Abstract
Interactions of RNA polymerase (RNAP) with nucleic acids must be tightly controlled to ensure precise and processive RNA synthesis. The RNAP β′-subunit Switch-2 (SW2) region is part of a protein network that connects the clamp domain with the RNAP body and mediates opening and closing of the active center cleft. SW2 interacts with the template DNA near the RNAP active center and is a target for antibiotics that block DNA melting during initiation. Here, we show that substitutions of a conserved Arg339 residue in the Escherichia coli RNAP SW2 confer diverse effects on transcription that include defects in DNA melting in promoter complexes, decreased stability of RNAP/promoter complexes, increased apparent KM for initiating nucleotide substrates (2- to 13-fold for different substitutions), decreased efficiency of promoter escape, and decreased stability of elongation complexes. We propose that interactions of Arg339 with DNA directly stabilize transcription complexes to promote stable closure of the clamp domain around nucleic acids. During initiation, SW2 may cooperate with the σ3.2 region to stabilize the template DNA strand in the RNAP active site. Together, our data suggest that SW2 may serve as a key regulatory element that affects transcription initiation and RNAP processivity through controlling RNAP/DNA template interactions.
INTRODUCTION
The efficient synthesis of RNA by bacterial RNAP depends on a complex and dynamic network of its interactions with the nucleic acids. The catalytically competent core RNAP is a five-subunit (α2ββ′ω) complex, in which the two largest, β and β′, subunits jointly form the active site and establish most of the key contacts to the DNA and RNA chains. Structural and functional studies (1–6) identified several mobile β and β′ domains that are proposed to undergo concerted transitions at different steps of transcription in RNAPs across all domains of life. However, the detailed understanding of the functional role of these elements is just beginning to emerge.
Transcription initiation is carried out by RNAP holoenzyme, a complex of core RNAP and a σ subunit, which plays the key role in the specific recognition of the promoter elements and the DNA strand separation. Most promoters are recognized by a holoenzyme species containing the ‘housekeeping’ σ subunit (σ70 in Escherichia coli and σA in other bacteria); recognition is followed by melting of the DNA, which commences at the second position of the −10 element and propagates downstream to include the start of transcription (7,8 and references therein). During initiation, RNAP reiteratively synthesizes and releases short RNA products but remains bound to the promoter, a process known as abortive initiation (9,10); only a fraction of RNAP molecules clears the promoter to escape into the productive synthesis mode at each cycle (11). The σ conserved region 3.2 (σ3.2) has been implicated in both the initiation of RNA synthesis, by stimulating binding of the initiating nucleotides in the RNAP active center (12), and the promoter clearance, by clashing with the growing RNA transcript in the RNA exit channel (12–14).
The elongation complex is characterized by the very high stability, as demanded by the obligatory processivity of the RNAP that must remain bound to DNA and RNA throughout elongation. The β′ clamp domain was proposed to play a pivotal role in transcription complex stabilization. Based on its apparent conformational mobility and different positions in different structures, the β′ clamp was proposed to close the active site cleft around nucleic acids during transcription (2–4,6). In the Thermus thermophilus elongation complex structure, the clamp directly interacts with the downstream DNA duplex and the DNA/RNA hybrid inside the main RNAP cleft (Figure 1A–C). In the T. thermophilus holoenzyme structure, the clamp is open in comparison with the elongation complex (Figure 1D; 3); however, its interactions with DNA upon a promoter complex formation may favor a more closed state.
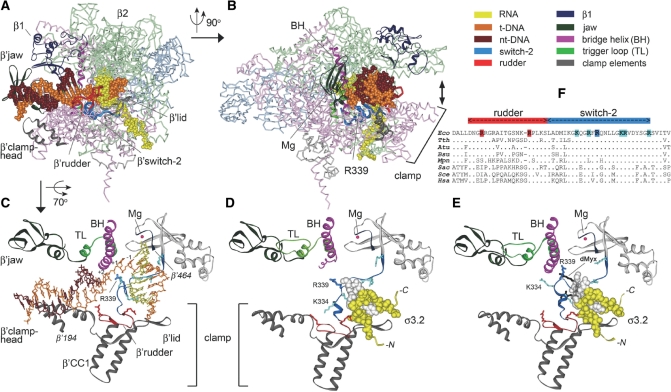
Figure 1.
RNAP–nucleic acids contacts in transcription complexes. (A) The structure of the elongation complex of T. thermophilus RNAP (4). RNAP subunits are shown as Cα-backbone stick models; α-subunits are light blue, β—light green, β′—light red, ω—light gray. β′SW2 and β′rudder are shown in blue and red, respectively; β′lid and β′clamphead are gray. DNA and RNA are shown as CPK models. Core RNAP domains that interact with downstream DNA duplex and substitutions in which affect stability of transcription complexes are shown as solid ribbons. The color scheme is shown at the upper right part of the figure. (B) The same structure but rotated 90° relative to (A). The direction of clamp movement is shown by a double arrowhead. The R339 residue is shown as a CPK model. The active site Mg ion is shown as a red sphere. (C) A close-up view of the RNAP active center in the elongation complex. The color scheme is the same as in (A) and (B). The bridge helix (BH) and trigger loop (TL) at the RNAP active center are shown in violet and light green, respectively. The β′-segment between amino acids 194 and 464 (E. coli numbering) shown on the figure includes the clamphead, β′ coiled-coil 1, lid and rudder elements of the clamp, SW2 and the active site Mg ion. Conserved positively charged residues in SW2 are shown in blue (R339 is dark blue). Conserved positively charged residues from β′rudder that interact with the downstream DNA duplex and the DNA/RNA hybrid are shown in red. (D) The same view in the holoenzyme of T. thermophilus RNAP (3). The σ3.2 region is shown in yellow as a CPK model; the σ513–519 region is in white. (E) The same view in the complex of the T. thermophilus holoenzyme RNAP with desmethyl-myxopyronin B (dMyx) (27). dMyx is shown in black, the part of the molecule that is located behind σ3.2 and SW2 is translucent. A loop including SW2 residues K334 and R339 blocks the path of the template DNA strand downstream of the RNAP active center. (F) Alignment of the rudder and SW2 regions in various RNAPs: Eco, Escherichia coli; Tth, Thermus thermopilus; Atu, Agrobacterium tumefaciens; Bsu, Bacillus subtilis; Mpn, Mycoplasma pneumoniae; Sac, Sulfolobus acidocaldarius; Sce, Saccharomyces cerevisiae RNAPII; Hsa, Homo sapiens RNAPII. Amino acid residues shown in (C and D) are colored.
Mutations in several clamp elements that interact with DNA/RNA hybrid and downstream DNA duplex were shown to affect the stability of RNAP–nucleic acids complexes at different steps of transcription. In particular, deletions of two clamp loops, the β′ lid and β′ rudder (Figure 1A and C), destabilize elongation complexes of E. coli and Thermus aquaticus RNAPs, respectively (15–17), whereas deletions in the β′ clamphead (18,19) and the β′ lid (17) dramatically decrease the E. coli promoter complex stability. In addition, mutations in other E. coli RNAP elements interacting with nucleic acids in the main cleft, including the β1 and β′ jaw domains (Figure 1A), also affect the open complex stability (18,20–22).
The clamp domain is connected to the main RNAP body through several evolutionary conserved ‘switch’ regions. These switches were proposed to couple DNA binding with the clamp movement and the closure of the main RNAP cleft around the nucleic acids (1,2). The focus of this study is the β′ SW2 (amino acid residues 327–352, E. coli numbering is used throughout the article unless otherwise indicated) that occupies a prominent position within the transcription elongation complex—it directly contacts the template DNA strand at the RNAP active center (Figure 1C; 2,4,5,23–25). In the holoenzyme, SW2 also interacts with σ3.2 (3). Deletion of amino acids 513–519 in this region in E. coli σ70 (shown in white in Figure 1D) has been shown to impair initiating nucleotide binding and promoter escape by RNAP (12).
This raises a possibility that SW2 may have specific functions in transcription initiation. Indeed, a number of substitutions in SW2 in E. coli RNAP decreased stability of open promoter complexes (26,27) and affected regulation by DksA, a protein that alters the pathway of the initiation complex formation (26). Furthermore, analysis of SW2 substitutions in eukaryotic (28) and archaeal RNAPs (29) suggested that this region may be involved in start site selection, abortive initiation, promoter escape and RNA chain elongation. Finally, a group of antibiotics that target bacterial RNAP, including myxopyronin, corallopyronin and ripostatin, were recently shown to stabilize SW2 in inactive conformation (27,30), thereby altering the path of the template DNA strand and blocking DNA melting near the transcription start site (Figure 1E). SW2 substitutions in E. coli RNAP designed to mimic the antibiotic-stabilized state conferred similar effects on DNA melting, but did not block transcription irreversibly (27), suggesting that SW2 may alternate between different conformational states, acting as a gate that specifically controls the downstream propagation of the transcription bubble. However, the role of SW2 at subsequent steps of transcription by bacterial RNAP was not investigated further.
In this study, we set out to probe the role of the SW2/DNA contacts throughout transcription. SW2 contains several highly conserved positively charged residues that contact the template DNA strand (K334, R337, R339, K345, R346 and R352; Figure 1F). Among these, R339 is particularly interesting because (i) it is absolutely conserved among bacterial, archaeal and eukaryotic lineages (Figure 1F; 31); (ii) contacts DNA phosphates near the active center (Figure 1B and C); (iii) is differently positioned relative to the DNA phosphates in different elongation complex structures: the closest DNA phosphates are at −1 in bacterial elongation complexes (4,5), +2 in most yeast elongation complexes (2,23,24), and +1 in the yeast elongation complex inhibited by α-amanitin (25); and (iv) substitutions of the corresponding residue in archaeal RNAP from Pyrococcus furiosis (R313A in the largest subunit) resulted in gross defects in transcription initiation and elongation (29). We created three variants of the E. coli RNAP with substitutions of R339 to A (lacking charge and the side chain), K (preserving positive charge) and E (reversing charge). We report that these substitutions perturb different steps of the transcription cycle, including promoter binding and opening, initiation of RNA synthesis, promoter escape, and RNA elongation.
MATERIALS AND METHODS
RNAPs and promoters
Wild-type E. coli core RNAP bearing a hexahistidine tag at the C-terminus of the β′ subunit was purified from E. coli BL21(DE3) cells overproducing all four core RNAP subunits from the plasmid pVS10 as described previously (32). Plasmids with mutant rpoC fragments encoding the β′ R339A, R339E and R339K substitutions (pIA827, pIA828 and pIA829, respectively) were obtained by PCR mutagenesis of pIA458 (33). The SbfI-BsmI fragments containing the mutant alleles were sequenced and recloned into the same sites of pVS10, resulting in pIA830, pIA831 and pIA832 plasmids, respectively. Mutant RNAPs were overexpressed and purified similarly to the wild-type enzyme. Wild-type σ70 and σ70 with the 513–519 deletion in region 3.2 were obtained as described in ref. (12).
Promoter-containing DNA fragments for in vitro transcription assays were obtained as follows. The T7A1 promoter (positions from −85 to +53 relative to the starting point of transcription) used in the run-off transcription, promoter complex stability, permanganate footprinting and promoter escape assays was obtained by PCR from synthetic oligonucleotide template. The T7A1cons promoter (see Supplementary Figure S1; 12), containing a consensus −10 element with two substitutions in comparison with wild-type T7A1 (G-13T and C-9A) was obtained in a similar way. The partially melted T7A1 promoter (Supplementary Figure S1) was prepared by annealing of two 66-mer synthetic oligonucleotides (Syntol, Moscow). The T7A1 promoter-containing template (positions −100 to 1225) used for the elongation rate measurements was obtained by PCR from the pIA146 plasmid encoding the E. coli rpoB gene (22). The lacUV5 promoter fragment (positions −69 to +23) was obtained by PCR from pFW11 (a gift from Bryce Nickels). The λPR promoter (positions −81 to +387) was obtained by PCR from the pIA226 plasmid (27).
In vitro transcription
Most transcription assays were performed in transcription buffer containing 40 mM Tris–HCl, pH 7.9, 40 mM KCl and 10 mM MgCl2, unless otherwise indicated. Holoenzyme RNAPs were prepared by incubating the corresponding core RNAP (100 nM final concentration) and either the wild type or the Δ513–519 σ70 subunit (500 nM) in the transcription buffer for 5 min at 37°C. The DNA template was added (10–30 nM) and the samples were incubated for another 5 min at 37°C. In the run-off transcription assay, all four nucleotide substrates were added (100 µM of ATP, CTP, GTP and 10 µM of UTP with addition of [α32P]-UTP, 3000 Ci/mmol, NEN), either in the absence or in the presence of the CpA primer (100 µM). The transcription reactions were proceeded for the desired time intervals and stopped by addition of an equal volume of buffer containing 8 M urea and 20 mM EDTA. RNA products were separated by denaturing PAGE and analyzed by using PhosphorImager (GE Healthcare).
Apparent KMs for the initiating substrates were measured on the wild type or synthetic bubble T7A1 promoter in reactions containing either the ATP and UTP nucleotides or the CpA primer and UTP. One of the two substrates was taken at 1 mM (with the addition of the corresponding [α32P]-labeled nucleotide), and the concentration of the other was varied from 1 µM to 6 mM. The reaction was proceeded for 1 minute at 37°C, RNA products were analyzed by either 30% or 23% PAGE. The data were fit to the hyperbolic equation R = Rmax[NTP]/(KM+[NTP]), where R is the observed amount of the RNA product and Rmax is the amount of RNA product synthesized at saturating substrate concentration, using GraFit software (Erithacus Software).
The promoter complex stability measurements were performed in buffer containing 20 mM Tris-acetate, pH 8.0, 20 mM Na-acetate, 10 mM Mg-acetate, 5% glycerol, 14 mM 2-mercaptoethanol, 0.1 mM EDTA. RNAP–promoter complexes were obtained as described above and heparin was added to 10 µg/ml. Following incubation of the samples for different time intervals at 37°C, the transcription reactions were initiated by the addition of the dinucleotide primer ApU (200 µM) and CTP (for the T7A1 promoter) or GTP (for λPR promoter) nucleotides (20 µM). The reactions were stopped after 3 min and trinucleotide RNA products were analyzed by 23% denaturing PAGE.
For elongation complex stability measurements, elongation complexes stalled at position +26 of the λPR promoter were obtained by incubating promoter complexes with a limited set of substrates containing 5 µM ATP, 5 µM GTP, 100 µM ApU and 1 µM UTP with addition of α-[32P]-UTP for 5 min at 37°C. Five microliters of Ni-NTA-agarose (Qiagen) was added and the samples were incubated for 5 min at 20°C with shaking. The sorbent was washed three times with the transcription buffer to remove unbound reactants and the bound elongation complexes were incubated in transcription buffer containing 1 M NaCl at 37°C for different time intervals. The dissociated RNAs were removed by washing with the low-salt transcription buffer and the bound transcripts were analyzed by 15% PAGE.
KMnO4 footprinting
For the KMnO4 footprinting experiments, the T7A1 and lacUV5 promoters were labeled at the 5′-end of the nontemplate strand by using 5′-[32P]-labeled primers during the PCR. In the case of the T7A1 promoter, holoenzyme RNAPs (100 nM core plus 500 nM σ70) were incubated with the labeled promoter fragment (10 nM) in transcription buffer containing 40 mM Tris–HCl, pH 7.9, 40 mM KCl and 10 mM MgCl2 for 15 min at 37°C. KMnO4 was added to 2 mM and the reaction was stopped after 15 s by addition of solution containing 1 M β-mercaptoethanol and 1 M sodium acetate (pH 4.8). DNA was processed as described (12) and analyzed on 10% denaturing polyacrylamide gel. In the case of lacUV5 promoter, transcription complexes were assembled in 20 μl of buffer containing 40 mM HEPES, pH7.5, 100 mM NaCl, 10 mM MgCl2, 2.5% glycerol and 0.025% DMSO using holoenzyme RNAP at 100 nM and lacUV5 promoter fragments at 100 nM for 15 min at 37°C. KMnO4 was added to 15 mM and the reaction was stopped after 30 s. DNA was processed in a similar way and analyzed on 8% denaturing gel.
Cell viability assay
For analysis of the effects of the R339 substitutions on cell viability, the mutant R339A, R339E and R339K rpoC alleles were recloned into the pRL662 plasmid (AmpR) under the control of the Ptrc promoter, resulting in plasmids pNB1, pNB2 and pNB3, respectively. Escherichia coli strain RL602 bearing a temperature-sensitive chromosomal rpoC allele (34) was transformed with plasmid pRL662 encoding for wild-type β′ subunit, pNB1, pNB2 and pNB3 plasmids, or control plasmid pBR322, and grown in the presence of Amp at 32°C to OD values 0.4–0.6. IPTG was added to 0.5 mM to induce expression of plasmid-encoded rpoC genes, and after growth for 45 min at 32°C the cells were put on IPTG containing LB plates. The plates were incubated overnight at 32°C or 42°C.
RESULTS
RNAPs with substitutions of R339 require a primer for efficient transcription
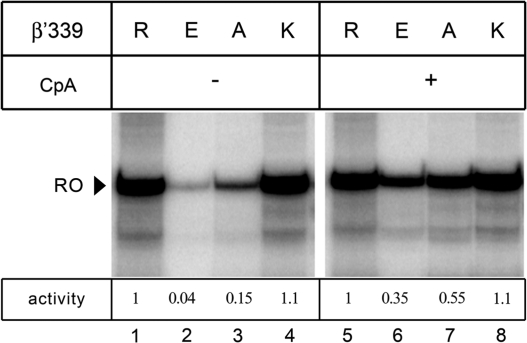
Activities of the mutationally altered E. coli RNAPs were tested in a multiple-round transcription assay on a template encoding the T7A1 promoter. In this assay, synthesis of a full-length ‘run-off’ RNA transcript (53 nt) requires sequential binding to the promoter, promoter escape, elongation and RNA release. In the presence of all four NTP substrates, the wild-type and the R339K RNAPs synthesized comparable amounts of the run-off transcript (Figure 2, lanes 1 and 4), whereas R339A and R339E variants displayed a significantly lower activity (lanes 2 and 3). However, in the presence of a dinucleotide primer corresponding to positions −1 and +1 of the promoter (CpA, see Figure 3A), the activity of the R339A and R339E RNAPs was greatly stimulated (Figure 2, lanes 5–8), suggesting that their major defect may be associated with the initiation step of transcription, including the promoter complex formation and/or first phosphodiester bond synthesis. On the other hand, even in the presence of the CpA primer, the overall activity of the R339A and R339E RNAPs was still 2- to 3-fold lower than that of the wild-type RNAP, suggesting that these substitutions may also compromise the later steps of transcription, i.e. promoter escape and RNA elongation.
Figure 2.
Transcription activity of RNAPs with substitutions of β′R339. Transcription was performed either in the absence (lanes 1–4) or in the presence (lanes 5–8) of the CpA primer (100 µM) as described in ‘Materials and Methods’ section. The position of the 53 nt run-off RNA (RO) is indicated by an arrowhead. The ratio of activity of each mutant RNAP to that of the wild-type enzyme is shown below the gel.
Figure 3.
Stability of promoter complexes formed by RNAPs with substitutions of β′ R339. (A) Stability of the T7A1 promoter complexes. Promoter complexes were incubated in the presence of 10 µg/ml heparin for time intervals varying from 10 s to 4 min, and the remaining activity was measured in the reaction of trinucleotide synthesis. (B) Stability of the λPR promoter complexes. The incubation time was varied from 30 s to 120 min. Promoter complex half-lives are indicated below each gel panel.
Substitutions of R339 increase apparent KMs for initiating substrates
The dramatic stimulation of the R339A and R339E RNAPs activity by the dinucleotide primer (Figure 2) suggested that these substitutions may specifically affect the formation of the first phosphodiester bond during initiation. Previously, we demonstrated that a deletion in σ3.2 (Δ513–519), which had a similar effect on the first bond synthesis, significantly increased the apparent KM values for initiating nucleotides, in particular, for the 3′-initiating NTP (12). To test the possibility of the functional interplay, we compared effects of the β′ R339 substitutions and the σ70 Δ513–519 deletion on the initiating NTP binding.
We measured apparent KM values in the reaction of dinucleotide synthesis from two initiating NTPs on the T7A1 promoter (ATP+UTP→pppApU). We found that the mutations had only modest effect on the apparent KM value for the 5′-initiating ATP that was increased 1.5-4 fold for different RNAP variants (Table 1). The mutations had more pronounced effects on the apparent KMs for 3′-initiating UTP that were increased about 3-, 6- and 13-fold for R339K, R339A and R339E variants, respectively (Table 1). The strength of the effect correlated with the nature of the substituted residue, with the conserved R339K substitution having the smallest and the charge-reversing R339E substitution—the strongest effect. In accordance with the published data, the deletion Δ513–519 in σ3.2 dramatically increased apparent KM for 3′-UTP (∼50-fold) while having only a subtle effect on the 5′-ATP binding (Table 1). Combination of the σ70 Δ513–519 and R339E mutations in the same holoenzyme RNAP did not have an additive effect on the apparent KM values (∼80-fold increase in KM for 3′-UTP in comparison with ∼50-fold increase for the σ70 Δ513–519 RNAP, Table 1). Thus, the observed defects in the first phosphodiester bond formation by the mutant enzymes may be explained by their defects in binding of the 3′-initiating NTP in the presence of a single-nucleotide triphosphate primer (ATP).
Table 1.
Apparent KM values for the initiating substrates on the T7A1 promoter for wild-type and mutant RNAPs
| RNAP | Reaction |
|||
|---|---|---|---|---|
| ATP+UTP |
CpA+UTP |
|||
| KM ATP, µM | KM UTP, µM | KM CpA, µM | KM UTP, µM | |
| WT | 190 ± 4 1 | 9.2 ± 0.5 1 | 90 ± 24.8 1 | 4.8 ± 1.1 1 |
| R339K | 272 ± 28 1.4 | 25 ± 2.8 2.7 | 116 ± 41 1.3 | 3.2 ± 2.6 0.7 |
| R339A | 700 ± 100 3.7 | 54 ± 6 5.9 | 185 ± 68 2.1 | 1.2 ± 0.2 0.25 |
| R339E | 410 ± 10 2.2 | 121 ± 20 13.2 | 174 ± 45 1.9 | 2.2 ± 0.5 0.4 |
| σ70Δ513-519 | 700 ± 28 3.7 | 490 ± 30 54 | 118 ± 16 1.3 | 9.8 ± 0.5 2.0 |
| σ70Δ+R339E | 584 ± 11 3.1 | 762 ± 183 83.2 | 92 ± 6 1.0 | 2.4 ± 0.8 0.5 |
Average values and standard deviations from three independent experiments are shown. The numbers in bold indicate the ratios of the KM values for different RNAPs to the KM value for wild-type RNAP in the same reaction.
This interpretation predicts that the initiation defects will be alleviated at higher 3′-initiating UTP concentrations or in the presence of a dinucleotide primer. Indeed, we found that the efficiency of full-length RNA synthesis by the mutant RNAPs relative to the wild-type RNAP increases with increasing the UTP concentration (Supplementery Figure S2). Furthermore, when we repeated KM measurements in the reaction of trinucleotide synthesis (CpA+UTP→CpApU), the apparent KM values for the 3′-UTP nucleotide for all mutant RNAPs, including the most defective R339E and the σ70 Δ3.2 enzymes, were comparable or even lower than the value for the wild-type RNAP (Table 1). In particular, in the case of the R339A and R339E mutants, the UTP KMs were decreased about 45- and 55-fold, respectively. The mutant and wild-type RNAPs also had comparable apparent KM values for the CpA primer (Table 1). Together, these data indicate that the defects of the mutant RNAPs in transcription initiation may be largely attributed to their defects in the binding of the initiating NTPs and are suppressed after the first bond formation (or in the presence of short RNA primers).
Mutant RNAPs form unstable promoter complexes
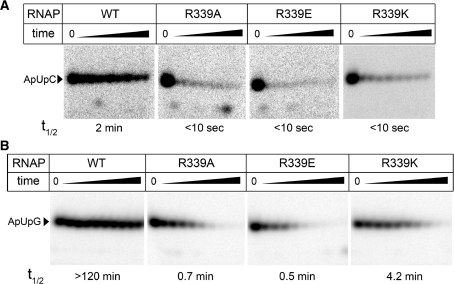
Mutations in the clamp domain and in SW2 have been shown to decrease the stability of transcription complexes (see ‘Introduction’ section). We therefore tested whether substitutions of R339 affect promoter complex stability. We measured the stability of T7A1 promoter complexes formed by RNAPs with R339 substitutions in the presence of heparin, a competitive inhibitor that would trap any free RNAP that has dissociated from the promoter DNA. For the wild-type E. coli RNAP, about half of the complexes dissociated within four minutes following the addition of heparin (Figure 3A). Stability of complexes formed by all three mutant RNAPs was dramatically reduced, with almost complete dissociation within just 10 s (Figure 3A).
To quantitatively evaluate the effects of different substitutions in SW2 on promoter complex stability, we utilized the λPR promoter that mediates formation of very stable open complexes. Complexes formed by the wild-type E. coli RNAP were stable for >2 h (half-life >120 min; Figure 3B), whereas those formed by mutant RNAPs were very sensitive to heparin challenge, with half-lives varying from less than a minute for R339A and R339E (0.7 ± 0.2 and 0.5 ± 0.1 min, respectively) to about four minutes for R339K RNAP (4.2 ± 1.1 min). Thus, while the removal of the positive charge led to the strongest defect, even a conservative lysine substitution resulted in a >30-fold decrease in the promoter complex stability.
Substitutions R339 inhibit downstream propagation of the transcription bubble
The β′ SW2 has been recently implicated in regulation of the initiation complex formation by the E. coli RNAP by a transcription factor DksA and antibiotics (26,27,30). Both DksA and antibiotic myxopyronin were shown to affect RNAP interactions with downstream promoter DNA, and myxopyronin was shown to stabilize a partially melted, inactive intermediate complex and to prevent its isomerization into a fully melted, transcriptionally competent open promoter complex. A deletion of the β’ residues 338–341 designed to block the SW2 refolding apparently ‘froze’ an intermediate state in which the DNA strand separation had commenced but stopped upstream from the start site (27). The R339 residue lies in the region that undergoes refolding, and its contacts to the DNA (Figure 1) may be particularly important for the hypothesized transition. We thus wanted to test if R339 alone affects the propagation of the transcription bubble.
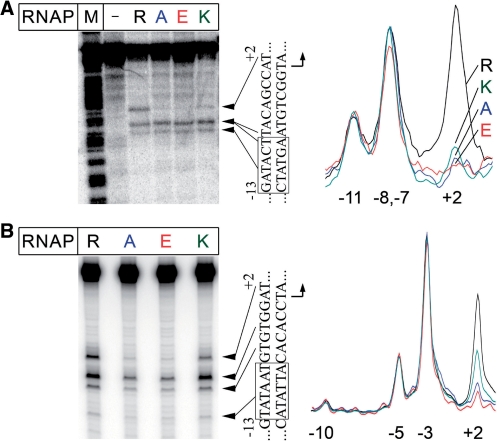
To monitor a melting state of promoter DNA in complexes formed by R339 RNAP variants at two model promoters, we performed KMnO4 footprinting. KMnO4 modifies single-stranded or unstacked T residues, and is thus commonly used to probe the extent of DNA melting. In the T7A1 promoter complex formed with the wild-type E. coli RNAP, efficient modification of thymine residues at positions −11, −8/−7 and +2 of the non-template DNA strand was observed (Figure 4A). Similarly, in complexes formed by the wild-type RNAP at the lacUV5 promoter, positions −10, −5, −3 and +2 were modified (Figure 4B). These patterns are indicative of the formation of the open promoter complex, in which the DNA bubble encompasses the transcription start point. In contrast, while RNAPs with substitutions of R339 were able to initiate the DNA strand separation in the upstream part of the bubble, melting did not extend to the start site. In particular, on both promoters, thymine at position +2 was much less reactive with the R339K and R339A RNAPs, and undetectable with the R339E RNAP (Figure 4A and B). Thus, substitutions of R339 impair melting of the downstream end of the transcription bubble during initiation.
Figure 4.
Promoter melting by RNAPs with substitutions of β′ R339. (A) KMnO4 footprinting of the non-template strand in the T7A1 promoter complexes of the wild-type and mutant E. coli RNAPs. The experiment was performed at 37°C as described in ‘Materials and Methods’ section. The ‘M’-lane is an A+G cleavage marker. The sequence of the melted promoter region is shown on the right of the gel. The −10 element is boxed. Scanned profiles of modification of thymine residues in the melted region are shown on the right of the figure. The numbers indicate promoter positions relative to the starting point of transcription. (B) KMnO4 footprinting of the non-template strand of the λPR promoter complexes.
It should be noted that, despite their defects in DNA melting, all three RNAPs with substitutions of R339 displayed considerable activity in the presence of a short RNA primer (Figure 2). These results suggest that partially melted complexes formed by the mutant enzymes can isomerize into the open complex to allow substrate binding. Other SW2 variants were also ‘rescued’ by the primer (27). To visualize downstream promoter melting in the initiation complexes formed by the mutant R339A RNAP variant, we performed permanganate footprinting in the presence of two initiating substrates, the dinucleotide primer CpA and UTP. At these conditions, we observed efficient permanganate modification of the +2 thymine in promoter complexes formed by both the wild-type and the R339A enzymes (Supplementary Figure S3). Thus, the initiating substrates stabilize melting of the downstream end of the transcription bubble by the mutant RNAPs.
To further test whether defects of the mutant RNAPs in transcription initiation can be connected to their inability to melt downstream DNA in promoter complexes, we analyzed transcription initiation on a T7A1 template containing an artificial bubble encompassing promoter positions −12 to +3 (Supplementary Figure S1). We observed that the differences between the wild-type and the mutant RNAPs in transcription initiation were partially compensated on this template (Supplementary Table S1 and Figure S4). In particular, the mutant RNAPs were able to initiate transcription on the bubble template even in the absence of a primer (Supplementary Figure S4). However, we found that the presence of the artificial bubble in the promoter region itself leads to defects in initiating nucleotide binding and promoter escape even in the case of the wild-type RNAP. Thus, transcription initiation on bubble templates may be significantly different from initiation on fully double-stranded promoters, limiting the use of this model in the analyses of transcription initiation (see Supplementary Data for discussion).
Mutant RNAPs are defective in promoter escape
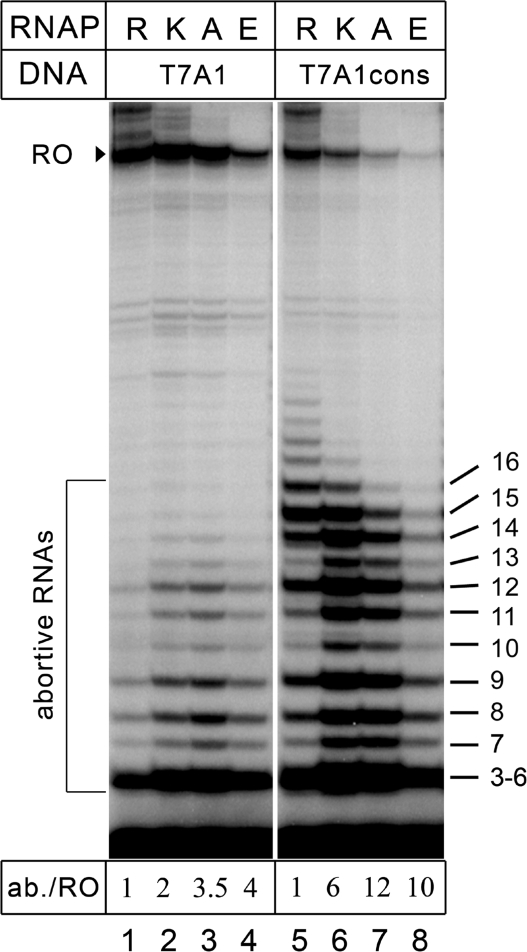
Changes in stability of promoter complexes may significantly affect the efficiency of promoter escape by RNAP (9,35). We thus tested the influence of the R339 substitutions on promoter escape from two T7A1 promoter variants, the wild-type and the T7A1cons promoter (12) in which the two positions in the −10 element (GATACT) have been changed to create the consensus TATAAT sequence (Supplementary Figure S1). The T7A1cons promoter displays a much higher efficiency of abortive synthesis and a correspondingly lower efficiency of productive synthesis, likely due to stronger σ interactions with the −10 region (12); on this promoter, the wild-type RNAP synthesizes large amounts of abortive RNAs up to 16 nt in length (Figure 5).
Figure 5.
Analysis of promoter escape by RNAPs with substitutions of β′ R339. Activities of the wild-type and mutant RNAPs were measured on the T7A1 and T7A1cons promoters in the presence of the CpA primer. Positions of the run-off and abortive RNAs are indicated on the left, the lengths of abortive RNAs are shown on the right. For each RNAP, the ratio of total radioactivity in abortive and run-off RNA products (ab./RO) is shown relative to the wild-type RNAP below the gel.
Transcription was performed in the presence of CpA primer that allowed initiation by all mutant RNAPs. On the wild-type promoter, the mutant RNAPs synthesized 2- to 4-fold higher amounts of short abortive RNAs (relative to the full-length RNA product) than the wild-type enzyme. In agreement with the previous data, the amounts of the full-length transcripts were comparable for all RNAPs except the R339E mutant (Figure 5, lanes 1–4) . On the T7A1cons promoter, the fraction of abortive RNAs was dramatically increased for all three mutant RNAPs (up to 10- to 12-fold for the R339A and R339E mutants; Figure 5, lanes 6–8), with a concomitant decrease in the full-length transcript, indicative of a serious defect in promoter escape.
Substitutions in the SW2 dramatically destabilize elongation complexes
The observed effects of the R339 substitutions on transcription initiation suggested that these substitutions may also affect RNA elongation by RNAP, through changes in the RNAP–DNA interactions network. We therefore tested how R339 substitutions would affect the elongation properties of RNAP.
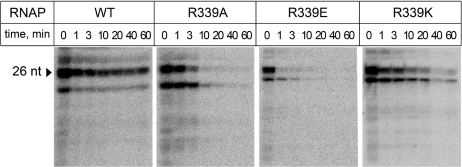
We measured average elongation rates of the mutant RNAPs on a pIA146 template that encodes the E. coli rpoB gene devoid of strong pause and terminator signals (22) resulting in a 1225-nt long run-off RNA transcript. We found that all RNAPs reached the end of the template in the time course of the experiment, but the rates of RNA synthesis by the mutant RNAPs were somewhat slower than that of the wild-type enzyme (Supplementary Figure S1). This effect was especially pronounced in the case of the R339E RNAP, which synthesized half of the RNA product (as calculated at 30 min) in >15 min, as compared with <10 min in the case of wild-type RNAP (Supplementary Figure S1B). We then tested whether the mutations affect the elongation complex stability; these assays were carried out at high ionic strength because elongation complexes are exceedingly stable at low ionic strength (36). We obtained the elongation complexes stalled at +26 position downstream from the λPR promoter by incubating promoter complexes with a subset of NTP substrates, lacking CTP (the first C residue is encoded by the 27th template position). The complexes were immobilized on a Ni-NTA affinity resin via a His6-tag present at the C-terminus of the β′-subunit and incubated in a buffer containing 1M NaCl. The RNA transcripts released from the enzyme were removed at the desired time points by washing the sorbent with the transcription buffer, and the RNAs that remained bound to the sorbent were analyzed by gel electrophoresis. The complexes formed by the wild-type RNAP were stable for >1 h at these conditions. In contrast, the mutant complexes completely dissociated within this time (Figure 6). As in the case of promoter complexes (see above), the rate of dissociation depended on the nature of the substitution: elongation complexes formed by R339K, R339A and R339E RNAPs dissociated within ∼30 min, 10 min and 1 min, respectively. Thus, substitutions of R339 have dramatic effects on the elongation complex stability.
Figure 6.
Stability of elongation complexes formed by RNAPs with substitutions of β′ R339. Halted A26 elongation complexes were formed at the λPR promoter and incubated in the presence of 1M NaCl. The RNA transcripts that remained bound to RNAP after different time intervals were analyzed by gel electrophoresis. The 26-mer RNA transcript is shown by an arrowhead. Mutant RNAPs also synthesized considerable amounts of a shorter RNA transcript, likely as a result of defects in promoter escape and/or RNA elongation.
Substitutions in the SW2 affect cell viability
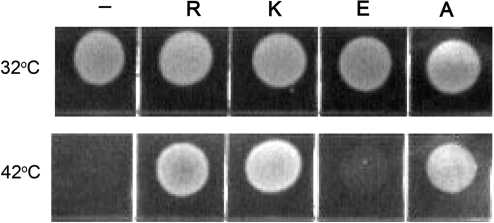
When analyzed in vitro, the three R339 substitutions had distinct effects on various steps of the transcription cycle, the strengths of the effects being dependent on the nature of the substituted residue. To test whether these substitutions would also affect RNAP function in vivo, we analyzed the ability of the mutant rpoC alleles to suppress temperature-dependent growth defects of the E. coli RL602 cells, bearing a temperature-sensitive rpoC allele (34). This strain is viable at 32°C but is unable to grow at 42°C (Figure 7). Transformation of the cells with plasmid encoding for the wild-type β′-subunit suppressed the growth defect at 42°C, while control pBR322 plasmid did not. The R339 substitutions differed in their effects on cell growth. The cells expressing the β′-subunits with the R339K and R339A substitutions were viable at 42°C, while the R339E allele did not support cell growth (Figure 7). Thus, substitution of the positively charged R339 in SW2 with the negatively charged glutamic acid residue inactivates RNAP function in vivo. At the same time, the in vitro defects of the R339K and R339A RNAPs are apparently compensated at the in vivo conditions, which likely favor stable open complex formation and efficient transcription initiation by the mutant RNAPs.
Figure 7.
The effect of the R339 substitutions on cell viability. The growth of the RL602 cells containing a temperature sensitive rpoC allele was assayed at either permissive (32°C, upper row) or restrictive (42°C, lower row) temperature as described in ‘Materials and Methods’ section. The cells expressed either wild-type (R) or mutant β′ subunits (K, E and A) from corresponding plasmids, or contained a control pBR322 plasmid (‘−’).
DISCUSSION
SW2 is a dynamic structural element that connects the clamp domain to the main part of the RNAP molecule and directly contacts the template DNA strand at the active center of the enzyme. In this work, we show that substitutions of a highly conserved SW2 residue R339 confer defects at different stages of transcription, including promoter opening, initiating substrate binding, promoter escape and RNA chain elongation. Comparison of bacterial and yeast elongation complex structures reveals that R339 makes direct contacts to phosphates in the −1 to +2 region of the template strand, but these contacts differ from one complex to another (2,23–25). Together with the relatively mild effect of the lysine substitution (as compared with other substitutions, see below), these results suggest that electrostatic interactions between R339 and DNA may play the key role in transcription and may change at different stages of transcription and during each nucleotide addition cycle. Below, we discuss possible functions of SW2 and, in particular, the role of R339/DNA interactions at different steps of RNA synthesis.
Role of SW2 in the promoter complex formation
Substitutions at position R339 in the E. coli RNAP prevented formation of the fully melted promoter complex. While the DNA strands became separated at the upstream edge of the transcription bubble, the DNA melting stopped upstream from the +1 position (Figure 4). Together with high sensitivity of these promoter complexes to heparin challenge, this phenotype is characteristic of an intermediate along the open complex formation pathway (37,38) and resembles the pattern reported for myxopyronin, the antibiotic that binds to SW2 and inhibits bubble propagation by stabilizing SW2 conformation in which R339 is unable to interact with DNA and is located in a loop that blocks the normal path of the template DNA in the active center (Figure 1E). This alternative SW2 state captured in the T. thermophilus holoenzyme-myxopyronin structure was hypothesized to correspond to a naturally occurring intermediate during the open complex formation (27) and is probably stabilized by the R339 substitutions, similarly to previously studied Δ338–341 and F338A mutations (27). However, in contrast to myxopyronin, changes in SW2 allow initiation in the presence of substrates, implying that a trapped intermediate can isomerize into the fully melted open complex.
Thus, the SW2 can adopt different conformations, and its contacts with the template DNA may stabilize the active conformation, which promotes DNA melting around the start site. Our observations that the R339 substitutions for alanine and glutamate produced stronger effects than that for lysine support the key role of electrostatic interactions of a positively charged R339 with the template DNA phosphate(s) in DNA melting and promoter complex stabilization. The key support for this proposal comes from the in vivo experiment that demonstrated that only the charge-reversing R339E substitution completely disrupted cellular RNAP function. Several other studied SW2 variants also mediated the formation of altered promoter complexes in vitro (26,27); these variants include changes in SW2 that alter its conformation (F338A, G333D), its contacts with DNA (R337S) or both (a short deletion including R339, Δ338–341). Thus, both the overall conformation of SW2 and its contacts to the template DNA are likely essential for the formation of transcriptionally competent promoter complexes; furthermore, changes in one would affect the other, in turn likely resulting in changes in the position or mobility of the clamp domain, which encloses the nucleic acids inside the main RNAP channel during initiation. Indeed, insertions and deletions in two regions of the clamp domain, the clamphead (18,19) and the lid (17), that interact with the downstream DNA duplex and the upstream part of the transcription bubble, respectively (Figure 1C), were also shown to destabilize the open complexes.
Role of the SW2 in binding of initiating nucleotides
Substitutions of R339 resulted in a notable increase in apparent KMs for initiating substrates, especially for the 3′-initiating NTP. This increase is most likely explained by changes in the position and/or conformation of the template DNA base at the i+1 site of the RNAP active center, resulting in defects in substrate binding. Indeed, the R339 substitutions also impaired downstream DNA melting, and their defects in transcription initiation were partially compensated on an artificially melted promoter template. The mutations may also affect the translocation state of RNAP in the promoter complex and change the register of the DNA template in the active center, which would also manifest in the increase in the apparent KM values. The smaller effect of the mutations on KM for the 5′-initiating NTP is consistent with the hypothesis that the binding site for the 5′-priming substrate is partially preformed in the core enzyme, and that the DNA template is not absolutely required for its formation (39).
Remarkably, the defects of the mutant RNAPs in the downstream DNA melting, initiating substrate binding and the first bond formation were suppressed in the presence of a dinucleotide primer. Thus, the presence of the nascent RNA in the active center likely stabilizes the template strand in the active conformation even in the absence of its contacts with R339. Interestingly, the mutant RNAPs had even lower apparent KM values (2- to 4-fold) for the 3′-UTP nucleotide in the presence of the primer, which may reflect better substrate binding after the first bond formation (Table 1). However, the functional significance of the improved UTP binding remains unknown and this effect is likely unrelated to the defects of the mutant RNAPs during the first bond formation and transcription initiation.
The observed effects of R339 substitutions on binding of initiating substrates is similar to that of the σ3.2 deletion (Δ513–519), which also increased KM for the 3′-initiating NTP in the absence, but not in the presence of a primer (Table 1; 12). Since σ3.2 interacts with SW2 near R339 (Figure 1D), one can speculate that this effect is connected to changes in the SW2 conformation and in the template strand positioning. The functional interplay between SW2 and σ3.2 is further supported by the fact that a combination of the σΔ513–519 with R339 substitutions did not lead to additional defects in substrate binding (Table 1). Furthermore, changes in the SW2 structure may be coupled with those at the tip of the σ3.2 loop, as revealed by comparison of the myxopyronin-bound and apo-holoenzyme RNAP structures (compare Figure 1D and E). Thus, σ3.2 may cooperate with SW2 to stimulate the initiating nucleotide binding, by stabilizing the proper conformation of the template DNA strand during initiation. However, in contrast to the SW2 substitutions, the σ3.2 deletion does not affect downstream DNA melting and promoter complex stability (12) suggesting that the functions of SW2 in stabilization of the open promoter complex may be relatively independent of its role in substrate binding. In further support of this notion, the R339K substitution that significantly destabilized promoter complexes had only a minor effect on initiating substrate binding (Table 1).
Strikingly, in initiating complexes of Saccharomyces cerevisiae RNAP II, a B-reader region of the general transcription factor TFIIB, consisting of a short B-reader helix and a B-reader loop, occupies a position similar to that of σ3.2 in bacterial RNAP, contacts SW2 and approaches the active center of RNAP (Supplementary Figure S6; 40–42). Substitutions at several positions in the B-reader helix and loop were shown to alter the transcription start site selection by the S. cerevisiae and human RNAPII (41,43 and references therein), whereas substitutions and deletions at the tip of the B-reader loop impaired the first bond formation by human RNAPII (44). Remarkably, substitutions in SW2 in S. cerevisiae RNAPII (Supplementary Figure S6) were shown to affect the start site selection by RNAP (28), a phenotype similar to the TFIIB-reader mutations, suggesting that TFIIB and SW2 may cooperate during transcription initiation.
Furthermore, mutations in the B-reader loop in the archael homologue of TFIIB, factor TFB, were shown to impair downstream propagation of the transcription bubble during promoter melting by Pyrococcus furiosis RNAP (Supplementary Figure S6; 41). This effect is similar to the effects of mutations in SW2 in bacterial RNAP on promoter melting described by us and others (26,27). One may therefore speculate that TFIIB stimulates downstream promoter melting and/or DNA template strand positioning at the RNAP active center through its contacts with SW2. Analysis of a possible functional interplay between SW2 and TFIIB would be an interesting goal of future studies.
Quite intriguingly, human TFIIB was shown to be phosphorylated at a Ser65 residue (corresponding to Ser77 in S. cerevisiae) in the B-reader loop, which likely contacts SW2, during transcription initiation (45). The phosphorylation of TFIIB was shown to be required for recruitment of RNA cleavage and polyadenylation factors to the promoter and for efficient transcription initiation by RNAPII. Since Ser65 is buried in the S. cerevisiae TFIIB-RNAP complex structure (Supplementary Figure S6), this suggests that the B-reader undergoes structural rearrangements during initiation that may be coupled with conformational changes of SW2.
Role of the SW2 in promoter escape
R339 substitutions increased the fraction of short abortive RNAs, especially on the consensus promoter that is characterized by strong RNAP–promoter interactions mediated by the σ70 subunit (12). Previous studies indicated that the lesser stability of promoter complex (determined by the strength of specific RNAP–promoter interactions) usually correlates with the higher efficiency of the initiation-to-elongation transition (9,35). In contrast to these observations, destabilization of the promoter complexes by the R339 substitutions was paralleled by decrease in the efficiency of promoter escape by the mutant RNAPs. Thus, efficient disruption of specific RNAP-promoter contacts likely depends on stable interactions of SW2 with template DNA. We propose that impairment of RNAP interactions with the DNA by the SW2 substitutions weakens the RNAP hold on the downstream DNA segment during ‘scrunching’ of the transcription bubble that is thought to be a prerequisite for promoter escape (46,47). This may result in a premature relaxation of the scrunched complex and the concurrent release of short abortive RNAs. SW2 may therefore be a key structural element involved in the regulation of scrunching and promoter escape by RNAP. Further experiments are required to establish whether scrunching is indeed impaired in the mutant RNAPs.
The effects of the R339 substitutions on abortive initiation contrast those of the σ3.2 deletion, whose defects in promoter escape were reflected in the preferential accumulation of longer abortive transcripts (12). The σ3.2 loop lies in the path of the nascent RNA (Figure 1C and D), whose extension has been proposed to trigger a steric clash with, and the subsequent displacement of σ3.2 from the main RNAP channel (3,13). Thus, promoter clearance defects of the Δ513–519 enzyme likely result from the loss of RNA-σ3.2 clash rather than from the altered SW2-σ3.2 contacts.
Role of R339 in transcript elongation
Previous work suggested that closing of the β′-clamp domain around the nucleic acids inside the main RNAP channel is a crucial factor that determines the high stability of elongation complexes (see ‘Introduction’ section). In this work, we demonstrated that SW2 and, in particular, positively charged R339 residue from this region, also critically contributes to the elongation complex stability. While substitutions of R339 had only moderate defects on elongation at low ionic strength conditions, they dramatically decreased the stability of elongation complexes at high salt. Since the strength of the effect was dependent on the charge of the substituted residue, we hypothesize that the R339 substitutions disrupt local SW2–DNA contacts and disfavor the β′-clamp closure around the DNA/RNA framework, thereby destabilizing the elongation complex. This interpretation is consistent with the reports that the high stability of elongation complexes is mediated by the clamp interactions with the downstream DNA duplex and the DNA/RNA hybrid (16,17,19,36,48–50). Our results thus suggest that contacts of the SW2 and, in particular, the R339 residue with the DNA may favor the closed clamp conformation during elongation.
RNAP from archaeon Pyrococcus furiosis with an alanine substitution of R313, which corresponds to R339 in the E. coli RNAP, was essentially inactive in RNA elongation (29). It was therefore suggested that SW2 is required for the separation of the DNA strands in front of the active site. However, since we found that E. coli RNAP with the same substitution can readily elongate RNA on double-stranded DNA templates at moderate ionic strength, SW2 is unlikely to be involved in the downstream DNA separation. We propose that, because the experiments with P. furiosis RNAP were performed on synthetic RNA/DNA scaffolds and at elevated salt concentrations (≥250 mM), the observed elongation defects could be explained by the inability of the mutant enzyme to stably bind to the scaffolds.
Perspective
Our results demonstrate that β′ SW2 plays a key role during transcription initiation and elongation and inactivation of its function by mutations disrupts RNAP activity both in vitro and in vivo. SW2 is a dynamic module whose conformational transitions can be controlled in response to regulatory signals of various types. During initiation, SW2 may serve as a gate controlling transcription start opening and nucleotide binding. The DNA melting activity of SW2 can be controlled by a number of protein factors such as DksA, which binds within the RNAP secondary channel and transmits an allosteric signal to SW2, preventing DNA melting (26). The σ3.2 region that directly interacts with SW2 may modulate the SW2–DNA contacts, thereby affecting the initiating nucleotide binding. In eukaryotic RNAP, TFIIB may play an analogous role in the transcription start selection and RNA initiation through contacts with SW2 (41). Other initiation factors, including alternative σ subunits, various bacterial secondary channel binding proteins, and eukaryotic transcription factors may also control the conformational state of SW2 directly or allosterically. Some antibiotics that block transcription initiation by RNAP also act through SW2. Myxopyronin and several related compounds freeze SW2 in an inactive conformation (27,30). Lipiarmycin may also target SW2, since some mutations conferring resistance to lipiarmycin have been mapped to this region in Enterococcus faecalis, Mycobacterium tuberculosis and Bacillus subtilis (51). Finally, other, as yet uncharacterized factors may affect SW2 conformation and its contacts with DNA during elongation, thus affecting the elongation complex stability, pausing and transcription termination. Analysis of possible functions of SW2 in transcription regulation is an important goal of our future studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Russian Academy of Sciences Presidium Program in Molecular and Cellular Biology (to A.K.); National Institutes of Health (GM67153 to I.A., GM074840 to D.G. Vassylyev); Russian Foundation for Basic Research (10-04-00925); President of Russian Federation MK-4743.2009.4. Funding for open access charge: National Institutes of Health (GM67153 to I.A.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Mustaev for inspiration of this work, K. Brodolin for helpful discussions, V. Svetlov for help in protein purification and R. Landick for the RL602 strain.
REFERENCES
- 1.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 2.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 3.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 4.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 5.Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 6.Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 7.Schroeder LA, Gries TJ, Saecker RM, Record M.T., Jr, Harris ME, DeHaseth PL. Evidence for a tyrosine-adenine stacking interaction and for a short-lived open intermediate subsequent to initial binding of Escherichia coli RNA polymerase to promoter DNA. J. Mol. Biol. 2009;385:339–349. doi: 10.1016/j.jmb.2008.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schroeder LA, Choi AJ, DeHaseth PL. The -11A of promoter DNA and two conserved amino acids in the melting region of sigma70 both directly affect the rate limiting step in formation of the stable RNA polymerase-promoter complex, but they do not necessarily interact. Nucleic Acids Res. 2007;35:4141–4153. doi: 10.1093/nar/gkm431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu LM. Promoter clearance and escape in prokaryotes. Biochim. Biophys. Acta. 2002;1577:191–207. doi: 10.1016/s0167-4781(02)00452-9. [DOI] [PubMed] [Google Scholar]
- 10.Goldman SR, Ebright RH, Nickels BE. Direct detection of abortive RNA transcripts in vivo. Science. 2009;324:927–928. doi: 10.1126/science.1169237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Susa M, Sen R, Shimamoto N. Generality of the branched pathway in transcription initiation by Escherichia coli RNA polymerase. J. Biol. Chem. 2002;277:15407–15412. doi: 10.1074/jbc.M112481200. [DOI] [PubMed] [Google Scholar]
- 12.Kulbachinskiy A, Mustaev A. Region 3.2 of the sigma subunit contributes to the binding of the 3′-initiating nucleotide in the RNA polymerase active center and facilitates promoter clearance during initiation. J. Biol. Chem. 2006;281:18273–18276. doi: 10.1074/jbc.C600060200. [DOI] [PubMed] [Google Scholar]
- 13.Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 14.Nickels BE, Garrity SJ, Mekler V, Minakhin L, Severinov K, Ebright RH, Hochschild A. The interaction between sigma70 and the beta-flap of Escherichia coli RNA polymerase inhibits extension of nascent RNA during early elongation. Proc. Natl Acad. Sci. USA. 2005;102:4488–4493. doi: 10.1073/pnas.0409850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuznedelov K, Minakhin L, Niedziela-Majka A, Dove SL, Rogulja D, Nickels BE, Hochschild A, Heyduk T, Severinov K. A role for interaction of the RNA polymerase flap domain with the sigma subunit in promoter recognition. Science. 2002;295:855–857. doi: 10.1126/science.1066303. [DOI] [PubMed] [Google Scholar]
- 16.Naryshkina T, Kuznedelov K, Severinov K. The role of the largest RNA polymerase subunit lid element in preventing the formation of extended RNA-DNA hybrid. J. Mol. Biol. 2006;361:634–643. doi: 10.1016/j.jmb.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 17.Toulokhonov I, Landick R. The role of the lid element in transcription by E. coli RNA polymerase. J. Mol. Biol. 2006;361:644–658. doi: 10.1016/j.jmb.2006.06.071. [DOI] [PubMed] [Google Scholar]
- 18.Bartlett MS, Gaal T, Ross W, Gourse RL. RNA polymerase mutants that destabilize RNA polymerase-promoter complexes alter NTP-sensing by rrn P1 promoters. J. Mol. Biol. 1998;279:331–345. doi: 10.1006/jmbi.1998.1779. [DOI] [PubMed] [Google Scholar]
- 19.Kulbachinskiy AV, Ershova GV, Korzheva NV, Brodolin KL, Nikiforov VG. Mutations in β′ subunit of the Escherichia coli RNA polymerase influence interaction with the downstream DNA duplex in the elongation complex. Genetika. 2002;38:1207–1211. [PubMed] [Google Scholar]
- 20.Severinov K, Darst SA. A mutant RNA polymerase that forms unusual open promoter complexes. Proc. Natl Acad. Sci. USA. 1997;94:13481–13486. doi: 10.1073/pnas.94.25.13481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nechaev S, Chlenov M, Severinov K. Dissection of two hallmarks of the open promoter complex by mutation in an RNA polymerase core subunit. J. Biol. Chem. 2000;275:25516–25522. doi: 10.1074/jbc.M002511200. [DOI] [PubMed] [Google Scholar]
- 22.Ederth J, Artsimovitch I, Isaksson LA, Landick R. The downstream DNA jaw of bacterial RNA polymerase facilitates both transcriptional initiation and pausing. J. Biol. Chem. 2002;277:37456–37463. doi: 10.1074/jbc.M207038200. [DOI] [PubMed] [Google Scholar]
- 23.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 24.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brueckner F, Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat. Struct. Mol. Biol. 2008;15:811–818. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- 26.Rutherford ST, Villers CL, Lee JH, Ross W, Gourse RL. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev. 2009;23:236–248. doi: 10.1101/gad.1745409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belogurov GA, Vassylyeva MN, Sevostyanova A, Appleman JR, Xiang AX, Lira R, Webber SE, Klyuyev S, Nudler E, Artsimovitch I, et al. Transcription inactivation through local refolding of the RNA polymerase structure. Nature. 2009;457:332–335. doi: 10.1038/nature07510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majovski RC, Khaperskyy DA, Ghazy MA, Ponticelli AS. A functional role for the switch 2 region of yeast RNA polymerase II in transcription start site utilization and abortive initiation. J. Biol. Chem. 2005;280:34917–34923. doi: 10.1074/jbc.M502932200. [DOI] [PubMed] [Google Scholar]
- 29.Naji S, Bertero MG, Spitalny P, Cramer P, Thomm M. Structure-function analysis of the RNA polymerase cleft loops elucidates initial transcription, DNA unwinding and RNA displacement. Nucleic Acids Res. 2008;36:676–687. doi: 10.1093/nar/gkm1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukhopadhyay J, Das K, Ismail S, Koppstein D, Jang M, Hudson B, Sarafianos S, Tuske S, Patel J, Jansen R, et al. The RNA polymerase “switch region” is a target for inhibitors. Cell. 2008;135:295–307. doi: 10.1016/j.cell.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lane WJ, Darst SA. Molecular evolution of multisubunit RNA polymerases: sequence analysis. J. Mol. Biol. 2010;395:671–685. doi: 10.1016/j.jmb.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, Grishin NV, Vassylyev DG, Artsimovitch I. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol. Cell. 2007;26:117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svetlov V, Vassylyev DG, Artsimovitch I. Discrimination against deoxyribonucleotide substrates by bacterial RNA polymerase. J. Biol. Chem. 2004;279:38087–38090. doi: 10.1074/jbc.C400316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weilbaecher R, Hebron C, Feng G, Landick R. Termination-altering amino acid substitutions in the beta' subunit of Escherichia coli RNA polymerase identify regions involved in RNA chain elongation. Genes Dev. 1994;8:2913–2927. doi: 10.1101/gad.8.23.2913. [DOI] [PubMed] [Google Scholar]
- 35.Hsu LM. Monitoring abortive initiation. Methods. 2009;47:25–36. doi: 10.1016/j.ymeth.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sidorenkov I, Komissarova N, Kashlev M. Crucial role of the RNA:DNA hybrid in the processivity of transcription. Mol. Cell. 1998;2:55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- 37.Saecker RM, Tsodikov OV, McQuade KL, Schlax P.E., Jr, Capp MW, Record M.T., Jr Kinetic studies and structural models of the association of E. coli sigma(70) RNA polymerase with the lambdaP(R) promoter: large scale conformational changes in forming the kinetically significant intermediates. J. Mol. Biol. 2002;319:649–671. doi: 10.1016/S0022-2836(02)00293-0. [DOI] [PubMed] [Google Scholar]
- 38.Kontur WS, Saecker RM, Capp MW, Record M.T., Jr Late steps in the formation of E. coli RNA polymerase-lambda PR promoter open complexes: characterization of conformational changes by rapid [perturbant] upshift experiments. J. Mol. Biol. 2008;376:1034–1047. doi: 10.1016/j.jmb.2007.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naryshkina T, Mustaev A, Darst SA, Severinov K. The beta' subunit of Escherichia coli RNA polymerase is not required for interaction with initiating nucleotide but is necessary for interaction with rifampicin. J. Biol. Chem. 2001;276:13308–13313. doi: 10.1074/jbc.M011041200. [DOI] [PubMed] [Google Scholar]
- 40.Bushnell DA, Westover KD, Davis RE, Kornberg RD. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science. 2004;303:983–988. doi: 10.1126/science.1090838. [DOI] [PubMed] [Google Scholar]
- 41.Kostrewa D, Zeller ME, Armache KJ, Seizl M, Leike K, Thomm M, Cramer P. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462:323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 42.Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science. 2010;327:206–209. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng W, Roberts SG. TFIIB and the regulation of transcription by RNA polymerase II. Chromosoma. 2007;116:417–429. doi: 10.1007/s00412-007-0113-9. [DOI] [PubMed] [Google Scholar]
- 44.Tran K, Gralla JD. Control of the timing of promoter escape and RNA catalysis by the transcription factor IIb fingertip. J. Biol. Chem. 2008;283:15665–15671. doi: 10.1074/jbc.M801439200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Fairley JA, Roberts SG. Phosphorylation of TFIIB links transcription initiation and termination. Curr. Biol. 2010;20:548–553. doi: 10.1016/j.cub.2010.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kapanidis AN, Margeat E, Ho SO, Kortkhonjia E, Weiss S, Ebright RH. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314:1144–1147. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Revyakin A, Liu C, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nudler E, Avetissova E, Markovtsov V, Goldfarb A. Transcription processivity: protein-DNA interactions holding together the elongation complex. Science. 1996;273:211–217. doi: 10.1126/science.273.5272.211. [DOI] [PubMed] [Google Scholar]
- 49.Kireeva ML, Komissarova N, Waugh DS, Kashlev M. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J. Biol. Chem. 2000;275:6530–6536. doi: 10.1074/jbc.275.9.6530. [DOI] [PubMed] [Google Scholar]
- 50.Kuznedelov K, Korzheva N, Mustaev A, Severinov K. Structure-based analysis of RNA polymerase function: the largest subunit's rudder contributes critically to elongation complex stability and is not involved in the maintenance of RNA-DNA hybrid length. EMBO J. 2002;21:1369–1378. doi: 10.1093/emboj/21.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gualtieri M, Tupin A, Brodolin K, Leonetti JP. Frequency and characterisation of spontaneous lipiarmycin-resistant Enterococcus faecalis mutants selected in vitro. Int. J. Antimicrob. Agents. 2009;34:605–606. doi: 10.1016/j.ijantimicag.2009.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.