 3.
HincII restriction digestions of PCR amplifications of four segments cloned into E. coli. Identical bands are seen for the three transposons for the first, second and fourth amplification products. Digestion of the third PCR product of Tn6084 yields two bands due to the insertion of ISEfa11 (Lane H). Size standard sizes are listed to the right of the figure. Lanes A, D, G, J: Tn6085a; Lanes B, E, H, K: Tn6084; Lanes C, F, I, L: Tn6085b. Lane SS: 1 kb size standard. PCR amplifications Lanes A,B,C: nt 7 TO 4449; Lanes D,E,F: nt 4228 to 10083; Lanes G,H,I: nt 9961 to 14956; Lanes J,K,L: nt 14865 to 17978.
Functional analysis of excision of the three transposons
We performed excision assays in E. coli to determine whether each element was capable of excision. The excision assay (Rice et al., 2007) measured the formation of the classical circular intermediate by amplifying the transposon termini and the “joint” region that connects the two ends when the transposon has circularized. Joint regions were readily amplified from each of the cloned transposons, indicating that each of the elements had a functional excision mechanism (data not shown).
Transferability of the Tn916-like elements
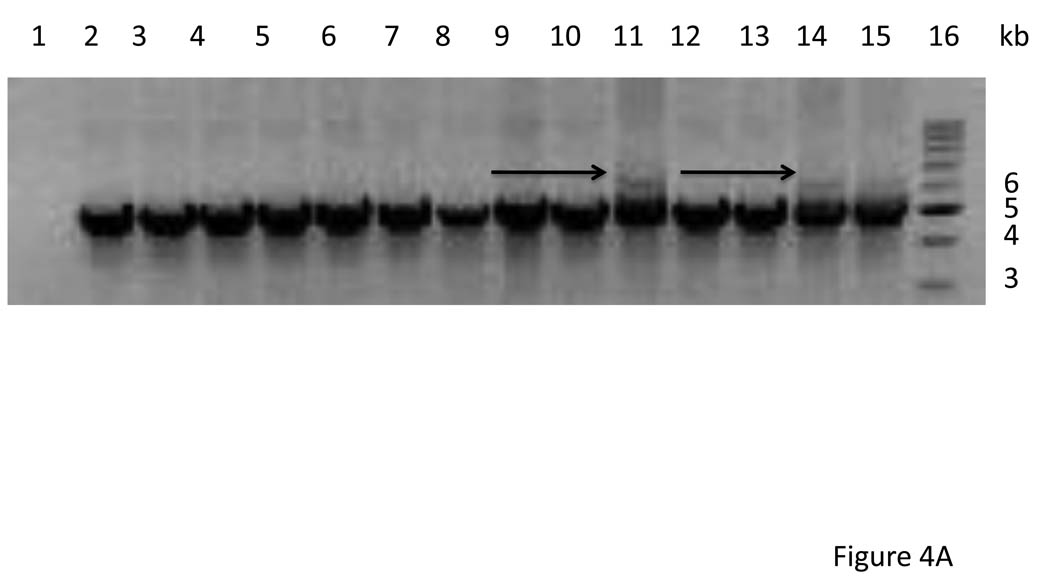
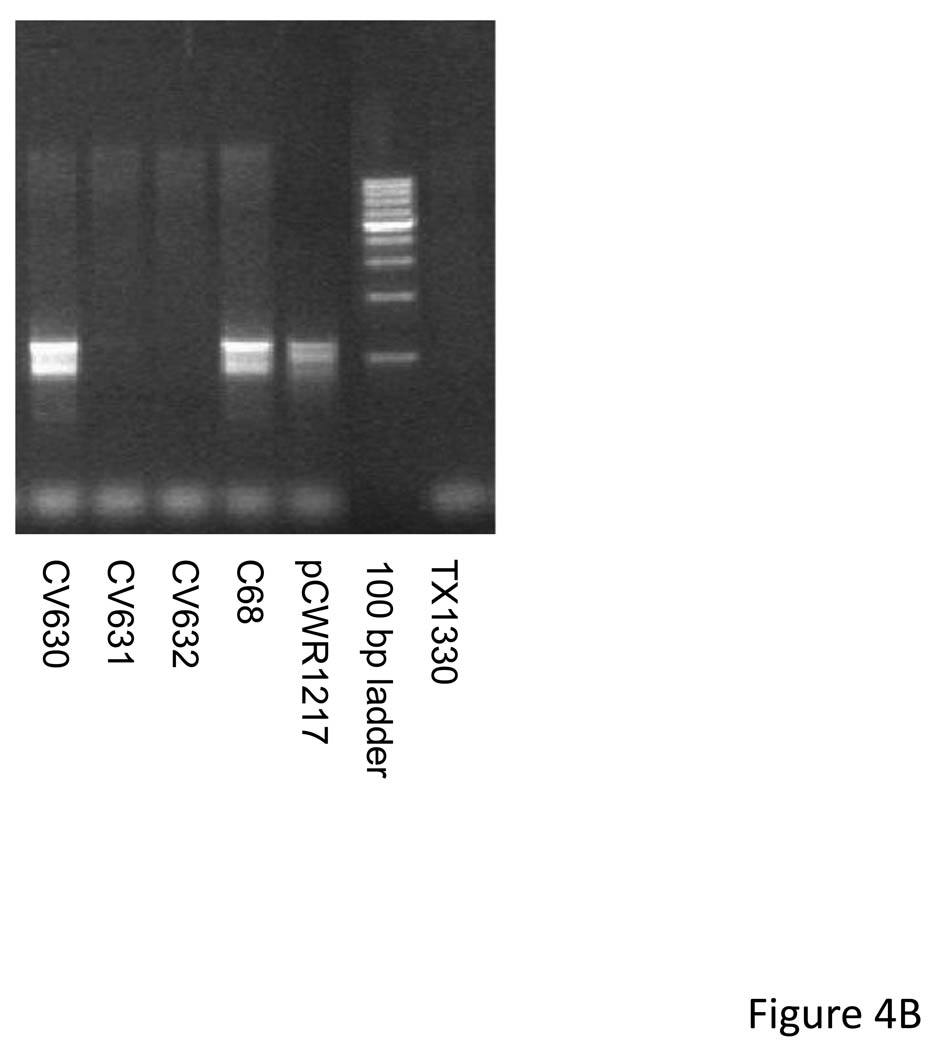
In order to determine whether any of these transposons was conjugative, we performed matings between C68 and E. faecium 1330 (kindly provided by Barbara E. Murray)(Nallapareddy et al., 2006). Matings were performed overnight on filters as previously described (Christie et al., 1987) with selection on BHI agar plates containing tetracycline (15 µg/ml), rifampin (100 µg/ml) and fusidic acid (25 µg/ml). Transconjugants were recovered at a low frequency (ca. 10−7/recipient CFU) indicating that at least one of the transposons was conjugative. Growth of the donor strain in tetracycline (15 µg/ml) increased the frequency of transfer 3-fold. PCR amplification designed to amplify the HincII fragment containing tet(M) revealed that all of the transconjugants had received a copy of the either Tn6085a or Tn6085b (Figure 4A). One transconjugant (CV63) revealed a second, fainter band that was larger in size and consistent with the expected amplification product from Tn6084, with the ISEfa11 inserted. Amplification with primers designed to amplify from upstream of tet(M) to within the ISEfa11 confirmed that Tn6084 was also present in CV630 but not in CV631 or CV632 (Figure 4B).
Figure 4.
A. PCR amplification of tet(M) region of 12 transconjugants resulting from a mating between E. faecium C68 and E. faecium TX1330. Lane 1 is the recipient strains (negative control). LANES 2–13: Transconjugants CV621-632. Lane 14: E. faecium C68 grown without tetracycline; Lane 15: C68 grown with tetracycline; Lane 16: 1 kb size standard. Sizes are marked to the right of the figure. The arrows indicate the additional band resulting from amplifying this segment in Tn6084, which has the insertion of ISEfa11 in the region. B. PCR amplification using primers ACJQ_149_7990 and 6773_B designed to amplify a segment from within ISEfa11 to a flanking region. The results confirm that transconjugant CV630 has a copy of Tn6084 while transoconjugants CV631 and CV632 do not.
Discussion
The wealth of genome sequence data that has accumulated over the past few years has provided an opportunity to better appreciate the variety and prevalence of mobile elements in the bacterial genome. However, the number of repeated IS elements in many of these genomes makes it very difficult to complete and close these sequences, so the majority of them are left as incomplete assemblies. Under these circumstances, it is not possible to determine precisely how many repeated elements exist within the genome. As such, the question of how many copies of a large transposon such as Tn916 exist within a specific genome cannot be answered easily by reference to publicly available genome sequences.
The data presented in this paper confirm that E. faecium C68 has three functional copies of nearly identical Tn916-like elements, all containing tet(M), in its genome. This would seem to be a rather inefficient arrangement if the purpose of acquiring Tn916 is to express resistance to tetracycline. The level of tetracycline resistance expressed by C68 does not differ significantly from enterococcal strains with just one copy of Tn916 (data not shown). The presence of these three elements in a single strain may confer some other selective advantage. It may be that one or more of the insertions activates or inactivates nearby genes in a manner that provides a selective advantage in an environment. Alternatively, the presence of three such elements in the genome may provide for genetic flexibility through interaction between individual elements. We have previously shown deletion of a large E. faecium genome segment that occurred through an interaction between non-identical but related Tn916-like elements (Rice et al., 2005). It is reasonable to presume that the rates of such interactions between identical elements would occur at an even higher frequency than among non-identical elements and that this flexibility could be advantageous in the challenging environment of the mammalian gastrointestinal tract. The presence of multiple elements may also increase the frequency of excision and circularization of each element (as has been shown in E. faecalis)(Flannagan and Clewell, 1991) which would then increase the frequency of genetic exchange through read-through transcription of the conjugation genes which are downstream of the integrase and excisionase only in the circular form (Celli and Trieu-Cuot, 1998). Such increased genetic exchange could also be advantageous in a challenging environment.
We can only speculate about the mechanism by which E. faecium acquired three copies of Tn916-like element. As noted above, transfer of Tn916 in the laboratory is frequently associated with the appearance of multiple copies in the transconjugant genome (Scott et al., 1994). It is conceivable that the three copies all arrived during a single mating event, with Tn6084 subsequently acquiring the ISEfa11 from elsewhere in the genome (there are one or two additional copies of ISEfa11 in the E. faecium C68 genome, based on analysis of individual trace files of the whole genome sequence), or that Tn6085a and Tn6085b arrived in one mating event, with the subsequent arrival of the ISEfa11-containing transposon during a different conjugation. Previous work in E. faecalis suggested that the presence of Tn916 in a genome did not confer immunity to subsequent acquisitions of similar elements (Norgren and Scott, 1991). It seems unlikely that the number was reached by the arrival of a single transposon with subsequent duplication, as Tn916 transposes via a conservative mechanism. However, recent work by Lee, et al (Lee et al.) indicates that some ICE elements may replicate after excision. If, contrary to current beliefs, Tn916 replicates after excision, then the multiplication of insertions could occur. The complete discordance of the junction sequences of the three elements would argue against one element being the immediate progenitor of the other or that the three copies resulted from a mating event from a donor with a single copy.
E. faecium C68 is a member of the highly effective clonal complex 17, a group of ampicillin-resistant E. faecium strains that has spread worldwide and is responsible for the majority of vancomycin-resistant E. faecium outbreaks around the world (Top et al., 2008). C68 itself is a representative of the only vancomycin-resistant E. faecium clone found in all 11 Cleveland hospitals studied in the 1990’s, suggesting it is well-adapted to survival in the hospital (Donskey et al., 1999). As noted above, C68 also contains a fourth Tn916-like element, the 30 kb VanB transposon Tn5382 (Carias et al., 1998). Taken together, these data indicate that 90 kb, or roughly three percent of the 3.08 megabase C68 genome consists of Tn916-like elements, constituting a large number of duplicated genes. If this is not an accident, then the presence of these elements must confer significant advantages to these opportunistic pathogens in the clinical setting.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Agerso Y, et al. Identification of Tn5397-like and Tn916-like transposons and diversity of the tetracycline resistance gene tet(M) in enterococci from humans, pigs and poultry. J Antimicrob Chemother. 2006;57:832–839. doi: 10.1093/jac/dkl069. [DOI] [PubMed] [Google Scholar]
- Burrus V, et al. Conjugative transposons: the tip of the iceberg. Mol Microbiol. 2002;46:601–610. doi: 10.1046/j.1365-2958.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- Carias LL, et al. Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J Bacteriol. 1998;180:4426–4434. doi: 10.1128/jb.180.17.4426-4434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli J, Trieu-Cuot P. Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol Microbiol. 1998;28:103–117. doi: 10.1046/j.1365-2958.1998.00778.x. [DOI] [PubMed] [Google Scholar]
- Christie PJ, et al. Two conjugation systems associated with plasmid pCF10: identification of a conjugative transposon that transfers between Streptococcus faecalis and Bacillus subtilis. J Bacteriol. 1987;169:2529–2536. doi: 10.1128/jb.169.6.2529-2536.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell DB, et al. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 1995;3:229–236. doi: 10.1016/s0966-842x(00)88930-1. [DOI] [PubMed] [Google Scholar]
- Dahl KH, et al. Genetic linkage of the vanB2 gene cluster to Tn5382 in vancomycin-resistant enterococci and characterization of two novel insertion sequences. Microbiol. 2000;146:1469–1479. doi: 10.1099/00221287-146-6-1469. [DOI] [PubMed] [Google Scholar]
- Donskey CJ, et al. A polyclonal outbreak of predominantly VanB vancomycin-resistant enterococci in northeast Ohio. Northeast Ohio Vancomycin-Resistant Enterococcus Surveillance Program. Clin Infect Dis. 1999;29:573–579. doi: 10.1086/598636. [DOI] [PubMed] [Google Scholar]
- Flannagan SE, Clewell DB. Conjugative transfer of Tn916 in Enterococcus faecalis: trans activation of homologous transposons. J Bacteriol. 1991;173:7136–7141. doi: 10.1128/jb.173.22.7136-7141.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, Clewell DB. Evidence for conjugal transfer of a Streptococcus faecalis transposon (Tn916) from a chromosomal site in the absence of plasmid DNA. Cold Spring Harbor Symp Quant Biol. 1980;45:77–80. doi: 10.1101/sqb.1981.045.01.014. [DOI] [PubMed] [Google Scholar]
- Hill C, et al. Cloning and characterization of the tetracycline resistance determinant of and several promoters from within the conjugative transposon Tn919. Appl Environ Microbiol. 1988;54:1230–1236. doi: 10.1128/aem.54.5.1230-1236.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CA, et al. Autonomous plasmid-like replication of a conjugative transposon. Mol Microbiol. 75:268–279. doi: 10.1111/j.1365-2958.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallapareddy SR, et al. Construction of improved temperature-sensitive and mobilizable vectors and their use for constructing mutations in the adhesin-encoding acm gene of poorly transformable clinical Enterococcus faecium strains. Appl Environ Microbiol. 2006;72:334–345. doi: 10.1128/AEM.72.1.334-345.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgren M, Scott JR. The presence of conjugative transposon Tn916 in the recipient strain does not impede transfer of a second copy of the element. J Bacteriol. 1991;173:319–324. doi: 10.1128/jb.173.1.319-324.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LB, Carias LL. Studies on excision of conjugative transposons in enterococci: evidence for joint sequences composed of strands with unequal numbers of nucleotides. Plasmid. 1994;31:312–316. doi: 10.1006/plas.1994.1034. [DOI] [PubMed] [Google Scholar]
- Rice LB, Carias LL. Transfer of Tn5385, a composite, multiresistance element from Enterococcus faecalis. J Bacteriol. 1998;180:714–721. doi: 10.1128/jb.180.3.714-721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LB, et al. Interaction of related Tn916-like transposons: analysis of excision events promoted by Tn916 and Tn5386 integrases. J Bacteriol. 2007;189:3909–3917. doi: 10.1128/JB.00859-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LB, et al. Tn5386, a novel Tn916-like mobile element in Enterococcus faecium D344R that interacts with Tn916 to yield a large genomic deletion. J Bacteriol. 2005;187:6668–6677. doi: 10.1128/JB.187.19.6668-6677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LB, et al. Tn5381, a conjugative transposon identifiable as a circular form in Enterococcus faecalis. J Bacteriol. 1992;174:7308–7315. doi: 10.1128/jb.174.22.7308-7315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JR, et al. Conjugative transposition of Tn916: preferred targets and evidence for conjugative transfer of a single strand and for a double stranded circular intermediate. Mol Microbiol. 1994;11:1099–1108. doi: 10.1111/j.1365-2958.1994.tb00386.x. [DOI] [PubMed] [Google Scholar]
- Spigaglia P, et al. New variants of the tet(M) gene in Clostridium difficile clinical isolates harbouring Tn916-like elements. J Antimicrob Chemother. 2006;57:1205–1209. doi: 10.1093/jac/dkl105. [DOI] [PubMed] [Google Scholar]
- Top J, et al. Emergence of CC17 Enterococcus faecium: from commensal to hospital-adapted pathogen. FEMS Immunol Med Microbiol. 2008;52:297–308. doi: 10.1111/j.1574-695X.2008.00383.x. [DOI] [PubMed] [Google Scholar]