Abstract
The Nobel Prize in Physiology or Medicine was recently awarded to Elizabeth Blackburn, Carol Greider and Jack Szostak for their pioneering studies on chromosome termini (telomeres) and their discovery of telomerase, the enzyme that synthesizes telomeres. Telomerase is a unique cellular reverse transcriptase that contains an integral RNA subunit, the telomerase RNA and a catalytic protein subunit, the telomerase reverse transcriptase (TERT), as well as several species-specific accessory proteins. Telomerase is essential for genome stability and is associated with a broad spectrum of human diseases including various forms of cancer, bone marrow failure and pulmonary fibrosis. A better understanding of telomerase structure and function will shed important insights into how this enzyme contributes to human disease. To this end, a series of high-resolution structural studies have provided critical information on TERT architecture and may ultimately elucidate novel targets for therapeutic intervention. In this review, we discuss the current knowledge of TERT structure and function, revealed through the detailed analysis of TERT from model organisms. To emphasize the physiological importance of telomeres and telomerase, we also present a general discussion of the human diseases associated with telomerase dysfunction.
INTRODUCTION
Telomeres are essential nucleoprotein structures that define the terminal segments of linear chromosomes. In eukaryotes, telomeres are essential for genome stability, functioning to prevent chromosome ends from being recognized and processed as bona fide DNA double-strand (ds) breaks. Importantly, telomeres also provide a solution to the ‘end-replication problem’, which was first proposed by Olovnikov and Watson in the early 1970s (1–3). This model predicts that during the process of DNA replication, a small amount of DNA from the 3′-ends of linear chromosomes is left unreplicated. As a result, chromosome 3′-ends progressively shorten during consecutive cell divisions, which limits cellular lifespan (1–3). Chromosome ends that lack sufficient telomeric repeats are prone to recombination and fusion with other pieces of genomic DNA, events that can interfere with normal cell cycle progression and promote genetic instability. Thus, telomeres provide a protective cap for the ends of linear chromosomes.
One universal feature of telomeric DNA is the organization into a C/A-rich strand and a complementary G/T-rich strand. Telomeric DNA almost always contains tandem repeats of simple, species-specific sequences that are 6–8 nucleotides (nt) long [e.g. (TTAGGG)n in mammals]. Another conserved feature of telomeric DNA is the organization into a ds segment with a single-stranded (ss) 3′-overhang. Electron microscopic analysis of psoralen cross-linked human and mouse telomeric DNA revealed large lariat-like structures containing thousands of TTAGGG repeats (4). These structures are known as telomere-loops (t-loops) and are postulated to be formed and stabilized by invasion of the telomeric 3′-overhang into the duplex repeat array (4). Telomere loops have also been detected in trypanosomes (5), ciliates (6), plants (7), nematodes (8) and some strains of yeast (9,10). The existence of t-loops provides an attractive model that could explain how ss chromosome ends are protected from degradation, recombination and fusion (11). However, the molecular mechanism(s) that regulate t-loop formation in vivo remain to be elucidated. If living cells do contain t-loops, one interesting possibility is that telomeres may adopt alternative conformations at specific stages of the cell cycle (12). The identification of novel, cell-cycle-specific telomere-associated proteins will provide important insights into telomere dynamics and length regulation in vivo.
Various proteins have been shown to associate with telomeric DNA and are implicated in the formation and maintenance of telomere architecture. These proteins are the focus of excellent reviews by Linger and Price (13) and Palm and de Lange (11), and will not be discussed in detail in this article. One general concept, however, relates to the evolution of telomere-associated proteins. Although telomere function is conserved in diverse organisms, the architecture and composition of telomere-associated proteins is remarkably varied and seems to have changed rapidly during evolution (13). For example, human telomeres are bound by a six-protein complex called shelterin, comprised of TRF1, TRF2, POT1, RAP1, TIN2 and TPP1, which interacts with ss and ds telomeric DNA (11). In contrast, budding yeast telomeres are bound by two separate protein complexes (13). One contains the Cdc13, Stn1 and Ten1 proteins and binds the G-rich 3′-overhang (14). The other, which binds duplex telomeric DNA, is minimally composed of Rap1, Rif1 and Rif2 (15). Despite these differences in composition, however, the telomere-associated proteins have a conserved function, which is to assemble a protective cap that ensures telomeres are maintained at an appropriate length and are protected from being recognized and processed as broken DNA. The ability to perform this task is facilitated by additional species-specific protein–protein interactions (11,13).
De novo synthesis of telomeric DNA in most eukaryotes is performed by the cellular ribonucleoprotein reverse transcriptase (RT) telomerase. Originally discovered by Carol Greider and Elizabeth Blackburn in the ciliate Tetrahymena thermophila (16–18), telomerase is a unique RT that contains a catalytic protein subunit, the telomerase RT (TERT), the telomerase RNA (TR) and species-specific accessory proteins. Telomere synthesis involves TERT-catalyzed reverse transcription of a small template region within TR and telomerase activity can be reconstituted in rabbit reticulocyte lysates by co-expressing the TERT and TR subunits (19–21). Importantly, species-specific accessory proteins regulate telomerase biogenesis, subcellular localization and function in vivo. For example, mass spectrometric analysis of affinity-purified telomerase from HeLa cells has identified integral protein components of human telomerase: dyskerin, NHP2, NOP10, pontin/reptin and TCAB1 (22–25). Dyskerin, NHP2 and NOP10 are required for the stability and accumulation of human telomerase RNA (hTR) in vivo (22). Similarly, pontin and reptin are two closely related ATPases necessary for the stability of dyskerin and hTR in vivo (24). The current model is that dyskerin, pontin and reptin form a scaffold that recruits and stabilizes hTR, and assembles the telomerase ribonucleoprotein particle. Once this complex is formed, pontin and reptin are thought to dissociate from the complex and yield the catalytically active enzyme (24). The subcellular localization of telomerase appears to be regulated by the recently identified protein TCAB1 (25). Further studies are needed to elucidate the biochemical and molecular significance of the intricate network of protein–protein and protein–nucleic acid interactions within the telomerase holoenzyme. Moreover, it will be important to investigate whether the composition of the holoenzyme changes in specific stages of the cell cycle. This information will provide important insight into how human telomerase functions in vivo. Notably, the composition of the telomerase holoenzyme varies significantly between species and the reader is referred to Collins (26) and Gallardo and Chartrand (27) for excellent reviews of this topic.
THE TELOMERASE RT SUBUNIT
The catalytic core of telomerase is composed of the RNA subunit (TR) and the catalytic protein subunit (TERT), whereas the holoenzyme contains additional species-specific accessory proteins. The first TERT subunits were identified through genetic screens in yeast (28) and the biochemical purification of Euplotes aediculatus telomerase (29,30). The E. aediculatus protein was identified as a homolog of the yeast protein and sequence comparison with prototypical RTs revealed an evolutionarily conserved RT domain in both proteins (30). The first direct evidence that TERT was the catalytic subunit of telomerase came from in vivo studies showing that substitution of evolutionarily conserved residues within the RT catalytic triad of the yeast protein caused telomere shortening and cellular senescence in vivo and eliminated enzymatic activity in vitro (30). TERT homologs were subsequently identified in humans, rodents and ciliates (21,31–35). Further evidence that TERT was the catalytic subunit of human and T. thermophila telomerase came from studies showing that telomerase activity could be reconstituted in vitro by co-expressing wild-type TERT and TR in rabbit reticulocyte lysates (19–21). These studies were also the first to demonstrate that TERT and TR were sufficient to reconstitute telomerase activity in vitro (19–21), although it is important to note that rabbit reticulocyte lysates contain endogenous proteins that interact with telomerase to facilitate its assembly and activity [e.g. heat shock (36) and chaperone (37) proteins].
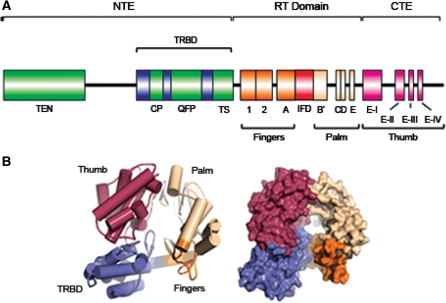
Bioinformatics and mutational studies have collectively established that TERT contains three main structural elements: (i) a long N-terminal extension that contains conserved DNA- and RNA-binding domains; (ii) a central catalytic RT domain; and (iii) a short C-terminal extension (38). To exemplify this domain organization, the predicted linear architecture of human TERT (hTERT) is illustrated in Figure 1A. Although this organization defines almost all TERT proteins, there are a few notable exceptions. First, certain insect and nematode TERTs harbor a truncated N-terminus that does not contain the telomerase essential N-terminal (TEN) domain (39,40). Second, the C-terminal region is absent from Giardia lamblia and nematode TERTs (40). Third, Plasmodium falciparum TERT contains an abundance of hypervariable insertions between the conserved domains and is at least three times larger than all other TERT proteins (41). The biological significance of this divergent architecture is not clear and it will be interesting to determine if other TERT domains and/or cellular proteins compensate for the missing domains in vivo.
Figure 1.
Structural organization of the TERT. (A) Predicted linear architecture of hTERT. In most organisms, TERT contains a long N-terminal extension (NTE), a central catalytic RT domain and a short C-terminal extension (CTE). Green boxes indicate the predicted locations of the telomerase essential N-terminal (TEN) domain and the telomerase-specific motifs CP, QFP and TS. Blue boxes indicate the TRBD, containing the CP motif, QFP motif and part of the TS motif. An unstructured linker region connects the TEN domain and TRBD. Orange and tan boxes represent the seven evolutionarily conserved motifs in the RT domain (1, 2, A, B′, C, D, E) and a red box illustrates the telomerase-specific IFD. The CTE contains four blocks of conserved amino acids, which are shown as pink boxes (E-I, E-II, E-III, E-IV). (B) Domain organization of the protein predicted to be T. castaneum TERT (cartoon and surface representation), reprinted with permission from ref. (42). The protein is organized into a ring-shaped structure, containing hallmark ‘thumb’ (red), ‘palm’ (tan) and ‘fingers’ (orange) motifs. The telomerase-specific TRBD is shown in violet. The color scheme used in (A) corresponds to that shown in (B).
A potential advance in our understanding of telomerase architecture came from the recent atomic-resolution structure of a protein thought to represent full-length Tribolium castaneum (flour beetle) TERT, in complex with an 18-nt telomeric oligonucleotide (42). However, whether this protein truly represents a telomerase is currently controversial. Formal proof that this T. castaneum protein represents the catalytic TERT subunit remains to be demonstrated, which is an important question to address given the complex evolution of telomerase and telomere-maintenance in insects (39,43). For example, Dipteran insects (e.g. mosquitoes) lack telomerase activity and maintain their telomeres via recombination-based mechanisms, whereas Arthropods (e.g. silkworm) have pentanucleotide (TTAGG)n telomeric repeats and contain detectable levels of telomerase activity (39,43). Interestingly, the predicted T. castaneum TERT lacks a significant portion of the N-terminus that is critical for telomerase activity in ciliates, yeast and humans (42). One possibility is that T. castaneum TERT may be inactive, which would be supported by the absence of canonical telomeric repeats in this insect. Another prediction, which cannot be excluded in the absence of biochemical support for telomerase activity, is that the putative TERT represents a non-telomerase RT. Despite these potential caveats, the crystal structure reported by Skordalakes and colleagues (42) represents an important snapshot of a TERT-like protein and provides the basis for structural comparisons with other TERTs.
The T. castaneum TERT structure suggests a ring-shaped protein held together by extensive hydrophobic interactions between the N- and C-terminal regions (Figure 1B) (42). This architecture is congruent with previous biochemical studies of human and Euplotes crassus TERT, which revealed physical interactions between N- and C-terminal domains (44–46). The ring-like structure of TERT is akin to that of other nucleic acid polymerases, including that of human immunodeficiency virus (HIV), and contains the hallmark ‘thumb’, ‘palm’ and ‘fingers’ motifs (Figure 1A and B) (42). Notably, the interior dimensions of the TERT ring structure predict that it can accommodate seven or eight base pairs of ds nucleic acid (42). This is consistent with previous biochemical studies demonstrating that the RNA–DNA duplex in the telomerase active site is maintained at a length of seven to eight base pairs (47,48). The ring’s interior is lined with amino acids that mediate protein–nucleic acid interactions, nucleotide binding and DNA synthesis in yeast and hTERT (49–54). An important future challenge will be to determine the high-resolution structure of TERT in complex with its integral RNA subunit and/or additional protein components. Towards this goal, recent studies allowed the determination of the crystal structure of the T. castaneum TERT in complex with a short RNA–DNA hybrid designed to resemble the putative RNA-templating region and complementary DNA sequence (55). One potential caveat to this study, however, is that the RNA component of this telomerase is not known and therefore, the physical contacts observed in this structure may be significantly different than those that occur in the presence of the biological RNA subunit and telomeric chromatin. An equally important challenge will be to solve the crystal structure of full length TERT from an organism with well-characterized telomerase activity.
The TERT N-terminal extension
The N-terminal extension of most TERTs contains two conserved domains, the TEN domain and telomerase RNA-binding domain (TRBD) (Figure 1A). High-resolution structures of the T. thermophila TEN domain and TRBD indicate that both of these regions represent novel protein folds involved in binding ss nucleic acids (56,57). In between these domains is a relatively long and unstructured linker region that may be important for conformational flexibility within the holoenzyme (Figure 1A). The N-terminal extension also contains several conserved telomerase-specific motifs, including the CP, QFP and TS motifs (Figure 1A) (58,59). These regions are important for TERT-TR binding interactions and the rate of template copying during telomere synthesis (19,58,60–69), and may thus be amenable to therapeutic interventions that inhibit or augment telomerase activity.
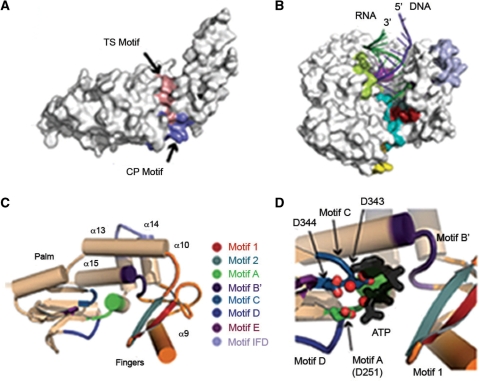
The crystal structure of the T. thermophila TRBD reveals that it is organized into two asymmetric lobes that contain residues from the CP and TS motifs (Figure 2A) (56). The two halves are connected by extended loops that impart the domain with structural flexibility (56). Hydrophilic and hydrophobic CP and TS residues form an extended RNA-binding groove on the surface of the domain. The dimensions of this cavity reveal a relatively wide hydrophilic pocket that could accommodate dsRNA and a narrow hydrophobic pocket that could accommodate ssRNA (56). In the context of full-length T. castaneum TERT, the RNA-binding groove is located on the side of the ring and faces the interior of the ring in close proximity to the active site (Figure 2B) (42). This is believed to permit the RNA 5′-end to enter the ring’s interior where the active site is located. The structural data is supported by biochemical studies with ciliate, yeast and human telomerase that have identified the CP and TS motifs as being critical for TR-binding and telomerase activity in vitro and in vivo (19,58,60–68).
Figure 2.
Molecular models of T. thermophila (A) and T. castaneum (B–D) TERT. (A) Surface representation of the isolated TRBD from T. thermophila TERT, reprinted with permission from ref. (56). This cartoon shows the two asymmetric lobes that comprise the TRBD. Amino acids that form the TS motif are shown in pink and those that comprise the CP motif are indicated in blue. (B) Model of the protein suggested to be T. castaneum TERT (surface representation) in complex with the telomerase RNA subunit (dark green) and single-stranded telomeric DNA (dark purple), reprinted with permission from ref. (42). The hallmark motifs of the RT domain are illustrated as follows: motif 1 (red), 2 (grey), A (green), B′ (dark purple), C (blue), D (dark blue), E (magenta) and IFD (light blue). The TRBD comprises residues from the CP (yellow) and TS (cyan) motifs. Structural elements of the TERT C-terminal extension (light green) are predicted to stabilize and orientate the TERT–RNA–DNA complex. (C) Domain folds of the predicted T. castaneum TERT RT domain, colored as in (B) and reprinted with permission from ref. (42). Elements that form the palm domain are colored in tan and those that form the fingers domain are shown in orange. (D) The active site and nucleotide-binding pocket of T. castaneum TERT, reprinted with permission from ref. (42). This figure shows the predicted location of three invariant aspartic acids (D251, D343 and D344) that form the catalytic triad of the active site in complex with a modeled nucleotide of ATP (black stick). The RT motifs are colored as in (B).
The most N-terminal domain in TERT (the TEN domain) exhibits binding affinity for ss telomeric DNA and contains residues that are essential for telomerase activity (38,57,70–75). Furthermore, a specific leucine in the TEN domain of ciliate TERT (L14) has been identified as a critical determinant of the enzyme’s ability to add multiple telomeric repeats to a single telomeric primer (74). These observations indicate that the TEN domain has an important role in DNA substrate recognition and elongation. Interestingly, the TEN domain also displays a weak non-specific RNA-binding activity (57). Although the significance of this interaction is presently unclear, it is possible that in the context of the full-length protein, the TEN domain and TRBD co-operate to ensure sequence-specific TERT–TR binding interactions and optimal template positioning during telomerase assembly and/or telomere synthesis.
The TERT RT domain
The catalytic domain of TERT is the most characterized region of the protein and contains seven evolutionarily conserved RT motifs (see Figures 1A and 2C) essential for enzymatic activity (19,20,30,31,33,51,76–78). The TERT RT domain is organized into two subdomains that resemble the ‘fingers’ and ‘palm’ subdomains of prototypical RT enzymes (Figure 2C) (30,31,79). These regions are connected by a loop that contains the conserved ‘primer grip’ region of motif E (42). Molecular models predict that the loop makes direct contacts with the RNA–DNA hybrid and the 3′-end of the ssDNA, suggesting that the primer grip region could be involved in positioning the 3′-end within the enzyme active site (Figure 2B) (42). This model is supported by biochemical studies of yeast and hTERT that have identified an important role for this region in ssDNA-binding and processive telomere synthesis in vitro and in vivo (80,81). Further evidence for contacts between the TERT primer grip and DNA primer have been provided by structural studies of T. castaneum TERT in complex with an RNA–DNA hairpin designed to resemble the putative telomerase RNA template and telomeric DNA (55).
One unique structural feature of the telomerase RT domain is a large insertion between motifs A and B′, called the ‘insertion in fingers’ domain (IFD) (Figure 1A) (30,31). The IFD is comprised of two antiparallel α-helices that are sandwiched between the fingers and palm subdomains (Figure 2C) (42). The crystal structure reveals that the IFD is engaged in intramolecular protein-protein interactions that organize and stabilize this region of the RT domain (42). Importantly, the IFD makes extensive contacts with an α-helix that is implicated in making direct contacts with the backbone of the incoming DNA primer (42). Mutations that alter the structural integrity of the IFD are expected to indirectly compromise protein–DNA and RNA–DNA interactions, thus impairing enzyme activity. Consistent with this, alanine substitutions of moderately conserved residues in the IFD of Est2p (yeast TERT) were found to impair telomerase activity and processivity in vitro and prevented telomere maintenance in vivo (52).
The TERT active site contains three invariant aspartic acids, one of which resides in motif A and the other two within motif C (Figure 2D) (30,31). These residues are conserved in RTs and form a catalytic triad that participates directly in nucleotide addition via a two-metal ion mechanism (30,31). Alanine substitution of these amino acids abrogates telomerase activity (19,33,64,76,77). Conserved residues from motifs 1, 2, A, B′, C and D form the nucleotide-binding pocket, located at the interface between the palm and fingers subdomains (Figure 2D) (42). Two conserved surface-exposed residues from motifs A and C (tyrosine and valine, respectively) form a hydrophobic patch that is predicted to bind the base of the nucleotide substrate. This interaction would facilitate nucleotide positioning in the active site for co-ordination with a metal ion and the 3′-end of the DNA primer (42). This model is supported by biochemical studies of ciliate, yeast and hTERT that have implicated these residues in nucleotide insertion rate, polymerase fidelity and enzyme processivity (78,80,82).
Sequence alignment of the TERT RT domain revealed a novel telomerase-specific motif (motif 3) between motifs 2 and A (Figure 1A) (83,84). Interestingly, although the predicted secondary-structure of motif 3 is evolutionarily conserved, the primary sequence is only conserved in vertebrate and ciliate telomerases. This suggests that motif 3 may regulate species-specific aspects of telomerase biochemistry, such as repeat addition processivity (84). Alanine substitution screening of this motif in hTERT identified mutations that either abrogated telomerase activity or caused a significant increase in the rate or processivity of telomere synthesis in vitro (84). These biochemical characterizations have provided new insight into the telomerase reaction mechanism by identifying specific amino acids in hTERT that independently regulate two critical aspects of telomere synthesis. An important area of future research will be to elucidate how mutations in motif 3 affect the biogenesis, activity and regulation of telomerase.
The TERT C-terminal extension
In contrast to the N-terminus and RT domain, the C-terminus of TERT shows only weak sequence conservation suggesting that it may have species-specific functions or that different amino acid sequences have evolved to fold into similar structural domains. The C-terminal extension of TERT adopts a novel protein fold, although structural comparison of TERT with the closely related HIV RT indicates that this region represents the thumb domain of telomerase (42). The C-terminus is a helical bundle that contains several surface-exposed loops that are thought to contribute to the formation and stabilization of an RNA–DNA heteroduplex in the enzyme active site (Figure 2B) (42,55). Mutations in this region affect the nucleotide and repeat addition processivity of human and yeast telomerase, telomere length maintenance in human cells and the subcellular localization of hTERT (49,53,54,80,85). Furthermore, it was shown that the addition of an epitope tag to the C-terminus of hTERT abolishes telomere elongation in vivo, suggesting that the conformation of this region is important for hTERT function (86). Interestingly, the C-terminal extension is essential for telomerase activity in T. thermophila and humans, but is dispensable for telomerase activity in yeast (58,62,63,65) and is completely absent in other organisms (40). These observations further suggest that the C-terminal extension has species-specific roles in vivo. Additional structure–function studies are needed to understand how this region contributes to the biochemical and molecular properties of telomerase from different organisms. In this regard, the in vitro and in vivo characterization of disease-associated hTERT mutants will likely provide invaluable insight these aspects of human telomerase (87,88).
THE TELOMERASE RNA SUBUNIT
A unique feature of telomerase is that the RNA template for DNA synthesis is an integral component of the holoenzyme. Although a detailed discussion of the RNA subunit is beyond the scope of this review, it is necessary to comment on some of the key properties of the vertebrate TR. The reader is referred to Theimer and Feigon (89) for a comprehensive review of the structure and function of TR subunits from various organisms.
Phylogenetic comparative analysis of vertebrate TR predicts three conserved domains: (i) the pseudoknot/template core domain; (ii) the CR4/CR5 domain (for conserved regions 4 and 5, respectively); and (iii) a box H/ACA domain (90). The core domain is essential for telomerase activity in vitro and in vivo (89). This region contains the template for telomere addition, the 5′- and 3′-boundary elements that prevent the incorporation of non-template nucleotides, a putative TERT binding site and a conserved pseudoknot structure (90,91). Human telomerase activity can be reconstituted in vitro by co-expressing the pseudoknot/template domain and the CR4/CR5 domain in the presence of hTERT (92,93). The TR domains work together to mediate TR-TERT interactions, nucleotide and repeat addition processivity, and enzyme fidelity (89). The 3′-end of vertebrate TR contains two conserved motifs, the box H and ACA elements (box H/ACA domain), which serve as binding sites for proteins involved in RNA processing, stability and subcellular localization (89). The box H/ACA domain is essential for TR stability, processing, nuclear localization and telomerase activity in vivo (89,94,95). Mutations in the gene encoding TR have been linked to the multi-system disorder called dyskeratosis congenita, highlighting the clinical importance of this molecule. Furthermore, the TR gene is amplified in several human cancers (96–98), thus making it a potential target for therapeutic inhibitors of telomerase activity in cancer cells (99).
THE TELOMERASE REACTION CYCLE
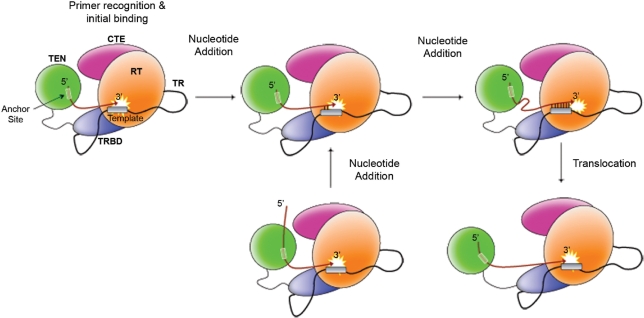
Telomerase has the unique ability to catalyze multiple rounds of template copying and add hundreds of nucleotides to the same DNA primer. This contrasts with prototypical RTs, which copy a relatively large RNA genome into a single molecule of complementary DNA. As illustrated in Figure 3 and discussed in the following sections, the telomerase reaction cycle can be divided into three basic steps: (i) primer recognition and binding; (ii) synthesis of the first telomeric repeat; and (iii) translocation and realignment of the new DNA 3′-end to initiate the next round of telomere synthesis.
Figure 3.
Schematic diagram of the telomerase reaction cycle. This figure summarizes the fundamental steps of the telomerase reaction cycle: initial primer recognition and binding, nucleotide addition and translocation. The telomerase RNA (TR) subunit is represented by a black line (not drawn to scale). The TR supplies the template (grey rectangle) for telomere synthesis. DNA synthesis is catalyzed by the TERT. TERT contains a TEN domain (pink sphere), a TRBD (purple oval), RT domain (blue sphere) and CTE (green oval). The TEN domain contains a unique ssDNA-binding region, called the telomerase anchor site (transparent rectangle). The red line represents telomeric ssDNA. Telomerase binds the telomeric ssDNA such that the 3′-end is aligned with the TR template in the active site (white star) and the 5′-end is positioned within the telomerase anchor site. Telomerase reverse transcribes the template region, 1 nt at a time (nucleotide addition), until reaching the 5′-template boundary element (top right). At this point, a translocation step repositions the new DNA 3′-end within the template for a second round of telomere synthesis (bottom). Conformational changes within the telomerase holoenzyme are believed to facilitate nucleotide and repeat addition processivity.
During telomere elongation, the RNA template is reverse transcribed using canonical Watson–Crick base-pairing to specify the telomeric sequence. Telomere synthesis proceeds by the sequential addition of deoxynucleotide triphosphates (dNTPs) to the free 3′-hydroxyl group of the telomeric ssDNA primer. The telomeric ssDNA overhang is believed to be the natural primer for telomerase-mediated DNA synthesis in vivo. However, telomerase can elongate almost any G-rich linear ssDNA primer that contains a free 3′-end in vitro (16, 100). Parallel intermolecular G-quadruplex substrates are also efficient substrates for ciliate telomerase in vitro (101–103).
Telomerase incorporates consecutive dNTPs without dissociating from the primer by a reaction known as nucleotide addition processivity (38). During telomere elongation, the RNA–DNA hybrid is kept at a constant length of seven to eight base pairs by melting bonds at the distal end of the template as new bonds are formed at the proximal end (48). When the 5′-template boundary element is reached, a translocation step repositions the new DNA 3′-end within the template for a second round of telomere synthesis. The ability of telomerase to catalyze more than one round of DNA synthesis while bound to the same telomeric primer is referred to as repeat addition processivity. As discussed below, interactions between the 5′-end of the primer and regions of TERT outside the RT domain are required for repeat addition processivity and have been termed ‘anchor sites’ to emphasize their predicted role in anchoring TERT to the DNA primer during telomere synthesis (38).
Repeat addition processivity
Repeat addition processivity was first observed with T. thermophila telomerase and subsequently with human telomerase (104,105). Many rodent and fungal telomerases, however, are relatively non-processive and catalyze short elongation products in vitro and in vivo (106–108). Furthermore, genetic studies with mutant T. thermophila telomerase RNAs indicate that telomerase has limited processivity in vivo (109,110). These studies raise questions regarding the significance of repeat addition processivity in vivo. Nonetheless, biochemical and genetic studies indicate that processive repeat addition is a biologically relevant property of telomerase. First, telomerase-specific residues that mediate processivity have been located in the RT domain (52,78,84), the N-terminal extension (71,74,111–113) and the C-terminal extension (53). Mutations in these regions in human, ciliate or yeast TERT selectively alters repeat addition processivity in vitro and causes defects in telomere length maintenance in vivo (52,69,74,84,111–113). Second, these regions are important for telomerase-dependent extension of partially or completely nontelomeric primers, which provides indirect evidence for a role in primer binding (74,111–113). Third, mutations that specifically increase repeat addition processivity in vitro and cause telomere over-elongation in vivo have been identified in ciliate and yeast TERT (71,78). Finally, in vivo analyses of budding yeast telomerase show that repeat addition processivity is significantly increased at extremely short telomeres, suggesting that this unique feature of telomerase may enable cells to rapidly elongate critically short telomeres (114,115). This model predicts that the extent of processive repeat addition depends upon telomere length and thus, could partially explain the variance observed between telomerase repeat addition processivity in vitro and in vivo. Further studies will be required to completely understand this unique aspect of telomerase biochemistry.
Telomerase anchor sites
A long-standing notion in telomerase biochemistry is that the enzyme contains DNA-binding regions outside the RT domain that are required for processive telomere elongation. The existence of one or more telomerase ‘anchor sites’ was precipitated by studies demonstrating that ciliate and human telomerase could elongate non-telomeric G-rich ssDNA primers in vitro (16,17,105,116). Furthermore, telomerase was shown to add telomeric repeats to chromosome breaks that contained non-telomeric DNA during in vivo chromosome healing (110,117). It was subsequently shown that primers containing almost any sequence of ssDNA could be extended in vitro if telomeric repeats were added to the 5′-end (100,118). These observations suggested that hybridization between the DNA 3′-end and RNA template was not required for primer elongation. This hypothesis was further supported by biochemical studies of human and yeast telomerase, which showed that the enzyme/primer complex was mainly stabilized by contacts with the catalytic protein subunit (119,120).
The first direct evidence for a telomerase anchor region was provided by photo-cross-linking studies performed with E. aediculatus telomerase (121). Specifically, a TERT-sized protein was cross-linked to the 5′-end of photo-reactive ssDNA primers that were aligned in the enzyme active site (121). The most efficient protein–DNA cross-links were established with bases 20–22 nt upstream of the primer 3′-end and required the telomerase RNA subunit. More recent studies have shown that human and T. thermophila TERT can interact sequence-specifically with telomeric ssDNA in the absence of TR (75,81). These studies used biotinylated ssDNA primers to investigate TERT–DNA interactions in vitro. One important finding was that a fragment of hTERT encompassing amino acids 1–300 bound telomeric ssDNA as efficiently as the full-length protein, providing the first physical evidence for an anchor site in the N-terminus of telomerase (81). An equally important finding was that the T. thermophila TERT (tTERT) C-terminus exhibited telomeric ssDNA-binding activity in vitro (75). Furthermore, it has been recently shown that a purified human TEN domain spanning amino acids 1–200 directly interacts with telomeric DNA and this interaction is dependent on the length and register of the telomeric repeat (73). These studies indicate that TERT contains multiple anchor sites, which are believed to co-ordinate and regulate various stages of primer binding and extension. Importantly, some studies indicate that TERT-DNA interactions are enhanced by the telomerase RNA subunit, suggesting that TR has an important role in regulating anchor site interactions (57,75,111,121).
Genetic and biochemical studies with ciliate, yeast and mammalian telomerase suggest that the anchor site may be comprised of a template-proximal and a template-distal component (57,71,74,81,105,112,116,120,122–125). In this model, both regions contribute to telomere binding and elongation, but only relatively long primers (≥20 nt) extend into the template-distal anchor region (47). The existence of two separate anchor sites indicates a tripartite mode of DNA-binding: (i) the primer 3′-end hybridizes with the RNA template in the active site; (ii) the primer nucleotides adjacent to the template-hybridizing region interact with a template-proximal anchor site; and (iii) the primer nucleotides at the 5′-end interact with the template-distal anchor site (123).
The template-proximal telomerase anchor region has been physically and functionally mapped to the N-terminus of ciliate, yeast and hTERT (38,71,72,74,75,81). The crystal structure of the tTERT TEN domain revealed a previously-unrecognized ssDNA-binding groove on the surface of this domain. Photo-cross-linking studies showed that the tTERT TEN domain possessed telomere-specific ssDNA-binding activity in vitro (57,72). Mutagenesis of key residues thought to be involved in ssDNA-binding, such as an invariant glutamine (tTERT Q168), significantly reduced the interaction between tTERT and telomeric ssDNA, and impaired enzyme activity in vitro (57,72). Furthermore, mutation of the corresponding residue in yeast Est2p (Q146A) caused severe growth defects and telomere loss in vivo (71). Most recently, hTERT Q169 was shown to be important for the structure of the human telomerase N-terminal extension, primer binding and nucleotide incorporation in vitro, as well as telomere length maintenance in vivo (70,73). Interestingly, Q169 appears to be particularly important for incorporating the second nucleotide during telomere extension in vitro (70). Collectively, the above studies indicate that this glutamine residue forms part of an evolutionarily conserved anchor site that facilitates primer recognition and orientation in the active site for telomere synthesis.
Telomerase structural mobility
DNA polymerases undergo a series of conformational changes during DNA synthesis (79,126). Structural mobility confers polymerases with the ability to discriminate against nucleotide misinsertion and incorporate multiple nucleotides without dissociating from the DNA primer. With respect to telomerase, it has been long proposed that conformational changes mediate the transition between telomere synthesis initiation, elongation and translocation (104).
Recent work with T. thermophila telomerase has provided new insights into the structural mobility of the telomerase anchor site (74). The crystal structure of the tTERT TEN domain revealed that an evolutionarily-conserved leucine (L14), in close proximity of Q168, was involved in forming one edge of the ssDNA-binding groove (57). The branched, hydrophobic side-chain of leucine can theoretically engage in intra- or inter-molecular protein–protein interactions. Structure–function studies of tTERT identified an essential role for L14 in the repeat addition processivity of ciliate telomerase in vitro (74). Importantly, L14 mutants displayed normal nucleotide addition processivity, thus identifying a specific role for L14 in the telomerase translocation step (74). An attractive model has been proposed in which L14 represents an intramolecular ‘door latch’, which holds the TEN domain in close proximity of the catalytic site and facilitates processive nucleotide addition (Figure 3) (74). After synthesizing a complete telomeric repeat, a conformational change occurs within telomerase that releases the door latch (‘open’ state) and allows the new DNA 3′-end to be repositioned within the RNA template and alignment regions for the next round of telomere synthesis. This model is supported by cross-linking studies indicating that the tTERT TEN domain is displaced relative to the catalytic site during telomere synthesis in vitro (72). A second prediction of this model is that the 5′-end of the elongated DNA slides through the ssDNA-binding groove on the surface of the TEN domain (Figure 3). This movement is believed to signal telomerase to re-establish the intramolecular protein–protein interaction (‘closed’ state) and initiate the next round of telomere synthesis (74).
Processivity factors
In addition to anchor sites located within telomerase itself, accumulating evidence suggests that telomerase-associated proteins facilitate the enzyme’s repeat addition processivity in vivo. For example, the TPP1–POT1 heterodimer stimulates the activity and processivity of human telomerase in vitro (127–129). TPP1 has been shown to interact with hTERT in vitro (128) and mutation of a conserved glycine residue in the hTERT TEN domain (G100) was shown to suppress the stimulatory effect of TPP1–POT1 on human telomerase processivity in vitro (129). These results are important because they identify a physical link between the human shelterin complex and telomerase, and provide new insight into the mechanism of processive telomere synthesis. One interpretation of these observations is that telomere-bound TPP1–POT1 interacts with hTERT to transfer the telomeric ssDNA 3′-end to telomerase for subsequent elongation. It will be interesting to elucidate whether the interaction between hTERT and TPP1 is direct or indirect (i.e. mediated by the telomeric ssDNA primer) and to investigate the conformational changes that occur in hTERT upon binding the TPP1–POT1 and TPP1–POT1–ssDNA complexes. Identification of the TERT binding site in TPP1 should also help clarify the relationships between telomerase, shelterin and telomeric ssDNA.
How else might telomere-associated proteins contribute to telomerase activity and repeat addition processivity in vivo? It is possible that ssDNA-binding proteins, such as RPA or POT1, coat the elongating telomere as it slides through the TEN domain. This protein–DNA interaction could stabilize and protect the otherwise naked ssDNA and facilitate primer movement through telomerase. In this model, telomere-binding proteins fulfill the template-distal anchor site function. Another possibility is that one or more telomerase-associated proteins mediate anchor site function in vivo. In this regard, a recent study of T. thermophila telomerase identified an RPA1-like DNA-binding protein p82 in stable association with the endogenous holoenzyme (130). Interestingly, p82 exhibits telomere sequence-specific ssDNA-binding activity and confers ciliate telomerase with high levels of repeat addition processivity in vitro (130,131). Recent observations suggest that the function of p82 may be to bind the telomeric primer and stabilize its association with telomerase as well as to suppress the formation of DNA structures that could impede processive telomere synthesis (131). It will be important to determine if repeat addition processivity is regulated by similar proteins and mechanisms in other organisms. To this end, it will be useful to devise purification strategies that can be used to characterize the composition of the telomerase holoenzyme at various stages of the cell cycle and in different organisms and cell types. This would provide new insight into specific factors that regulate telomerase biogenesis and activity in vivo.
TELOMERASE, TELOMERES AND HUMAN DISEASE
Many studies have illustrated the molecular complexity of telomerase and telomere biology by implicating these processes in a broad spectrum of human diseases, ranging from cancer to premature aging disorders. This area has been the focus of several excellent reviews (132–134), and therefore, we will only briefly summarize the relationships between telomerase and human disease to emphasize the clinical importance of understanding telomerase biochemistry.
Telomerase, chromosome instability and cancer
Telomerase is active in germ line and stem cells but repressed in most somatic cells and tissues, which is achieved through various transcriptional and post-transcriptional mechanisms (32,135). In the absence of telomerase activity, telomeres shorten during each round of cell division until they reach a critically short-length threshold (136–138). This triggers a p53 and pRB-dependent DNA damage response and induces a non-proliferative state called replicative senescence, which is an important barrier to tumorigenesis (139–142). Cells with defective p53 and pRB pathways can bypass this barrier and undergo additional 20–30 cell divisions (143,144). During this extended period of proliferation, telomeres continue to shorten and eventually undergo chromosome end-to-end fusions (i.e. the breakage-fusion-bridge cycle), which promotes genome instability by generating loss of heterozygosity or the amplification of genetic loci (132). This usually triggers a second proliferative blockade called crisis, which is characterized by massive genome instability and apoptosis (145). However, rare clones (∼1 in 107 human cells) can emerge from crisis by up-regulating telomerase or homologous recombination-based telomere maintenance mechanisms called alternative lengthening of telomeres (ALT), all of which stabilize critically short telomeres and permit further rounds of cell division (146). Telomerase activation is the most common pathway for cellular immortalization and transformation; at least 85% of human cancer cells constitutively express the telomerase catalytic subunit and utilize telomerase-dependent telomere elongation to attain unlimited growth potential (147). Thus, telomerase is an attractive target for the development of anti-cancer therapeutics that specifically target cancer versus healthy cells (99). However, some mammalian tumors and immortalized cell lines lack telomerase activity and use ALT pathways for telomere maintenance (146). A better understanding of these non-telomerase telomere-maintenance pathways could lead to novel anti-cancer therapies.
Diseases associated with telomerase deficiency
The first disease-associated mutations in human telomerase were identified in patients afflicted with a rare, multi-system disorder called dyskeratosis congenita [see Walne (148) for a historical review of dyskeratosis congenita]. The clinical manifestations of dyskeratosis congenita generally appear during childhood and include a monocutaneous triad of abnormal skin pigmentation, nail dystrophy and oral leukoplakia. These symptoms are accompanied by a spectrum of other somatic abnormalities, such as developmental delay, premature hair loss and organ failure. Bone marrow failure is the principal cause of premature mortality. More recently, telomerase mutations have been detected in the context of aplastic anemia (149–151), Hoyeraal–Hreidarsson syndrome (152–155), idiopathic pulmonary fibrosis (156,157) and liver disease (158). Aplastic anemia is a hematological disorder characterized by reduced red blood cell counts, bone marrow failure, and liver and lung disease. Hoyeraal–Hreidarsson syndrome is a multisystem disorder characterized by bone marrow failure, immunodeficiency and severe growth retardation. Idiopathic pulmonary fibrosis is a chronic, progressive, and fatal disease that is defined by irreversible lung fibrosis. The unifying molecular characteristic of these diseases is that patients harbor telomeres that are significantly shorter than age-matched control subjects (133). This indicates that mutations in the telomerase holoenzyme compromise its ability to maintain telomere length, which is thought to impair stem cell function and limit the renewal capacity of highly proliferative cells. Indeed, inherited forms of dyskeratosis congenita exhibit disease anticipation, whereby successive generations exhibit earlier disease onset and present with more severe phenotypes (159–161). The mechanism of disease anticipation is thought to be caused by the inheritence of short telomeres that continue to shorten at an accelerated rate in the afflicted offspring. Furthermore, patients afflicted with dyskeratosis congenita are predisposed to developing carcinomas, lymphomas and leukemias. This is believed to result from the accelerated rate of telomere attrition, which promotes genetic instability and tumor formation.
Mutations have been detected in five subunits of the human telomerase holoenzyme (TERT, TR, dyskerin, NHP2 and NOP10) and one shelterin protein (TIN2) [the reader is referred to the telomerase database (http://telomerase.asu.edu/diseases.html) for a current list of disease-linked mutations]. Dyskerin, NHP2 and NOP10 are essential for the biogeneisis of human telomerase and disease-linked mutant proteins have been shown to compromise enzyme stability (162–165). Similarly, mutations in hTR can either impair its stability or disrupt the catalytic activity of human telomerase (88,159,166–168). hTERT mutants exhibit catalytic defects in vitro and fail to maintain telomere length in human cells (84,87,88,150,161). Importantly, telomere maintenance defects observed in cultivated human cells can be rescued by the ectopic expression of wild-type hTR or hTERT, providing direct evidence that these mutations impair telomerase-mediated telomere maintenance (150,163,169). However, a comprehensive study that describes how these mutations influence the biochemical and cellular properties of human telomerase is not yet available. One possibility that has not been explored is that some of these mutations interfere with enzyme activity by disrupting critical protein–protein interactions within the holoenzyme. It will be interesting to isolate human telomerase from healthy and diseased cells and compare the composition of the holoenzyme in these different cellular contexts. These experiments will provide novel insights into the mechanism of telomerase regulation in healthy and diseased human cells.
CONCLUSIONS
Telomerase has emerged as a central player in several devastating human disorders, including various forms of cancer, bone marrow failure and pulmonary fibrosis. This emphasizes the need for a detailed understanding of telomerase structure and function. High-resolution crystal structures of ciliate and insect TERTs provide much-needed insight into the intricate details of this unique RT. An important future challenge will be to determine the crystal structure of TERT in complex with TR and integral protein components of the telomerase holoenzyme. This structural information will form the basis for biochemical and molecular studies that could provide important insights into telomerase assembly and regulation. An equally important challenge will be to solve the crystal structure of the full-length TERT from an organism with well-characterized enzyme activity, especially in light of the controversy regarding the identity of T. castaneum TERT. Finally, structure–function studies of disease-associated TERT mutants will be particularly informative since these mutations have abrogated amino acids that are required for telomerase function in vivo. The information obtained from these studies may ultimately translate into novel therapeutic strategies that improve the diagnosis, treatment and management of human diseases associated with telomerase dysfunction.
FUNDING
Alberta Cancer Research Institute and Canadian Institute of Health Research (T.L.B.); Cancer Research UK, the Louis-Jeantet Foundation and Swiss Bridge (S.C.W.). H.D.M.W. was a recipient of a doctoral student scholarship from the Alberta Cancer Research Institute and National Science and Engineering Research Council. Funding for open access charge: Cancer Research UK.
Conflict of interest statement. None declared.
REFERENCES
- 1.Olovnikov AM. Principle of marginotomy in template synthesis of polynucleotides. Dokl. Akad. Nauk. SSSR. 1971;201:1496–1499. [PubMed] [Google Scholar]
- 2.Watson JD. Origin of concatemeric T7 DNA. Nat. New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 3.Olovnikov A. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- 4.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 5.Munoz-Jordan JL, Cross GA, de Lange T, Griffith JD. T-loops at trypanosome telomeres. EMBO J. 2001;20:579–588. doi: 10.1093/emboj/20.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murti KG, Prescott DM. Telomeres of polytene chromosomes in a ciliated protozoan terminate in duplex DNA loops. Proc. Natl Acad. Sci. USA. 1999;96:14436–14439. doi: 10.1073/pnas.96.25.14436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cesare AJ, Quinney N, Willcox S, Subramanian D, Griffith JD. Telomere looping in P. sativum (common garden pea) Plant J. 2003;36:271–279. doi: 10.1046/j.1365-313x.2003.01882.x. [DOI] [PubMed] [Google Scholar]
- 8.Raices M, Verdun RE, Compton SA, Haggblom CI, Griffith JD, Dillin A, Karlseder J. C. elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell. 2008;132:745–757. doi: 10.1016/j.cell.2007.12.039. [DOI] [PubMed] [Google Scholar]
- 9.Tomaska L, Willcox S, Slezakova J, Nosek J, Griffith JD. Taz1 binding to a fission yeast model telomere: formation of telomeric loops and higher order structures. J. Biol. Chem. 2004;279:50764–50772. doi: 10.1074/jbc.M409790200. [DOI] [PubMed] [Google Scholar]
- 10.Cesare AJ, Groff-Vindman C, Compton SA, McEachern MJ, Griffith JD. Telomere loops and homologous recombination-dependent telomeric circles in a Kluyveromyces lactis telomere mutant strain. Mol. Cell Biol. 2008;28:20–29. doi: 10.1128/MCB.01122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 12.Tomaska L, Nosek J, Kramara J, Griffith JD. Telomeric circles: universal players in telomere maintenance? Nat. Struct. Mol. Biol. 2009;16:1010–1015. doi: 10.1038/nsmb.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linger BR, Price CM. Conservation of telomere protein complexes: shuffling through evolution. Crit. Rev. Biochem. Mol. Biol. 2009;44:434–446. doi: 10.3109/10409230903307329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 15.Marcand S, Gilson E, Shore D. A protein-counting mechanism for telomere length regulation in yeast. Science. 1997;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- 16.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 17.Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 18.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 19.Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, Trager JB, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 20.Beattie TL, Zhou W, Robinson MO, Harrington L. Reconstitution of human telomerase activity in vitro. Curr. Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- 21.Collins K, Gandhi L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc. Natl Acad. Sci. USA. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu D, Collins K. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol. Cell. 2007;28:773–785. doi: 10.1016/j.molcel.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen SB, Graham ME, Lovrecz GO, Bache N, Robinson PJ, Reddel RR. Protein composition of catalytically active human telomerase from immortal cells. Science. 2007;315:1850–1853. doi: 10.1126/science.1138596. [DOI] [PubMed] [Google Scholar]
- 24.Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–648. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins K. The biogenesis and regulation of telomerase holoenzymes. Nat. Rev. Mol. Cell Biol. 2006;7:484–494. doi: 10.1038/nrm1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gallardo F, Chartrand P. Telomerase biogenesis: the long road before getting to the end. RNA Biol. 2008;5:212–215. doi: 10.4161/rna.7115. [DOI] [PubMed] [Google Scholar]
- 28.Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lingner J, Cech TR. Purification of telomerase from Euplotes aediculatus: requirement of a primer 3′ overhang. Proc. Natl Acad. Sci. USA. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 32.Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergen RL, Davidoff MJ, Liu Q, et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 33.Harrington L, Zhou W, McPhail T, Oulton R, Yeung DS, Mar V, Bass MB, Robinson MO. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bryan TM, Sperger JM, Chapman KB, Cech TR. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc. Natl Acad. Sci. USA. 1998;95:8479–8484. doi: 10.1073/pnas.95.15.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greenberg RA, Allsopp RC, Chin L, Morin GB, DePinho RA. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene. 1998;16:1723–1730. doi: 10.1038/sj.onc.1201933. [DOI] [PubMed] [Google Scholar]
- 36.Holt SE, Aisner DL, Baur J, Tesmer VM, Dy M, Ouellette M, Trager JB, Morin GB, Toft DO, Shay JW, et al. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 1999;13:817–826. doi: 10.1101/gad.13.7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bachand F, Boisvert FM, Cote J, Richard S, Autexier C. The product of the survival of motor neuron (SMN) gene is a human telomerase-associated protein. Mol. Biol. Cell. 2002;13:3192–3202. doi: 10.1091/mbc.E02-04-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 39.Osanai M, Kojima KK, Futahashi R, Yaguchi S, Fujiwara H. Identification and characterization of the telomerase reverse transcriptase of Bombyx mori (silkworm) and Tribolium castaneum (flour beetle) Gene. 2006;376:281–289. doi: 10.1016/j.gene.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 40.Malik HS, Burke WD, Eickbush TH. Putative telomerase catalytic subunits from Giardia lamblia and Caenorhabditis elegans. Gene. 2000;251:101–108. doi: 10.1016/s0378-1119(00)00207-9. [DOI] [PubMed] [Google Scholar]
- 41.Figueiredo LM, Rocha EP, Mancio-Silva L, Prevost C, Hernandez-Verdun D, Scherf A. The unusually large Plasmodium telomerase reverse-transcriptase localizes in a discrete compartment associated with the nucleolus. Nucleic Acids Res. 2005;33:1111–1122. doi: 10.1093/nar/gki260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 43.Sasaki T, Fujiwara H. Detection and distribution patterns of telomerase activity in insects. Eur. J. Biochem. 2000;267:3025–3031. doi: 10.1046/j.1432-1033.2000.01323.x. [DOI] [PubMed] [Google Scholar]
- 44.Wang L, Dean SR, Shippen DE. Oligomerization of the telomerase reverse transcriptase from Euplotes crassus. Nucleic Acids Res. 2002;30:4032–4039. doi: 10.1093/nar/gkf513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arai K, Masutomi K, Khurts S, Kaneko S, Kobayashi K, Murakami S. Two independent regions of human telomerase reverse transcriptase are important for its oligomerization and telomerase activity. J. Biol. Chem. 2002;277:8538–8544. doi: 10.1074/jbc.M111068200. [DOI] [PubMed] [Google Scholar]
- 46.Beattie TL, Zhou W, Robinson MO, Harrington L. Functional multimerization of the human telomerase reverse transcriptase. Mol. Cell Biol. 2001;21:6151–6160. doi: 10.1128/MCB.21.18.6151-6160.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hammond PW, Cech TR. Euplotes telomerase: evidence for limited base-pairing during primer elongation and dGTP as an effector of translocation. Biochemistry. 1998;37:5162–5172. doi: 10.1021/bi972988o. [DOI] [PubMed] [Google Scholar]
- 48.Forstemann K, Lingner J. Telomerase limits the extent of base pairing between template RNA and telomeric DNA. EMBO Rep. 2005;6:361–366. doi: 10.1038/sj.embor.7400374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hossain S, Singh S, Lue NF. Functional analysis of the C-terminal extension of telomerase reverse transcriptase. A putative “thumb” domain. J. Biol. Chem. 2002;277:36174–36180. doi: 10.1074/jbc.M201976200. [DOI] [PubMed] [Google Scholar]
- 50.Bosoy D, Lue NF. Functional analysis of conserved residues in the putative “finger” domain of telomerase reverse transcriptase. J. Biol. Chem. 2001;276:46305–46312. doi: 10.1074/jbc.M108168200. [DOI] [PubMed] [Google Scholar]
- 51.Haering CH, Nakamura TM, Baumann P, Cech TR. Analysis of telomerase catalytic subunit mutants in vivo and in vitro in Schizosaccharomyces pombe. Proc. Natl Acad. Sci. USA. 2000;97:6367–6372. doi: 10.1073/pnas.130187397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lue NF, Lin YC, Mian IS. A conserved telomerase motif within the catalytic domain of telomerase reverse transcriptase is specifically required for repeat addition processivity. Mol. Cell Biol. 2003;23:8440–8449. doi: 10.1128/MCB.23.23.8440-8449.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huard S, Moriarty TJ, Autexier C. The C-terminus of the human telomerase reverse transcriptase is a determinant of enzyme processivity. Nucleic Acids Res. 2003;31:4059–4070. doi: 10.1093/nar/gkg437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Banik SS, Guo C, Smith AC, Margolis SS, Richardson DA, Tirado CA, Counter CM. C-terminal regions of the human telomerase catalytic subunit essential for in vivo enzyme activity. Mol. Cell Biol. 2002;22:6234–6246. doi: 10.1128/MCB.22.17.6234-6246.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat. Struct. Mol. Biol. 2010;17:513–518. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- 56.Rouda S, Skordalakes E. Structure of the RNA-binding domain of telomerase: implications for RNA recognition and binding. Structure. 2007;15:1403–1412. doi: 10.1016/j.str.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 57.Jacobs SA, Podell ER, Cech TR. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 2006;13:218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- 58.Friedman KL, Cech TR. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev. 1999;13:2863–2874. doi: 10.1101/gad.13.21.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xia J, Peng Y, Mian IS, Lue NF. Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol. Cell Biol. 2000;20:5196–5207. doi: 10.1128/mcb.20.14.5196-5207.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller MC, Liu JK, Collins K. Template definition by Tetrahymena telomerase reverse transcriptase. EMBO J. 2000;19:4412–4422. doi: 10.1093/emboj/19.16.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bosoy D, Peng Y, Mian IS, Lue NF. Conserved N-terminal motifs of telomerase reverse transcriptase required for ribonucleoprotein assembly in vivo. J. Biol. Chem. 2003;278:3882–3890. doi: 10.1074/jbc.M210645200. [DOI] [PubMed] [Google Scholar]
- 62.Bachand F, Autexier C. Functional regions of human telomerase reverse transcriptase and human telomerase RNA required for telomerase activity and RNA-protein interactions. Mol. Cell Biol. 2001;21:1888–1897. doi: 10.1128/MCB.21.5.1888-1897.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beattie TL, Zhou W, Robinson MO, Harrington L. Polymerization defects within human telomerase are distinct from telomerase RNA and TEP1 binding. Mol. Biol. Cell. 2000;11:3329–3340. doi: 10.1091/mbc.11.10.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bryan TM, Goodrich KJ, Cech TR. Telomerase RNA bound by protein motifs specific to telomerase reverse transcriptase. Mol. Cell. 2000;6:493–499. doi: 10.1016/s1097-2765(00)00048-4. [DOI] [PubMed] [Google Scholar]
- 65.Lai CK, Mitchell JR, Collins K. RNA binding domain of telomerase reverse transcriptase. Mol. Cell Biol. 2001;21:990–1000. doi: 10.1128/MCB.21.4.990-1000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lai CK, Miller MC, Collins K. Template boundary definition in Tetrahymena telomerase. Genes Dev. 2002;16:415–420. doi: 10.1101/gad.962602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moriarty TJ, Huard S, Dupuis S, Autexier C. Functional multimerization of human telomerase requires an RNA interaction domain in the N-terminus of the catalytic subunit. Mol. Cell Biol. 2002;22:1253–1265. doi: 10.1128/MCB.22.4.1253-1265.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O'Connor CM, Lai CK, Collins K. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J. Biol. Chem. 2005;280:17533–17539. doi: 10.1074/jbc.M501211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Drosopoulos WC, Prasad VR. The telomerase-specific T motif is a restrictive determinant of repetitive reverse transcription by human telomerase. Mol. Cell Biol. 30:447–459. doi: 10.1128/MCB.00853-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wyatt HD, Tsang AR, Lobb DA, Beattie TL. Human telomerase reverse transcriptase (hTERT) Q169 is essential for telomerase function in vitro and in vivo. PLoS One. 2009;4:e7176. doi: 10.1371/journal.pone.0007176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lue NF, Li Z. Modeling and structure function analysis of the putative anchor site of yeast telomerase. Nucleic Acids Res. 2007;35:5213–5222. doi: 10.1093/nar/gkm531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Romi E, Baran N, Gantman M, Shmoish M, Min B, Collins K, Manor H. High-resolution physical and functional mapping of the template adjacent DNA binding site in catalytically active telomerase. Proc. Natl Acad. Sci. USA. 2007;104:8791–8796. doi: 10.1073/pnas.0703157104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sealey DC, Zheng L, Taboski MA, Cruickshank J, Ikura M, Harrington LA. The N-terminus of hTERT contains a DNA-binding domain and is required for telomerase activity and cellular immortalization. Nucleic Acids Res. 2010;38:2019–2035. doi: 10.1093/nar/gkp1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zaug AJ, Podell ER, Cech TR. Mutation in TERT separates processivity from anchor-site function. Nat. Struct. Mol. Biol. 2008;15:870–872. doi: 10.1038/nsmb.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Finger SN, Bryan TM. Multiple DNA-binding sites in Tetrahymena telomerase. Nucleic Acids Res. 2008;36:1260–1272. doi: 10.1093/nar/gkm866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Counter CM, Meyerson M, Eaton EN, Weinberg RA. The catalytic subunit of yeast telomerase. Proc. Natl Acad. Sci. USA. 1997;94:9202–9207. doi: 10.1073/pnas.94.17.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nakayama J, Tahara H, Tahara E, Saito M, Ito K, Nakamura H, Nakanishi T, Ide T, Ishikawa F. Telomerase activation by hTRT in human normal fibroblasts and hepatocellular carcinomas. Nat. Genet. 1998;18:65–68. doi: 10.1038/ng0198-65. [DOI] [PubMed] [Google Scholar]
- 78.Bryan TM, Goodrich KJ, Cech TR. A mutant of Tetrahymena telomerase reverse transcriptase with increased processivity. J. Biol. Chem. 2000;275:24199–24207. doi: 10.1074/jbc.M003246200. [DOI] [PubMed] [Google Scholar]
- 79.Cote ML, Roth MJ. Murine leukemia virus reverse transcriptase: structural comparison with HIV-1 reverse transcriptase. Virus Res. 2008;134:186–202. doi: 10.1016/j.virusres.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peng Y, Mian IS, Lue NF. Analysis of telomerase processivity: mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Mol. Cell. 2001;7:1201–1211. doi: 10.1016/s1097-2765(01)00268-4. [DOI] [PubMed] [Google Scholar]
- 81.Wyatt HD, Lobb DA, Beattie TL. Characterization of physical and functional anchor site interactions in human telomerase. Mol. Cell Biol. 2007;27:3226–3240. doi: 10.1128/MCB.02368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Drosopoulos WC, Prasad VR. The active site residue Valine 867 in human telomerase reverse transcriptase influences nucleotide incorporation and fidelity. Nucleic Acids Res. 2007;35:1155–1168. doi: 10.1093/nar/gkm002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li Y, Yates JA, Chen JJ. Identification and characterization of sea squirt telomerase reverse transcriptase. Gene. 2007;400:16–24. doi: 10.1016/j.gene.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 84.Xie M, Podlevsky JD, Qi X, Bley CJ, Chen JJ. A novel motif in telomerase reverse transcriptase regulates telomere repeat addition rate and processivity. Nucleic Acids Res. 2010;38:1982–1996. doi: 10.1093/nar/gkp1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Seimiya H, Sawada H, Muramatsu Y, Shimizu M, Ohko K, Yamane K, Tsuruo T. Involvement of 14-3-3 proteins in nuclear localization of telomerase. EMBO J. 2000;19:2652–2661. doi: 10.1093/emboj/19.11.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Counter CM, Hahn WC, Wei W, Caddle SD, Beijersbergen RL, Lansdorp PM, Sedivy JM, Weinberg RA. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc. Natl Acad. Sci. USA. 1998;95:14723–14728. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vulliamy TJ, Walne A, Baskaradas A, Mason PJ, Marrone A, Dokal I. Mutations in the reverse transcriptase component of telomerase (TERT) in patients with bone marrow failure. Blood Cells Mol. Dis. 2005;34:257–263. doi: 10.1016/j.bcmd.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 88.Robart AR, Collins K. Investigation of human telomerase holoenzyme assembly, activity, and processivity using disease-linked subunit variants. J. Biol. Chem. 2009;285:4375–4386. doi: 10.1074/jbc.M109.088575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Theimer CA, Feigon J. Structure and function of telomerase RNA. Curr. Opin. Struct. Biol. 2006;16:307–318. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 90.Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 91.Chen JL, Greider CW. Template boundary definition in mammalian telomerase. Genes Dev. 2003;17:2747–2752. doi: 10.1101/gad.1140303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Autexier C, Pruzan R, Funk WD, Greider CW. Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J. 1996;15:5928–5935. [PMC free article] [PubMed] [Google Scholar]
- 93.Tesmer VM, Ford LP, Holt SE, Frank BC, Yi X, Aisner DL, Ouellette M, Shay JW, Wright WE. Two inactive fragments of the integral RNA cooperate to assemble active telomerase with the human protein catalytic subunit (hTERT) in vitro. Mol. Cell Biol. 1999;19:6207–6216. doi: 10.1128/mcb.19.9.6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Theimer CA, Jady BE, Chim N, Richard P, Breece KE, Kiss T, Feigon J. Structural and functional characterization of human telomerase RNA processing and Cajal body localization signals. Mol. Cell. 2007;27:869–881. doi: 10.1016/j.molcel.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 95.Cristofari G, Adolf E, Reichenbach P, Sikora K, Terns RM, Terns MP, Lingner J. Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol. Cell. 2007;27:882–889. doi: 10.1016/j.molcel.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 96.Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J, et al. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 97.Nowak T, Januszkiewicz D, Zawada M, Pernak M, Lewandowski K, Rembowska J, Nowicka K, Mankowski P, Nowak J. Amplification of hTERT and hTERC genes in leukemic cells with high expression and activity of telomerase. Oncol. Rep. 2006;16:301–305. [PubMed] [Google Scholar]
- 98.Andersson S, Wallin KL, Hellstrom AC, Morrison LE, Hjerpe A, Auer G, Ried T, Larsson C, Heselmeyer-Haddad K. Frequent gain of the human telomerase gene TERC at 3q26 in cervical adenocarcinomas. Br. J. Cancer. 2006;95:331–338. doi: 10.1038/sj.bjc.6603253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Phatak P, Burger AM. Telomerase and its potential for therapeutic intervention. Br. J. Pharmacol. 2007;152:1003–1011. doi: 10.1038/sj.bjp.0707374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Morin GB. Recognition of a chromosome truncation site associated with alpha-thalassaemia by human telomerase. Nature. 1991;353:454–456. doi: 10.1038/353454a0. [DOI] [PubMed] [Google Scholar]
- 101.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 102.Oganesian L, Moon IK, Bryan TM, Jarstfer MB. Extension of G-quadruplex DNA by ciliate telomerase. EMBO J. 2006;25:1148–1159. doi: 10.1038/sj.emboj.7601006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oganesian L, Graham ME, Robinson PJ, Bryan TM. Telomerase recognizes G-quadruplex and linear DNA as distinct substrates. Biochemistry. 2007;46:11279–11290. doi: 10.1021/bi700993q. [DOI] [PubMed] [Google Scholar]
- 104.Greider CW. Telomerase is processive. Mol. Cell Biol. 1991;11:4572–4580. doi: 10.1128/mcb.11.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Morin GB. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 106.Cohn M, Blackburn EH. Telomerase in yeast. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- 107.Prowse KR, Avilion AA, Greider CW. Identification of a nonprocessive telomerase activity from mouse cells. Proc. Natl Acad. Sci. USA. 1993;90:1493–1497. doi: 10.1073/pnas.90.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Prescott J, Blackburn EH. Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes Dev. 1997;11:528–540. doi: 10.1101/gad.11.4.528. [DOI] [PubMed] [Google Scholar]
- 109.Yu GL, Bradley JD, Attardi LD, Blackburn EH. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature. 1990;344:126–132. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]
- 110.Yu GL, Blackburn EH. Developmentally programmed healing of chromosomes by telomerase in Tetrahymena. Cell. 1991;67:823–832. doi: 10.1016/0092-8674(91)90077-c. [DOI] [PubMed] [Google Scholar]
- 111.Lue NF. A physical and functional constituent of telomerase anchor site. J. Biol. Chem. 2005;280:26586–26591. doi: 10.1074/jbc.M503028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Moriarty TJ, Ward RJ, Taboski MA, Autexier C. An anchor site-type defect in human telomerase that disrupts telomere length maintenance and cellular immortalization. Mol. Biol. Cell. 2005;16:3152–3161. doi: 10.1091/mbc.E05-02-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Moriarty TJ, Marie-Egyptienne DT, Autexier C. Functional organization of repeat addition processivity and DNA synthesis determinants in the human telomerase multimer. Mol. Cell Biol. 2004;24:3720–3733. doi: 10.1128/MCB.24.9.3720-3733.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 115.Chang M, Arneric M, Lingner J. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 2007;21:2485–2494. doi: 10.1101/gad.1588807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Collins K, Greider CW. Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev. 1993;7:1364–1376. doi: 10.1101/gad.7.7b.1364. [DOI] [PubMed] [Google Scholar]
- 117.Wang H, Blackburn EH. De novo telomere addition by Tetrahymena telomerase in vitro. EMBO J. 1997;16:866–879. doi: 10.1093/emboj/16.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Harrington LA, Greider CW. Telomerase primer specificity and chromosome healing. Nature. 1991;353:451–454. doi: 10.1038/353451a0. [DOI] [PubMed] [Google Scholar]
- 119.Prescott J, Blackburn EH. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 1997;11:2790–2800. doi: 10.1101/gad.11.21.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wallweber G, Gryaznov S, Pongracz K, Pruzan R. Interaction of human telomerase with its primer substrate. Biochemistry. 2003;42:589–600. doi: 10.1021/bi026914a. [DOI] [PubMed] [Google Scholar]
- 121.Hammond PW, Lively TN, Cech TR. The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol. Cell Biol. 1997;17:296–308. doi: 10.1128/mcb.17.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lee MS, Blackburn EH. Sequence-specific DNA primer effects on telomerase polymerization activity. Mol. Cell Biol. 1993;13:6586–6599. doi: 10.1128/mcb.13.10.6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lue NF, Peng Y. Negative regulation of yeast telomerase activity through an interaction with an upstream region of the DNA primer. Nucleic Acids Res. 1998;26:1487–1494. doi: 10.1093/nar/26.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Baran N, Haviv Y, Paul B, Manor H. Studies on the minimal lengths required for DNA primers to be extended by the Tetrahymena telomerase: implications for primer positioning by the enzyme. Nucleic Acids Res. 2002;30:5570–5578. doi: 10.1093/nar/gkf676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee SR, Wong JM, Collins K. Human telomerase reverse transcriptase motifs required for elongation of a telomeric substrate. J. Biol. Chem. 2003;278:52531–52536. doi: 10.1074/jbc.M311359200. [DOI] [PubMed] [Google Scholar]
- 126.Berdis AJ. DNA polymerases as therapeutic targets. Biochemistry. 2008;47:8253–8260. doi: 10.1021/bi801179f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 128.Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 129.Zaug AJ, Podell ER, Nandakumar J, Cech TR. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 2010;24:613–622. doi: 10.1101/gad.1881810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Min B, Collins K. An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol. Cell. 2009;36:609–619. doi: 10.1016/j.molcel.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Min B, Collins K. Multiple mechanisms for elongation processivity within the reconstituted Tetrahymena telomerase holoenzyme. J. Biol. Chem. 2010 doi: 10.1074/jbc.M110.119172. doi:10.1074/jbc.M110.119172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Armanios M. Syndromes of telomere shortening. Annu. Rev. Genomics Hum. Genet. 2009;10:45–61. doi: 10.1146/annurev-genom-082908-150046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Calado RT, Young NS. Telomere diseases. N. Engl. J. Med. 2009;361:2353–2365. doi: 10.1056/NEJMra0903373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW. Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 1996;18:173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 136.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 137.Allsopp RC, Vaziri H, Patterson C, Goldstein S, Younglai EV, Futcher AB, Greider CW, Harley CB. Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl Acad. Sci. USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hemann MT, Strong MA, Hao LY, Greider CW. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- 139.Smogorzewska A, de Lange T. Different telomere damage signaling pathways in human and mouse cells. EMBO J. 2002;21:4338–4348. doi: 10.1093/emboj/cdf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a) Mol. Cell. 2004;14:501–513. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- 141.d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 142.Beausejour CM, Krtolica A, Galimi F, Narita M, Lowe SW, Yaswen P, Campisi J. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J. 2003;22:4212–4222. doi: 10.1093/emboj/cdg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shay JW, Pereira-Smith OM, Wright WE. A role for both RB and p53 in the regulation of human cellular senescence. Exp. Cell Res. 1991;196:33–39. doi: 10.1016/0014-4827(91)90453-2. [DOI] [PubMed] [Google Scholar]
- 144.Hara E, Tsurui H, Shinozaki A, Nakada S, Oda K. Cooperative effect of antisense-Rb and antisense-p53 oligomers on the extension of life span in human diploid fibroblasts, TIG-1. Biochem. Biophys. Res. Commun. 1991;179:528–534. doi: 10.1016/0006-291x(91)91403-y. [DOI] [PubMed] [Google Scholar]
- 145.Wright WE, Shay JW. The two-stage mechanism controlling cellular senescence and immortalization. Exp. Gerontol. 1992;27:383–389. doi: 10.1016/0531-5565(92)90069-c. [DOI] [PubMed] [Google Scholar]
- 146.Cesare AJ, Reddel RR. Telomere uncapping and alternative lengthening of telomeres. Mech. Ageing Dev. 2008;129:99–108. doi: 10.1016/j.mad.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 147.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 148.Walne AJ, Dokal I. Dyskeratosis Congenita: a historical perspective. Mech. Ageing Dev. 2008;129:48–59. doi: 10.1016/j.mad.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 149.Yamaguchi H, Calado RT, Ly H, Kajigaya S, Baerlocher GM, Chanock SJ, Lansdorp PM, Young NS. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N. Engl. J. Med. 2005;352:1413–1424. doi: 10.1056/NEJMoa042980. [DOI] [PubMed] [Google Scholar]
- 150.Xin ZT, Beauchamp AD, Calado RT, Bradford JW, Regal JA, Shenoy A, Liang Y, Lansdorp PM, Young NS, Ly H. Functional characterization of natural telomerase mutations found in patients with hematologic disorders. Blood. 2007;109:524–532. doi: 10.1182/blood-2006-07-035089. [DOI] [PubMed] [Google Scholar]
- 151.Vulliamy T, Marrone A, Dokal I, Mason PJ. Association between aplastic anaemia and mutations in telomerase RNA. Lancet. 2002;359:2168–2170. doi: 10.1016/S0140-6736(02)09087-6. [DOI] [PubMed] [Google Scholar]
- 152.Knight SW, Heiss NS, Vulliamy TJ, Aalfs CM, McMahon C, Richmond P, Jones A, Hennekam RC, Poustka A, Mason PJ, et al. Unexplained aplastic anaemia, immunodeficiency, and cerebellar hypoplasia (Hoyeraal-Hreidarsson syndrome) due to mutations in the dyskeratosis congenita gene, DKC1. Br. J. Haematol. 1999;107:335–339. doi: 10.1046/j.1365-2141.1999.01690.x. [DOI] [PubMed] [Google Scholar]
- 153.Marrone A, Walne A, Tamary H, Masunari Y, Kirwan M, Beswick R, Vulliamy T, Dokal I. Telomerase reverse-transcriptase homozygous mutations in autosomal recessive dyskeratosis congenita and Hoyeraal-Hreidarsson syndrome. Blood. 2007;110:4198–4205. doi: 10.1182/blood-2006-12-062851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Walne AJ, Vulliamy T, Beswick R, Kirwan M, Dokal I. TINF2 mutations result in very short telomeres: analysis of a large cohort of patients with dyskeratosis congenita and related bone marrow failure syndromes. Blood. 2008;112:3594–3600. doi: 10.1182/blood-2008-05-153445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Savage SA, Giri N, Baerlocher GM, Orr N, Lansdorp PM, Alter BP. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am. J. Hum. Genet. 2008;82:501–509. doi: 10.1016/j.ajhg.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Marrone A, Sokhal P, Walne A, Beswick R, Kirwan M, Killick S, Williams M, Marsh J, Vulliamy T, Dokal I. Functional characterization of novel telomerase RNA (TERC) mutations in patients with diverse clinical and pathological presentations. Haematologica. 2007;92:1013–1020. doi: 10.3324/haematol.11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Armanios MY, Chen JJ, Cogan JD, Alder JK, Ingersoll RG, Markin C, Lawson WE, Xie M, Vulto I, Phillips J.A., 3rd, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N. Engl. J. Med. 2007;356:1317–1326. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 158.Calado RT, Regal JA, Kleiner DE, Schrump DS, Peterson NR, Pons V, Chanock SJ, Lansdorp PM, Young NS. A spectrum of severe familial liver disorders associate with telomerase mutations. PLoS One. 2009;4:e7926. doi: 10.1371/journal.pone.0007926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, Dokal I. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 160.Vulliamy T, Marrone A, Szydlo R, Walne A, Mason PJ, Dokal I. Disease anticipation is associated with progressive telomere shortening in families with dyskeratosis congenita due to mutations in TERC. Nat. Genet. 2004;36:447–449. doi: 10.1038/ng1346. [DOI] [PubMed] [Google Scholar]
- 161.Armanios M, Chen JL, Chang YP, Brodsky RA, Hawkins A, Griffin CA, Eshleman JR, Cohen AR, Chakravarti A, Hamosh A, et al. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc. Natl Acad. Sci. USA. 2005;102:15960–15964. doi: 10.1073/pnas.0508124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 163.Wong JM, Collins K. Telomerase RNA level limits telomere maintenance in X-linked dyskeratosis congenita. Genes Dev. 2006;20:2848–2858. doi: 10.1101/gad.1476206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Walne AJ, Vulliamy T, Marrone A, Beswick R, Kirwan M, Masunari Y, Al-Qurashi FH, Aljurf M, Dokal I. Genetic heterogeneity in autosomal recessive dyskeratosis congenita with one subtype due to mutations in the telomerase-associated protein NOP10. Hum. Mol. Genet. 2007;16:1619–1629. doi: 10.1093/hmg/ddm111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Vulliamy T, Beswick R, Kirwan M, Marrone A, Digweed M, Walne A, Dokal I. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc. Natl Acad. Sci. USA. 2008;105:8073–8078. doi: 10.1073/pnas.0800042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Fu D, Collins K. Distinct biogenesis pathways for human telomerase RNA and H/ACA small nucleolar RNAs. Mol. Cell. 2003;11:1361–1372. doi: 10.1016/s1097-2765(03)00196-5. [DOI] [PubMed] [Google Scholar]
- 167.Errington TM, Fu D, Wong JM, Collins K. Disease-associated human telomerase RNA variants show loss of function for telomere synthesis without dominant-negative interference. Mol. Cell Biol. 2008;28:6510–6520. doi: 10.1128/MCB.00777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Trahan C, Dragon F. Dyskeratosis congenita mutations in the H/ACA domain of human telomerase RNA affect its assembly into a pre-RNP. RNA. 2009;15:235–243. doi: 10.1261/rna.1354009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kirwan M, Beswick R, Vulliamy T, Nathwani AC, Walne AJ, Casimir C, Dokal I. Exogenous TERC alone can enhance proliferative potential, telomerase activity and telomere length in lymphocytes from dyskeratosis congenita patients. Br. J. Haematol. 2009;144:771–781. doi: 10.1111/j.1365-2141.2008.07516.x. [DOI] [PubMed] [Google Scholar]