Abstract
Previous studies have shown that the translation level of in vitro transcribed messenger RNA (mRNA) is enhanced when its uridines are replaced with pseudouridines; however, the reason for this enhancement has not been identified. Here, we demonstrate that in vitro transcripts containing uridine activate RNA-dependent protein kinase (PKR), which then phosphorylates translation initiation factor 2-alpha (eIF-2α), and inhibits translation. In contrast, in vitro transcribed mRNAs containing pseudouridine activate PKR to a lesser degree, and translation of pseudouridine-containing mRNAs is not repressed. RNA pull-down assays demonstrate that mRNA containing uridine is bound by PKR more efficiently than mRNA with pseudouridine. Finally, the role of PKR is validated by showing that pseudouridine- and uridine-containing RNAs were translated equally in PKR knockout cells. These results indicate that the enhanced translation of mRNAs containing pseudouridine, compared to those containing uridine, is mediated by decreased activation of PKR.
INTRODUCTION
In vitro transcribed messenger RNA (mRNA) has many advantages as a vehicle for gene delivery. Transfection of mRNA is very efficient (1), and rapid expression of the encoded protein can be achieved. Unlike viral vectors or plasmid DNA, cell-delivered mRNA does not introduce the risk of insertional mutagenesis (2,3). Previous studies have shown that RNA can activate a number of innate immune receptors, including Toll-like receptor (TLR)3, TLR7, TLR8 and retinoic acid-inducible gene I (RIG-I). However, activation of these receptors can be avoided by incorporating modified nucleosides, e.g. pseudouridine (Ψ) or 2-thiouridine (s2U), into the RNA (4,5).
RNA-dependent protein kinase (PKR) is a ubiquitous mammalian enzyme with a variety of cellular functions, including regulation of translation during conditions of cell stress. During viral infection, PKR binds viral double-stranded (ds)RNA, autophosphorylates and subsequently phosphorylates the alpha subunit of translation initiation factor 2 (eIF-2α), thus repressing translation (6,7). Originally, potent activation of PKR was thought to require >30-bp-long dsRNA (8). It has subsequently been shown that PKR can be activated by a variety of RNA structures that include single-stranded (ss)RNA forming hairpins (9,10), imperfect dsRNA containing mismatches (10), short dsRNA with ss tails (11), stem–loop structures with 5′-triphosphates (12,13), and unique elements present in interferon gamma (IFN-γ) and tumor necrosis factor-alpha mRNAs (14). Viral (15,16) and cellular RNAs (17–20) transcribed as ssRNA but containing secondary structure can also be potent PKR activators. PKR activation by short dsRNA, such as siRNA, has also been demonstrated (21–26). These reports indicate that a wide variety of RNA structures can activate PKR, provided they contain some dsRNA element. Modified nucleosides present in homopolymeric RNAs (27–30) or in short transcripts (25,31,32) can influence activation of PKR. However, it has not been investigated whether modified nucleosides present in long, protein-encoding mRNAs impact activation of PKR.
Previously, we demonstrated that in vitro transcribed mRNAs containing Ψ are translated at significantly higher levels than those containing unmodified uridines (33). However, the molecular mechanism underlying this enhancement has not been identified. Here, we show that one cause of this translational difference is that Ψ-containing mRNA activates PKR less efficiently than uridine-containing mRNA. This reduced PKR activation also mitigates general translational inhibition of cellular proteins that is induced when unmodified in vitro transcribed mRNAs are delivered to cells. Since replacing uridines with pseudouridines also abrogates innate immune activation by RNA, Ψ-modified mRNAs are attractive vectors for gene delivery or replacement, vaccine antigen delivery or other RNA-based therapeutic applications.
MATERIALS AND METHODS
Cells and reagents
Human embryonic kidney (HEK) 293T cells were obtained from the American Type Culture Collection and were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 2 mM l-glutamine (Life Technologies), 100 U/ml penicillin and 100 µg/ml streptomycin (Invitrogen) and 10% fetal calf serum (HyClone). Immortalized wild-type (WT) and PKR knockout (PKR−/–) mouse embryonic fibroblasts (MEFs) were generously provided by Robert Silverman (Cleveland Clinic Foundation) and were maintained in RPMI medium supplemented with 2 mM l-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin and 10% fetal calf serum. Polyinosinic:polycytidylic acid (poly(I:C)) was purchased from Sigma and polydeoxycytidylic acid (poly(dC)) was purchased from Midland Certified Reagent Co.
mRNA synthesis
RNAs were transcribed as previously described (4), using linearized plasmids encoding firefly luciferase (pT7TS-fLuc and pTEVluc) or Renilla luciferase (pT7TS-Ren) and T7 RNA polymerase (Megascript, Ambion). Except where otherwise specified, capped mRNA was generated by performing transcription in the presence of cap analog 3′-O-Me-m7G(5′)ppp(5′)G (New England Biolabs). All mRNAs were transcribed to contain 30 or 50-nt-long 3′ poly(A) tails. Triphosphate-derivatives of Ψ, s2U, m5C, m6A and m5U (TriLink) were used in place of their cognate unmodified NTP to generate modified nucleoside-containing RNA. Following transcription, the template plasmids were digested with Turbo DNase and RNAs were precipitated with 2.5 M lithium chloride at −20°C for 4 h. RNAs were pelleted by centrifugation, washed with 75% ethanol and then reconstituted in nuclease-free water. The concentration of RNA was determined by measuring the optical density at 260 nm. All RNA samples were analyzed by denaturing agarose gel electrophoresis for quality assurance. Each RNA type was synthesized in 4–10 independently performed transcription experiments and all experiments were performed with at least two different batches of mRNA. Enzymatic capping was performed using ScriptCap m7G capping kit (Epicentre) on mRNA transcribed with guanosine 5′-[γ-32P]-triphosphate (GE Healthcare). Efficiency of capping was verified by monitoring the elimination of γ-32P from the mRNA. Biotinylated mRNA was transcribed with the addition of 1:5 biotinylated CTP (Roche Applied Sciences) in the transcription reaction.
Detection of reporter proteins in RNA-transfected cells
Cells were seeded into 96-well plates at a density of 5.0 × 104 cells/well 1 day prior to transfection. RNA was complexed with lipofectin (Invitrogen) as described previously (4). Cells were exposed to 50 µl DMEM containing lipofectin-complexed RNA (0.25 µg) for 1 h, which was then replaced with complete medium and further cultured. Cells were lysed in 25 µl firefly, Renilla, or dual-luciferase specific lysis reagents (Promega). Aliquots of 2 µl were assayed with the corresponding enzyme substrates and a LUMAT LB 950 luminometer (Berthold) at a 10-s measuring time.
Assessment of total protein synthesis
HEK293T cells were seeded into 96-well plates at a density of 5.0 × 104 cells/well with 1000 U/ml interferon-αA/D (Sigma) 1 day prior to transfection. Cells were incubated in methionine/cysteine-free medium (Invitrogen) for 1 h, then pulsed with complete medium supplemented with 35S-methionine/cysteine (140 mCi/ml) (PerkinElmer) for 1–3 h. Cells were lysed in RIPA lysis buffer supplemented with protease inhibitor cocktail (Sigma). Lysate was diluted in 0.1% bovine serum albumin (BSA), and macromolecules were precipitated by the addition of trichloroacetic acid (TCA) and 30 min incubation on ice. Precipitates were filtered onto glass microfiber filters (Whatman) and washed with 10% TCA and 100% ethanol. Incorporated 35S-methionine/cysteine was quantified using Ecolite(+) scintillation cocktail (MP Biomedicals) and a Beckman LS 6000IC scintillation counter.
PKR activation in vitro
Purified PKR prepared as described (11) was dephosphorylated using lambda protein phosphatase (New England Biolabs). Final concentrations of 0.75 µM dephosphorylated PKR, 0.1 mM ATP and 0.15 µCi/µl adenosine 5′-[γ-32P]-triphosphate (γ-32P-ATP) (PerkinElmer) were mixed with the indicated concentration of RNA for 10 min at 30°C in a buffer consisting of 4 mM MgCl2, 100 mM KCl and 20 mM HEPES, pH 7.5. The reaction was stopped by the addition of NuPage LDS sample buffer and reducing agent (Invitrogen) and heating for 10 min at 70°C. Unincorporated γ-32P-ATP was separated from radiolabeled PKR by running samples on a 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) gel. Phosphorylated PKR was imaged in dried gels using a phosphor storage screen (Molecular Dynamics) and detected using Storm or Typhoon Phosphorimagers (GE Healthcare). Band densities were quantified using ImageQuant software (GE Healthcare).
Western blotting
HEK293T cells were seeded into 96-well plates at a density of 5.0 × 104 cells/well, with 1000 U/ml interferon-αA/D 1 day prior to transfection. At the indicated time following RNA transfection, cells were lysed in RIPA lysis buffer supplemented with protease inhibitor cocktail and HALT phosphatase inhibitor (Pierce). Equal mass of protein (10−30 µg per sample) was loaded onto a 12% SDS–PAGE gel. Proteins were subsequently transferred to a Hybond-P polyvinylidene fluoride (PVDF) membrane (GE Amersham), blocked with 2.5% non-fat milk in TBS containing 0.05% Tween-20, and probed with antibodies for PKR-pT446 and PKR (Epitomics), eIF-2α-pS51 and eIF-2α (Cell Signaling Technologies), or PABP (Abcam). Membranes were stripped by agitating gently in a buffer of 2% SDS, 100 mM β-mercaptoethanol, 62.5 mM Tris pH 6.7 for 30 min at 50°C, then subsequently re-blocked and re-probed. Image was captured using the Fujifilm LAS1000 digital imaging system. Linear brightness and contrast were adjusted using GIMP 2.6 software.
Biotinylated RNA pull-down
HEK293T cells were lysed in RIPA lysis buffer supplemented with protease inhibitor cocktail and RNase inhibitor (RNasin, Promega). Biotinylated mRNA (2 µg) was added to 25 µl lysate and incubated on ice for 2 h. Subsequently, 50 µl of streptavidin-agarose bead 50% slurry (Invitrogen) was added and incubated on ice for 1 h. Beads with bound RNA and proteins were centrifuged and washed, and proteins were released from RNA by heating samples at 70°C for 10 min in the presence of NuPage LDS sample buffer and reducing agent. Samples were separated by 10% SDS–PAGE and transferred to PVDF membranes. PKR and poly(A)-binding protein (PABP) were detected by western blotting.
Statistical analysis
All data are reported as mean ± standard error of the mean (SEM). Statistical differences between treatment groups were calculated by the Student’s t-test using Microsoft Excel. For all statistical testing, a P-value <0.05 was considered significant.
RESULTS
Conventional in vitro transcribed mRNA induces translational repression
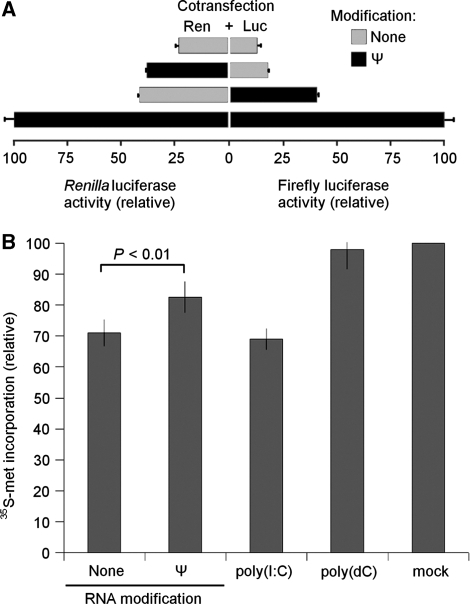
We previously reported that mRNA transcribed in vitro containing Ψ in place of uridine is translated more efficiently than mRNA containing unmodified nucleosides (33). In order to determine whether the translational enhancement exerted by Ψ incorporated into RNA is restricted to the modified transcript or also extends to unmodified transcripts, we performed co-transfection experiments delivering equal amounts of Renilla and firefly luciferase-encoding mRNAs to cells. As expected, the mRNAs were translated much more efficiently when both contained Ψ as compared to when both were unmodified (Figure 1A). However, when only one of the mRNAs contained Ψ modification, the translation level of the Ψ-containing RNA decreased (∼50%) relative to the level measured when both contained Ψ. One explanation for these findings could be that unmodified RNA inhibits the translation of the co-delivered RNA, while Ψ-containing RNA has no such inhibitory effect. To explore whether translation of endogenous cellular mRNAs are similarly influenced by exogenously delivered in vitro transcribed mRNAs, total cellular protein synthesis was monitored in cells transfected with mRNA containing Ψ modification or no modification. Both types of mRNA reduced cellular protein translation; however, the suppression of protein synthesis was greater with unmodified RNA than with Ψ-containing RNA (Figure 1B). PKR-activating poly(I:C) and non-activating poly(dC) were used as controls. Mock transfected cells were treated with the transfection reagent (lipofectin) only, without nucleic acid.
Figure 1.
Translational inhibition by unmodified in vitro transcribed mRNA. (A) In vitro transcribed mRNAs encoding Renilla luciferase (Ren) and firefly luciferase (Luc) were synthesized with and without Ψ modifications then mixed (1 : 1 mass ratio) as indicated. The mixed mRNA was complexed with lipofectin and added to HEK293T cells seeded in 96-well plates (0.25 µg RNA/well). Cells were lysed 4 h after transfection and dual luciferase measurements were performed in aliquots (1/20th) of the lysates. Values presented are normalized to cells transfected with Ren and Luc mRNAs when both contained Ψ modifications. Error bars indicate the standard error of n = 3 samples. (B) Unmodified or pseudouridine-containing RNA was delivered to HEK293T cells by lipofection. Cells were subsequently incubated with 35S-methionine/cysteine supplemented medium, lysed, and proteins were TCA precipitated. Data are presented as percentage of counts obtained from mock transfected cells. Data shown are mean values from three independent experiments ± SEM.
Conventional in vitro transcribed mRNA activates PKR
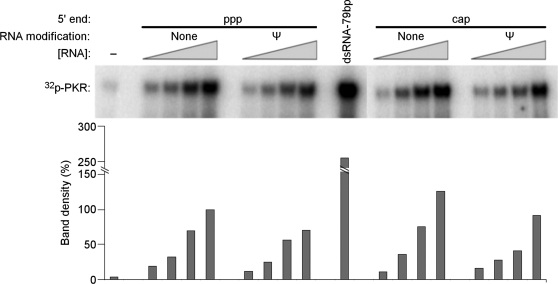
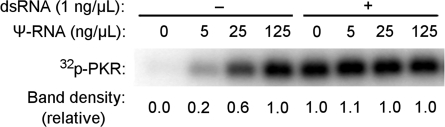
To determine whether the inhibition of translation by unmodified mRNA is mediated by PKR, in vitro transcribed mRNAs were first analyzed in a cell-free system using purified PKR. Four different mRNAs were tested: unmodified and Ψ-modified mRNA, each with either a cap or a triphosphate at their 5′-end (5′ppp). In vitro transcribed mRNA with 5′ppp and containing uridines activated PKR to a greater extent than those containing Ψ (Figure 2). This reduced activation of PKR by Ψ-containing transcripts is consistent with the previously observed enhancement of in vitro translation from Ψ-containing RNA in rabbit reticulocyte lysates (33). Since the presence of 5′ppp on short RNAs has previously been shown to enhance the activation of PKR (12,13), it was important to determine whether the 5′ppp present on long mRNAs also contributed to PKR activation. To remove 5′ppp, in vitro transcripts were capped enzymatically (Supplementary Figure S1), which completely removed the 5′ppp, and then tested. As Figure 2 demonstrates, the presence or absence of 5′ppp on unmodified and Ψ-modified transcripts did not significantly alter their ability to activate PKR. It has been shown that a variety of nucleoside modifications in RNA can influence the activation of RNA sensors (4,5,32); therefore, the effect of incorporating the modified nucleosides s2U, 5-methylcytidine (m5C), 6-methyladenosine (m6A) or 5-methyluridine (m5U) into mRNA was also analyzed. mRNA containing s2U, m5C or m6A activated PKR to a lesser extent than unmodified RNA, while RNA with m5U activated PKR to the greatest extent (Supplementary Figure S2).
Figure 2.
Activation of purified PKR by in vitro transcribed RNA. Purified PKR was incubated with γ-32P-ATP and in vitro transcribed mRNA for 10 min. Reaction products were separated by SDS–PAGE and imaged using phosphor storage radiography. Unmodified or Ψ-containing mRNAs encoding firefly luciferase contained triphosphates (ppp) or cap at their 5′-ends. Complete capping of RNA was achieved post-transcriptionally using vaccinia capping enzyme. Concentration of mRNA in reactions was 3.1, 6.2, 12.5 and 25 µg/ml. Quantified phosphorylation is presented as a bar graph below each band. Values were normalized to those obtained with 25 µg/ml uncapped, unmodified RNA. No RNA (−) and 79 bp dsRNA were used as negative and positive controls.
Pseudouridine-containing mRNA does not activate PKR in cells
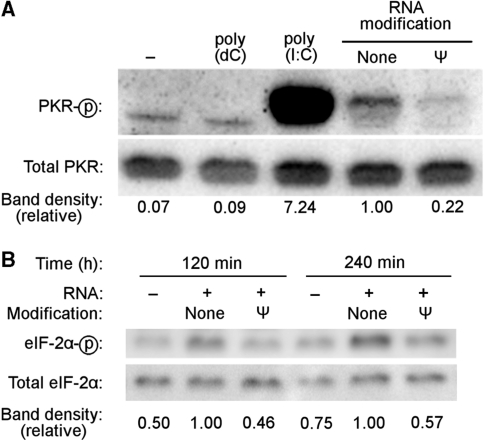
Next, we investigated the impact of Ψ-containing mRNA on PKR activation in the complex cellular environment. Following control studies demonstrating that RNAs with or without nucleoside modification can be delivered to cells with the same efficiency (data not shown), unmodified or Ψ-containing mRNA was complexed with lipofectin and delivered into HEK293T cells. PKR activation was assessed by western blot using an antibody specific for PKR phosphorylated on Thr446, a site at which phosphorylation is requisite for PKR activation (34). Consistent with the results observed using purified PKR, transfection of unmodified transcript induced PKR phosphorylation, which was dramatically reduced if the transfected RNA contained Ψ (Figure 3A). Similarly, incorporation of s2U or m5C into RNA reduced the level of PKR phosphorylation relative to that induced by unmodified RNA, while m5U incorporation into RNA enhanced PKR phosphorylation (Supplementary Figure S3A). Incorporation of m6A into RNA also enhanced PKR phosphorylation in cells, despite reducing PKR activation in vitro.
Figure 3.
PKR activation by in vitro transcribed mRNA in cells. Unmodified or Ψ-containing in vitro transcribed firefly luciferase mRNA was delivered to cells by lipofection. Following RNA transfection, cells were lysed at 4 h (A) or at the indicated time (B), proteins were separated by SDS–PAGE, and assayed for phosphorylation of PKR (A) or eIF-2α (B) by western blotting. No RNA (−), poly(dC) and poly(I:C) were used as controls. Relative phosphorylation is indicated below each gel lane, calculated as phosphorylated band density divided by total band density and then normalized to the phosphorylation induced by unmodified RNA.
Phosphorylation of eIF-2α, a substrate of PKR, was induced in HEK293T cells by transfection with unmodified RNA but not with Ψ-containing RNA (Figure 3B). Incorporation of modified nucleosides other than Ψ into mRNA altered the phosphorylation of eIF-2α in direct parallel to their alterations of PKR phosphorylation (Supplementary Figure S3B).
Translation of unmodified mRNA is enhanced upon inhibiting or eliminating PKR
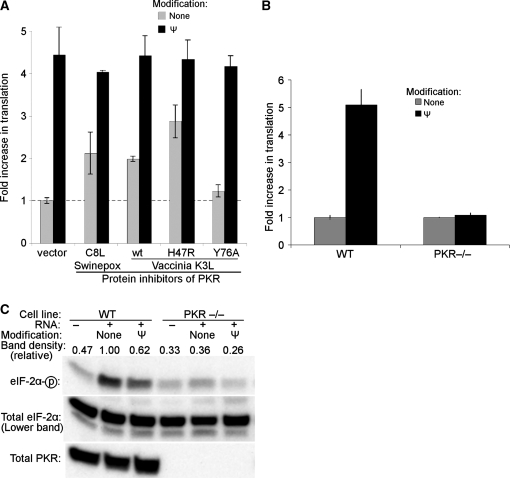
Viral proteins C8L of swinepox and K3L of vaccinia are inhibitors of PKR and have been shown to reverse PKR-mediated inhibition of translation in mammalian cells (35). Thus, to confirm the role of PKR in the translational differences observed between uridine- and Ψ-containing transcripts, we utilized C8L, K3L and two K3L mutants: hyperactive K3L-H47R and inactive K3L-Y76A (35,36). Based on the premise that PKR is activated by in vitro transcribed mRNAs that contain uridine but not by those with Ψ, inhibition of PKR would be expected to increase the translation of unmodified mRNA but to have no effect on the translation of Ψ-containing RNA. Indeed, in the presence of PKR inhibitors, the amount of translation increased from unmodified transcripts but not from Ψ-modified transcripts (Figure 4A).
Figure 4.
Translation of in vitro transcribed mRNA in the absence of PKR activity. (A) HEK293T cells were transfected with plasmids encoding protein inhibitors of PKR: swinepox C8L protein, wt vaccinia K3L, hyperactive K3L-H47R, inactive K3L-Y76A, or pG5 empty vector. Twenty-four hours later, unmodified or Ψ-modified in vitro transcribed mRNAs encoding firefly luciferase were delivered by lipofection, and luciferase activity was measured 4 h later. Data were normalized to values obtained when cells were first transfected with empty vector then with unmodified RNA. Presented data are mean values from three replicates ± SEM. (B) MEF cell lines derived from wild-type (WT) or transgenic mice that do not express functional PKR (PKR−/−) were transfected with unmodified or Ψ-containing in vitro transcribed mRNAs encoding firefly luciferase. Data were normalized to values obtained when cells were transfected with unmodified RNA and expressed as fold increase in translation of Ψ-containing mRNA over unmodified RNA. Values are from three replicate wells ± SEM, and are representative of at least three independently performed experiments. (C) WT and PKR−/– MEF cells were transfected with unmodified or Ψ-containing in vitro transcribed mRNAs encoding firefly luciferase, or mock transfected with no RNA (–). Cells were lysed 2 h following RNA transfection; proteins were then separated by SDS–PAGE and assayed for eIF-2α phosphorylation by western blotting. Relative phosphorylation is indicated above each gel lane, calculated as phosphorylated band density divided by total band density and then normalized to the phosphorylation induced by unmodified RNA in wild-type cells. Absence of PKR was also confirmed by western blotting.
Further evidence confirming the role of PKR in suppressing translation of unmodified mRNAs was obtaining using mouse embryonic fibroblasts (MEFs) derived from PKR-knockout animals. In wild-type MEFs, translation of Ψ-containing transcripts was 4–5-fold greater than that of unmodified transcripts (Figure 4B). In PKR-deficient MEFs, however, the extent of translation of Ψ-modified mRNA was not different from that of unmodified mRNA. Additionally, RNA transfection does not induce phosphorylation of eIF-2α in PKR-deficient MEFs, as it does in WT cells (Figure 4C). These results demonstrate that the activity of PKR is necessary for the decreased translation of unmodified transcripts relative to Ψ-containing transcripts.
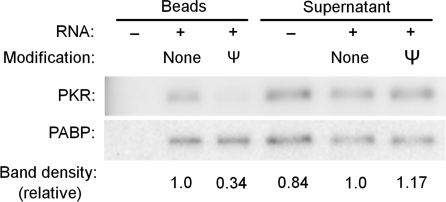
Pseudouridine-containing mRNA is not bound by PKR
To test whether Ψ-modified mRNA is a competitive inhibitor of PKR, a 200-bp dsRNA known to activate PKR was mixed with a 5–125-fold mass excess ofΨ-modified RNA. All concentrations of Ψ-modified RNA tested failed to inhibit the activation of PKR by the 200-bp dsRNA (Figure 5). Similarly, a 125-fold mass excess of mRNA containing s2U, m5C or m6A did not inhibit PKR activation by dsRNA (Figure S4). The results were the same using lower mass excess, equal mass or equal molar mixes (data not shown), demonstrating that RNAs containing modified nucleosides are not competitive inhibitors of PKR. The lack of PKR inhibition by transcripts containing modified nucleosides suggests a lack of binding between PKR and modified RNAs. To directly test binding, biotinylated transcripts having 30-nt-long poly(A) tails and containing either Ψ or uridines were mixed with HEK293T cell lysates, and complexes were then precipitated using streptavidin-agarose beads. Western blots of the precipitates indicated that PKR bound to unmodified RNA, but bound poorly to Ψ-modified RNA (Figure 6), consistent with reduced activation of PKR by Ψ-containing RNA. By contrast, poly(A)-binding protein (PABP) bound equally well to both transcripts. These results indicate that unmodified RNA, but not Ψ-modified RNA, binds to and activates PKR.
Figure 5.
Ψ-containing mRNA does not inhibit PKR activation. An activating 200 bp dsRNA was mixed with a 5–125-fold mass excess of Ψ-containing in vitro transcribed firefly luciferase mRNA prior to incubation with purified PKR. Reaction products were separated by SDS–PAGE. Relative band densities are presented below each gel lane and normalized to dsRNA only. Data shown are representative of three independent experiments.
Figure 6.
Ψ-containing mRNA does not pull-down PKR. Biotinylated in vitro transcribed unmodified or Ψ-containing RNAs were incubated with HEK293T cell lysates for 2 h. The RNA and bound proteins were pulled down using streptavidin-agarose beads. An aliquot of lysate that was incubated only with beads but without RNA (−) was also processed. Aliquots of pull-down proteins as well as the supernatants were separated by SDS–PAGE. PKR and PABP were detected by western blotting. Relative band densities of PKR divided by PABP compared to unmodified RNA are presented below each gel lane.
DISCUSSION
We demonstrate that modified nucleosides in mRNA reduce PKR activation and identify a mechanism by which Ψ-incorporation in mRNA enhances translation of the encoded protein. Our data show that conventional in vitro transcribed RNA inhibits translation of reporter and cellular mRNAs, in part through the activation of PKR. However, this inhibitory activity is not induced by Ψ-containing mRNA. Using multiple lines of investigation, our studies demonstrate that unmodified in vitro transcribed mRNA activates PKR, resulting in phosphorylation of eIF-2α and inhibition of translation. Replacement of 5′ppp with 5′cap structure on the mRNA does not substantially alter this PKR activation. Examining translation in the context of PKR inhibitors and in PKR-deficient cells confirmed that enhanced translation of Ψ-containing mRNA is a consequence of diminished PKR activation. Mechanistically, modified nucleoside incorporation reduces RNA recognition by PKR. This is supported by data demonstrating that RNAs containing modified nucleosides do not inhibit PKR activation by dsRNA and that PKR binds poorly to Ψ-containing RNA.
PKR activation by unmodified RNA has a more pronounced impact on translation of the transfected reporter mRNA than on total cellular translation (Figure 1). A similar local translation effect has been observed with PKR activation by IFN-γ mRNA (19,37). The pronounced local inhibition is likely due to the kinetics of phosphorylation and dephosphorylation of PKR. Activated PKR most dramatically inhibits local translation because rapid dephosphorylation of PKR limits the impact on more distant translation. Therefore, translation of a PKR-activating mRNA is more severely impacted than total cellular translation. Furthermore, the observation that Ψ-containing RNA also causes some reduction in total protein synthesis suggests that there are additional effects on cellular translation which are not mediated by PKR.
Ψ-containing RNA activates PKR more effectively in vitro as compared to in vivo (Figures 2 and 3). One possible reason for this difference is that PKR activation in vivo occurs in the presence of competing factors such as phosphatases, components of the translational system and other proteins affecting the structure and accessibility of the RNA to PKR. In contrast, in vitro assays lack such competing factors that would limit or reverse PKR phosphorylation.
Although mRNA is normally transcribed without a complementary antisense transcript or long stretches of self-complementarity, it contains many short ds regions and other intramolecular secondary structures (Figure S5). In addition to long perfectly dsRNA, PKR is activated by RNA that contains either hairpins (9), bulges, mismatched base-pairing (10), short internal dsRNA regions (11) or unique structures naturally present in selected cellular mRNAs (17–20). As previously demonstrated for TLR3 (38), it is likely that the activation of PKR by in vitro transcribed mRNA is due to the formation of intra- and intermolecular secondary structures. PKR is then activated upon binding to these structures, similar to the classical dsRNA-mediated mechanism of PKR activation. Nucleoside modifications influence base pairing and secondary structure formation (39–46), which likely contribute to their effects on PKR activation. Alterations to the shape of the helix formed and interruptions to the minor groove, which is presumed to be the principal location of PKR interaction with RNA (32,47,48), are also likely to play significant roles in determining how each modified nucleoside will impact RNA-mediated PKR activation.
Unlike short ssRNAs (12), PKR activation by long in vitro transcribed mRNA is not dependent on the presence of a 5′-triphosphate, as mRNA containing complete replacement of 5′ppp with cap structure also activates PKR (Figures 2 and S1). The difference between these findings might reflect the amount of 5′ppp in the RNAs being compared. Forty-seven nucleotide-long ssRNA induced 100-fold more PKR activation when the 5′-end contained triphosphates (12), while our data did not show any significant effect of removing the 5′ppp from 1976-nt-long mRNA, which contains ∼40-fold less 5′ppp. Our finding is more consistent with the result reported for 47-bp-long dsRNA wherein PKR activation did not depend on 5′ppp (12).
Previous reports indicate that PKR activation is altered by the presence of modified nucleosides in homopolymeric RNA (27,28,30) and short ssRNA and dsRNA (32). Our data extend these findings by demonstrating that incorporation of modified nucleosides into long in vitro transcribed mRNA also alters activation of PKR, and subsequent translation of the RNA. We observe substantial PKR activation by in vitro transcribed mRNA, which is reduced by incorporation of Ψ. Additionally, our studies show reduced PKR activation by mRNA that contains m5C, enhanced PKR activation by mRNA containing m5U and elimination of PKR activation by s2U-containing mRNAs. These results vary from those obtained when testing PKR activation by short 47 nt ssRNA: a low level of PKR activation by unmodified RNA, which was dependent on the presence of a 5′-triphosphate, and near-complete elimination of PKR activation by incorporation of modified nucleosides (32). However, when testing short 47-bp dsRNA, the effects observed were similar to those reported here: PKR activation by unmodified RNA, which is reduced by Ψ incorporation, increased by m5U incorporation, and eliminated by s2U incorporation. This similarity to short dsRNA, and dissimilarity to ssRNA, supports our model that PKR activation by long in vitro transcribed mRNA is due to regions of secondary structure formed within the RNA and is independent of the 5′-end.
Unlike the other nucleoside modifications tested, the presence of m6A in mRNA impacted PKR activation differently in vivo than in vitro. In vitro, mRNA containing m6A activated PKR only moderately (Figure S2) whereas in vivo, m6A-containing mRNA activated PKR more potently than unmodified RNA (Figure S3). Although the significance of this observation is not fully understood, the discrepancy may be explained by the presence of additional factors in cells that facilitate increased ds formation in m6A-containing mRNA in vivo.
Nucleic acids containing modified nucleosides can act as antagonists of nucleic acid-sensing TLRs (49–52). Therefore, we asked whether mRNAs containing modified nucleosides inhibit activation of PKR by its cognate ligand, dsRNA. PKR is still activated by dsRNA in the presence of a 125-fold excess of mRNA containing Ψ or other modified nucleosides (s2U, m5C or m6A), indicating that mRNAs containing modified nucleosides are not inhibitors of PKR (Figures 5 and S4). This extends previous data demonstrating that short ssRNAs containing modified nucleosides do not inhibit PKR (32). Furthermore, in cell lysates, RNA containing Ψ pulls down less PKR than RNA containing uridine (Figure 6). This reduction in PKR binding is consistent with prior in vitro data demonstrating small reductions in PKR binding to short dsRNA and ssRNA that contain modified nucleosides (32). From these data we conclude that the mechanism of reduced PKR activation is reduced recognition and binding to RNA containing modified nucleosides.
It is possible that mRNAs with different nucleoside modifications have different optimal concentrations for activating PKR. Figures S2 and S3 indicate that none of the modified nucleosides tested, with the exception of s2U, completely eliminate PKR activation. Rather, each modified nucleoside might alter the ability of RNA to bind and activate PKR (Figure 6).
PKR plays an integral part in the cellular response to viral RNA. However, mechanisms to avoid PKR activation by cellular RNAs are required, as constitutive PKR activation and translational inhibition would obstruct normal cellular function. Here, our data show that PKR activation is reduced when RNAs contain nucleoside modifications that are naturally present in many cellular RNAs, including piRNA (53), snRNA, tRNA, mRNA and rRNA (54). Activation of TLRs (4) and RIG-I (5) is also influenced by modified nucleosides in RNA, and most commonly RNA modifications decrease the immunogenicity of RNA. Together, these data support a general interpretation that modified nucleosides supply a pattern for differential recognition by RNA-binding proteins. One purpose of common natural modifications may be avoiding activation of PKR and other RNA sensors by self-RNA.
Using mRNA for gene delivery has the benefits of efficient transfection and rapid protein expression without the risk of insertional mutagenesis. The potential of mRNA as a delivery vehicle is enhanced further by incorporating modified nucleosides that reduce host defense responses initiated by PKR, TLRs, and RIG-I (4,5,32). We recently reported the additional benefit of increased translation from Ψ-containing mRNA (33). In vitro transcribed mRNA is regularly delivered to cells in a research setting and has entered clinical trials as a cancer vaccine. As the interest in non-coding RNA continues, the delivery of RNA is likely to continue expanding. In most cases, activating PKR is an unwanted side effect. High translation and low immunogenicity make mRNA containing Ψ or m5C applicable to express therapeutic proteins, whereas s2U-modified RNA is best suited for applications where avoiding nonspecific immunogenicity is desirable but where translation is unnecessary (33), such as delivering antisense RNA (55) or stimulating RNA interference.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institutes of Health (R01AI50484, R21DE019059 to D.W., T32GM07229, T32DK07748, T32RR007063 to B.R.A., R42HL87688 to K.K. and R01GM058709 to P.C.B.). Funding for open access charge: National Institutes of Health grant R42HL87688.
Conflict of interest statement. K.K. and D.W. have formed a small biotech company RNARx that receives funding from the National Institutes of Health to explore the use of nucleoside-modified mRNA for gene therapy.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Tom Dever (NIH) for providing plasmids encoding swinepox C8L and vaccinia K3L, K3L-H47R and K3L-Y76A, and pG5 empty vector. We thank Robert Silverman (Cleveland Clinic Foundation) for providing PKR-deficient and wild-type MEF cell lines.
REFERENCES
- 1.Weissman D, Ni H, Scales D, Dude A, Capodici J, McGibney K, Abdool A, Isaacs SN, Cannon G, Karikó K. HIV gag mRNA transfection of dendritic cells (DC) delivers encoded antigen to MHC class I and II molecules, causes DC maturation, and induces a potent human in vitro primary immune response. J. Immunol. 2000;165:4710–4717. doi: 10.4049/jimmunol.165.8.4710. [DOI] [PubMed] [Google Scholar]
- 2.Nienhuis AW, Dunbar CE, Sorrentino BP. Genotoxicity of retroviral integration in hematopoietic cells. Mol. Ther. 2006;13:1031–1049. doi: 10.1016/j.ymthe.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Donsante A, Miller DG, Li Y, Vogler C, Brunt EM, Russell DW, Sands MS. AAV vector integration sites in mouse hepatocellular carcinoma. Science. 2007;317:477. doi: 10.1126/science.1142658. [DOI] [PubMed] [Google Scholar]
- 4.KarikóK, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann K.-K, Schlee M, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 6.Hunt T, Ehrenfeld E. Cytoplasm from poliovirus-infected HeLa cells inhibits cell-free haemoglobin synthesis. Nature New Biol. 1971;230:91–94. doi: 10.1038/newbio230091a0. [DOI] [PubMed] [Google Scholar]
- 7.Ehrenfeld E, Hunt T. Double-stranded poliovirus RNA inhibits initiation of protein synthesis by reticulocyte lysates. Proc. Natl Acad. Sci. U.S.A. 1971;68:1075–1078. doi: 10.1073/pnas.68.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minks MA, West DK, Benvin S, Baglioni C. Structural requirements of double-stranded RNA for the activation of 2′,5′-oligo(A) polymerase and protein kinase of interferon-treated HeLa cells. J. Biol. Chem. 1979;254:10180–10183. [PubMed] [Google Scholar]
- 9.Hunter T, Hunt T, Jackson RJ, Robertson HD. The characteristics of inhibition of protein synthesis by double-stranded ribonucleic acid in reticulocyte lysates. J. Biol. Chem. 1975;250:409–417. [PubMed] [Google Scholar]
- 10.Bevilacqua PC, George CX, Samuel CE, Cech TR. Binding of the protein kinase PKR to RNAs with secondary structure defects: role of the tandem A-G mismatch and noncontiguous helixes. Biochemistry. 1998;37:6303–6316. doi: 10.1021/bi980113j. [DOI] [PubMed] [Google Scholar]
- 11.Zheng X, Bevilacqua PC. Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA. 2004;10:1934–1945. doi: 10.1261/rna.7150804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318:1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- 13.Dauber B, Martinez-Sobrido L, Schneider J, Hai R, Waibler Z, Kalinke U, Garcia-Sastre A, Wolff T. Influenza B virus ribonucleoprotein is a potent activator of the antiviral kinase PKR. PLoS Pathog. 2009;5:e1000473. doi: 10.1371/journal.ppat.1000473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaempfer R. RNA sensors: novel regulators of gene expression. EMBO Rep. 2003;4:1043–1047. doi: 10.1038/sj.embor.7400005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edery I, Petryshyn R, Sonenberg N. Activation of double-stranded RNA-dependent kinase (dsl) by the TAR region of HIV-1 mRNA: a novel translational control mechanism. Cell. 1989;56:303–312. doi: 10.1016/0092-8674(89)90904-5. [DOI] [PubMed] [Google Scholar]
- 16.SenGupta DN, Silverman RH. Activation of interferon-regulated, dsRNA-dependent enzymes by human immunodeficiency virus-1 leader RNA. Nucleic Acids Res. 1989;17:969–978. doi: 10.1093/nar/17.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis S, Watson JC. In vitro activation of the interferon-induced, double-stranded RNA-dependent protein kinase PKR by RNA from the 3′ untranslated regions of human alpha-tropomyosin. Proc. Natl Acad. Sci. U.S.A. 1996;93:508–513. doi: 10.1073/pnas.93.1.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian B, White RJ, Xia T, Welle S, Turner DH, Mathews MB, Thornton CA. Expanded CUG repeat RNAs form hairpins that activate the double-stranded RNA-dependent protein kinase PKR. RNA. 2000;6:79–87. doi: 10.1017/s1355838200991544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben-Asouli Y, Banai Y, Pel-Or Y, Shir A, Kaempfer R. Human interferon-gamma mRNA autoregulates its translation through a pseudoknot that activates the interferon-inducible protein kinase PKR. Cell. 2002;108:221–232. doi: 10.1016/s0092-8674(02)00616-5. [DOI] [PubMed] [Google Scholar]
- 20.Nussbaum JM, Gunnery S, Mathews MB. The 3′-untranslated regions of cytoskeletal muscle mRNAs inhibit translation by activating the double-stranded RNA-dependent protein kinase PKR. Nucleic Acids Res. 2002;30:1205–1212. doi: 10.1093/nar/30.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Armstrong ME, Gantier M, Li L, Chung WY, McCann A, Baugh JA, Donnelly SC. Small interfering RNAs induce macrophage migration inhibitory factor production and proliferation in breast cancer cells via a double-stranded RNA-dependent protein kinase-dependent mechanism. J. Immunol. 2008;180:7125–7133. doi: 10.4049/jimmunol.180.11.7125. [DOI] [PubMed] [Google Scholar]
- 22.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z, Weinschenk T, Guo K, Schluesener HJ. siRNA binding proteins of microglial cells: PKR is an unanticipated ligand. J. Cell. Biochem. 2006;97:1217–1229. doi: 10.1002/jcb.20716. [DOI] [PubMed] [Google Scholar]
- 24.Schlee M, Hornung V, Hartmann G. siRNA and isRNA: two edges of one sword. Mol. Ther. 2006;14:463. doi: 10.1016/j.ymthe.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 25.Puthenveetil S, Whitby L, Ren J, Kelnar K, Krebs JF, Beal PA. Controlling activation of the RNA-dependent protein kinase by siRNAs using site-specific chemical modification. Nucleic Acids Res. 2006;34:4900–4911. doi: 10.1093/nar/gkl464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorina R, Santalucia T, Petegnief V, Ejarque-Ortiz A, Saura J, Planas AM. Astrocytes are very sensitive to develop innate immune responses to lipid-carried short interfering RNA. Glia. 2009;57:93–107. doi: 10.1002/glia.20738. [DOI] [PubMed] [Google Scholar]
- 27.Torrence PF, Friedman RM. Are double-stranded RNA-directed inhibition of protein synthesis in interferon-treated cells and interferon induction related phenomena? J. Biol. Chem. 1979;254:1259–1267. [PubMed] [Google Scholar]
- 28.Minks M, West D, Benvin S, Greene J, Ts′o P, Baglioni C. Activation of 2′,5′-oligo(A) polymerase and protein kinase of interferon-treated HeLa cells by 2′-O-methylated poly (inosinic acid):poly(cytidylic acid) J. Biol. Chem. 1980;255:6403–6407. [PubMed] [Google Scholar]
- 29.Content J, Lebleu B, De Clercq E. Differential effects of various double-stranded RNAs on protein synthesis in rabbit reticulocyte lysates. Biochemistry. 1978;17:88–94. doi: 10.1021/bi00594a012. [DOI] [PubMed] [Google Scholar]
- 30.Torrence PF, Johnston MI, Epstein DA, Jacobsen H, Friedman RM. Activation of human and mouse 2-5A synthetases and mouse protein P1 kinase by nucleic acids. Structure-activity relationships and correlations with inhibition of protein synthesis and interferon induction. FEBS Lett. 1981;130:291–296. doi: 10.1016/0014-5793(81)81142-8. [DOI] [PubMed] [Google Scholar]
- 31.Puthenveetil S, Veliz EA, Beal PA. Site-specific modification of Epstein-Barr virus-encoded RNA 1 with N2-benzylguanosine limits the binding sites occupied by PKR. Chembiochem. 2004;5:383–386. doi: 10.1002/cbic.200300816. [DOI] [PubMed] [Google Scholar]
- 32.Nallagatla SR, Bevilacqua PC. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008;14:1201–1213. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kariko K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romano PR, Garcia-Barrio MT, Zhang X, Wang Q, Taylor DR, Zhang F, Herring C, Mathews MB, Qin J, Hinnebusch AG. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2alpha kinases PKR and GCN2. Mol. Cell. Biol. 1998;18:2282–2297. doi: 10.1128/mcb.18.4.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawagishi-Kobayashi M, Cao C, Lu J, Ozato K, Dever TE. Pseudosubstrate inhibition of protein kinase PKR by swine pox virus C8L gene product. Virology. 2000;276:424–434. doi: 10.1006/viro.2000.0561. [DOI] [PubMed] [Google Scholar]
- 36.Kawagishi-Kobayashi M, Silverman JB, Ung TL, Dever TE. Regulation of the protein kinase PKR by the vaccinia virus pseudosubstrate inhibitor K3L is dependent on residues conserved between the K3L protein and the PKR substrate eIF2alpha. Mol. Cell. Biol. 1997;17:4146–4158. doi: 10.1128/mcb.17.7.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen-Chalamish S, Hasson A, Weinberg D, Namer LS, Banai Y, Osman F, Kaempfer R. Dynamic refolding of IFN-gamma mRNA enables it to function as PKR activator and translation template. Nat. Chem. Biol. 2009;5:896–903. doi: 10.1038/nchembio.234. [DOI] [PubMed] [Google Scholar]
- 38.Karikó K, Weissman D. Naturally-occurring nucleoside modifications suppress the immunostimulatory activity of RNA: Implication for therapeutic RNA development. Curr. Opin. Drug Discov. Devel. 2007;10:523–532. [PubMed] [Google Scholar]
- 39.Szer W, Ochoa S. Complexing ability and coding properties of synthetic polynucleotides. J. Mol. Biol. 1964;12:823–834. doi: 10.1016/s0022-2836(64)80163-7. [DOI] [PubMed] [Google Scholar]
- 40.Ziomek K, Kierzek E, Biala E, Kierzek R. The influence of various modified nucleotides placed as 3′-dangling end on thermal stability of RNA duplexes. Biophys. Chem. 2002;97:243–249. doi: 10.1016/s0301-4622(02)00073-x. [DOI] [PubMed] [Google Scholar]
- 41.Ziomek K, Kierzek E, Biala E, Kierzek R. The thermal stability of RNA duplexes containing modified base pairs placed at internal and terminal positions of the oligoribonucleotides. Biophys. Chem. 2002;97:233–241. doi: 10.1016/s0301-4622(02)00074-1. [DOI] [PubMed] [Google Scholar]
- 42.Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 43.Kumar RK, Davis DR. Synthesis and studies on the effect of 2-thiouridine and 4-thiouridine on sugar conformation and RNA duplex stability. Nucleic Acids Res. 1997;25:1272–1280. doi: 10.1093/nar/25.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nair TM, Myszka DG, Davis DR. Surface plasmon resonance kinetic studies of the HIV TAR RNA kissing hairpin complex and its stabilization by 2-thiouridine modification. Nucleic Acids Res. 2000;28:1935–1940. doi: 10.1093/nar/28.9.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luyten I, Herdewijn P. Hybridization properties of base-modified oligonucleotides within the double and triple helix motif. Eur. J. Med. Chem. 1998;33:515–576. [Google Scholar]
- 46.Kierzek E, Kierzek R. The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res. 2003;31:4472–4480. doi: 10.1093/nar/gkg633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bevilacqua PC, Cech TR. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry. 1996;35:9983–9994. doi: 10.1021/bi9607259. [DOI] [PubMed] [Google Scholar]
- 48.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robbins M, Judge A, Liang L, McClintock K, Yaworski E, Maclachlan I. 2′-O-methyl-modified RNAs act as TLR7 antagonists. Mol. Ther. 2007;15:1663–1669. doi: 10.1038/sj.mt.6300240. [DOI] [PubMed] [Google Scholar]
- 50.Yu D, Wang D, Zhu FG, Bhagat L, Dai M, Kandimalla ER, Agrawal S. Modifications incorporated in CpG motifs of oligodeoxynucleotides lead to antagonist activity of toll-like receptors 7 and 9. J. Med. Chem. 2009;52:5108–5114. doi: 10.1021/jm900730r. [DOI] [PubMed] [Google Scholar]
- 51.Wang D, Bhagat L, Yu D, Zhu FG, Tang JX, Kandimalla ER, Agrawal S. Oligodeoxyribonucleotide-based antagonists for Toll-like receptors 7 and 9. J. Med. Chem. 2009;52:551–558. doi: 10.1021/jm8014316. [DOI] [PubMed] [Google Scholar]
- 52.Hamm S, Latz E, Hangel D, Müller T, Yu P, Golenbock D, Sparwasser T, Wagner H, Bauer S. Alternating 2′-O-ribose methylation is a universal approach for generating non-stimulatory siRNA by acting as TLR7 antagonist. Immunobiology. 2009 doi: 10.1016/j.imbio.2009.09.003. In Press, doi:10.1016/j.imbio.2009.1009.1003. [DOI] [PubMed] [Google Scholar]
- 53.Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat. Struct. Mol. Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- 54.Rozenski J, Crain P, McCloskey J. The RNA Modification Database: 1999 update. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prakash TP, Naik N, Sioufi N, Bhat B, Swayze EE. Activity of siRNAs with 2-thio-2′-O-methyluridine modification in mammalian cells. Nucleosides Nucleotides Nucleic Acids. 2009;28:902–910. doi: 10.1080/15257770903316145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.