Abstract
HIV-1 infection and antiretroviral therapy associate with a dyslipidemia marked by low HDL levels and increased cardiovascular disease, but it is obscure whether virion replication plays a causative role in these changes. The HIV-1 Nef protein can impair ABCA1 cholesterol efflux from macrophages, a potentially pro-atherosclerotic effect. This viral inhibition of efflux was correlated with a direct interaction between ABCA1 and Nef. Here, we defined the ABCA1 domain required for the Nef-ABCA1 protein-protein interaction and tested if this interaction mediates the ability of Nef to down-regulate ABCA1. Nef expressed in HEK293 cells strongly inhibited ABCA1 efflux and protein levels, but did not alter levels of cMIR, another transmembrane protein. Analysis of a panel of ABCA1 C-terminal mutants showed Nef binding required the ABCA1 C-terminal amino acids between 2225 and 2231. However, the binding of Nef to ABCA1 was not required for inhibition since the C-terminal ABCA1 mutants that did not bind Nef were still down-regulated by Nef. Given this discordance, the mechanism of downregulation was investigated and was found to involve the acceleration of ABCA1 protein degradation but did not to depend upon the ABCA1 PEST sequence, which mediates the calpain proteolysis of ABCA1. Furthermore, it did not associate with a Nef dependent induction of signaling through the unfolded protein response, but did have a significant dependence upon proteasomal function and could act on an ABCA1 mutant that fails to exit the endoplasmic reticulum. In summary, we show that Nef down-regulates ABCA1 function by a post-translational mechanism that stimulates ABCA1 degradation but does not require the ability of Nef to bind ABCA1.
INTRODUCTION
HIV infection is associated with pro-atherosclerotic changes in lipid metabolism and an increase in the risk of cardiovascular disease (CVD) (1-7). Whereas numerous studies have investigated the role of antiretroviral therapy (ART) in driving this dyslipidemia (8), the effect of primary HIV infection on cellular cholesterol metabolism remains poorly characterized (9). Understanding the role of infection versus treatment is important since recent clinical trials have associated treatment interruption with a higher incidence of adverse cardiovascular events (10-12). These results suggests that early and persistent inhibition of viral replication may reduce the risk of CVD in infected individuals and indicates viral replication itself may induce certain aspects of the HIV associated dyslipidemia (13). In this regard, it is noteworthy that both treatment-naïve patients and HIV infected individuals receiving ART have significantly lower levels of circulating high-density lipoprotein (HDL), a change that is predicted to increase atherosclerosis and CVD (14-19).
HIV replication can disrupt host lipid homeostasis (20) and this effect could be due to the critical role that cholesterol plays in the HIV life cycle (21). HIV assembly, budding, and infection of new target cells all depend on plasma membrane cholesterol (22-27). Depletion of virion-associated cholesterol attenuates HIV-1 fusion with the host cell membrane and cholesterol-sequestering compounds such as β-cyclodextrin permeabilize and inactivate virions, thus rendering them incompetent for cell entry (28-32). Additionally, monoclonal anti-cholesterol antibodies that remodel the plasma membrane of HIV-1 permissive human T-cells and macrophages, result in inhibition of HIV-1 infection and production in vitro (33). Moreover, HIV-1 budding from the host cell is thought to occur at lipid rafts and the cholesterol-to-phospholipid molar ratio of the viral envelope is approximately 2.5 times that of the host cell surface membranes (34). Recently, mass spectrometry was used to quantitate the lipid constituents of the HIV envelope and compare this to the host cell membrane. The composition of HIV-1 envelope lipid was similar to that of lipid rafts being enriched in cholesterol and sphingolipids, which further supports the hypothesis that HIV-1 buds from membrane microdomains (35). Thus, HIV may reprogram cellular lipid metabolism to maximize lipid rafts, the sites of virus assembly, budding and release. Supporting this notion are studies that demonstrate HIV-1 Nef, a 27-29 kDa myristoylated viral protein, alters cellular cholesterol metabolism so as to maximize virion production. In particular, Nef expression increases cholesterol biosynthesis by up-regulating CYP51, which encodes a sterol 14α-demethylase that catalyzes the demethylation of lanosterol during cholesterol biosynthesis (36). Nef also up-regulates other genes involved in de novo cholesterol biosynthesis, including HMG-CoA reductase, the rate-limiting enzyme for this process (37). Finally, Nef contains a C-terminal cholesterol recognition motif, preserved in all clades of HIV, which binds cholesterol both in vitro and in vivo (36). Since lipid raft-associated cholesterol is critical for HIV assembly and infectivity, it is thought that Nef serves as a carrier to enrich cholesterol at the sites of virion budding in the plasma membrane of infected cells (26, 38). Thus, by up-regulating its biosynthesis and binding cholesterol, Nef may ensure an adequate delivery of cholesterol to nascent virions that bud from infected cells (36).
To date, all of the pathways by which Nef is thought to maximize virion lipid content involve the synthesis of cholesterol. However, that HIV infection lowers HDL levels suggests the virus may also disrupt pathways by which cells rid themselves of cholesterol, such as by ATP-binding cassette transporter A1 (ABCA1)-dependent cholesterol efflux (39). Indeed, we have found that Nef impairs ABCA1-dependent cholesterol efflux from human macrophages, a key step in the reverse cholesterol transport process by which the body rids itself of excess cholesterol. This viral-mediated inhibition of efflux was critical for HIV-1 to form infectious particles and correlated with reduced ABCA1 protein levels and a direct interaction between ABCA1 and Nef (40).
Here we determined the domain on ABCA1 that allows it to physically interact with Nef and assessed whether this interaction mediates the ability of Nef to inhibit ABCA1 efflux function. Using a panel of ABCA1 C-terminal mutants (41), the binding of ABCA1 by Nef was found to depend upon a motif in the ABCA1 C-terminus between amino acids 2225 and 2231. Although Nef binding to the ABCA1 C-terminal mutants was ablated, Nef was still able to significantly reduce the efflux activity and protein levels of the mutant ABCA1 transporters. Given this discordance, we further investigated the mechanism by which Nef was able to reduce ABCA1 protein levels. This Nef effect was associated with an acceleration of ABCA1 protein degradation, but did not depend upon the ABCA1 PEST sequence mediated calpain degradation of the transporter (42, 43) and was not associated with activation of unfolded protein response (UPR) signaling pathways (44). In contrast, the Nef downregulation of ABCA1 was significantly dependent upon proteasomal function and repressed an ABCA1 mutant that fails to exit the endoplasmic reticulum. Thus, we conclude that Nef down-regulates ABCA1 function by a novel post-translational mechanism that stimulates ABCA1 degradation but does not require the ability of Nef to bind the ABCA1 C-terminus.
MATERIALS AND METHODS
Reagents
The following reagents were purchased from the indicated suppliers: Metafectene (Biontex); Lipofectamine LTX (Invitrogen); LXR-agonist TO-901317 (Calbiochem); [1,2- 3H]-cholesterol (PerkinElmer); human lipid free Apolipoprotein A-I (apoA-I, Biodesign); Calpeptin (BIOMOL); Cycloheximide, Lactacystin, Epoxomicin, Thapsigargin, Tunicamycin, FLAG peptide; anti-FLAG M2 affinity gel (Sigma); anti-ABCA1 rabbit polyclonal antibody (Abcam); anti-ABCA1 mouse monoclonal antibody (GenWay); anti-β-actin mouse monoclonal antibody (Millipore); anti-β-actin rabbit polyclonal antibody (Sigma); anti-CHOP mouse monoclonal antibody (ABR); anti-XBP-1 rabbit polyclonal antibody (Santa Cruz Biotechnology) and anti-HA mouse monoclonal antibody (Covance). The following reagents were obtained through the AIDS Research and Reference Reagent Program, AIDS Program, NIAID, NIH: HIV-1JR-CSF Nef monoclonal antibody from Dr. Kai Krohn and Dr. Vladimir Ovod (Ovod V, Lagerstedt A, Ranki A, Gombert FO, Spohn R, Tähtinen M, Jung G, Krohn KJE. AIDS 6: 25-34, 1992) and HIV-1 Nef polyclonal antibody from Dr. Ronald Swanstrom (Shugars DC, Smith MS, Glueck DH, Nantermet PV, Seillier-Moiseiwitsch F, Swanstrom R. J Virol 67: 4639-4650, 1993). We are grateful to Dr. Matija Peterlin (UCSF) for pHSF2Nef, pHSF2NefΔXho, SF2-NefWT, and SF2-NefG2A constructs; to Dr. Olivier Schwartz (Institut Pasteur) for AD8, AD8ΔNef viruses, and LAI-NefWT and LAI-NefG2A expression constructs and Dr. Giovanna Chimini (Centre d’Immunologie de Marseille-Luminy) for the YFP-ABCA1 construct. The FLAG-tagged ABCA1 (ABCA1WT, ABCA1Δ40 and ABCA1Δ46) constructs and the anti-ABCA1 polyclonal antibody have been previously described (41, 45). The ABCA1DDDHLK→AAAAAA and ABCA1ΔPEST mutants were generated and sequence verified as previously described (41-43, 45).
Cell culture and Transfection assays
HeLa, HEK 293, HEK 293T (ATCC) and HEK 293-EBNA-T (Gift of Dr. Brian Seed, MGH) cells were transfected using Metafectene reagent (Biontex, Munich, Germany) following the manufacturer’s protocol. In brief, the cells were seeded into 24-well poly-D-lysine-coated tissue culture plates at a density of 100,000 cells/well and 24h later were transfected with 0.5μg cDNA (in 30 μl of serum and antibiotic-free medium or PBS) with 2 μl of metafectene (in 30 μl of serum and antibiotic-free medium or PBS). RAW 264.7 murine macrophages were transfected using Lipofectamine LTX reagent (Invitrogen) following the manufacturer’s protocol. In brief, the cells were seeded into 12-well poly-D-lysine-coated tissue culture plates at a density of 100,000 cells/well and 24h later were treated with 1μM TO-901317 (LXR agonist). After 24h, the cells were transfected with 1 μg cDNA (in 120 μl Opti-MEM 1 reduced serum medium with 1 μl PLUS Reagent and 3 μl Lipofectamine LTX Reagent).
Immunoprecipitations
Immunoprecipitations were used to analyze the physical interaction of FLAG-tagged ABCA1WT and NefWT in lysates prepared from transfected HEK 293 EBNA-T cells seeded in a 6-well tissue culture plates. 24 h after co-transfection, the cells were washed with ice cold PBS and lysed in 1ml TX-100 buffer (1% Triton X-100, 140 mm NaCl, 3 mm MgCl2, 10% glycerol, 50 mm HEPES, pH 7.0 and a protease inhibitor cocktail). The cell lysates were centrifuged (10 min, 2000 X g, 4 °C), the supernatants were transferred to fresh 1.5 ml eppendorf tubes, and the ABCA1/Nef complexes were co-precipitated using the anti-FLAG M2 Affinity Gel (1 mg total cell protein lysate determined by Bradford Assay incubated with 40 μl of anti-FLAG M2 Affinity Gel overnight at 4 °C with rotation). After centrifugation, the anti-FLAG M2 Affinity Gel was washed 4 times with 1 ml TX-100 buffer and transferred to fresh tubes. The FLAG-ABCA1 complexes were eluted from the affinity gel with 30 μl of FLAG peptide (0.5 mg/ml, 1 h, 4 °C), and the resulting supernatants were separated on 4-20% SDS PAGE gradient gels and immunoblotted for the amount of precipitated Nef and ABCA1.
Cholesterol Efflux Assays
Cholesterol efflux assays were carried out as previously described (41, 45). In brief, HEK 293-EBNA-T cells were seeded into 24-well poly-D-lysine-coated tissue culture plates at a density of 100,000 cells/well and 72 h later were transfected in triplicate with empty vector or the indicated cDNAs (ABCA1WT, ABCA1Δ40, ABCA1Δ46, ABCA1DDDHLK→AAAAAA, ABCA1ΔPEST with NefAS or NefWT) using Metafectene (Biontex). In assays involving transfection of multiple cDNAs, empty vector was used to maintain an equal amount of transfected DNA. 24 h post-transfection, the cells were incubated with 0.5 μCi/mL [3H]-cholesterol in complete medium (10% FBS/DMEM) for an additional 24 h. Non-cell-associated cholesterol was removed by two washes with 1× PBS followed by a 2 h incubation in 2 mg/mL fatty acid-free BSA/DMEM at 37 °C and two additional washes in 1× PBS. The cells were further incubated in medium alone (2 mg/mL fatty acid-free BSA/DMEM) or in this medium with 10 μg/mL lipid free apoA-I for 20 h. Medium was collected from the cells and cleared of debris by a 5 min 1000× g spin. To measure total cholesterol uptake and efflux, cell layers were dissolved in 0.1 N NaOH for 1 h and the amount of radioactivity in the media and cell lysates was measured by scintillation counting. ApoA-I-dependent cholesterol efflux was expressed as the difference in the percentage of efflux [medium counts per minute/(medium + cell counts per minute) × 100] for the apoA-I-treated cells minus the percentage of efflux from the cells treated with medium alone.
Immunoblotting and Protein Degradation Assays
HEK 293-EBNA-T cells were seeded into 24-well poly-D-lysine-coated tissue culture plates at a density of 100,000 cells/well and 24 h later were transfected in triplicate with empty vector or the indicated cDNAs (ABCA1WT with NefAS or NefWT). 24 h after transfection and culture in complete media (10% FBS/DMEM), the cells were treated with 100 μg/ml cycloheximide and cell lysates were subsequently collected at 1h, 4h, 6h and 8h after cycloheximide treatment using RIPA buffer (0.1 M Tris HCl, pH 7.5, 0.15 M NaCl, 0.5% sodium deoxycholate, 1% IGEPAL CA360 and protease inhibitor cocktail). The amount of ABAC1, Nef and β-actin in the lysates was assessed by immunoblotting and enhanced chemiluminescence using a Biorad Molecular Imager ChemiDoc XRS+ System. Likewise, RAW 264.7 murine macrophages were seeded into 12-well poly-D-lysine-coated tissue culture plates at a density of 100,000 cells/well and 24h later were treated with 1 μM TO-901317 (LXR agonist). After 24h, the cells were transfected with the indicated cDNAs (NefAS or NefWT). 24 h after transfection and culture in complete media (10% FBS/DMEM), the cell were treated with 100 μg/ml cycloheximide and cell lysates were subsequently collected at 1h, 2h, 4h, 6h and 8h after cycloheximide treatment using RIPA buffer (0.1 M Tris HCl, pH 7.5, 0.15 M NaCl, 0.5% sodium deoxycholate, 1% IGEPAL CA360 and protease inhibitor cocktail). The amount of ABCA1, Nef andd β-actin in the lysates was assessed by immunoblotting and enhanced chemiluminescence using a Biorad Molecular Imager ChemiDoc XRS+ System. The ABCA1 protein half-life was estimated by logarithmic graphs the β-actin normalized ABCA1 values versus time.
ABCA1 localization by Confocal Microscopy and Ultracentrifugation
The cellular distribution of YFP- or GFP-tagged ABCA1 in 293 cells was determined using a Leica TCS SP confocal microscope and a 63X oil immersion lens. The effect of Nef expression on the cellular localization of ABCA1 was determined by co-transfection of GFP-Nef and YFP-ABCA1, or untagged Nef, GFP-ABCA1 and pDsRed2-ER. The cellular distribution of ABCA1 was further determined by ultracentrifugation of 293 cell lysates expressing ABCA1 with or without Nef derived by douncing the cells in buffer (250 mM sucrose, 10 mM HEPES, pH 7.5), and obtaining a 30,000 x g membrane pellet (30P). The pellet was then suspended in 0.450 ml of 12.5% sucrose buffer and overlain onto a discontinuous sucrose gradient (0.319ml of 20%, 0.319ml of 26%, 0.638ml of 32%, 1.275ml of 36%, 0.85ml of 40%, 0.638ml of 46% and 0.425ml of 60% sucrose in buffer). The gradient was spun at 33,000 rpm for 3hr with SW55 Beckman rotor, and fractions were collected by needle puncture from the bottom of the tubes. Fractions were diluted by 3 volumes of HEPES buffer, centrifuged for 2.5h at 100,000 x g, and membrane pellets were suspended in a constant volume of SDS buffer for analysis by immunoblotting. Fractions enriched in endoplasmic reticulum membranes were identified by the presence of the ER resident protein SPTLC endogenously expressed in these cells.
Statistical Analysis
Data from the cholesterol efflux assays and cycloheximide assays were found to have equal variance and were further compared by two-tailed Student’s t tests and ANOVA, respectively, using SigmaStat software. A value of p<0.05 was defined as statistically significant.
RESULTS
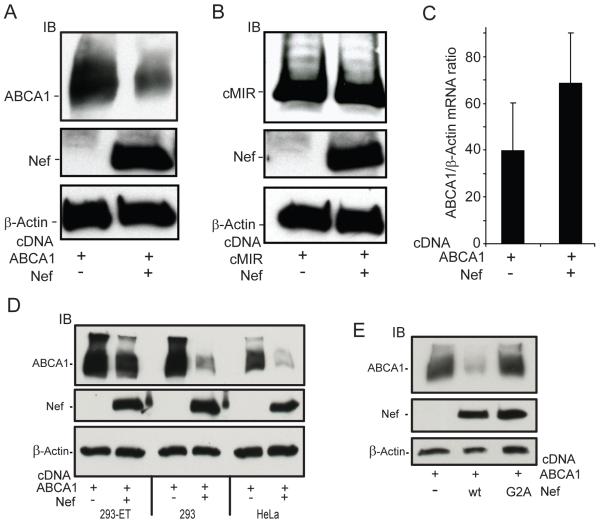
Previously, HIV-1 infection of macrophages was found to impair ABCA1 efflux activity through a process that depended upon Nef, a virus-encoded accessory protein. This effect lowered ABCA1 protein levels and was associated with a physical interaction between Nef and ABCA1 (40). Since we wanted to explore the structure-function relationship of the Nef-ABCA1 interaction and the ability of Nef to down-regulate the transporter, we first tested whether this activity could be recapitulated in a non-macrophage cell line that is readily transfected. To this end, we co-transfected cDNAs for Nef and ABCA1 into 293ET cells, which are a human embryonic kidney derived 293 cell line that also express the SV40 T and Epstein-Barr viral nuclear antigens. We favor these cells because they support robust ABCA1 expression and efflux activity when transfected with pcDNA based vectors expressing the ABCA1 cDNA driven by the cytomegalovirus (CMV) promoter. Next, protein lysates collected from the cells 24h post-transfection were assessed for ABCA1 expression by immunoblotting. We found that co-expression of Nef strongly reduced protein levels of ABCA1 (Fig. 1A). In contrast, co-transfection of the Nef cDNA with a pcDNA CMV based vector encoding another transmembrane domain protein, cMIR (46), did not result in the downregulation of the cMIR protein (Fig. 1B). This indicates that the effect of Nef on ABCA1 protein expression can be recapitulated in a non-macrophage cell line and is not due to a non-specific downregulation of transmembrane proteins by Nef.
Figure 1. The HIV-1 viral protein Nef down-regulates ABCA1 protein expression by a post-transcriptional mechanism.
A, Human embryonic kidney cells (293-ET) were transfected with an ABCA1 cDNA in the presence or absence of vectors expressing the HIV-1 Nef protein either in the anti-sense orientation (−) or in the sense orientation (+). 24 hours post-transfection, protein lysates were prepared from the cells and the amount of ABCA1, Nef and β-actin in the lysates was determined by immunoblotting (IB). B, Co-expression of Nef with cMIR, another transmembrane protein, in 293ET cells does not effect cMIR protein expression. C, Co-expression of Nef does not inhibit ABCA1 mRNA levels as determined by RT-QPCR assays indicating Nef is down-regulating ABCA1 protein levels by a post-transcriptional mechanism (n=4, ±SEM, p=0.37). The Nef mediated suppression of ABCA1 protein levels also occurs in HEK293 and HeLa cells (D), and is abrogated by the NefG2A mutation, which blocks the myristoylation and membrane association of Nef (E).
Since Nef can alter transcriptional pathways in infected cells, forcing an environment conducive to dynamic viral production, we wanted to test if Nef expression results in the suppression of ABCA1 mRNA levels (47, 48). To address this, 293ET cells were again co-transfected with ABCA1 and Nef (or a control vector containing the Nef sequence in an antisense orientation, NefAS) and quantitative RT-PCR was performed. This experiment showed that levels of ABCA1 mRNA were not being reduced by co-expression of Nef (Fig. 1C). This result confirmed our previous findings that Nef also did not reduce the level of endogenous ABCA1 mRNA in macrophages (40). Additionally, we tested whether the ability of Nef to down-regulate ABCA1 abundance was unique to the 293ET line because of SV40-T and EBNA viral antigen expression. This was not found to be the case as Nef was able to down-regulate ABCA1 protein expression in HEK 293 cells, which do not express the SV40 T and EBNA antigens (Fig. 1D). Moreover, when HeLa cells were co-transfected with the Nef and ABCA1 cDNAs, ABCA1 expression was also reduced, indicating that this effect of Nef was not restricted to the HEK 293 line (Fig.1D). Finally, we tested whether the myristoylation of Nef was important for this ABCA1 downregulation. To address this question, we tested the ability of a Nef mutant (NefG2A), which blocks the myristoylation of Nef and prevents it from associating with lipid bilayers to reduce ABCA1 expression (49). In contrast to Nefwt, the NefG2A mutant showed little or no ability to down-regulate ABCA1 in spite of being expressed at levels similar to Nefwt (Fig. 1E). In aggregate, these results indicate that Nef is able to down-regulate ABCA1 expression in the 293ET cells, and mutation of the Nef myristoylation site blocks the ability of Nef to down-regulate ABCA1.
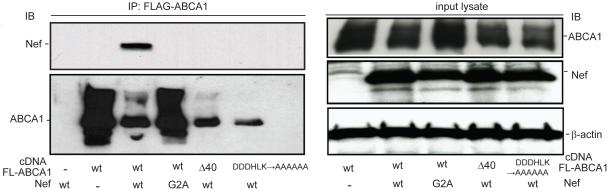
Since the downregulation of CD4 and MHC I have been shown to depend on a physical interaction between Nef and these receptors (50-52), we next tested whether the Nef-mediated downregulation of ABCA1 was acting through the reported ability of Nef to bind ABCA1. To address this, 293ET cells were co-transfected with Nef or the NefG2A myristoylation mutant and FLAG-tagged ABCA1. Our previous study showed that insertion of the FLAG epitope into the first large N-terminal extracellular loop of ABCA1 preserves the ability of the transporter to bind and transfer cholesterol and phospholipid to apoA-I (45). FLAG-ABCA1 from the transfected cells was immunoprecipitated using anti-FLAG antibody beads and after elution of FLAG-ABCA1 from the beads with free FLAG peptide, the amount of Nef that co-precipitated with ABCA1 was assessed by immunoblotting. This analysis revealed that Nefwt co-precipitated with ABCA1, whereas NefG2A did not (Fig. 2 top left panel, IP), despite similar expression levels of the NefG2A mutant in the cell lysates used for the immunoprecipitations (Fig. 2 middle right panel, input lysate). Thus, disrupting myristoylation of Nef blocked both its ability to physically interact with ABCA1 and to down-regulate protein levels of the transporter.
Figure 2. Physical interaction between Nef and ABCA1 depends upon Nef myristoylation and is disrupted by mutation of sequences between 2225 and 2231 of the ABCA1 C-terminus.
293ET cells were transfected with cDNAs that express wild type FLAG-ABCA1 (FLAG tag located in the N-terminal first extracellular loop of ABCA1) or the indicated FLAG-ABCA1 C-terminal mutants in the presence of the indicated Nef constructs (ABCA1-Δ40: a 40-AA C-terminal deletion, ABCA1-DDDHLK-AAAAAA: sequences between −31 and −36 from the C-terminus selectively mutated to alanines) (NefG2A, alanine mutation of myristoylation motif). FLAG-ABCA1 was immunoprecipitated and the amount of co-precipitated Nef and ABCA1 was determined by immunoblotting (left panels). The amount of ABCA1, Nef and β-actin expressed in the transfected cells was further determined by immunoblotting the input protein lysates (right panels).
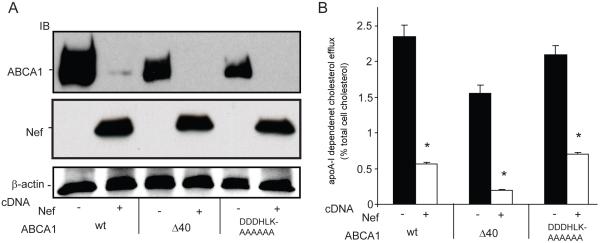
Previously, it has been shown that the Nef mediated post-translational downregulation of the mannose receptor (MMR) required the presence of a motif (SDTKDLV) in the MMR cytoplasmic tail (53). Such motifs are present in other membrane proteins susceptible to Nef-mediated downregulation including CD4, CCR5 and CXCR4. We found a similar sequence in the ABCA1 C-terminus (SDDDHLK) and tested whether Nef loses its ability to bind an ABCA1Δ40 mutant, which lacks the last 40 amino acids of the ABCA1 C-terminus including the SDDDHLK residues (41). Interestingly, the ABCA1Δ40 mutant did not co-immunoprecipitate Nef (Fig. 2, top left panel, IP) indicating that the last 40 amino acids of the ABCA1 C-terminus were required for Nef binding. To test whether mutation of just the ABCA1 residues between −30 and −40 that encompass the SDDDHLK motif disrupted the ability of ABCA1 to co-immunoprecipitate Nef, we generated an ABCA1 mutant that selectively changed these sequences to alanines (ABCA1DDDHLK→AAAAAA). Like the ABCA1Δ40 mutant, ABCA1DDDHLK→AAAAAA also failed to co-precipitate Nef, indicating that mutation of just the SDDDHLK motif in the C-terminus of ABCA1 could disrupt binding of Nef (Fig. 2, top left panel, IP). Functional assessment of the ABCA1Δ40 mutant showed that it possessed significantly less efflux activity compared to wild type ABCA1, while the ABCA1DDDHLK→AAAAAA mutant displayed activity that did not differ significantly from ABCA1 (supplemental Fig. 1). Thus, the inability of these ABCA1 mutants to bind Nef could not be ascribed to a complete failure of their ability to efflux cholesterol to apoA-I. However, probing the anti-FLAG immunoprecipitates from cells co-transfected with the FLAG-ABCA1 mutants and Nef for the amount of the mutant ABCA1 transporters showed levels that were more comparable to the amount of ABCA1wt precipitated in the presence of Nefwt than to the much greater amount of ABCA1wt that was precipitated in the presence of the NefG2A mutant that did not downregulate ABCA1 protein levels (Fig. 2, bottom left panel, IP). This result raised the possibility that in spite of not physically interacting with Nef, the ABCA1 C-terminal mutants might still have been downregulated by Nef, as was further suggested by assessing the amount of total ABCA1 expressed in these cells (Fig. 2 top right panel, input lysate). To investigate this possibility directly, additional 293ET cells were co-transfected with Nef or with control vector (NefAS) along with ABCA1WT, ABCA1Δ40 or ABCA1DDDHLK→AAAAAA. Confirming the results of the co-immunoprecipitation experiment, the expression of ABCA1Δ40 or ABCA1DDDHLK→AAAAAA was still strongly inhibited by Nef co-expression (Fig. 3A), which indicates that binding of Nef to ABCA1 was not a prerequisite for inhibiting protein expression of the ABCA1 mutants. Consistent with inhibition of mutant ABCA1 expression, cholesterol efflux to apoA-I mediated by the ABCA1Δ40 or ABCA1DDDHLK→AAAAAA mutants was also significantly inhibited by Nef, as was the activity of ABCA1wt (Fig. 3B). In sum, these results indicate that the Nef-mediated impairment of apoA-I-mediated cholesterol efflux does not have a critical dependence upon the ABCA1-Nef protein-protein interaction.
Figure 3. Disruption of the Nef/ABCA1 protein complex does not block the ability of Nef to down-regulate ABCA1 expression.
293ET cells were transfected with cDNAs that express wild type FLAG-ABCA1 or the indicated C-terminal FLAG-ABCA1 mutants as described in Figure 2. A, Cell lysates were immunoblotted for the amount of ABCA1 and Nef, which shows Nef was still able to down-regulate the expression of the ABCA1 C-terminal mutants that did not bind Nef. B, Nef also maintains the ability to suppress cholesterol efflux activity of the ABCA1 C-terminal mutants that did not bind Nef. Cells were transfected as in A, loaded with 3H-cholesterol for 24 hours, washed and incubated overnight with media alone (DMEM, 1mg/ml fatty acid free BSA) or with media and lipid free apolipoprotein A-I (apoA-I, 10 μg/ml). scintillation counting was used to quantitate media and cell associated 3H-cholesterol and the amount of apoA-I dependent cholesterol efflux was graphed (% of total cell associated cholesterol) (N=3, ±SD, *p<0.05, ABCA1 versus ABCA1 & Nef). Results are representative of two or more experiments.
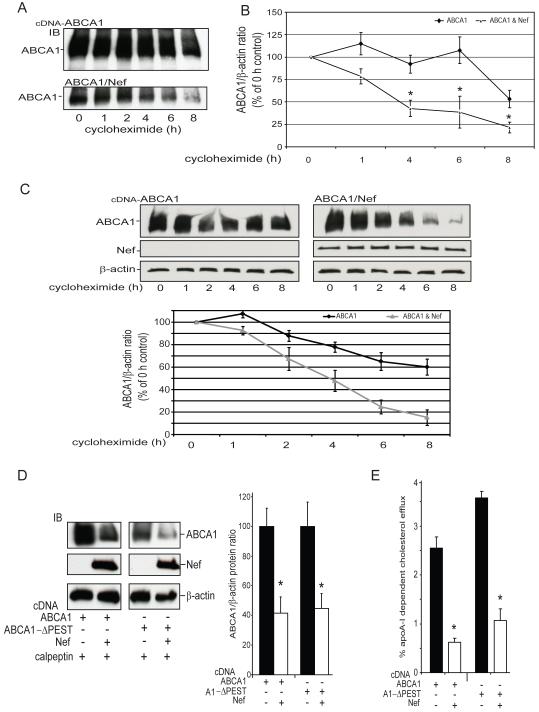
Given this discordance, we further explored the mechanism by which Nef was downregulating ABCA1 protein levels by testing whether it was due to accelerated degradation of the transporter. To address this question, 293ET cells expressing Nef and ABCA1 were treated with cycloheximide to inhibit protein synthesis and lysates of the treated cells were collected at a series of time points (2h to 8h). ABCA1 protein expression was assessed in these lysates by immunoblotting (Fig. 4A) and after probing the membranes for the level of β-actin protein, the amount of ABCA1 protein was quantified and expressed relative the level in the vehicle treated cells, thus providing an estimate of ABCA1 degradation (Fig.4B). Compared to cells expressing the control vector, degradation of ABCA1 was significantly enhanced in the cells expressing Nef, which reduced the half-life of ABCA1 protein by over 50% (ABCA1 ½ life 6.7 ± 1.5 h, ABCA1 & Nef ½ life 2.9 ± 0.4 h, p< 0.05). We further tested if Nef expression could accelerate the degradation of ABCA1 endogenously expressed in a macrophage environment. ABCA1 expression in RAW267 mouse macrophages was transcriptionally induced using the Liver-X-Receptor (LXR) agonist T0901317 (10 uM, 24h), the cells were tranfected with the antisense vector or with the wild type Nef vector and 24h later were treated with cycloheximide. Again expression of Nef was found to significantly reduce ABCA1 protein stability (Fig. 4C, ABCA1 ½ life 7.0 ± 1.0 h, ABCA1 & Nef ½ life 3.9 ± 0.4 h, p< 0.01). These results indicate that Nef was suppressing ABCA1 protein levels by a mechanism that stimulated the degradation rate of the transporter.
Figure 4. Nef down-regulates ABCA1 protein expression by a mechanism that stimulates degradation of the transporter but does not depend upon the ABCA1 PEST motif.
To determine the effect of Nef expression on the degradation of ABCA1, 293ET cells expressing ABCA1 alone or in the presence of Nef were treated with the protein synthesis inhibitor cycloheximide for the indicated time periods. A, Immunoblots are shown of ABCA1 expression in the presence and absence of Nef. B, The ABCA1β-actin protein ratio was quantitated using BioRad ECL imaging and expressed as a percentage of the ABCA1 level in cells treated with vehicle only (0 h) [n=3 (1 and 6 h), n=5 (0, 4, 8 h), ±SEM, *p<0.05, ABCA1 versus ABCA1 & Nef]. C, Nef expression in RAW264.7 macrophages also significantly accelerates the degradation of endogenously expressed ABCA1 induced by treatment with an LXR agonist (TO-901217, 1uM, 24h). Representative immunoblots of ABCA1, Nef and β-actin are shown in the top panels and the ABCA1/β-actin protein is graphed below (n=2, ±SEM, p<0.01, ANOVA) D, Nef is able to significantly suppress the expression of ABCA1 and the ABCA1ΔPEST mutant and this effect is not block by calpain protease inhibitor calpeptin (N=3, ±SD, *p<0.05, ABCA1 versus ABCA1 & Nef). E, Nef is able to significantly suppress the cholesterol efflux activity of ABCA1 and the ABCA1ΔPEST mutant (N=3, ±SD, *p<0.05, ABCA1 versus ABCA1 & Nef). Results in A-E are representative of two or more experiments.
ABCA1 contains a ‘PEST’ sequence (a motif rich in proline, glutamic acid, serine, and threonine residues) in its central intracellular cytoplasmic loop, and phosphorylation of this motif has been associated with enhancement of ABCA1 proteolytic degradation by calpain (42), whereas apoA-I binding to ABCA1 inhibits this PEST mediated degradation of the transporter (43). We thus tested whether the Nef-mediated ABCA1 downregulation required the PEST sequence of ABCA1 by generating and analyzing an ABCA1ΔPEST mutant that has previously been shown to disrupt the calpain mediated degradation of ABCA1. HEK 293ET cells were co-transfected with ABCA1ΔPEST and Nef, or with the NefAS control vector, and protein levels (Fig. 4C), and efflux activity (Fig. 4D) of the ABCA1ΔPEST mutant were assessed. As with ABCA1, Nef expression significantly repressed protein levels and efflux activity of the ABCA1ΔPEST mutant. Moreover, the ability of Nef to repress ABCA1 and the ABCA1ΔPEST mutant was not blocked by calpeptin, an inhibitor of the calpain protease that induces the PEST-mediated degradation of ABCA1. Also, since Nef accelerated the degradation of ABCA1 in the cyclohexamide assays where saturating amounts of apoA-I were provided by the added fetal bovine serum this indicates the binding of apoA-I to ABCA1 was not capable of blocking the Nef mediated down-regulation of ABCA1. In aggregate, these results indicate that Nef was increasing ABCA1 turnover by a mechanism that did not depend upon the PEST/calpain degradation pathway.
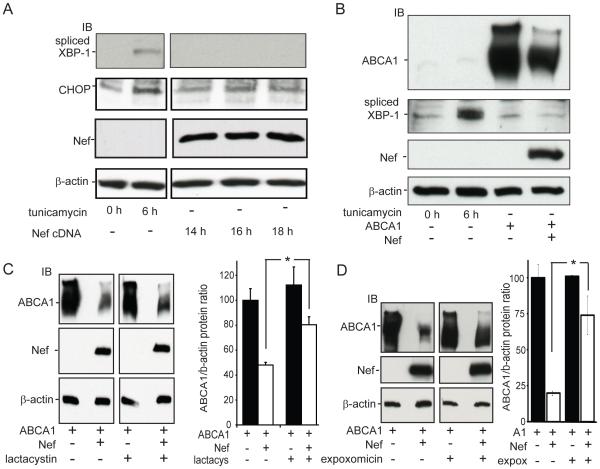
Since HIV-1 Tat, another viral accessory protein, has been reported to induce signaling through the Unfolded Protein Response (UPR) (54), a pathway that can lead to the degradation of poorly folded proteins via endoplasmic reticulum associated degradation (ERAD) (55) and another ATP-binding cassette transporter, ABCG2 is subject to ERAD (56-58), we questioned whether expression of Nef may also have the capacity to induce UPR signaling and thus explain its ability to enhance the degradation of ABCA1. To address this question, 293ET cells were transfected with Nef or with the NefAS control vector and cell lysates were collected at 14h, 16h and 18h post-transfection, a timeframe where Nef expression plateaus in this cell system. In parallel, additional cells were treated with tunicamycin, an inhibitor of protein glycosylation and a known inducer of UPR signaling. Lysates from these cells were then analyzed for activation of UPR signaling by immunoblotting for the level of spliced XBP-1 or CHOP/GADD153, two proteins whose up-regulation during the UPR mediates transcriptional and degradative responses of this pathway (44). In cells treated with tunicamycin for 6 hours, an increase in the levels of both spliced XBP-1 and CHOP was noted. However, in the cells transfected with Nef, no increase in the expression of these two UPR signaling components was detected (Fig. 5A). These results indicated that the 293ET cells were able to respond to signals that induce the UPR response, but that expression of Nef alone does not induce this signaling as monitored by changes in the levels of spliced XBP-1 and CHOP.
Figure 5. Nef inhibition of ABCA1 protein expression depends upon proteasomal activity but is not associated with signaling through the Unfolded Protein Response Pathway.
A, To test if expression of Nef alone stimulated UPR signaling 293ET cells expressing Nef for the indicate time periods or treated with tunicamycin, an inhibitor a protein glycosylation that induces UPR signaling were analyzed for the expression of spliced XBP-1 and CHOP, two factors that mediate the UPR response and whose protein levels are induced upon UPR activation. In the tunicamycin treated cells levels of both spliced XBP-1 and CHOP increased, whereas in the Nef expressing cells no such induction was apparent as determined by immunoblotting for the indicated proteins. B, Likewise in cells expressing ABCA1 alone, or with Nef, levels of spliced XBP-1 was not induced. C, Treatment of 293ET cells expressing ABCA1 and Nef with the proteasomal inhibitor lactacystin significantly blocks the ability of Nef to suppress ABCA1 protein levels (N=3, ±SD, *p<0.05, ABCA1& Nef versus ABCA1, Nef & lactacystin). D, Treatment of 293ET cells expressing ABCA1 and Nef with the proteasomal inhibitor expoxomicin significantly blocks the ability of Nef to suppress ABCA1 protein levels (N=3, ±SD, *p<0.05, ABCA1& Nef versus ABCA1, Nef & expoxomicin). Results in A-D are representative of two or more experiments.
Since ABCA1 is a large protein (2262 amino acids) with twelve transmembrane domains, it represents a complex target for chaperones of the endoplasmic reticulum to properly fold. Indeed, it has been reported that ER stress signals and UPR activation can lead to the degradation of ABCA1 (59, 60). Thus, it was possible that the combined expression of ABCA1 and Nef could induce UPR signaling, whereas expression of Nef alone did not. To address this possibility, we first verified that induction of UPR signaling leads to suppression of endogenous full length ABCA1 and the appearance of proteolytic fragments in THP-1 macrophages treated with tunicamycin (Supplemental Fig. 2A). Moreover, treatment of 293ET cells with tunicamycin also markedly suppressed the amount of ABCA1 protein after transfection of the ABCA1 cDNA (Supplemental Fig. 2B). These results indicated that 293ET cells were capable of mounting an unfolded protein response that suppresses ABCA1 expression. Thus, to test if the combined expression of ABCA1 and Nef induced UPR signaling, we co-expressed ABCA1 and Nef in 293ET cells and treated additional cells with tunicamycin in parallel, to serve as a positive control for induction of UPR signaling. Protein lysates prepared from these cells were immunoblotted for ABCA1 and Nef, which confirmed that co-expression of Nef reduced ABCA1 protein levels (Fig. 5B). Additionally, when these membranes were stripped and probed for levels of spliced XBP-1, the cells treated with tunicamycin showed increased levels of spliced XBP-1, but in the cells expressing ABCA1 alone, or in the presence of Nef, little or no spliced XBP-1 was detected (Fig. 5B). This result indicated that Nef stimulates the degradation of ABCA1 without inducing a detectable increase in the levels of spliced XBP-1, and further probing of these lysates also showed no induction of CHOP expression when Nef and ABCA1 were co-expressed.
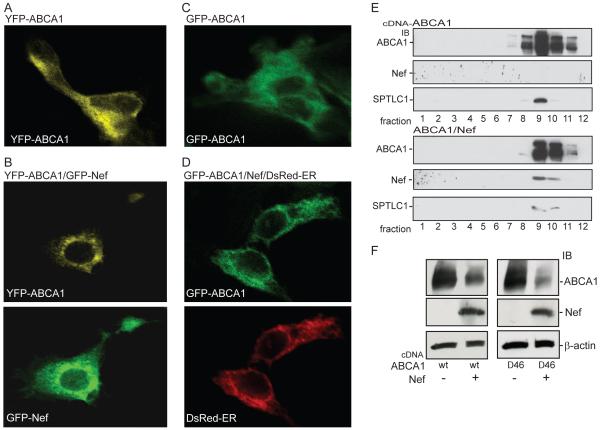
The above results indicate that the Nef mediated degradation of ABCA1 was not associated with induction of ER stress as detected by increased expression of CHOP or spliced XBP-1. However, these experiments did not rule out the possibility that Nef was triggering a degradative process that was dependent upon activity of the cytoplasmic proteasome. To test this possibility we treated cells expressing ABCA1 and Nef with the proteasomal inhibitor lactacystin (61). As expected, in vehicle treated cells, Nef significantly reduced ABCA1 protein abundance by well over 50%, whereas treatment of the cells with lactacystin significantly blocked the ability of Nef to down-regulate ABCA1 resulting in only a 25% reduction of ABCA1 expression (Fig. 5C). Likewise, in the presence of the expoxomicin proteasomal inhibitor the effect of Nef on ABCA1 protein expression was again significantly blocked by 75% (Fig. 5D). Thus, the mechanism by which Nef was suppressing ABCA1 function depends, in part, on a proteasomal degradation process. Typically, the proteasomal turnover of integral membrane proteins occurs through ERAD, wherein the targeted protein is dislocated from the ER membrane and ubiquitinated before being degraded by the proteasome (55). To begin to address whether Nef may be triggering the degradation of ABCA1 through this process we first used confocal microscopy to co-localize GFP-Nef and YFP-ABCA1 in HEK293 cells. In contrast to the normal prominent cell surface localization seen for YFP-ABCA1 in the absence of Nef (Fig. 6A), the fluorescence of YPF-ABCA1 in cells also expressing Nef was largely confined to an intracellular location with a reticulate ER-like pattern (Fig. 6B). This suggested that Nef might re-localize or trap ABCA1 in the ER. To test whether this was the case we co-transfected HEK293 cells with a non-fluorescent Nef construct, GFP-ABCA1 and a DsRed fusion protein that localizes to the ER by virtue of a KDEL ER targeting motif (62). In these cells, the GFP-ABCA1 construct expressed alone was again found to prominently localize to the cell surface (Fig. 6C), whereas when expressed with Nef, GFP-ABCA1 was found to strongly co-localize with the DsRed-ER marker (Fig. 6B). This result suggested that the mechanism by which Nef was triggering the degradation of ABCA1 was associated with a build-up of the transporter in the ER. A result that was further supported by sucrose gradient fractionation of the 293ET cells expressing ABCA1 alone or in the presence of Nef (Fig. 6E). When expressed along with Nef, ABCA1 was found to more prominently localize to the fractions that also contained SPTLC1, an ER resident protein endogenously expressed by the 293ET cells.
Figure 6. Nef causes a prominent trapping of ABCA1 in the ER.
Confocal microscopy was used to image 293 cells expressing GFP-Nef and YFP-ABCA1. As opposed to the strong cell surface distribution of YFP-ABCA1 seen when not expressed with Nef (A), the transporter was predominantly localized to an internal organelle with a reticulate-like pattern reminiscent of the endoplasmic reticulum in cells expressing GFP-Nef (B). Likewise GFP-ABCA1 expressed alone is prominently expressed at the cell surface (C), whereas when expressed with un-tagged Nef the transporter is prominently re-localized to the endoplasmic reticulum as shown by the strong co-localization of GFP-ABCA1 and a DsRed-ER fusion protein that marks the ER (D). E, The relocalization of ABCA1 by Nef is further demonstrated as analyzed by ultracentrifugation of 293 cell lysates using sucrose gradients. Immunoblots of the resulting fractions show that in cells expressing Nef, ABCA1 is more prominently localized to fractions that also contain the resident ER protein SPTLC1 endogenously expressed by these cells. F, In 293ET cells Nef is able to suppress the expression of an ABCA1−Δ46 mutant transporter that is trapped in the ER (shown are immunoblots of the indicated proteins).
Given these results, we questioned whether Nef would be able to down-regulate an ABCA1 transporter that fails to exit the ER. To test this possibility Nef was co-expressed with an ABCA1 mutant lacking the last 46 amino acids of the transporter (ABCA1Δ46), which we have previously shown is prominently retained in the ER (63). As with ABCA1, co-expression of Nef markedly suppressed expression of the ABCA1Δ46 mutant (Fig. 6F). Together, these results suggest that the mechanism by which Nef suppresses the expression of ABCA1 involve retention of ABCA1 in the ER and a triggering of the ER-associated degradation of ABCA1.
DISCUSSION
We have investigated the mechanism by which Nef, a HIV-1 viral accessory protein, interacts with and suppresses the function of ABCA1, a cholesterol efflux transporter. HIV-1 infection of human macrophages has previously been shown to inhibit ABCA1-mediated cholesterol efflux, an effect that is important for HIV-1 viral infectivity and that may contribute to the increased atherosclerosis seen in HIV infected individuals (64). Here we defined what domain of ABCA1 allowed Nef to physically interact with the transporter and tested if this interaction was critical for the ability of Nef to suppress ABCA1 function. By analyzing a series of ABCA1 mutants, we found that the physical interaction between Nef and ABCA1 required a motif in the cytoplasmic C-terminal domain of ABCA1 between residues 2225 and 2231 (SDDDHLK). Deletion of the C-terminal 40 amino acids of ABCA1 including the SDDDHLK motif, or selective mutation of residues within this motif to alanines disrupted the physical interaction of Nef with ABCA1. However, these mutations did not block the ability of Nef to down-regulate the efflux activity and protein expression of ABCA1. This indicates that a close physical association of Nef and ABCA1 is not essential for the Nef-mediated downregulation of ABCA1 efflux activity. In contrast, we did find that the NefG2A mutation, which blocks the myristoylation of Nef and its association with membrane bilayers, did block the ability of Nef to both interact and downregulate ABCA1, a result that recapitulates what has been reported for the Nef inhibition of ABCA1 in macrophages (40). Finally, we show that Nef suppression of ABCA1 in both 293ET cells and RAW264.7 macrophages is acting at a post-translational level by a process that increases the degradation of ABCA1.
How may Nef be acting to increase the degradation of ABCA1? To date a number of post-translational and proteolytic processes have been shown to regulate ABCA1 function. The most thoroughly described of these is the turnover of cell surface ABCA1 by a process that involves a calpain-mediated degradation of ABCA1 (42, 43). The ability of calpain to degrade ABCA1 is regulated by a PEST sequence located in the central intracellular loop of ABCA1 near the first ATP binding cassette of the transporter. Given this degradative pathway can be modulated by the phosphorylation status of the ABCA1 PEST sequence, it was possible that Nef was stimulating the degradation of ABCA1 by modulating the phosphorylation of the ABCA1 PEST sequence and thus the calpain degradation of ABCA1. Our work indicates that this is not the case because Nef still down-regulated the ABCA1ΔPEST mutant, which is not subject to proteolysis mediated by calpain. Furthermore, treatment of cells expressing ABCA1wt and Nef with calpeptin, a specific inhibitor of the calpain-mediated degradation of ABCA1, did not block the ability of Nef to inhibit ABCA1 protein expression. Finally, since Nef accelerated the degradation of ABCA1 in the presence of saturating amounts of apoA-I this indicates the ability of apoA-I to modulate the degradation of ABCA1 through the PEST motif was not capable of blocking the Nef mediated down-regulation of ABCA1 (43). In aggregate, these results indicate that the simulation of ABCA1 degradation mediated by Nef was not proceeding through the PEST/calpain pathway.
An alternative pathway described that stimulates degradation of ABCA1 is through the induction of ER stress (59). Since Nef has been reported to induce cholesterol synthesis (36, 37), this could induce ER stress and the unfolded protein response through the accumulation of excess cholesterol levels (60). We therefore tested whether Nef was triggering the degradation of ABCA1 through the UPR pathway. We first verified that tunicamycin, a known inducer of ER stress and UPR signaling, can lead to the induction of ABCA1 degradation in macrophages, and that 293ET cells when treated with tunicamycin, can initiate UPR signaling as detected by increased levels of spliced XBP-1 and CHOP. These results indicated that 293ET cells have the ability to mount a UPR that suppresses ABCA1 protein expression, yet the expression of Nef alone, or co-expression of Nef and ABCA1 were not associated with increased UPR signaling. This indicates that the Nef-mediated stimulation of the degradation of ABCA1 was likely not proceeding through a UPR mechanism.
However, the ER contains machinery that can dislocate large integral membrane proteins for degradation by the proteasome. Indeed, the cystic fibrosis ABC transporter can be degraded by such a mechanism (65) and our analysis demonstrated that Nef increases the association of ABCA1 with ER. We therefore tested whether the Nef-mediated suppression of ABCA1 was blocked by the proteasomal inhibitors lactacystin and expoxomicin (61). Both of these inhibitors significantly reduced the ability of Nef to downregulate ABCA1 expression. Moreover, Nef was able to suppress the expression of an ABCA1 mutant lacking the C-terminal 46 residues that we have previously shown to be largely trapped in the ER (63). This result suggests a two-tier mechanism whereby Nef increases ABCA1 association with ER and also triggers the ER associated degradation machinery in order to suppress ABCA1 efflux function. However, these inhibitors were only able to block from approximately 50 to 75% of the Nef-mediated downregulation of ABCA1 expression. This suggests that Nef may also trigger the degradation of ABCA1 through alternative mechanisms that do not depend upon proteasomal activity. Since Nef has been reported to re-route cell surface CD4 and MHC1 molecules to the lysosome for degradation (66), and because ABCA1 can traffic to a late endosomal/lysosomal compartment (67), it is possible that Nef also stimulates ABCA1 degradation by re-routing the transporter from the cell surface to the lysosome. Indeed, our previous imaging experiments suggest that Nef may also be able to cause at least a transitory build-up of ABCA1 at the cell surface of macrophages (40). These divergent effects on ABCA1 localization indicate Nef may modulate multiple trafficking pathways in order to block ABCA1 function. Finally, it is noted that our estimate of the baseline ABCA1 protein half-life is greater by approximately 4h compared to what has been previously published (42, 43). Why this is the case remains to be investigated. However, it may be that in our assays where we have included fetal bovine sera and LXR agonist this reduces ABCA1 protein turnover. That Nef is able to accelerate ABCA1 degradation under these conditions suggest it is able to override the processes that sera and LXR agonists may trigger to stabilize ABCA1.
In summary, we have confirmed that the myristoylation status of Nef is critical for its ability to suppress the function of ABCA1 and physically interact with ABCA1. Although the ability of Nef to physically interact with the transporter may play additional regulatory roles not revealed by our assays, our results indicate that the interaction of Nef with the ABCA1 C-terminal domain is not required for Nef to suppress ABCA1 protein abundance. Although we cannot exclude the possibility that mutation of the ABCA1 C-terminus preserves a weak physical interaction between Nef and ABCA1 not detected in our assays, our work suggests that Nef is acting at a distance to stimulate ABCA1 degradation, at least in part through a process depending upon proteasomal activity that may act on ABCA1 located in the endoplasmic reticulum.
A critical question remains as to whether the effect of Nef on ABCA1 is specific? Indeed, if Nef increases proteasomal activity in general, then it should increase the general catabolism of proteins affected by this pathway. However, we previously demonstrated that ABCG1 and SR-BI are not affected by Nef (40), and results in this report show that another transmembrane protein, c-MIR, is not down-regulated by Nef. One possible explanation is that these proteins are not metabolized through the mechanisms described here for ABCA1. This possibility appears unlikely given the high similarity between ABCA1 and ABCG1 regulation (68). Another possibility is that Nef, by increasing formation of lipid rafts, dislodges ABCA1 from the cell surface making it susceptible for degradation. Yet another possibility is that weaker interactions of Nef with sites in other domains of ABCA1 may provide specificity for the effects of Nef. Clearly, additional studies are warranted to address these questions.
Supplementary Material
Acknowledgements
This work was supported by NIH NHLBI grants HL074136 (MLF) and HL093818 (DS and MB), an American Heart Association grant 09GRNT2260352 (MLF), fellowship training grants from the American Heart Association (ZM) and the Uehara Memorial Foundation, the Nagai Foundation of Tokyo and the Japan Heart Foundation (NT) and a pre-doctoral training grant from the American Heart Association (AG).
ABBREVIATIONS
- ABCA1
ATP cassette binding transporter A1
- apoA-I
apolipoproteinA-I
- ER
endoplasmic reticulum
- ERAD
ER associated degradation
- FBS
fetal bovine serum
- HDL
high density lipoprotein
- UPR
unfolded protein response
Footnotes
Supporting Information Available: Supplementary Figures 1 & 2 are available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Carr A. Pathogenesis of cardiovascular disease in HIV infection. Curr Opin HIV AIDS. 2008;3:234–239. doi: 10.1097/COH.0b013e3282fb7be0. [DOI] [PubMed] [Google Scholar]
- 2.Hsue PY, Hunt PW, Schnell A, Kalapus SC, Hoh R, Ganz P, Martin JN, Deeks SG. Role of viral replication, antiretroviral therapy, and immunodeficiency in HIV-associated atherosclerosis. AIDS. 2009;23:1059–1067. doi: 10.1097/QAD.0b013e32832b514b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lo J, Abbara S, Shturman L, Soni A, Wei J, Rocha-Filho JA, Nasir K, Grinspoon SK. Increased prevalence of subclinical coronary atherosclerosis detected by coronary computed tomography angiography in HIV-infected men. AIDS. 2010;24:243–253. doi: 10.1097/QAD.0b013e328333ea9e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lorenz MW, Stephan C, Harmjanz A, Staszewski S, Buehler A, Bickel M, von Kegler S, Ruhkamp D, Steinmetz H, Sitzer M. Both long-term HIV infection and highly active antiretroviral therapy are independent risk factors for early carotid atherosclerosis. Atherosclerosis. 2008;196:720–726. doi: 10.1016/j.atherosclerosis.2006.12.022. [DOI] [PubMed] [Google Scholar]
- 5.Maggi P, Quirino T, Ricci E, De Socio GV, Gadaleta A, Ingrassia F, Perilli F, Lillo A, Bonfanti P. Cardiovascular risk assessment in antiretroviral-naive HIV patients. AIDS Patient Care STDS. 2009;23:809–813. doi: 10.1089/apc.2009.0102. [DOI] [PubMed] [Google Scholar]
- 6.Oliviero U, Bonadies G, Apuzzi V, Foggia M, Bosso G, Nappa S, Valvano A, Leonardi E, Borgia G, Castello G, Napoli R, Sacca L. Human immunodeficiency virus per se exerts atherogenic effects. Atherosclerosis. 2009;204:586–589. doi: 10.1016/j.atherosclerosis.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 7.van Vonderen MG, Smulders YM, Stehouwer CD, Danner SA, Gundy CM, Vos F, Reiss P, Agtmael MA. Carotid intima-media thickness and arterial stiffness in HIV-infected patients: the role of HIV, antiretroviral therapy, and lipodystrophy. J Acquir Immune Defic Syndr. 2009;50:153–161. doi: 10.1097/QAI.0b013e31819367cd. [DOI] [PubMed] [Google Scholar]
- 8.Mondy K, Tebas P. Cardiovascular risks of antiretroviral therapies. Annu Rev Med. 2007;58:141–155. doi: 10.1146/annurev.med.58.072905.180040. [DOI] [PubMed] [Google Scholar]
- 9.Lo J, Grinspoon S. Cardiovascular disease in HIV-infected patients: does HIV infection in and of itself increase cardiovascular risk? Curr Opin HIV AIDS. 2008;3:207–213. doi: 10.1097/COH.0b013e3282fb7ba6. [DOI] [PubMed] [Google Scholar]
- 10.Calmy A, Gayet-Ageron A, Montecucco F, Nguyen A, Mach F, Burger F, Ubolyam S, Carr A, Ruxungtham K, Hirschel B, Ananworanich J. HIV increases markers of cardiovascular risk: results from a randomized, treatment interruption trial. AIDS. 2009;23:929–939. doi: 10.1097/qad.0b013e32832995fa. [DOI] [PubMed] [Google Scholar]
- 11.El-Sadr WM, Lundgren JD, Neaton JD, Gordin F, Abrams D, Arduino RC, Babiker A, Burman W, Clumeck N, Cohen CJ, Cohn D, Cooper D, Darbyshire J, Emery S, Fatkenheuer G, Gazzard B, Grund B, Hoy J, Klingman K, Losso M, Markowitz N, Neuhaus J, Phillips A, Rappoport C. CD4+ count-guided interruption of antiretroviral treatment. N Engl J Med. 2006;355:2283–2296. doi: 10.1056/NEJMoa062360. [DOI] [PubMed] [Google Scholar]
- 12.Kuller LH, Tracy R, Belloso W, De Wit S, Drummond F, Lane HC, Ledergerber B, Lundgren J, Neuhaus J, Nixon D, Paton NI, Neaton JD. Inflammatory and coagulation biomarkers and mortality in patients with HIV infection. PLoS Med. 2008;5:e203. doi: 10.1371/journal.pmed.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oh J, Hegele RA. HIV-associated dyslipidaemia: pathogenesis and treatment. Lancet Infect Dis. 2007;7:787–796. doi: 10.1016/S1473-3099(07)70287-6. [DOI] [PubMed] [Google Scholar]
- 14.Anastos K, Lu D, Shi Q, Tien PC, Kaplan RC, Hessol NA, Cole S, Vigen C, Cohen M, Young M, Justman J. Association of serum lipid levels with HIV serostatus, specific antiretroviral agents, and treatment regimens. J Acquir Immune Defic Syndr. 2007;45:34–42. doi: 10.1097/QAI.0b013e318042d5fe. [DOI] [PubMed] [Google Scholar]
- 15.Baker J, Ayenew W, Quick H, Hullsiek KH, Tracy R, Henry K, Duprez D, Neaton JD. High-density lipoprotein particles and markers of inflammation and thrombotic activity in patients with untreated HIV infection. J Infect Dis. 2010;201:285–292. doi: 10.1086/649560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernal E, Masia M, Padilla S, Gutierrez F. High-density lipoprotein cholesterol in HIV-infected patients: evidence for an association with HIV-1 viral load, antiretroviral therapy status, and regimen composition. AIDS Patient Care STDS. 2008;22:569–575. doi: 10.1089/apc.2007.0186. [DOI] [PubMed] [Google Scholar]
- 17.Currier J, Scherzer R, Bacchetti P, Heymsfield S, Lee D, Sidney S, Tien PC. Regional adipose tissue and lipid and lipoprotein levels in HIV-infected women. J Acquir Immune Defic Syndr. 2008;48:35–43. doi: 10.1097/QAI.0b013e318164227f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duprez DA, Kuller LH, Tracy R, Otvos J, Cooper DA, Hoy J, Neuhaus J, Paton NI, Friis-Moller N, Lampe F, Liappis AP, Neaton JD. Lipoprotein particle subclasses, cardiovascular disease and HIV infection. Atherosclerosis. 2009;207:524–529. doi: 10.1016/j.atherosclerosis.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rose H, Hoy J, Woolley I, Tchoua U, Bukrinsky M, Dart A, Sviridov D. HIV infection and high density lipoprotein metabolism. Atherosclerosis. 2008;199:79–86. doi: 10.1016/j.atherosclerosis.2007.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasheed S, Yan JS, Lau A, Chan AS. HIV replication enhances production of free fatty acids, low density lipoproteins and many key proteins involved in lipid metabolism: a proteomics study. PLoS One. 2008;3:e3003. doi: 10.1371/journal.pone.0003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waheed AA, Freed EO. Lipids and membrane microdomains in HIV-1 replication. Virus Res. 2009;143:162–176. doi: 10.1016/j.virusres.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carter GC, Bernstone L, Sangani D, Bee JW, Harder T, James W. HIV entry in macrophages is dependent on intact lipid rafts. Virology. 2009;386:192–202. doi: 10.1016/j.virol.2008.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Del Real G, Jimenez-Baranda S, Lacalle RA, Mira E, Lucas P, Gomez-Mouton C, Carrera AC, Martinez AC, Manes S. Blocking of HIV-1 infection by targeting CD4 to nonraft membrane domains. J Exp Med. 2002;196:293–301. doi: 10.1084/jem.20020308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao Z, Cimakasky LM, Hampton R, Nguyen DH, Hildreth JE. Lipid rafts and HIV pathogenesis: host membrane cholesterol is required for infection by HIV type 1. AIDS Res Hum Retroviruses. 2001;17:1009–1019. doi: 10.1089/088922201300343690. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen DH, Hildreth JE. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J Virol. 2000;74:3264–3272. doi: 10.1128/jvi.74.7.3264-3272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ono A, Freed EO. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc Natl Acad Sci U S A. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Popik W, Alce TM, Au WC. Human immunodeficiency virus type 1 uses lipid raft-colocalized CD4 and chemokine receptors for productive entry into CD4(+) T cells. J Virol. 2002;76:4709–4722. doi: 10.1128/JVI.76.10.4709-4722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell SM, Crowe SM, Mak J. Virion-associated cholesterol is critical for the maintenance of HIV-1 structure and infectivity. AIDS. 2002;16:2253–2261. doi: 10.1097/00002030-200211220-00004. [DOI] [PubMed] [Google Scholar]
- 29.Graham DR, Chertova E, Hilburn JM, Arthur LO, Hildreth JE. Cholesterol depletion of human immunodeficiency virus type 1 and simian immunodeficiency virus with beta-cyclodextrin inactivates and permeabilizes the virions: evidence for virion-associated lipid rafts. J Virol. 2003;77:8237–8248. doi: 10.1128/JVI.77.15.8237-8248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guyader M, Kiyokawa E, Abrami L, Turelli P, Trono D. Role for human immunodeficiency virus type 1 membrane cholesterol in viral internalization. J Virol. 2002;76:10356–10364. doi: 10.1128/JVI.76.20.10356-10364.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao Z, Graham DR, Hildreth JE. Lipid rafts and HIV pathogenesis: virion-associated cholesterol is required for fusion and infection of susceptible cells. AIDS Res Hum Retroviruses. 2003;19:675–687. doi: 10.1089/088922203322280900. [DOI] [PubMed] [Google Scholar]
- 32.Viard M, Parolini I, Sargiacomo M, Fecchi K, Ramoni C, Ablan S, Ruscetti FW, Wang JM, Blumenthal R. Role of cholesterol in human immunodeficiency virus type 1 envelope protein-mediated fusion with host cells. J Virol. 2002;76:11584–11595. doi: 10.1128/JVI.76.22.11584-11595.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beck Z, Balogh A, Kis A, Izsepi E, Cervenak L, Laszlo G, Biro A, Liliom K, Mocsar G, Vamosi G, Fust G, Matko J. New cholesterol-specific antibodies remodel HIV-1 target cells’ surface and inhibit their in vitro virus production. J Lipid Res. 2010;51:286–296. doi: 10.1194/jlr.M000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aloia RC, Tian H, Jensen FC. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc Natl Acad Sci U S A. 1993;90:5181–5185. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brugger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Krausslich HG. The HIV lipidome: a raft with an unusual composition. Proc Natl Acad Sci U S A. 2006;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng YH, Plemenitas A, Fielding CJ, Peterlin BM. Nef increases the synthesis of and transports cholesterol to lipid rafts and HIV-1 progeny virions. Proc Natl Acad Sci U S A. 2003;100:8460–8465. doi: 10.1073/pnas.1437453100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van ’t Wout AB, Swain JV, Schindler M, Rao U, Pathmajeyan MS, Mullins JI, Kirchhoff F. Nef induces multiple genes involved in cholesterol synthesis and uptake in human immunodeficiency virus type 1-infected T cells. J Virol. 2005;79:10053–10058. doi: 10.1128/JVI.79.15.10053-10058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng YH, Plemenitas A, Linnemann T, Fackler OT, Peterlin BM. Nef increases infectivity of HIV via lipid rafts. Curr Biol. 2001;11:875–879. doi: 10.1016/s0960-9822(01)00237-8. [DOI] [PubMed] [Google Scholar]
- 39.Fitzgerald ML, Mujawar Z, Tamehiro N. ABC transporters, atherosclerosis and inflammation. Atherosclerosis. 2010 doi: 10.1016/j.atherosclerosis.2010.01.011. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mujawar Z, Rose H, Morrow MP, Pushkarsky T, Dubrovsky L, Mukhamedova N, Fu Y, Dart A, Orenstein JM, Bobryshev YV, Bukrinsky M, Sviridov D. Human immunodeficiency virus impairs reverse cholesterol transport from macrophages. PLoS Biol. 2006;4:e365. doi: 10.1371/journal.pbio.0040365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fitzgerald ML, Okuhira K, Short GF, 3rd, Manning JJ, Bell SA, Freeman MW. ATP-binding cassette transporter A1 contains a novel C-terminal VFVNFA motif that is required for its cholesterol efflux and ApoA-I binding activities. J Biol Chem. 2004;279:48477–48485. doi: 10.1074/jbc.M409848200. [DOI] [PubMed] [Google Scholar]
- 42.Martinez LO, Agerholm-Larsen B, Wang N, Chen W, Tall AR. Phosphorylation of a pest sequence in ABCA1 promotes calpain degradation and is reversed by ApoA-I. J Biol Chem. 2003;278:37368–37374. doi: 10.1074/jbc.M307161200. [DOI] [PubMed] [Google Scholar]
- 43.Wang N, Chen W, Linsel-Nitschke P, Martinez LO, Agerholm-Larsen B, Silver DL, Tall AR. A PEST sequence in ABCA1 regulates degradation by calpain protease and stabilization of ABCA1 by apoA-I. J Clin Invest. 2003;111:99–107. doi: 10.1172/JCI16808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 45.Fitzgerald ML, Morris AL, Rhee JS, Andersson LP, Mendez AJ, Freeman MW. Naturally occurring mutations in the largest extracellular loops of ABCA1 can disrupt its direct interaction with apolipoprotein A-I. J Biol Chem. 2002;277:33178–33187. doi: 10.1074/jbc.M204996200. [DOI] [PubMed] [Google Scholar]
- 46.Ohmura-Hoshino M, Goto E, Matsuki Y, Aoki M, Mito M, Uematsu M, Hotta H, Ishido S. A novel family of membrane-bound E3 ubiquitin ligases. J Biochem. 2006;140:147–154. doi: 10.1093/jb/mvj160. [DOI] [PubMed] [Google Scholar]
- 47.Shaheduzzaman S, Krishnan V, Petrovic A, Bittner M, Meltzer P, Trent J, Venkatesan S, Zeichner S. Effects of HIV-1 Nef on cellular gene expression profiles. J Biomed Sci. 2002;9:82–96. doi: 10.1007/BF02256581. [DOI] [PubMed] [Google Scholar]
- 48.Simmons A, Aluvihare V, McMichael A. Nef triggers a transcriptional program in T cells imitating single-signal T cell activation and inducing HIV virulence mediators. Immunity. 2001;14:763–777. doi: 10.1016/s1074-7613(01)00158-3. [DOI] [PubMed] [Google Scholar]
- 49.Gerlach H, Laumann V, Martens S, Becker CF, Goody RS, Geyer M. HIV-1 Nef membrane association depends on charge, curvature, composition and sequence. Nat Chem Biol. 6:46–53. doi: 10.1038/nchembio.268. [DOI] [PubMed] [Google Scholar]
- 50.Geyer M, Fackler OT, Peterlin BM. Structure--function relationships in HIV-1 Nef. EMBO Rep. 2001;2:580–585. doi: 10.1093/embo-reports/kve141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia JV, Miller AD. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature. 1991;350:508–511. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- 52.Schwartz O, Marechal V, Le Gall S, Lemonnier F, Heard JM. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat Med. 1996;2:338–342. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]
- 53.Vigerust DJ, Egan BS, Shepherd VL. HIV-1 Nef mediates post-translational down-regulation and redistribution of the mannose receptor. J Leukoc Biol. 2005;77:522–534. doi: 10.1189/jlb.0804454. [DOI] [PubMed] [Google Scholar]
- 54.Norman JP, Perry SW, Reynolds HM, Kiebala M, De Mesy Bentley KL, Trejo M, Volsky DJ, Maggirwar SB, Dewhurst S, Masliah E, Gelbard HA. HIV-1 Tat activates neuronal ryanodine receptors with rapid induction of the unfolded protein response and mitochondrial hyperpolarization. PLoS One. 2008;3:e3731. doi: 10.1371/journal.pone.0003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Furukawa T, Wakabayashi K, Tamura A, Nakagawa H, Morishima Y, Osawa Y, Ishikawa T. Major SNP (Q141K) variant of human ABC transporter ABCG2 undergoes lysosomal and proteasomal degradations. Pharm Res. 2009;26:469–479. doi: 10.1007/s11095-008-9752-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakagawa H, Wakabayashi-Nakao K, Tamura A, Toyoda Y, Koshiba S, Ishikawa T. Disruption of N-linked glycosylation enhances ubiquitin-mediated proteasomal degradation of the human ATP-binding cassette transporter ABCG2. FEBS J. 2009;276:7237–7252. doi: 10.1111/j.1742-4658.2009.07423.x. [DOI] [PubMed] [Google Scholar]
- 58.Wakabayashi-Nakao K, Tamura A, Furukawa T, Nakagawa H, Ishikawa T. Quality control of human ABCG2 protein in the endoplasmic reticulum: ubiquitination and proteasomal degradation. Adv Drug Deliv Rev. 2009;61:66–72. doi: 10.1016/j.addr.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 59.Feng B, Tabas I. ABCA1-mediated cholesterol efflux is defective in free cholesterol-loaded macrophages. Mechanism involves enhanced ABCA1 degradation in a process requiring full NPC1 activity. J Biol Chem. 2002;277:43271–43280. doi: 10.1074/jbc.M207532200. [DOI] [PubMed] [Google Scholar]
- 60.Feng B, Yao PM, Li Y, Devlin CM, Zhang D, Harding HP, Sweeney M, Rong JX, Kuriakose G, Fisher EA, Marks AR, Ron D, Tabas I. The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nat Cell Biol. 2003;5:781–792. doi: 10.1038/ncb1035. [DOI] [PubMed] [Google Scholar]
- 61.Fujita K, Omura S, Silver J. Rapid degradation of CD4 in cells expressing human immunodeficiency virus type 1 Env and Vpu is blocked by proteasome inhibitors. J Gen Virol. 1997;78(Pt 3):619–625. doi: 10.1099/0022-1317-78-3-619. [DOI] [PubMed] [Google Scholar]
- 62.Teasdale RD, Jackson MR. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the golgi apparatus. Annu Rev Cell Dev Biol. 1996;12:27–54. doi: 10.1146/annurev.cellbio.12.1.27. [DOI] [PubMed] [Google Scholar]
- 63.Tamehiro N, Zhou S, Okuhira K, Benita Y, Brown CE, Zhuang DZ, Latz E, Hornemann T, von Eckardstein A, Xavier RJ, Freeman MW, Fitzgerald ML. SPTLC1 binds ABCA1 to negatively regulate trafficking and cholesterol efflux activity of the transporter. Biochemistry. 2008;47:6138–6147. doi: 10.1021/bi800182t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crowe SM, Westhorpe CL, Mukhamedova N, Jaworowski A, Sviridov D, Bukrinsky M. The macrophage: the intersection between HIV infection and atherosclerosis. J Leukoc Biol. 2009 doi: 10.1189/jlb.0809580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Younger JM, Chen L, Ren HY, Rosser MF, Turnbull EL, Fan CY, Patterson C, Cyr DM. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- 66.Roeth JF, Collins KL. Human immunodeficiency virus type 1 Nef: adapting to intracellular trafficking pathways. Microbiol Mol Biol Rev. 2006;70:548–563. doi: 10.1128/MMBR.00042-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boadu E, Bilbey NJ, Francis GA. Cellular cholesterol substrate pools for adenosine-triphosphate cassette transporter A1-dependent high-density lipoprotein formation. Curr Opin Lipidol. 2008;19:270–276. doi: 10.1097/MOL.0b013e3282feea99. [DOI] [PubMed] [Google Scholar]
- 68.Oram JF, Vaughan AM. ATP-Binding cassette cholesterol transporters and cardiovascular disease. Circ Res. 2006;99:1031–1043. doi: 10.1161/01.RES.0000250171.54048.5c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.