Abstract
CeR-2 RNA is one of the newly identified Caenorhabditis elegans noncoding RNAs (ncRNAs). The characterization of CeR-2 by RNomic studies has failed to classify it into any known ncRNA family. In this study, we examined the spatiotemporal expression patterns of CeR-2 to gain insight into its function. CeR-2 is expressed in most cells from the early embryo to adult stages. The subcellular localization of this RNA is analogous to that of fibrillarin, a major protein of the nucleolus. It was observed that knockdown of C/D small nucleolar ribonucleoproteins (snoRNPs), but not of H/ACA snoRNPs, resulted in the aberrant nucleolar localization of CeR-2 RNA. A mutant worm with a reduced amount of cellular CeR-2 RNA showed changes in its pre-rRNA processing pattern compared with that of the wild-type strain N2. These results suggest that CeR-2 RNA is a C/D snoRNA involved in the processing of rRNAs.
INTRODUCTION
The ribosome is an essential component of the cell and is deeply involved in the regulation of cell growth, proliferation and differentiation (1–7). Despite its significance, little is known about the biogenesis of the ribosome. In eukaryotes, hundreds of nonribosomal proteins and small nucleolar RNAs (snoRNAs) are involved in ribosome assembly, and it is thought that they are elaborately coordinated to form a functional ribosome in response to cell circumstances (8,9).
Four RNA species are involved in eukaryotic ribosomes: the 18S ribosomal RNA (rRNA), which is a component of the small subunit (40S), and the 5.8S rRNA, 28S rRNA (25S or 28S, depending on the organism) and 5S rRNA, which are components of the large subunit (60S). The 18S, 5.8S and 28S rDNAs are aligned in tandem and cotranscribed by RNA polymerase I in the nucleolus (10). The primary transcripts contain extra sequences designated the 5′ external transcribed spacer (ETS), 3′-ETS, internal transcribed spacer 1 (ITS1) and ITS2, which are removed to produce the mature 18S, 5.8S and 28S rRNAs (11). Studies of rRNA maturation in yeast, frogs and mammals have shown that there are many similarities and differences in the pathways of pre-rRNA processing among species (6,12–15).
Nine snoRNAs (U3, U14, U17/E1/snR30, snR10, U8, U22, MRP, E2 and E3) are related to pre-rRNA processing. U3, U14 and U17/E1/snR30 are evolutionarily conserved snoRNAs that are required for the maturation of 18S rRNA (16–18). MRP snoRNAs are found in a variety of eukaryotic organisms. According to yeast analyses, this RNA functions in the cleavage of ITS1. However, it is not yet clear whether MRP snoRNA is involved in rRNA maturation in mammals and other organisms too. Similarly, snR10 has been identified in several organisms, but it has not been confirmed that this RNA functions in the cleavage of pre-rRNAs other than in yeast. The remaining snoRNAs, U8, U22, E2 and E3, have only been found in vertebrates to date. U3, U14, U8 and U22 snoRNAs share some features with C/D snoRNAs, and U17/E1/snR30, snR10, E2 and E3 RNAs share features with H/ACA snoRNAs (6,17–23).
Caenorhabditis elegans is a good model to study how various physiological phenomena occur based on the molecular systems in cells. However, little is known about C. elegans ribosome biogenesis. Until recently, the cleavage sites of the pre-rRNAs remained unclear (24). U3 is the only snoRNA that probably functions in pre-rRNA cleavage in C. elegans (25). Although several RNomic studies have suggested candidates for snR10, U14, U17 and MRP RNA homologs in C. elegans (23,26,27), there is no biochemical or genetic evidence that they are involved in pre-rRNA processing. Moreover, it is unclear whether RNAs homologous to U8, U22, E2 or E3 are expressed in C. elegans.
In our previous study, we identified 19 novel ncRNA candidates in C. elegans (28,29). Seven showed the characteristic secondary structure of the modification-guiding C/D or H/ACA snoRNAs (28). None of the remaining 12 candidates showed marked similarity to any known ncRNA sequence in the database. Here, we show that one of these RNAs, designated CeR-2 RNA and also known as CeN21 or Ce9 (26,30), has several characteristics of a C/D snoRNA and is likely to function in rRNA processing.
MATERIALS AND METHODS
Caenorhabditis elegans strains and culture
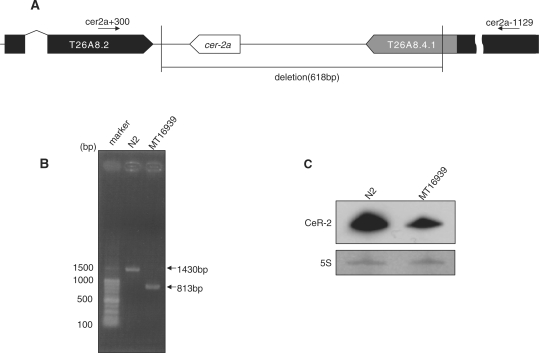
Worms were grown and maintained by standard procedures (31). Strain MT16939 containing a cer-2a mutant allele (n5007), which lacks a region encompassing nts 8 428 510–8 429 125 of chromosome IV, was generated by ethane methyl sulfonate mutagenesis. The cer-2a (n5007) worms were outcrossed to N2 animals six times before analysis. The cer-2a (n5007) worms were genotyped by polymerase chain reaction (PCR) using the primers cer-2a-1129 (5′-CCACAAGCTTTCATTTAGAGG-3′) and cer-2a+300 (5′-TTTACAATTGTTGATTACGTTTTTACCTC-3′). The positions and directions of these primers are shown in Figure 4A.
Figure 4.
CeR-2 RNA expression in MT16939. (A) Genomic map around cer-2a of MT16939 on chromosome IV. The primers cer-2a-1129 and cer-2a+300 refer to the PCR primers used for genotyping N2 and MT16939. There are two putative protein genes, T26A8.2 and T26A8.4.1, adjacent to cer-2a. (B) Genotyping of N2 and MT16939 worms by single-worm PCR. Marker, 100-bp DNA ladder; N2, PCR product amplified from the genomes of N2 worms; MT16939, PCR product amplified from the genomes of MT16939 worms. The PCR products are indicated by arrows with their putative lengths (bp). (C) Northern hybridization of total RNAs from N2 and MT16939 with the CeR-2 RNA antisense probe. The 5S rRNA band on the blot, stained with methylene blue, is shown subsequently.
Plasmids
The plasmid pT7CER2aSP6 was designed to express CeR-2 RNA from a T7 promoter and to express an antisense CeR-2 RNA from an SP6 promoter. The DNA fragments were amplified by nested PCR. The first PCR was performed with a C. elegans genomic DNA template and primers CeR2aT7F1 (5′-CGACTCACTATAGTCTTCAGTATGGGTCA-3′) and CeR2aSP6R (5′-AGGTGACACTATAGTTCAGAATCGGGCTGG-3′), which contain T7 promoter and SP6 promoter sequences (underlined), respectively. The PCR mixture was then used as the template for the second PCR, which was performed with the primers EcoRIT7 (5′-AAAGAATTCTAATACGACTCACTATA-3′) and PstISP6 (5′-AAACTGCAGATTTAGGTGACACTATA-3′). The resulting DNA fragment was digested with EcoRI and PstI and ligated into the same sites of pUC19. Clones pT7U18SP6 and pT7U17SP6 were prepared by the same procedures. The primers used for the first PCR were U18(T7)F 5′-CGACTCACTATAGTGGCAGTGATGATCACAAATC-3′, U18(SP6)R 5′-AGGTGACACTATAGTGGCTCAGCCGGTTTTC-3′, U17(T7)F 5′-CGACTCACTATAGCTCGACATGTGACTAGCG-3′ and U17(SP6)R 5′-AGGTGACACTATAGATTTGTAATTTGCATGGTTTG-3′. EcoRIT7 and PstISP6 were used as the second PCR primers. The clone containing part of the rRNA precursor sequence has been described previously (24).
Northern hybridization
Total RNAs from N2 and MT16939 were extracted with TRIzol Reagent or the PureLink RNA Mini Kit (Invitrogen). The RNAs were resolved on formaldehyde-containing 1.0% agarose gel or by 7 M urea/6% polyacrylamide gel electrophoresis and blotted onto Biodyne Plus membrane (Pall Corporation). The blot was hybridized with RNA probes prepared with the DIG RNA Labeling Kit (Roche). The templates for RNA synthesis were amplified by PCR from the clones as described earlier. The probes used for detecting the pre-rRNA intermediates have been described previously (24). Immunoprecipitation with anti-2,2,7-trimethylguanosine (TMG) antibody K121 was based on a previous work (25).
RNA fluorescence in situ hybridization of small RNAs and immunofluorescence analysis
Specimens for RNA fluorescence in situ hybridization and immunofluorescence analysis were prepared as described previously (32,33). The RNA probes were prepared using the DIG RNA Labeling Kit (SP6/T7) or with MEGAscript/SP6 (Ambion, Inc.) and fluorescein-12-UTP (Enzo Industries, Inc.). The DIG haptens were detected by Cy3-conjugated IgG fraction monoclonal mouse anti-DIG antibody (1:400 dilution; Jackson ImmunoResearch Laboratories, Inc.; lot 59998) or fluorescein-conjugated anti-DIG Fab fragment (1:25 dilution; Roche). Cy3-conjugated Affinipure goat anti-mouse IgG (H+L) antibody (1:400 dilution; Jackson ImmunoResearch Laboratories, Inc.) or Alexa-488-conjugated anti-fluorescein/Oregon Green rabbit polyclonal IgG (1:100 dilution; Molecular Probes, Inc.) was used as the secondary antibody. The signals of the fluorescein-labeled RNA probes were enhanced with Alexa-488-conjugated anti-fluorescein/Oregon Green rabbit antibody and anti-rabbit chicken IgG antibody (1:100; Molecular Probes, Inc.). Fibrillarin (FIB-1) was visualized with the anti-FIB-1 antibody 38F3 (1:400 dilution; EnCor Biotechnology Inc.) and Cy3-conjugated anti-mouse antibody. The nuclei were stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI). The signals were observed under a light fluorescence microscope (Olympus, BX60) or a confocal laser microscope (Leica, DMI6000, TCS SP5 and Olympus, FV1000D, FV10-ASW).
RNA interference
The template DNAs for in vitro transcription were generated by PCR using yk cDNA clones (nop10 [C25A1.6], yk1472f12; nop56 [K07C5.4], yk1604e03), which were kind gifts from Dr Yuji Kohara. Oligonucleotides containing the T7 promoter sequence were used as primers (T7ME774FW, 5′-TTTTAATAATACGACTCACTATAGCTTCTGCTCTAAAAGCTGCG-3′ and T7ME1250RV, 5′-TAAAGATAATACGACTCACTATAGTGTGGGAGGTTTTTTCTCTTAG-3′). Sense and antisense RNAs were synthesized using MEGAscript/T7 (Ambion). The resulting RNAs were annealed to generate double-stranded RNAs. The double-stranded RNAs (1 µg/µl) were injected into L4 worms.
RESULTS AND DISCUSSION
Isolation of CeR-2 RNA
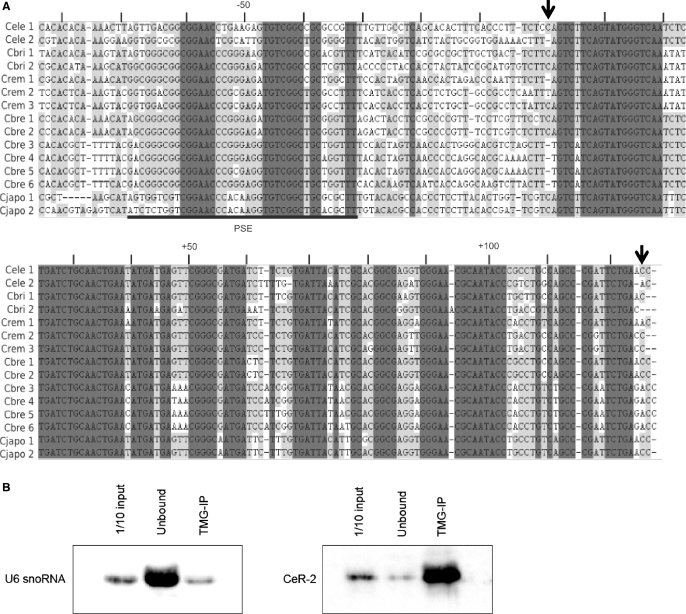
CeR-2 RNA is one of the 19 small novel candidate ncRNAs isolated from C. elegans (28,29). This RNA is encoded in the intergenic region of chromosome IV. A homologous gene is found on the same chromosome: the former is designated cer-2a and the latter is designated cer-2b (chromosome IV, nts 4 880 726–4 880 851, complement). A BLAST search showed that the genomes of nematodes closely related to C. elegans, C. briggsae (CB3; 2006), C. brenneri (6.0.1 contigs), C. remanei (15.0.1 supercontigs) and C. japonica (3.0.2 supercontigs), have two, six, three and two homologs of CeR-2, respectively, but no similar sequence was found in any other organism. Figure 1A shows the sequence alignment of the CeR-2 RNA gene with its homologs, constructed using Align X of the software package Vector NTI ver. 9 (Invitrogen). There are two highly conserved sequences: one corresponds to the sequence of CeR-2 RNA itself and the other is located ∼30–40-bp upstream from the CeR-2-RNA-coding region. The upstream sequence (−32 to −69 in Figure 1A) is similar to the proximal sequence element, which functions as a promoter in U snRNA genes (34–37). This suggests that CeR-2 RNA is transcribed by RNA polymerase II and has a TMG cap at its 5′ end. To examine this possibility, we performed immunoprecipitation with the anti-TMG antibody K121 against the total RNAs extracted from mixed-stage worms. The anti-TMG-precipitated RNAs were then subjected to Northern blot analysis (Figure 1B). As a control for immunoprecipitation specificity, U6 snRNA, which does not have a TMG cap (25), was also monitored by Northern blot analysis. Most of the CeR-2 RNA was detected in the K121 precipitate, whereas the majority of the U6 snRNA was detected in the supernatant. This indicates that most CeR-2 RNAs in the cell have a TMG cap at their 5′ ends, like other C. elegans small RNAs, such as the U snRNAs, SL RNAs (38) and U3 snoRNA (25).
Figure 1.
Sequence alignment of CeR-2 RNA genes. (A) Sequence alignment of cer-2a and its homologs. The sequences conserved among all 15 homologs are shaded in dark gray. Partially conserved sequences are shaded in light gray. Cele1, cer-2a; Cele2, cer-2b; Cbri1, C. briggsae homolog of cer-2a; Cbri2, C. briggsae homolog of cer-2b; Crem1-3, three homologs found in C. remanei; Cbre1-6, six homologs found in C. brenneri; Cjapo1 and Cjapo2, two homologs found in C. japonica. The putative 5′- and 3′-terminal nucleotides of the RNA-coding region in cer-2a are indicated by arrows. (B) Caenorhabditis elegans TMG-capped RNA was precipitated with the anti-TMG antibody K121. Northern hybridization was performed with antisense probes of CeR-2 RNA and U6 snRNA against the precipitate (TMG-IP) and the supernatant (Unbound). A 1/10 amount of total RNA input was also separated on the same denaturing gel and blotted onto a nylon membrane (1/10 input).
Spatiotemporal expression patterns and subcellular localization of CeR-2 RNA
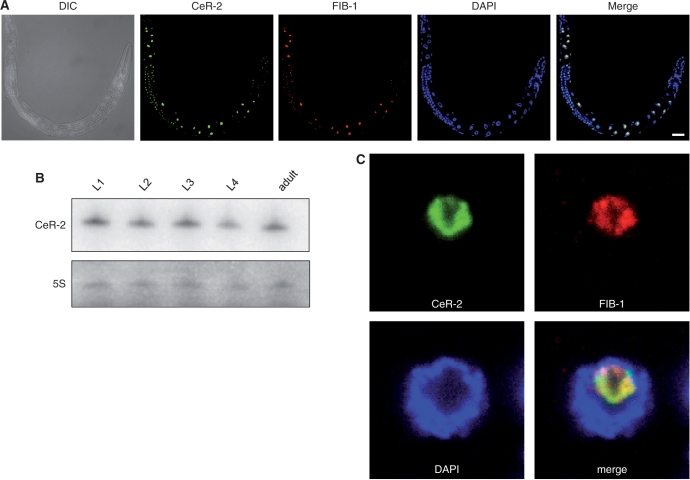
We examined the spatiotemporal expression patterns and subcellular localization of CeR-2 RNA by in situ hybridization. As shown in Figure 2A, CeR-2 is expressed in most cells in L1 larvae and it continues to be expressed until adulthood in both the somatic and germline cells. Consistently, Northern hybridization of total RNA revealed that CeR-2 is expressed constitutively during the four larval stages and the adult stage (Figure 2B).
Figure 2.
Spatiotemporal expression patterns of CeR-2 RNA. (A) CeR-2 RNA was detected by in situ hybridization (CeR-2, green). FIB-1 was costained with an anti-FIB antibody (FIB-1, red). DIC, Nomarski differential interference contrast microscopic image; DAPI, DNA visualized with DAPI staining; Merge, merged images of CeR-2, FIB-1, and DAPI. Scale bar, 5 µm. (B) Northern hybridization of CeR-2 RNA against total RNAs prepared from the larvae of each stage (L1 to L4) and adults. The 5S rRNA band on the blot was detected with methylene blue staining. (C) CeR-2 RNA foci in the nucleus overlapped with FIB-1 foci. The stained images of an intestinal nucleus are magnified. CeR-2 RNA (green) and FIB-1 (red) were stained with DAPI.
To determine the subcellular localization of CeR-2, we inspected the large intestinal nucleus (Figure 2C). Foci of CeR-2 RNA were detected inside the nucleus and completely overlapped with the signals for FIB-1 (Figure 2C). This indicates that CeR-2 RNA localizes in the nucleolus. Thus, CeR-2 RNA shares the most characteristic feature of the snoRNAs that function in pre-rRNA processing or rRNA modification.
Changes in the nucleolar localization of CeR-2 RNA by knockdown of the C/D snoRNP gene
There are two major snoRNA families, the C/D snoRNA and H/ACA snoRNA families. Four core proteins interact specifically with the RNAs of each family: fibrillarin/NOP1, NOP56, NOP58 and a 15.5-kDa protein with C/D snoRNAs, and dyskerin/NAP57, NHP2, NOP10 and GAR1 with H/ACA snoRNAs (39). We expected that some of these proteins would interact with CeR-2 RNA and contribute to its function and nucleolar localization. Therefore, we knocked down the expression of the C. elegans C/D snoRNP gene nop56 (K07C5.4) and the H/ACA snoRNP gene nop10 (C25A1.6). The effects of RNA interference (RNAi) were assayed by in situ hybridization and immunofluorescence.
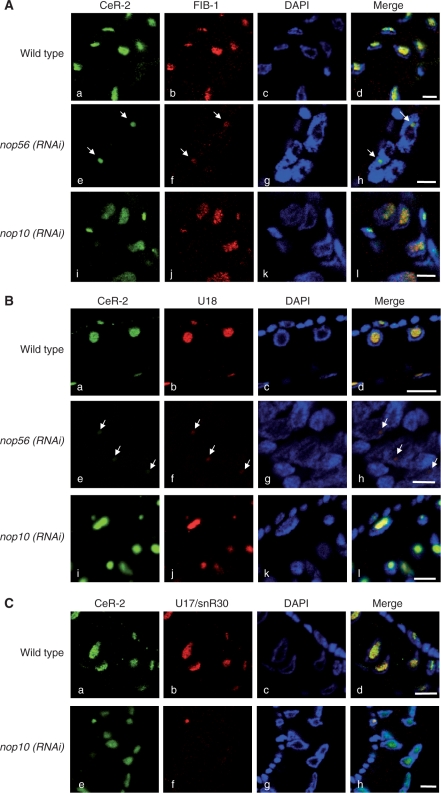
As expected, knockdown of nop56, which encodes a C/D snoRNP-specific protein, markedly reduced the signals of CeR-2 RNA and FIB-1 (Figure 3A). In several cells of the nop56 (RNAi) worm, both signals were observed in a limited region at the periphery of the nucleolus (Figure 3A and B, arrows). U18 snoRNA, a typical C/D snoRNA, was used as an internal control to monitor the knockdown effect of nop56. The level of nucleolar localization was reduced by the reduction of Nop56 but not by the reduction of Nop10. The effect of nop10 knockdown was confirmed by the reduction of the U17 H/ACA-type snoRNA (Figure 3C). It is likely that CeR-2 RNA is a member of the C/D snoRNA family and functions in the nucleolus together with C/D snoRNPs. A recent study based on a microarray indicated that the knockdown of nop58 or snu13 leads to a severe reduction in CeR-2 RNA (CeN21 RNA in refs. 26 and 40). This also supports our prediction that CeR-2 RNA is a C/D snoRNA.
Figure 3.
Knockdown of nop56 reduced the nucleolar localization of CeR-2 RNA. Each nucleolar factor was observed by in situ hybridization or immunofluorescence analysis. The white arrows indicate the accumulation of CeR-2 RNA, FIB-1 or U18 snoRNA in the foci, which newly appeared in the nucleoplasm after the knockdown of nop56. Scale bars, 5 µm. (A) Costaining of CeR-2 RNA (green, panels a, e and i) and FIB-1 (red, panels b, f and j) in nop56 (RNAi) and nop10 (RNAi) worms. (B) Costaining of CeR-2 RNA (green, panels a, e and i) and U18 C/D snoRNA (red, panels b, f and j) in nop56 (RNAi) worms and nop10 (RNAi) worms. (C) Costaining of CeR-2 RNA (green, panels a, e and i) and U17/snR30 H/ACA snoRNA (red, panels b, f and j) in nop10 (RNAi) worms. Panels c, g and k, DAPI staining; panels d, h and l, merged images.
A mutant lacking cer-2a shows an altered accumulation pattern of pre-rRNAs
To determine the function of CeR-2 RNA, we produced a mutant strain lacking cer-2a and designated it MT16939. This deletion mutant of cer-2a (n5007) lacks a 618-bp sequence on chromosome IV (Figure 4A and B). Northern blot analysis indicated that CeR-2 RNA was reduced by about half to one-third in cer-2a mutants compared with that in wild-type N2 worms (Figure 4C). The remaining CeR-2 RNA signal on Northern blots originated from cer-2b, a homolog of cer-2a, with a 98% identical sequence. The homozygous mutant of cer-2a showed a slow growth phenotype and abnormal fertilization, especially in old adults. We tried to generate a mutant of cer-2b, but were unsuccessful.
Because CeR-2 RNA exhibits the characteristics of a box C/D-type snoRNA, it was expected that CeR-2 RNA would function in guiding the 2′-O-methylation of rRNAs and/or in processing pre-rRNAs (1–6,41,42). One important structural feature of modification guiding C/D snoRNAs is that the region upstream from the D or D′ box encompasses 10–21 bp complementary to the target rRNA around the modification site. When the duplex is <9 bp or contains substantial AU or GU pairs, methylation becomes less efficient (42). Therefore, searching for a complementary sequence to an rRNA sequence longer than 10 bp is one way to assess the function of a C/D snoRNA in guiding the 2′-O-methylation of rRNA. We searched for a complementary sequence between CeR-2 RNA and C. elegans rRNAs that was longer than 10 bp. However, no such continuous sequences were found, which reduced our expectation that CeR-2 RNA functions in guiding the modification of rRNAs.
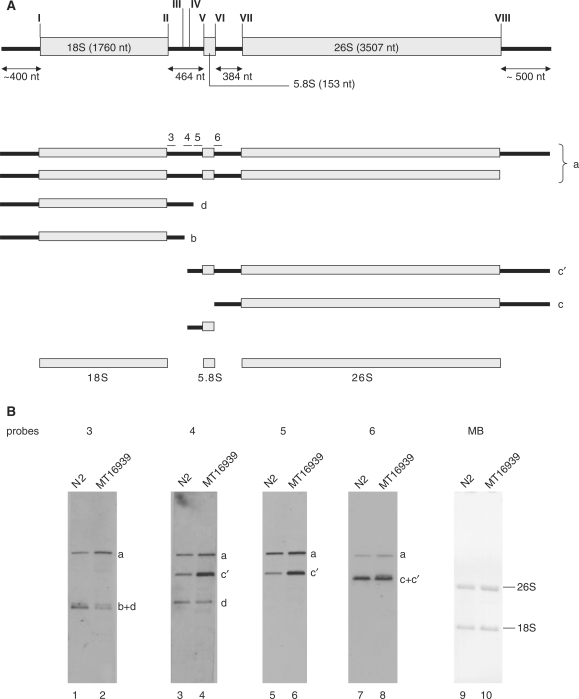
An outline of C. elegans rRNA processing was established in a previous study (Figure 5A) (24). In N2 worms, five pre-rRNA processing intermediates were detected and designated a, b, c, c′ and d. We designed four probes (probes 3, 4, 5 and 6), with reference to the study of Saijou et al. (24), to detect each intermediate. Figure 5B shows the results of Northern hybridization of RNA extracts from N2 (wild-type) worms and MT16939 (cer-2a [n5007]) worms with these probes. Intermediate c′ accumulated more in MT16939 than in N2, as shown in the results for probes 4, 5 and 6 (Figure 5B, lanes 4, 6 and 8, respectively). The accumulation of c′ indicates that the efficiency of processing the large subunit rRNA precursor into 5.8S and 26S rRNAs was reduced in the mutant after the cleavage of the pre-rRNA in ITS1. The results for probe 3 showed reductions in intermediates b and/or d in the mutant (Figure 5B, lanes 1 and 2, respectively). Intermediate d, detected with probe 4, did not differ significantly between N2 and MT16939 (Figure 5B, lanes 3 and 4, respectively), which indicates that intermediate b, which is a precursor of 18S rRNA, was reduced in the mutant. Therefore, MT16939 exhibited changes in the accumulation patterns of the rRNA precursors. This suggests that CeR-2 RNA is involved in the processing of pre-rRNAs, although it is still possible that CeR-2 RNA guides the modification of rRNAs.
Figure 5.
pre-rRNAs in MT16939. (A) Schematic representation of pre-rRNA processing pattern. Probes 3, 4, 5, and 6 used for the hybridization are indicated by bars above intermediate a. The cleavage sites (I–VIII) are indicated along the precursor a with reference to the study of Saijou et al. (24). The length of the rDNA is based on nucleotide data for GenBank accession number X03680. (B) Comparison of the pre-rRNA patterns of N2 and MT16939. Northern hybridization of the RNAs from N2 (lanes 1, 3, 5 and 7) and MT16939 (lanes 2, 4, 6 and 8) with each probe. Intermediates a, b, c, c′ and d indicate the pre-rRNAs shown in Figure 5A. The membrane was stained with methylene blue (lanes 9 and 10, MB), and the 26S and 18S rRNA bands are shown.
Because MT16939 lacks a part of the 3′ untranslated region (UTR) of the gene upstream from cer-2a (T26A8.4.1), it is possible that T26A8.4.1 affects the processing of rRNAs. The homozygous mutation of T26A8.4.1 is lethal and it is therefore difficult to analyze the pre-rRNA patterns. We tried to detect pre-rRNAs in worms in which T26A8.4.1 was knocked down. Although no obvious changes in the pre-rRNA pattern were observed, we cannot completely rule out the possibility that T26A8.4.1 is relevant to rRNA processing.
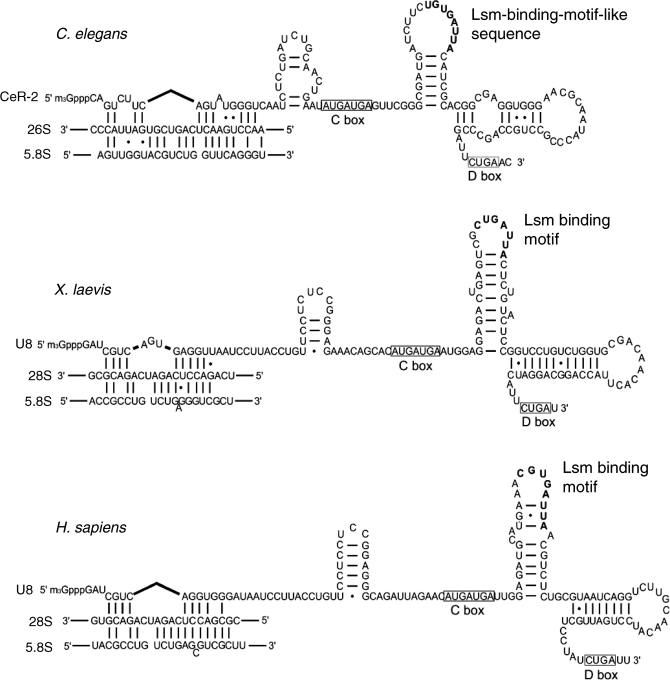
U8 and U22 snoRNAs are C/D-type snoRNAs related to the cleavage of rRNA processing. They have been identified only in vertebrates and their homologs have not been found in invertebrates to date. Some features of CeR-2 RNA shown here revealed similarity to those of U8 snoRNAs: both RNAs are TMG-capped, have features of C/D snoRNAs, and are involved in the cleavage of ITS2. In addition, the 5′-terminal sequence of CeR-2 RNA has the potential to base pair with that of C. elegans 26S rRNA, as U8 snoRNA base pairs with the 5′ terminus of 28S rRNA (Figure 6). There is a sequence similar to the conserved LSm binding motif of U8 snoRNA in the second stem-loop (Figure 6). Thus, CeR-2 RNA is an excellent candidate to be a U8 ortholog.
Figure 6.
Comparison of the predicted secondary structures of CeR-2 RNA and vertebrate U8 snoRNAs. Potential base pairing between the 5′ region of CeR-2 RNA and the 5′ region of 26S rRNA is shown and compared with that between U8 snoRNA and 28S rRNA of Xenopus laevis and Homo sapiens (20,43). There are three stem-loops in the remaining 3′ part of CeR-2 RNA, which appear in the secondary structure of U8 snoRNAs in similar regions: one upstream from the C-box and the other two between the C-box and the D-box (open rectangles). The LSm binding motif is a conserved octameric sequence located in the loop of the second stem-loop in U8 snoRNAs (44). Similar sequences (six of the conserved eight nucleotides, LSm-binding-motif-like sequence) are also found in the loop of the second stem-loop of CeR-2 RNA (bold letters).
FUNDING
Grant-in Aid for Scientific Research (C) 18570157 to C.U. from the Ministry of Education, Science, Sports and Culture (MEXT), Japan; a grant from the Functional RNA Project to C.U. from the Ministry of Economy, Trade and Industry; NEDO, Special Coordination Funds for Promoting Science and Technology; the Creation of Innovation Centers for Advanced Interdisciplinary Research Areas (Innovative Bioproduction Kobe) to T.F. from MEXT, Japan; and the 21st COE Program of Iwate University to Y.H.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dr Robert Horvitz and Dr Ezequiel Alvarez-Saavedra at the Department of Biology, Massachusetts Institute of Technology, Cambridge, for isolating the cer-2a (n5007) worm. They also thank Dr Katsuya Yamada at the School of Medicine, Hirosaki University, and the staff of the Gene Research Center of Hirosaki University for allowing us the use of their facilities.
REFERENCES
- 1.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 2.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 3.Granneman S, Baserga SJ. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 2004;296:43–50. doi: 10.1016/j.yexcr.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 4.Hage AE, Tollervey D. Why is ribosome assembly so much more complicated in eukaryotes than bacteria? RNA Biol. 2004;1:10–15. [PubMed] [Google Scholar]
- 5.Dinman J. The eukaryotic ribosome: current status and challenges. J. Biol. Chem. 2009;284:11761–11765. doi: 10.1074/jbc.R800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerbi SA, Borovjagin AV, Ezrokhi M, Lange TS. The Ribosome, Cold Spring Harbor Symposia on Quantitative Biology. Vol. LXVI. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. Ribosome biogenesis: role of small nucleolar RNA in maturation of eukaryotic rRNA; pp. 575–590. [DOI] [PubMed] [Google Scholar]
- 7.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yewdell JW, Nicchitta CV. The DRiP hypothesis decennial: support, controversy, refinement and extension. Trends Immunol. 2006;27:368–373. doi: 10.1016/j.it.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Kelleher RJ, III, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 10.Mougey EB, O’Reilly M, Osheim Y, Miller O.L., Jr, Beyer A, Sollner-Webb B. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev. 1993;7:1609–1619. doi: 10.1101/gad.7.8.1609. [DOI] [PubMed] [Google Scholar]
- 11.Winicov I. Alternate temporal order in ribosomal RNA maturation. J. Mol. Biol. 1976;100:141–155. doi: 10.1016/s0022-2836(76)80145-3. [DOI] [PubMed] [Google Scholar]
- 12.Dabeva MD, Dudov KP, Hadjiolov AA, Emanuilov I, Todorov BN. Intranuclear maturation pathways of rat liver ribosomal ribonucleic acids. Biochem. J. 1976;160:495–503. doi: 10.1042/bj1600495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loening UE, Jones KW, Birnstiel ML. Properties of the ribosomal RNA precursor in Xenopus laevis; comparison to the precursor in mammals and in plants. J. Mol. Biol. 1969;45:353–366. doi: 10.1016/0022-2836(69)90110-7. [DOI] [PubMed] [Google Scholar]
- 14.Udem SA, Warner JR. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J. Biol. Chem. 1973;248:1412–1416. [PubMed] [Google Scholar]
- 15.Borovjagin AV, Gerbi SA. U3 small nucleolar RNA is essential for cleavage at sites 1, 2 and 3 in pre-rRNA and determines which rRNA processing pathway is taken in Xenopus oocytes. J. Mol. Biol. 1999;286:1347–1363. doi: 10.1006/jmbi.1999.2527. [DOI] [PubMed] [Google Scholar]
- 16.Atzorn V, Fragapane P, Kiss T. U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production. Mol. Cell. Biol. 2004;24:1769–1778. doi: 10.1128/MCB.24.4.1769-1778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enright CA, Maxwell ES, Eliceiri GL, Sollner-Webb B. 5′ETS rRNA processing facilitated by four small RNAs: U14, E3, U17, and U3. RNA. 1996;2:1094–1099. [PMC free article] [PubMed] [Google Scholar]
- 18.Mishra RK, Eliceiri GL. Three small nucleolar RNAs that are involved in ribosomal RNA precursor processing. Proc. Natl Acad. Sci. USA. 1997;94:4972–4977. doi: 10.1073/pnas.94.10.4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peculis BA, Steitz JA. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993;73:1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- 20.Peculis BA. The sequence of the 5′ end of the U8 small nucleolar RNA is critical for 5.8S and 28S rRNA maturation. Mol. Cell. Biol. 1997;17:3702–3713. doi: 10.1128/mcb.17.7.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tycowski KT, Shu MD, Steitz JA. Requirement for intron-encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science. 1994;266:1558–1561. doi: 10.1126/science.7985025. [DOI] [PubMed] [Google Scholar]
- 22.Tycowski KT, Kolev NG, Conrad NK, Fok V, Steitz JA. The ever-growing world of small nuclear ribonucleoproteins. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 327–368. [Google Scholar]
- 23.Huang ZP, Chen CJ, Zhou H, Li BB, Qu LH. A combined computational and experimental analysis of two families of snoRNA genes from Caenorhabditis elegans, revealing the expression and evolution pattern of snoRNAs in nematodes. Genomics. 2007;89:490–501. doi: 10.1016/j.ygeno.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Saijou E, Fujiwara T, Suzaki T, Inoue K, Sakamoto H. RBD-1, a nucleolar RNA-binding protein, is essential for Caenorhabditis elegans early development through 18S ribosomal RNA processing. Nucleic Acids Res. 2004;32:1028–1036. doi: 10.1093/nar/gkh264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasano Y, Hokii Y, Inoue K, Sakamoto H, Ushida C, Fujiwara T. Distribution of U3 small nucleolar RNA and fibrillarin during early embryogenesis in Caenorhabditis elegans. Biochimie. 2008;90:898–907. doi: 10.1016/j.biochi.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Deng W, Zhu X, Skogerbø G, Zhao Y, Fu Z, Wang Y, He H, Cai L, Sun H, Liu C, et al. Organization of the Caenorhabditis elegans small non-coding transcriptome: genomic features, biogenesis, and expression. Genome Res. 2006;16:20–29. doi: 10.1101/gr.4139206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez MD, Rosenblad MA, Samuelsson T. Conserved and variable domains of RNase MRP RNA. RNA Biol. 2009;6:208–220. doi: 10.4161/rna.6.3.8584. [DOI] [PubMed] [Google Scholar]
- 28.Wachi M, Ogawa T, Yokoyama K, Hokii Y, Shimoyama M, Muto A, Ushida C. Isolation of eight novel Caenorhabditis elegans small RNAs. Gene. 2004;335:47–56. doi: 10.1016/j.gene.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Hokii Y, Kubo A, Ogasawara T, Nogi Y, Taneda A, Arai R, Muto A, Ushida C. Twelve novel C. elegans RNA candidates isolated by two-dimensional polyacrylamide gel electrophoresis. Gene. 2006;365:83–87. doi: 10.1016/j.gene.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 30.Zemann A, op de Bekke A, Kiefmann M, Brosius J, Schmitz J. Evolution of small nucleolar RNAs in nematodes. Nucleic Acids Res. 2006;34:2676–2685. doi: 10.1093/nar/gkl359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabara H, Motohashi T, Kohara Y. A multi-well version of in situ hybridization on whole mount embryos of Caenorhabditis elegans. Nucleic Acids Res. 1996;24:2119–2124. doi: 10.1093/nar/24.11.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitani S, Du H, Hall DH, Driscoll M, Chalfie M. Combinatorial control of touch receptor neuron expression in Caenorhabditis elegans. Development. 1993;119:773–783. doi: 10.1242/dev.119.3.773. [DOI] [PubMed] [Google Scholar]
- 34.Savino R, Hitti Y, Gerbi SA. Genes for Xenopus laevis U3 small nuclear RNA. Nucleic Acids Res. 1992;20:5435–5442. doi: 10.1093/nar/20.20.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peculis BA, DeGregorio S, McDowell K. The U8 snoRNA gene family: identification and characterization of distinct, functional U8 genes in Xenopus. Gene. 2001;274:83–92. doi: 10.1016/s0378-1119(01)00596-0. [DOI] [PubMed] [Google Scholar]
- 36.Thomas J, Lea K, Zucker-Aprison E, Blumenthal T. The spliceosomal snRNAs of Caenorhabditis elegans. Nucleic Acids Res. 1990;18:2633–2642. doi: 10.1093/nar/18.9.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li T, He H, Wang Y, Zheng H, Skogerbø G, Cheng R. In vivo analysis of Caenorhabditis elegans noncoding RNA promoter motifs. BMC Mol. Biol. 2008;9:71. doi: 10.1186/1471-2199-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas JD, Conrad RC, Blumenthal T. The C. elegans trans-spliced leader RNA is bound to Sm and has trimethylguanosine cap. Cell. 1988;54:533–539. doi: 10.1016/0092-8674(88)90075-x. [DOI] [PubMed] [Google Scholar]
- 39.Reichow SL, Hamma T, Ferré-D'Amaré AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aftab MN, He H, Skogerbø G, Chen R. Microarray analysis of ncRNA expression patterns in Caenorhabditis elegans after RNAi against snoRNA associated proteins. BMC Genomics. 2008;9:278. doi: 10.1186/1471-2164-9-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matera AG, Terns RM, Terns M. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 42.Cavaillé J, Bachellerie JP. SnoRNA-guided ribose methylation of rRNA: structural features of the guide RNA duplex influencing the extent of the reaction. Nucleic Acids Res. 1998;26:1576–1587. doi: 10.1093/nar/26.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peculis BA, Steitz JA. Sequence and structural elements critical for U8 snRNP function in Xenopus oocytes are evolutionarily conserved. Genes Dev. 1994;8:2241–2255. doi: 10.1101/gad.8.18.2241. [DOI] [PubMed] [Google Scholar]
- 44.Tomasevic N, Peculis BA. Xenopus LSm proteins bind U8 snoRNA via an internal evolutionarily conserved octamer sequence. Mol. Cell Biol. 2002;22:4101–4112. doi: 10.1128/MCB.22.12.4101-4112.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]