Abstract
We experimentally identified and characterized 97 novel, non-protein-coding RNA candidates (npcRNAs) from the human pathogen Salmonella enterica serovar Typhi (hereafter referred to as S. typhi). Three were specific to S. typhi, 22 were restricted to Salmonella species and 33 were differentially expressed during S. typhi growth. We also identified Salmonella Pathogenicity Island-derived npcRNAs that might be involved in regulatory mechanisms of virulence, antibiotic resistance and pathogenic specificity of S. typhi. An in-depth characterization of S. typhi StyR-3 npcRNA showed that it specifically interacts with RamR, the transcriptional repressor of the ramA gene, which is involved in the multidrug resistance (MDR) of Salmonella. StyR-3 interfered with RamR–DNA binding activity and thus potentially plays a role in regulating ramA gene expression, resulting in the MDR phenotype. Our study also revealed a large number of cis-encoded antisense npcRNA candidates, supporting previous observations of global sense–antisense regulatory networks in bacteria. Finally, at least six of the npcRNA candidates interacted with the S. typhi Hfq protein, supporting an important role of Hfq in npcRNA networks. This study points to novel functional npcRNA candidates potentially involved in various regulatory roles including the pathogenicity of S. typhi.
INTRODUCTION
Recent discoveries of a growing number of non-protein coding RNAs (npcRNAs) in bacteria have challenged the previous impression that proteins are the only relevant players in the control of bacterial gene expression. Bacterial small npcRNAs constitute a structurally diverse class of molecules that are typically 50–200 nt long and play a crucial role in many cellular networks, including responses to environmental stress, plasmid and viral replication, quorum sensing and bacterial virulence (1–9). The majority of known bacterial npcRNAs regulate gene expression by base pairing with mRNAs acting in trans- or cis-, and thereby either activating or repressing translation efficiency or the stability of the mRNA targets. Another subclass of cis-encoded riboregulators are located in the 5′-untranslated regions (UTR) of mRNA and are termed riboswitches (10), and facilitate feedback regulation at the transcription/translation level, following structure changes in the RNA in response to the binding of the metabolites.
Salmonella is categorized under the family of Enterobacteriaceae that includes the group of enteric bacteria. Based on DNA sequences, the Salmonella genus includes two species, S. enterica and S. bongori that are further subdivided into subspecies and serovars (>2500 serovars). Salmonella enterica serovar Typhi (hereafter referred to as S. typhi) is a Gram-negative, human-specific pathogen causing enteric typhoid fever, an acute life-threatening febrile illness that affects the reticuloendothelial system. According to the World Health Organization, 16–33 million cases and 500 000–600 000 deaths are reported around the world annually. S. typhi is characterized by its flagellar H-antigen, its lipopolysaccharidic O-antigen and its polysaccharide capsular virulence antigen found at the surface of freshly isolated strains.
The genome of S. typhi strain Ty2 was recently sequenced (11). It has a single 4.8-Mb chromosome with an average G+C content of ∼52% and encodes 4339 genes of which more than 200 are functionally inactive. Unlike the multidrug resistance (MDR) strain S. typhi CT18, S. typhi Ty2 has no plasmids and is sensitive to antibiotics. Comparisons of S. typhi isolates from around the globe indicated that they are highly related and can be traced to a single point of origin ∼30 000–50 000 years ago (12). Only ∼10% of the core genes are not shared between Escherichia coli and S. enterica, suggesting that they most likely branched from a common ancestor over 100 million years ago (12). A genome comparison with the S. typhi CT18 strain demonstrated a remarkable degree of conservation; one of the more interesting differences being an additional cluster of a few genes in Ty2 that may be a novel Salmonella Pathogenicity Island (SPI) (11). There are 10 SPIs (SPI-1–10) presently known in the S. typhi genome, all of which are thought to be involved in the virulence of the pathogen.
One of the key features of S. typhi is its restriction to human hosts, which has inhibited direct studies of its pathogenicity. Instead, much has been learned from comparative research with its close serovar, S. typhimurium in the mouse model. A number of npcRNAs involved in different regulatory pathways were identified in S. typhimurium, including SPI-1-derived InvR npcRNA. Surprisingly, InvR is not functionally linked to SPI-1 secretion or invasion pathways, but instead regulates the expression of OmpD porin (13). Padalon-Brauch et al. (14) identified and characterized 19 novel npcRNAs encoded by genes in the SPIs of S. typhimurium that possibly function in the pathogenicity of the bacterium. Using high-throughput pyrosequencing, Sittka et al. (15) detected 65 npcRNA candidates in S. typhimurium, many of which are conserved in S. typhi and related pathogenic enterobacterial species (16,17) and may function in complex with the Hfq protein. A number of characterized, trans-encoded RNAs in bacteria require the Sm-like RNA chaperone Hfq for regulating the translation of their mRNA targets. Hfq binds with high affinity to OxyS, DsrA, RprA, RyhB, Spot42, SgrS and other npcRNAs identified in E. coli and is frequently required for both their intracellular stability and interaction with target mRNAs (18–20), either to activate or predominantly to silence translation (21). Deletion of Hfq in S. typhimurium attenuates its ability to infect mice and especially to invade epithelial cells, indicating that Hfq and its ribonucleoprotein particle (RNP) complexes are essential for the virulence of the bacterium (5). Expression of Hfq is also essential for the virulence of Shigella flexneri (22).
Recently, the S. typhi transcriptome was analyzed using Illumina-platform high-throughput sequencing, whereby 40 putative npcRNAs were identified (23); surprisingly, only five of them were also found in common with those in S. typhimurium (15,23). Transcriptome analyses also revealed the presence of cis-encoded npcRNAs expressed in antisense orientation to open reading frames (ORFs) of bacterial genes (24–26). Although many of the reported examples negatively regulate expression of the complementary mRNAs, some of them (like GadY) increase the stability of the corresponding mRNAs (27).
To survey the population of npcRNAs in S. typhi, we generated a specialized cDNA library representing three different growth stages of the bacterium. We experimentally identified and characterized many novel npcRNA candidates. Their differential expression patterns were compared with potential homologs in E. coli. Many of the npcRNA candidates are restricted to Salmonella species or to S. typhi and some of the npcRNA candidates may be involved in the MDR of S. typhi.
MATERIALS AND METHODS
S. typhi culture conditions
S. typhi was cultured in Luria-Bertani medium (LB); 100 μl of a clinical isolate of S. typhi from glycerol stock was aseptically inoculated into 10 ml of LB medium and incubated overnight at 37°C with 220 rotations per minute. LB medium (250 ml) was then inoculated with a 2.5 ml aliquot of the overnight culture. Cells were harvested during the lag phase (OD600 0.3–0.4), log phase (OD600 0.6–0.7) and stationary phase (OD600 1.5). Total RNA was extracted by Trizol reagent (Gibco BRL) according to the manufacturer’s instructions.
cDNA library construction
To avoid unintended bias in our starting material, equal amounts of total RNA (50 μg) extracted from different growth stages of S. typhi were combined, size fractionated (20–500 nt) on 8% denaturing PAGE gels (7 M urea, 1 × TBE buffer) and passively eluted in 0.3 M NaOAc (pH 5.3) overnight at 4°C. Size-selected RNA was ethanol precipitated. All further steps of library construction were performed as described by Raabe et al. (28). The resulting 1700 cDNA clones were randomly sequenced and assembled into contigs. All cDNAs shorter then 18 nt were excluded from further analysis. The resulting 388 contigs were manually analyzed and mapped to the S. typhi Ty2 genome (AE014613) using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and BLAT (http://archaea.ucsc.edu/cgi-bin/hgBlat) genomic browsers.
Northern blot analysis
Total RNA (8–10 μg) was separated on 8% denaturing polyacrylamide gels and electro-transferred onto positively charged nylon membranes (Ambion Ltd). Northern blot analyses using specific oligonucleotides (Supplementary Table S1) complimentary to putative npcRNAs were performed as described by Khanam et al. (29).
Cloning and purification of recombinant RamR and Hfq proteins from S. typhi
DNA fragments representing the ORFs of ramR and hfq protein-coding genes from S. typhi were generated by PCR amplification of genomic DNA. The reaction was performed using a standard protocol (30), and oligonucleotides RamRndeIF and RamRxhoIR (for ramR), and HfqforvndeI and HfqrevxhoI (for hfq) (Supplementary Table S1). To facilitate cloning, NdeI and XhoI restriction sites were introduced into the oligonucleotide sequence. The pET28-b+ vector was used for cloning and expression of RamR and Hfq proteins. The expected clones were confirmed by sequence analysis. The recombinant RamR and Hfq proteins were purified from E. coli BL21 cells as instructed by the manufacturer (Qiagen). The proteins were dialyzed against buffer containing 20 mM HEPES–KOH (pH 7.4), 150 mM KCl, 1.5 mM MgCl2 and 5% glycerol.
RNA gel shift assay
For binding assays to RamR or Hfq protein, S. typhi novel npcRNA candidates were in vitro-transcribed from PCR templates containing a T7 promoter using 250 U of T7 RNA polymerase, and gel-purified npcRNAs were radioactively labeled by incorporating [α-32P] UTP during the in vitro transcription. Before incubating with recombinant RamR or Hfq protein, RNAs were dissolved in buffer containing 20 mM HEPES-KOH (pH 7.4), 80 mM NaCl, 10 mM KCl, 2.5 mM MgCl2, heat-denatured (2 min at 85°C) and subsequently placed in ice. RNP complexes of recombinant RamR or Hfq proteins with npcRNAs were formed in reaction volumes of 20 μl containing 20 mM HEPES–KOH (pH 7.4), 80 mM NaCl, 10 mM KCl, 2.5 mM MgCl2, 2 mM DTT, 10% glycerol, 1 μg BSA, 5 μg tRNA and 5 U of ribonuclease inhibitor (RNasin, Fermentas); 2 μg of sheared salmon sperm DNA was added for the RamR protein binding assay. Aliquots (0.05 pmol) of npcRNAs were incubated with increasing concentrations of RamR or Hfq for 30 min at room temperature. RNA and RNA–protein complexes were separated on native 8% (w/v) polyacrylamide gels containing 1 × TBE (90 mM Tris; 64.6 mM boric acid; 2.5 mM EDTA; pH 8.3). All further steps were performed as described by Rozhdestvensky et al. (31). The apparent K50 values of the RNA–protein complexes were defined as the protein concentrations for which 50% of the input RNA was shifted to an RNP complex.
DNA gel shift assay
For binding assays to RamR protein, two complementary 60-nt oligonucleotides harboring the promoter region of the S. typhi ramA gene were synthesized (MWG Biotech, Ebersberg, Germany) (Figure 3A; Supplementary Table S1). As controls, oligonucleotides harboring 9-nt and 2-nt deletions were also generated (Figure 3A, Supplementary Table S1; 32,33). Complementary single-stranded DNA fragments were end-labeled with [γ-32P]-ATP using T4 polynucleotide kinase and annealed. The resulting double-stranded DNA fragments were gel-purified from native 10% (w/v) polyacrylamide gels containing 1 × TBE and passively eluted in 0.3 M NaOAc (pH 7.0) and 1 mM EDTA buffer at 4°C overnight.
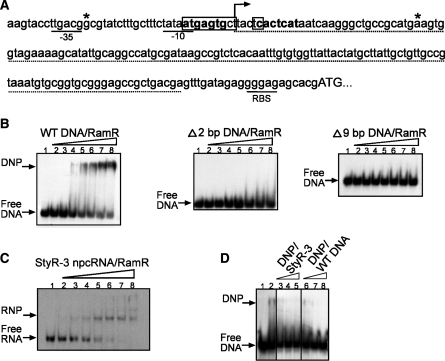
Figure 3.
Gel retardation analysis of the interaction of RamR protein with ramA gene regulatory region and StyR-3. (A) Sequence of ramA regulatory region. The −35, −10 and RBS sequences are underlined. Predicted RamR–DNA binding sites are indicated with bold letters. The 2-bp (Δ2 bp DNA) and 9-bp (Δ9 bp DNA) deletions within the ramA regulatory region are boxed. The transcriptional start site is indicated by an arrow. Asterisks indicate the 5′- and 3′-nt for the DNA fragment (WT DNA) used in the gel retardation assay. The StyR-3 sequence is indicated with a dotted line. (B) Gel retardation analysis of the interaction of RamR protein with the ramA gene regulatory region, showing wild-type DNA fragment (left), 2-bp (middle) and 9-bp (right) deletion fragments. (C) Interaction of RamR protein with StyR-3. (B and C) 32P-labeled DNA fragments or StyR-3 were incubated with S. typhi recombinant protein RamR as follows: lane 1, no protein added; (B and C) lanes 2–8: 50, 100, 150, 200, 400, 600 and 800 nM RamR protein was added, respectively; (D) Competition assay. 32P-labeled WT DNA was competed from RamR–DNP complex with increasing concentrations of unlabeled StyR-3 (lanes 3–5: 50, 250 and 500 nM, respectively) and WT DNA (lanes 6–8: 50, 250 and 500 nM, respectively). DNA: competitor molar ratios for lanes 3 and 6; 4 and 7; 5 and 8 were: 1:20; 1:100 and 1:200, respectively.
Complexes of recombinant RamR protein with DNA fragments (∼0.05 pmol per reaction) were formed in a volume of 20 μl containing 20 mM HEPES–KOH (pH 7.4), 80 mM NaCl, 10 mM KCl, 2.5 mM MgCl2, 2 mM DTT, 10% glycerol, 1 μg BSA, 5 μg tRNA, 2 μg of sheared salmon sperm DNA and 5 U of ribonuclease inhibitor (RNasin, Fermentas) (Figure 3B). The subsequent steps were performed as described above for RNA gel shift assays.
Gel shift competition assays
Competition assays were performed under the above binding conditions with 300 nM RamR protein and increasing concentrations of DNA fragments or S. typhi StyR-3 npcRNA as competitors (Figure 3D). All reactions were incubated for 30 min at room temperature. DNA and DNA-protein complexes were separated on native 10% (w/v) polyacrylamide gels containing 1 × TBE buffer. Electrophoresis and subsequent steps were performed as described above.
Oligonucleotides used in this work
All oligonucleotides used in this work are listed in Supplementary Table S1.
RESULTS AND DISCUSSION
cDNA library construction and analysis
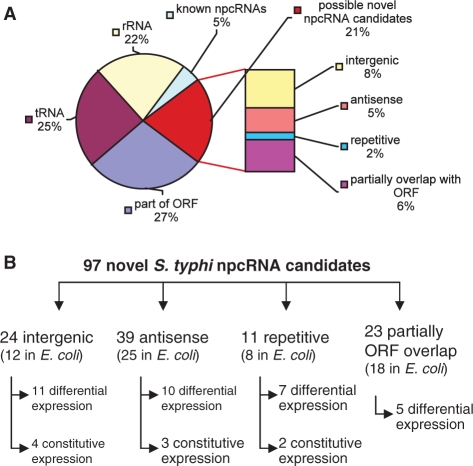
To survey the population of npcRNAs in S. typhi, we generated a specialized cDNA library representing the lag, log and stationary growth stages of the bacterium. A total of 1700 cDNA clones were randomly sequenced and assembled into contigs. All cDNAs shorter than 18 nt were excluded from further analysis. The resulting 388 contigs were manually analyzed and mapped to the S. typhi Ty2 genome (AE014613) using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and BLAT (http://archaea.ucsc.edu/cgi-bin/hgBlat) genomic browsers. To identify known npcRNAs in our data set, we screened cDNA sequences against known npcRNAs in Salmonella and E. coli. Nearly half of the cDNAs were derived from the highly abundant npcRNA species of rRNAs (22%) and tRNAs (25%) (Figure 1A). Roughly 5% of the cDNA sequences belonged to other known npcRNAs (Supplementary Table S3). cDNAs that mapped to the ORFs of known or hypothetical protein-coding genes constituted the largest subgroup (27%). Some of these cDNAs might represent potential npcRNA species that overlap with ORFs, similar to RNAIII, a small npcRNA in S. aureus (34). Moreover, the predicted ORFs of the hypothetical protein-coding genes matching some of the cDNAs may be wrongly annotated and may encode potential novel npcRNAs instead (Supplementary Table S2). Furthermore, some of those candidates overlap with putative 5′-UTR regions and may represent the products of transcriptional attenuation and correspond to potential candidates for novel riboswitches (Supplementary Table S2).
Figure 1.
Classification of cDNA sequences and identified novel npcRNA candidates. (A) Distribution of the cDNA sequences among various classes of RNA species; frequencies indicated as percentages. (B) Classification and expression profiles of novel npcRNA candidates in S. typhi and their E. coli homologs.
Interestingly, 21% of the cDNAs could not be assigned to any known RNAs, and thus represent candidates for novel npcRNAs in S. typhi (Figure 1A and B). Eight of these 97 npcRNA candidates overlap with those identified by Perkins et al. (23) via deep sequencing. As mentioned above, deep sequencing approaches in the closely related S. typhimurium and S. typhi species also revealed only 5 putative npcRNAs in common (15,23). Such a paucity of overlap was also observed in our laboratory in another instance where two different sequencing approaches were available for the same or similar RNA preparations (our unpublished data). Most likely, the differences are a consequence of alternative methodologies for preparing RNAs for cDNA libraries and for sequencing (e.g. pre-selections, tailing, linker ligation and PCR amplification). Hence, comparisons between two data sets are valid only when generated with virtually identical experimental approaches. These observations are a clear warning that even deep sequencing is unlikely to represent all RNA species, the identification of which may require a combination of various approaches.
Potential candidates for novel npcRNAs in S. typhi
The 97 npcRNA candidates that we identified experimentally (Figure 1B) have been assigned reference identifications based on the template, StyR-n (S. typhi npcRNA - number). Three of the npcRNA candidates are specific to S. typhi and 22 are restricted to Salmonella species; of these three are restricted to S. typhi and S. paratyphi C and one to S. typhi and S. paratyphi A. Most of our npcRNA candidates are transcribed complimentarily to ORFs of known or hypothetical protein-coding genes (39 candidates, Figure 1B), supporting previous experimental evidence of significant antisense transcription in bacteria (35,36). Twenty-four npcRNA candidates were mapped to intergenic regions of the S. typhi genome (Figure 1B). Given that most known npcRNAs in bacteria were previously identified and characterized in E. coli and the close phylogenetic relationship between S. typhi and E. coli (60–70% genomic similarity), we computationally predicted the existence of 63 putative homologous and novel npcRNAs that we designated as EcoR (Escherichia coli npcRNA- number).
Expression analysis of novel StyR npcRNAs
To determine the differential expression of our npcRNA candidates and to validate our computational analysis, we performed northern blot hybridizations on total RNA samples from three different bacterial growth stages. To ensure specific detection of npcRNA candidates, we designed a second oligonucleotide probe complementary to the respective RNA sequence in most of the cases (Supplementary Table S1). Approximately 45% of the npcRNA candidates were detected on northern blots (Figure 1B). Many of these transcripts exhibited the same length as the corresponding cDNA sequence contigs, leading us to conclude that our technique for constructing the bacterial cDNA library often yields full-length npcRNAs. However, in a few cases, the resulting northern blot signals suggested longer RNAs than the cDNAs predicted. Differential expression analyses of the npcRNA candidates indicated that the majority of them were regulated in a growth-dependent manner in S. typhi. To confirm the validity of the 63 putative npcRNAs orthologs predicted in E. coli, we carried out northern blot analysis with E. coli total RNA isolated from three different growth stages in parallel. Northern blot signals supported the phylogenetic predictions for 19 of these npcRNA homologs (Figures 2, 4, 5B and 6)
Figure 2.
Expression analysis of intergenic-derived npcRNA candidates at different growth stages of S. typhi and E. coli. Northern blot analysis was performed on total RNA isolated from S. typhi and E. coli cultures at different OD600 values, as shown across the top of the blots. OD600 0.3: lag phase-steady bacterial growth; OD600 0.7: log phase-exponential bacterial growth; and OD600 1.5: stationary growth phase. The npcRNA candidates and their sizes are indicated. As loading controls, a few examples of ethidium bromide-stained 5S rRNA are shown at the bottom of each panel.
Figure 4.
Expression analysis of cis-encoded antisense npcRNA candidates at different growth stages of S. typhi and E. coli. Northern blot analyses were performed on total RNA isolated from different growth stages of S. typhi and E. coli at different OD600 values, as shown at the top. OD600 0.3: lag phase-steady bacterial growth; OD600 0.7: log phase-exponential bacterial growth; and OD600 1.5: stationary growth phase. The npcRNA candidates and RNA lengths are indicated. As loading controls, a few examples of ethidium bromide-stained 5S rRNA are shown.
Figure 5.
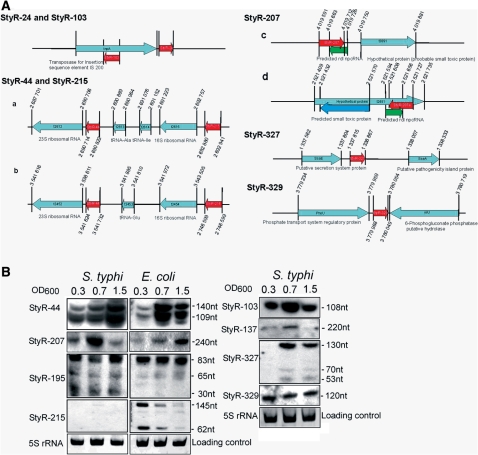
Schematic representations and expression analyses of S. typhi repetitive npcRNA candidates. (A) Schematic representations of S. typhi repetitive npcRNA candidates. Coordinates of depicted genes are based on the completed genome of S. typhi Ty2 (AE014613). Locations of npcRNA candidates are depicted by red arrows. Known or predicted hypothetical genes are indicated with light blue arrows. Drawings are not to scale. StyR-24 and StyR-103 npcRNA candidates are each repeated 24 times in the S. typhi genome; only a schematic representation is shown. StyR-44 and StyR-215 are each repeated seven times in the S. typhi genome; only two rRNA operons are shown (a, b). StyR-207 and its schematic representation is depicted in (c); green arrow indicates putative S. typhi rdl npcRNA gene (annotated in this study). StyR-207a npcRNA (d) was predicted based on sequence similarity, putative ldr and rdl genes (annotated in this study) are depicted by blue and green arrows, respectively. (B) Northern blots showing expression patterns of repetitive npcRNA candidates at different growth stages of S. typhi and E. coli. OD600 values are indicated across the tops of the blots. OD600 0.3: lag phase-steady bacterial growth; OD600 0.7: log phase-exponential bacterial growth; and OD600 1.5: stationary growth phase. The npcRNA candidates and RNA lengths are indicated. As loading controls, a few examples of ethidium bromide-stained 5S rRNA are shown.
Figure 6.
Northern blots showing growth-dependent expression patterns of S. typhi npcRNA candidates that partially overlap with ORFs of protein-coding genes. Growth stages of S. typhi and E. coli. OD600 values are indicated across the tops of the blots. OD600 0.3: lag phase-steady bacterial growth; OD600 0.7: log phase-exponential bacterial growth; and OD600 1.5: stationary growth phase. The npcRNA candidates and RNA lengths are indicated. As loading controls, a few examples of ethidium bromide-stained 5S rRNA are shown.
Based on their genomic localization and orientation, we grouped the novel npcRNA candidates into four classes: (i) intergenic npcRNAs; (ii) antisense npcRNAs; (iii) repetitive npcRNAs; and (iv) npcRNAs partially overlapping with ORFs (Figure 1B). To a large degree, however, these classifications were somewhat arbitrary, chiefly due to incomplete annotations of the genome. For example, with regard to the intergenic regions (Class 1), there are no annotations for 5′- and 3′-UTRs, because very few experimental data are available. An intergenic RNA might simply be derived from a UTR representing a more stable domain of an mRNA. Furthermore, the promoter-associated npcRNAs (Class 1.1) and terminator-associated npcRNAs (Class 1.2) are, strictly speaking, not intergenic and in many cases might be more appropriately classified as 5′- and 3′- UTR-associated or -derived RNAs, a definition that would also include riboswitches. The same difficulty arises for npcRNAs that map between genes in an operon (Class 1.3). Only the individual genes (Class 1.4) may be truly intergenic. On the other hand, npcRNAs located in a sense or antisense orientation to hypothetical ORFs might also be truly intergenic npcRNAs. The candidates located in a sense orientation to ORFs are listed in Supplementary Table S2 and are marked as being derived from hypothetical ORFs to discriminate them from sense RNAs located within proven ORFs, as the majority of the latter merely represent more stable degradation products of bona fide mRNAs.
Class 1: novel npcRNA candidates from intergenic regions of S. typhi
Obviously, intergenic regions remain the ‘hot spot’ for computational searches of npcRNAs. In fact, a number of previous predictions were focused solely on intergenic regions of bacteria (37–39). However, the intergenic npcRNA candidates that harbor low phylogenetic conservation or do not encode conserved secondary structures might not be detected in computational screens. Moreover, in some cases, the criterion of sequence conservation alone is insufficient and could be misleading. This might be particularly important for evolutionarily young and probably species-specific npcRNAs that nevertheless might exert important functions. Our experimental approach uncovered 24 npcRNA candidates derived from intergenic regions of S. typhi and the expressions of 15 of them were monitored by northern blot analysis. The corresponding RNAs range in size from 32 to 212 nt and many of them show growth stage-specific expression (Figure 2). We identified putative transcriptional regulatory elements (promoters and ρ-independent terminators) for five npcRNA genes from this category. However, their transcriptions might be more complex and some might be initiated from promoters of the adjacent genes. The intergenic npcRNAs were further subdivided into four categories depending on their location (Table 1).
Table 1.
Compilation of npcRNA candidates from intergenic regions in S. typhi
| npcRNAs | cDNA (nt) | Orient. | Adj. genes (ID) | Presence in E. coli | N. Blot S. typhi /E. coli | Growth reg. S. typhi /E. coli | Specificity | Comments | Acc. No. |
|---|---|---|---|---|---|---|---|---|---|
| Promoter-associated npcRNAs | |||||||||
| StyR-3 | 144 | < < > | 1067322/1069368 | No | +/− | −/− | Sal. sp | FJ746361 | |
| StyR-8 | 94 | > > > | 1069235/1069233 | Yes | −/− | −/− | FJ746362 | ||
| StyR-10 | 147 | < < > | 1067006/1067023 | Yes | +/+ | +/− | FJ746363 | ||
| StyR-88 | 109 | > > > | 1067919/1067920 | Yes | +/− | +/− | FJ746366 | ||
| StyR-99 | 32 | < > > | 1067609/1068774 | No | −/− | −/− | Sal. sp | FJ746367 | |
| StyR-134 | 72 | > > > | 1069120/1069122 | No | −/− | −/− | Sal. sp | FJ746369 | |
| StyR-241 | 152 | < < > | 1068271/1068273 | Yes | +/+ | +/+ | ρ-ind. term. | FJ746373 | |
| StyR-262 | 156 | < > > | 1069506/1069507 | Yes | −/− | −/− | FJ746374 | ||
| StyR-287 | 50 | > > > | 1067282/1069315 | No | −/− | −/− | Sal. sp | FJ746376 | |
| StyR-293 | 59 | < > > | 1070460/1070461 | Yes | −/− | −/− | FJ746378 | ||
| StyR-362 | 141 | < > > | 1070630/1070631 | No | −/− | −/− | Sal. sp | ρ-ind. term. | FJ746382 |
| StyR-383 | 22 | < < > | 1068808/1068821 | No | −/− | −/− | Sal. sp | FJ746384 | |
| Terminator-associated npcRNAs | |||||||||
| StyR-169 | 32 | < < < | 1069647/1067454 | Yes | +/+ | +/+ | FJ746371 | ||
| StyR-288 | 44 | > > < | 1070605/1070606 | Yes | +/+ | −/− | FJ746377 | ||
| Operon-derived npcRNAs | |||||||||
| StyR-55 | 77 | < < < | 1068343/1071526 | Yes | +/+ | +/+ | FJ746364 | ||
| StyR-126 | 34 | < < < | 1069335/1067249 | Yes | +/+ | +/+ | FJ746368 | ||
| StyR-175 | 77 | > > > | 1069764/1069765 | Yes | +/+ | +/+ | FJ746372 | ||
| StyR-280 | 102 | < > > | 1068036/1068041 | Yes | +/+ | +/+ | FJ746375 | ||
| StyR-306 | 28 | > > > | 1071397/1071398 | No | +/− | +/− | FJ746379 | ||
| StyR-333 | 40 | < < < | 1067027/1067676 | No | +/− | +/− | S. typhi | FJ746380 | |
| StyR-354 | 46 | < < < | 1067015/1068236 | No | −/− | −/− | Sal. sp | FJ746381 | |
| Possible individual intergenic npcRNA genes | |||||||||
| StyR-59 | 162 | > < < | 1068621/1068614 | No | +/− | −/− | S. typhi | ρ-ind. term. | FJ746365 |
| StyR-161 | 132 | > < > | 1068552/1069548 | No | +/− | +/− | S. typhi, S. para C | ρ-ind. term. | FJ746370 |
| StyR-381 | 73 | > > > | 1067420/1067421 | No | +/− | −/− | S. typhi, S. para C and S. dublin | ρ-ind. term. | FJ746383 |
npcRNAs, reference identifications of npcRNA candidates; cDNA (nt), length of cDNAs in nt; Orient., orientation of npcRNA (middle arrow) and adjacent genes (right and left arrows); Adj. Genes (ID), gene ID numbers of genes flanking the npcRNA candidates; Presence in E. coli, npcRNA homolog is present (Yes) or not present (No) in the E. coli K12 genome; N. blot S. typhi/E. coli, northern blots were positive (+) or negative (−) for the given npcRNAs; Growth reg. S. typhi/E. coli, npcRNA is (+) or is not (−) growth phase-regulated in S. typhi/E. coli; Specificity, species specific; Sal sp, Salmonella species specific; S. typhi, S. typhi specific; S. para C, npcRNA is present in S. paratyphi C; Comments: ρ-ind. term., probable ρ-independent/intrinsic terminator detected; Acc. No., accession number of npcRNA sequence in DDBJ/EMBL/GenBank databases.
Subclass 1.1: promoter-associated npcRNAs
The principle of transcriptional interference, also termed promoter occlusion, has been known for a long time in bacteria. It describes the regulatory interplay between two neighboring promoters. Transcription initiated by the upstream promoter inhibits transcriptional start of the downstream promoter (40,41). Recently, the mechanisms of transcriptional control involving small npcRNAs associated with transcription start sites in higher eukaryotes were described (42,43). In addition, there are reports indicating regulation of promoter activity by npcRNAs in yeast (44,45). Therefore, the control of transcription by promoter-associated RNA transcripts might be of broad importance.
We identified 12 npcRNA candidates that are transcribed within conserved mRNA promoter regions. In addition, five of the promoter-associated npcRNAs contain putative transcription factor binding sites within their primary sequences. Interestingly, there are reports indicating that transcription factors other than those with DNA binding activity also interact with RNA molecules (for review see 46). In this scenario, the npcRNA involves not only cis-mediated promoter occlusion but also regulates transcription in trans.
StyR-3 is an example of a promoter-associated npcRNA candidate. The RNA is co-transcribed by the promoter of the ramA gene and shares its putative transcriptional start site (Figures 2 and 3A). StyR-3 is likely a stable processing product derived from the RamA 5′-UTR. StyR-3 represents an abundant npcRNA species in our library, with 24 independent cDNAs. The northern blot signal is consistent with a stable 144-nt StyR-3 transcript (Figure 2).
RamA protein was recently identified as an alternative global regulator and an activator of the MDR regulation cascade in S. typhimurium. Overexpression of RamA was associated with MDR also in Enterobacter aerogenes, Klebsiella pneumonia and S. enterica serovar Paratyphi B (47–49). Recent reports identified two deletions in the promoter region of ramA that remarkably enhanced transcription of ramA, and consequently the MDR of bacteria (32,33). StyR-3 overlaps the location of these two deletions, suggesting the presence of important suppressive cis-acting elements within the region encoding the StyR-3. In addition, two inverted repeats, one of them overlapping with StyR-3, were predicted to be DNA binding sites for the local repressor, RamR (Figure 3A; 32,33). RamR belongs to the TetR family of proteins and inactivation of its gene in a susceptible serovar of the Typhimurium strain resulted in an MDR phenotype (32). Several transcription regulatory proteins interact both with DNA and RNA (46). Hence, we investigated whether the RamR transcriptional repressor protein specifically binds to StyR-3 npcRNA.
First, we conducted a gel retardation assay to examine whether the predicted binding sites within the ramA regulatory region specifically interact with and form a stable deoxyribonucleoprotein (DNP) complex with RamR protein. The in vitro-synthesized DNA fragments harboring the proposed RamR binding sites were radioactively labeled and a gel retardation assay was performed using increasing concentrations of RamR protein (see ‘Material and Methods’ section; Figure 3B). We detected formation of a stable DNP complex with an apparent K50 of 300 nM (complex formation was performed in the presence of vast excesses of non-specific DNA and RNA competitors Figure 3B). To provide further evidence of the specificity of RamR recognition, we assayed DNA fragments harboring the aforementioned 2- and 9-bp deletions within the ramA regulatory region. As expected, we did not observe a mobility shift with mutant DNA when incubated with increasing concentrations of RamR protein (Figure 3B). The loss of complex formation is consistent with overexpression of RamA and the MDR phenotype observed in S. typhimurium mutant strains (32,33).
Next, we examined via a gel retardation assay whether StyR-3 is specifically recognized by and forms an RNP complex with RamR protein. The in vitro binding assay detected the formation of an StyR-3/RamR–RNP complex with an apparent K50 of 300 nM (again, complex formation was done in the presence of vast excesses of non-specific DNA and RNA competitors Figure 3C). Thus, we demonstrated that RamR specifically interacts with DNA harboring the regulatory region for the ramA gene and with StyR-3 npcRNA. Moreover, both DNP and RNP complexes were formed with similar apparent K50 values (Figure 3B and C).
We therefore investigated whether StyR-3 interferes with RamR-DNA binding activity using a competition binding assay with increasing concentrations of unlabeled StyR-3. Even low concentrations (1 pmol) of competing StyR-3 abolished RamR–DNA binding (Figure 3D). Notably, similar results were observed when increasing concentrations of the same unlabeled wild-type DNA were added (Figure 3D). In contrast, when we competed RamR-DNA complex formation with unlabeled DNA fragments harboring the 9-bp deletion within the RamR binding site, no competition was detected (data not shown). Our data suggest an involvement of StyR-3 in the regulation of RamR-DNA binding activity, which might consequently effect ramA gene expression and the MDR of S. typhi. Given the obvious medical importance of MDR, StyR-3 might be considered a potential drug target.
We also identified other npcRNA candidates that overlap with DNA binding sites for regulatory proteins: StyR-88 with IscR, StyR-241 with Ntrc, StyR-293 with FNR and StyR-287 with GadX (Figure 2; Supplementary Figure S1). It will be interesting to examine whether these npcRNA candidates are specifically recognized by and form RNP complexes with the corresponding transcription factors. Alternatively, these npcRNA candidates may be involved in bacterial chromosome remodeling and consequently regulate promoter activity in cis.
Subclass 1.2: terminator-associated npcRNAs
Terminator-associated npcRNA candidates are a subclass of small stable npcRNAs located in regions overlapping ρ-independent terminator signals. The two candidates of this class are transcribed in a sense orientation with respect to the corresponding mRNAs and are likely derived from their 3′-UTRs. StyR-169 is associated with the terminator of the hypothetical protein gene yaiA, and StyR-288 overlaps with the ρ-independent terminator signal of the putative-exported protein-coding gene t0311 (Supplementary Figure S1). Northern blot analyses revealed that StyR-169 (32 nt) and StyR-288 (44 nt) are generated from longer primary transcripts that are subject to differential processing steps (Figure 2).
Subclass 1.3: operon-derived npcRNAs
This subgroup consists of npcRNA candidates that do not contain discernible transcription regulatory elements, are possibly transcribed as parts of operons, and undergo additional RNA processing steps for maturation. Seven such npcRNA candidates were identified and their expressions confirmed by northern blot hybridization. StyR-55 contains a sequence motif that is essential for translational feedback regulation and efficient translation of L10 and L12 ribosomal protein-coding mRNA from the L10 operon (Figure 2; Supplementary Figure S1). Proteins L10 and L12 form a complex that specifically interacts (if rRNA binding sites are saturated) with the 5′-leader sequence of the L10 operon mRNA and prevents translation (50). StyR-55 harbors the unique binding site for the inhibitory L10/L12 complex. Expression of this candidate npcRNA was detected only in the stationary phase of S. typhi growth (Figure 2). StyR-55, as an RNP with L10/L12 proteins, might participate in regulation of the L10 operon, be involved in a yet unknown function or both. Similarly, in E. coli, the t44 npcRNA was recently identified in an analogous region of the rps2 gene, encoding ribosomal protein S2. The t44 npcRNA was proposed to play a role in attenuating rps2 gene expression (51). An additional novel npcRNA candidate from S. typhi, StyR-8, originating from the leader sequence of the rmpB gene encoding ribosomal protein L28, was identified in our study (Supplementary Figure S1). This finding might provide new insight into the regulation of ribosomal protein-coding gene expression. Future experimentation will determine whether, due to their stable structures and bound proteins, such RNAs are simply more stable degradation products of the transcribed operons or true functional riboregulators.
Subclass 1.4: possible individual npcRNA genes
This category contains npcRNA candidates that are transcribed from intergenic regions of the S. typhi genome and for which associated regulatory elements (promoter and ρ-independent/intrinsic terminator) were predicted computationally. However, the situation might be more complex; for example, transcription might be initiated from promoters of adjacent genes. Three such npcRNA candidates were identified. Importantly, all of them were restricted to Salmonella serovars, representing evolutionarily young npcRNA species with potential strain-specific functions (Table 1; Figure 2; Supplementary Figure S1). Thus, StyR-381 is found in S. typhi and S. dublin genomes; StyR-161 is present in S. typhi and S paratyphi C; and StyR-59 is restricted to S. typhi.
Class 2: Antisense S. typhi npcRNAs
Small antisense npcRNAs have been studied in bacteria for over two decades and are classified according to their location on either chromosomal or plasmid DNA. Chromosomally encoded antisense npcRNAs are involved in diverse bacterial regulatory pathways. Generally, the functions of these RNAs are associated with transcriptional attenuation, inhibition of translation, or promotion of RNA degradation or cleavage (52,53). Most of the chromosomally encoded antisense npcRNA genes are expressed only under certain conditions, for example, as gadY in a growth rate-dependent manner (27) or as isrR under stress conditions (54). The antisense npcRNAs are cis-encoded with perfect complementarity to large stretches of the corresponding target RNAs.
We identified 39 npcRNAs candidates derived from the strands of DNA complementary to protein-encoding genes (i.e. cis-encoded). In fact, they constituted the largest number of novel npcRNAs identified (Table 2). Of these, 24 npcRNA candidates have homologs in E. coli (Table 2) and 10 show growth stage-specific expression patterns in S. typhi (Figures 1B and 4). Based on their location with respect to the ORF of the target genes, we established four subcategories.
Table 2.
Compilation of antisense npcRNA candidates in S. typhi
| npcRNAs | cDNA (nt) | Orient. | Gene (ID) | Presence in E. coli | N. blot S. typhi /E. coli | Growth reg, S. typhi /E. coli | Specificity | Comments | Acc. No. |
|---|---|---|---|---|---|---|---|---|---|
| npcRNA candidates antisense to the 3′-end of ORF | |||||||||
| StyR-143 | 144 | < | 1068407 | No | −/− | −/− | S. typhi, S. para A | ρ-ind. term. | FJ746389 |
| StyR-219 | 104 | > | 1068601 | Yes | +/+ | +/+ | ρ-ind. term. | FJ746397 | |
| StyR-250 | 222 | > | 1069193 | Yes | −/− | −/− | FJ746402 | ||
| npcRNA candidates antisense to the 5′-end of ORF | |||||||||
| StyR-341 | 49 | > | 1070547 | No | −/− | −/− | Sal. sp | FJ746419 | |
| npcRNA candidates antisense to both 5′- and 3′-end of single ORF | |||||||||
| StyR-281 | 253 | < | 1070369 | Yes | +/− | +/− | FJ746407 | ||
| StyR-47 | 216 | > | Predicted ORF | Yes | −/− | −/− | FJ746385 | ||
| npcRNA candidates antisense to both 5′- and 3′-end of ORFs | |||||||||
| StyR-328 | 78 | < | 1068197 | Yes | −/− | −/− | FJ746417 | ||
| StyR-150 | 46 | > | 1067798/ 1067800 | No | −/− | −/− | Sal. sp | FJ746390 | |
| cis-encoded antisense npcRNA candidates to ORFs of protein-coding genes | |||||||||
| StyR-101 | 45 | < | 1067667 | No | −/− | −/− | S. typhi | FJ746386 | |
| StyR-122 | 25 | > | 1069722 | Yes | +/− | −/− | FJ746387 | ||
| StyR-128 | 83 | < | 1069075 | Yes | −/− | −/− | FJ746388 | ||
| StyR-151 | 109 | > | 1067671 | Yes | +/− | −/− | FJ746391 | ||
| StyR-172 | 57 | < | 1069704 | Yes | −/− | −/− | FJ746392 | ||
| StyR-173 | 76 | < | 1068387 | No | +/− | +/− | Sal. sp | FJ746393 | |
| StyR-178 | 34 | < | 1067312 | Yes | −/− | −/− | FJ746394 | ||
| StyR-196 | 82 | < | 1069862 | Yes | +/− | +/− | FJ746395 | ||
| StyR-199 | 48 | > | 1067680 | Yes | +/− | +/− | FJ746396 | ||
| StyR-222 | 75 | < | 1069061 | Yes | −/− | −/− | FJ746398 | ||
| StyR-234 | 119 | > | 1069670 | Yes | +/+ | +/+ | FJ746399 | ||
| StyR-243 | 135 | > | 1069109 | Yes | +/− | +/− | FJ746400 | ||
| StyR-248 | 211 | > | 1070765 | Yes | +/− | +/− | FJ746401 | ||
| StyR-252 | 162 | < | 1071079 | Yes | −/− | −/− | FJ746403 | ||
| StyR-254 | 98 | < | 1071250 | Yes | +/+ | +/− | FJ746404 | ||
| StyR-264 | 85 | > | 1067864 | Yes | +/+ | −/− | ρ-ind. term. | FJ746405 | |
| StyR-277 | 99 | > | 1069782 | Yes | −/− | −/− | FJ746406 | ||
| StyR-285 | 110 | < | 1070539 | No | −/− | −/− | Sal. sp | FJ746408 | |
| StyR-291 | 61 | > | 1070390 | Yes | −/− | −/− | FJ746409 | ||
| StyR-292 | 38 | > | 1070899 | No | −/− | −/− | Sal. sp | FJ746410 | |
| StyR-295 | 109 | > | 1066997 | Yes | −/− | −/− | FJ746411 | ||
| StyR-302 | 22 | < | 1067039 | No | −/− | −/− | Sal. sp | FJ746412 | |
| StyR-305 | 57 | < | 1067199 | No | −/− | −/− | FJ746413 | ||
| StyR-309 | 32 | > | 1067690 | Yes | −/− | −/− | FJ746414 | ||
| StyR-315 | 69 | < | 1069646 | Yes | −/− | −/− | FJ746415 | ||
| StyR-322 | 90 | < | 1069020 | Yes | −/− | −/− | FJ746416 | ||
| StyR-336 | 199 | > | 1071148 | Yes | −/− | −/− | FJ746418 | ||
| StyR-358 | 113 | < | 1067275 | No | +/− | +/− | Sal. sp | FJ746420 | |
| StyR-369 | 22 | < | 1067888 | No | −/− | −/− | Sal. sp | FJ746421 | |
| StyR-377 | 23 | > | 1069015 | No | −/− | −/− | FJ746422 | ||
| StyR-378 | 23 | < | 1069059 | Yes | −/− | −/− | Sal. sp | FJ746423 | |
npcRNAs, reference identifications of npcRNA candidates; cDNA (nt), length of cDNA clones in nt; Orient, orientation of npcRNA; Gene (ID), ID number of protein-coding genes; Presence in E. coli, npcRNA homolog is present (Yes) or not present (No) in the E. coli K12 genome; N. blot S. typhi/E. coli, northern blots were positive (+) or negative (-) for the given transcript; Growth reg. S. typhi /E. coli, npcRNA was (+) or was not (-) growth phase-regulated; Specificity, species specific; Sal sp, Salmonella species-specific, S. typhi, S. typhi-specific, EPEC, enteropathogenic E. coli; EHEC, enterohemorrhagic E. coli;. Comments: ρ-ind. term., probable ρ-independent terminator detected; Acc. No., accession number of npcRNA sequence in DDBJ/EMBL/GenBank databases.
Subclass 2.1: npcRNA candidates antisense to the 3′-end of ORFs
Three of the antisense npcRNAs (StyR-143, StyR-219 and StyR-250) partially overlap the 3′-ends of ORFs (Table 2; Supplementary Figure S2). The cis-antisense candidate StyR-219 is complementary to the 3′-part of the ORF of prfB that encodes peptide chain release factor-2 (Supplementary Figure S2). StyR-219 features a ρ-independent/intrinsic terminator and is conserved in many other enterobacteria. We computationally predicted an E. coli K12 homolog (EcoR-219) that shares 89% sequence similarity. Interestingly, the expression of the E. coli prfB gene is upregulated during later growth stages (55,56), whereas the expression of StyR-219 is highest during the log phase of growth and expressions of both the S. typhi and E. coli homologs are reduced as the cultures reach their stationary phases (Figure 4). Phylogenetic conservation and the reversely correlated expression profiles of npcRNA and mRNA strongly indicate that StyR-219 is involved in regulation of prfB expression.
Subclass 2.2: npcRNA candidates antisense to the 5′-end of ORFs
The cis-encoded StyR-341 npcRNA candidate was located complementary to the ribosomal binding site (RBS) and translational start codon of the protein-coding gene t2574, which is restricted to Salmonella species and maps to SPI-6 (Figure 7; Supplementary Figure S2), suggesting a function in S. typhi pathogenicity. In bacteria, npcRNAs exhibiting complementarities to mRNA regions involved in translation initiation, such as RBSs and AUG start codons, are obvious candidates for control at the level of translation regulation. Therefore, StyR-341 might be an example of a riboregulator that is associated with virulence gene control.
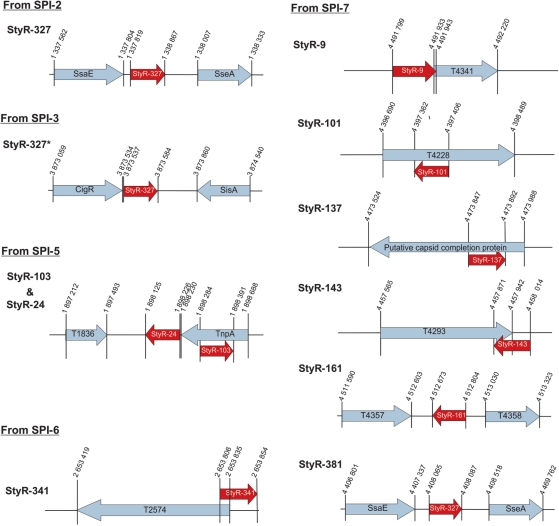
Figure 7.
Schematic representations of SPI-derived npcRNA candidates. Coordinates are based on the S. typhi Ty2 complete genome annotation (AE014613). Locations of npcRNA candidates are indicated by red arrows. ORFs of known or hypothetical genes are indicated by light blue arrows. SPI-numbers are indicated. Drawings are not to scale.
Subclass 2.3: npcRNA candidates antisense to the 5′- and 3′-ends of two different ORFs
Two cis-encoded antisense npcRNA candidates (StyR-150 and StyR-328) are transcribed such that they are partially complementary to the 5′- and 3′-ends of two neighboring ORFs, as well as to the corresponding UTRs and the entire intergenic region (Supplementary Figure S2). StyR-328 is complementary to the RBS plus AUG start codon of the fdhE gene and the stop codon of the fdoI gene. Both protein-coding genes are involved in formate metabolism. In addition to molecular oxygen, Salmonella can utilize formate dehydrogenase-O under aerobic conditions as an alternative terminal electron acceptor to generate a proton motive force (57). The physiological role of formate dehydrogenase-O is to ensure rapid adaptation during a sudden shift from aerobiosis to anaerobiosis to compensate for low levels of formate dehydrogenase-N (57). Formate dehydrogenase-O shares sequence similarity and immunological properties with the anaerobically expressed formate dehydrogenase-N. StyR-328 might possibly regulate bacterial adaptation to anaerobic conditions. This might partially explain our failure to detect the mature npcRNA during the examined growth stages of S. typhi under aerobic conditions.
Subclass 2.4: cis-encoded antisense npcRNA candidates complementary to ORFs
Thirty-one npcRNA candidates belong to a subclass of cis-encoded antisense RNA species that are complementary to internal parts of the ORFs of known or hypothetical protein-coding genes (Table 2; Supplementary Figure S2). StyR-243 (135 nt) is expressed in an antisense orientation to ORF of the oxyR gene, which encodes a DNA binding, transcriptional dual regulator of the LysR family (58). The gene encoding StyR-243 is highly conserved in all Salmonella species with a predicted putative homolog in E. coli (EcoR-243, 83% sequence similarity). OxyR participates in controlling several genes involved in the response to oxidative stress and the production of surface proteins by activating the expression of several genes (e.g. dps, fur and katG) whose protein products regulate intracellular hydrogen peroxide levels (59–61). oxyR is negatively autoregulated and its expression decreases towards the stationary growth phase (58). Interestingly, under aerobic growth conditions, StyR-243 expression significantly increases during the log phase and decreases in the stationary phase (Figure 4; Supplementary Figure S3). In contrast, we did not detect expression of the putative EcoR-243 homolog in E. coli.
StyR-248 (211 nt) is located antisense to part of the tus ORF, which encodes the DNA replication terminus (ter) site binding protein. Tus, also known as ter-binding protein (TBP), binds to ter sites, blocking the progress of DNA replication in a polar fashion by blocking the progress of the helicase DnaB, eventually leading to replication termination (62–64). The ter-bound Tus also blocks the activity of helicases PriA, Rep and UvrD (65). StyR-248 is predominantly expressed in the stationary phase (Figure 4). This might well reflect its possible role in the control of tus gene expression and replication of the S. typhi bacterial chromosome. The RNA is highly conserved in all published Salmonella species genomes with a putative homolog in E. coli (EcoR-248, 75% sequence similarity). Inexplicably, StyR-248 gives a signal of only ∼80 nt on northern blots (Figure 4). Interestingly, StyR-248 forms a complex with the S. typhi Hfq protein in vitro, implicating this npcRNA as a potential riboregulator (Supplementary Data and Supplementary Figure S4).
StyR-264 (85 nt) is expressed complementary to part of the ORF of the ung gene that encodes the ubiquitous uracil-DNA glycosylate, an enzyme that prevents mutagenesis by eliminating uracil from DNA molecules by cleaving the N-glycosylic bond and initiating the base-excision repair pathway (66). The expression of StyR-264 was slightly decreased during the late growth stage of S. typhi (Figure 4). Expression of the ung gene was reported to remain constant up to the early stationary phase of E. coli and to decline in the late stationary phase (67). Interestingly, the EcoR-264 homolog (85 nt) exhibits a similar expression pattern (Figure 4), suggesting that the npcRNA might play a role in stabilizing Ung mRNA.
To gain further insight into the functional significance of other members of this subclass of antisense npcRNAs, we investigated their differential expression profiles and those of the corresponding mRNAs at various growth stages of S. typhi using real-time PCR (Supplementary Figure S3). Most of the sense–antisense pairs exhibited positively co-regulated expression profiles, indicating a possible involvement of antisense npcRNA in stabilizing cis-encoded mRNAs, probably enhancing their translation (Supplementary Figure S3).
Class 3: multicopy npcRNAs in S. typhi
We considered npcRNA candidates that were represented by more than one copy in the S. typhi genome to be repetitive (Table 3). Repetitive npcRNAs have been reported in all three domains of life. For example, four dispersed long-direct-repeat (LDR) sequences express both mRNAs (ldrA-D, where the designations A to D represent four different locations in the E. coli genome) encoding a toxic peptide and their cis-encoded antisense riboregulators, rdlA-D (68). In addition, six regions in the E. coli genome display high sequence similarity to the plasmid hok/sok loci encoding Sok npcRNAs controlling the sense-orientated protein-coding hok genes (69). We identified 11 repetitive npcRNA candidates. Eight originated from intergenic regions and the other three were transcribed in antisense orientations to bona fide or hypothetical genes. Seven of these repetitive npcRNAs exhibited growth stage-specific expression patterns (Figure 5A and B).
Table 3.
Compilation of repetitive npcRNA candidates in S. typhi
| npcRNAs | cDNA (nt) | Repeats | Presence E. coli | N. Blot S. typhi /E. coli | Growth reg. S. typhi /E. coli | Comments | Acc. No. |
|---|---|---|---|---|---|---|---|
| Intergenic repetitive npcRNAs | |||||||
| StyR-24 | 102 | 26× | No | −/− | −/− | Downstream from ORF of IS200 transposase | FJ746424 |
| StyR-44 | 109 | 7× | Yes | +/+ | +/+ | Part of ribosomal operon | FJ746425 |
| StyR-184 | 84 | >22× | Yes | −/− | −/− | FJ746429 | |
| StyR-195 | 83 | >30× | Yes | +/+ | −/− | FJ746430 | |
| StyR-207 | 122 | 2× | Yes | +/+ | +/− | FJ746431 | |
| StyR-327 | 53 | 8× | No | +/− | +/− | FJ746433 | |
| StyR-329 | 120 | >20× | Yes | +/− | −/− | FJ746434 | |
| StyR-215 | 62 | 7× | Yes | +/+ | +/+ | Upstream to ribosomal operon | FJ746432 |
| Antisense repetitive npcRNAs | |||||||
| StyR-90 | 90 | 2× | Yes | +/− | +/− | FJ746426 | |
| StyR-103 | 108 | 26× | No | +/− | +/− | Antisense to ORF of IS200 transposase | FJ746427 |
| StyR-137 | 46 | 2× | Yes | +/− | +/− | Derived from SPI-7 | FJ746428 |
npcRNAs, reference identifications of S. typhi npcRNA candidates; cDNA (nt), length of cDNA clones in nt; Repeats, number of copies in the S. typhi TY2 genome (AE014613); Presence in E. coli, npcRNA is present (Yes) or not present (No) in E. coli K12 genome; N. blot S. typhi/E. coli, northern blots were positive (+) or negative (−) for the given transcript; Growth phase S. typhi/E. coli, npcRNA was (+) or was not (−) growth phase-regulated; Acc. No., accession number of npcRNA sequence in DDBJ/EMBL/GenBank databases.
StyR-207 perfectly matches an S. typhi locus upstream from hypothetical protein-coding gene t3891, and at the same time is complementary to the S. typhi t3892 pseudogene (Figure 5A-c). Based on sequence similarities to E. coli, these two S. typhi ORFs might represent S. typhi ldrD and rdlD genes, respectively (Figure 5A-c and B). Moreover, a sequence similar to StyR-207 was detected in the S. typhi genome that was complementary to the hypothetical ORF t2451 (Figure 5A-d). S. typhi ORF t2451 can be aligned to the E. coli ldrB and rdlB gene pair (71% sequence similarity; Figure 5A-d). In E. coli, StyR-207 homologs (EcoR-207) are present in four copies complementary to rdlA-rdlD npcRNA genes. Therefore, StyR-207 npcRNAs might represent cis-encoded riboregulators for rdl npcRNAs, and thereby play a role in the ldr/rdl toxin–antitoxin system.
StyR-44 and StyR-215 are derived from ribosomal operons and are each repeated seven times in the S. typhi and E. coli genomes. Based on the tRNA genes located in the internal spacer region between 16S and 23S rRNAs, there are two subtypes of ribosomal RNA operons defined in many bacterial species (Figure 5A-a and -b). One subtype features tRNA-Glu and the second tRNA-Ile and tRNA-Ala in the spacer (70). StyR-44 (109 nt) is located upstream of the respective 23S rRNA genes in all rRNA operons. StyR-215 (62 nt) is located 123 nt upstream from the 16S rRNA gene in all seven ribosomal RNA operons (Figure 5A-a and -b). StyR-44 and StyR-215 are both growth rate-regulated and processed from longer precursors (Figure 5B). At this point, we cannot rule out the possibility that both RNAs are merely metastable RNA intermediates generated during maturation of ribosomal RNA. Conspicuously, although StyR-215 was cloned from S. typhi, a northern blot signal was only detected in E. coli (Figure 5B).
StyR-24 and StyR-103 are both derived from the repetitive IS200 transposon (Figure 5A), of which there are 24 copies in S. typhi. StyR-24 is located 4 nt downstream of the ORF of the transposase gene in the same orientation, whereas StyR-103 is expressed antisense to the transposase ORF (Figures 5A, B and 7). RNA-OUT, a cis-endoded antisese npcRNA to the ORF of the IS10 transposase-encoded gene (RNA-IN), was previously shown to regulate transposase expression and control transposition activity in E. coli (71). StyR-103 represents an abundantly expressed RNA species with pronounced upregulation during the exponential growth phase (Figure 5B). Therefore, similar to RNA-OUT, StyR-103 npcRNA may act as a cis-encoding antisense npcRNA to regulate transposase expression and influence the rate of transposition of IS200 elements. This may be the reason for the extremely low transposition frequency of IS200 during S. typhi growth (72). Indeed, cis-encoded antisense npcRNA regulation of transposition activity is a widespread phenomenon in prokaryotes (52).
Class 4: S. typhi npcRNAs overlapping with ORFs in the sense orientation
Twenty-three npcRNA candidates were transcribed in the same orientation as ORFs and were partially congruent with the corresponding mRNAs (Table 4). The E. coli t44 npcRNA (135 nt) (51) maps upstream of the 30S ribosomal protein S2 gene, rpS2, suggesting its role in attenuating rpS2 expression. StyR-7 appears to be a putative S. typhi counterpart of the E. coli t44 npcRNA; however, cDNAs for StyR-7 are up to 238 nt long (103 nt longer than t44) covering the S. typhi rpS2 translational start codon and part of the N-terminus of the ORF (Supplementary Table S3).
Table 4.
Compilation of npcRNA candidates overlapping with ORFs in sense orientation
| npcRNAs | cDNA (nt) | Orient. | Overlap (nt) | Gene ID | Presence in E. coli | N. blot S. typhi / E. coli | Growth reg., S. typhi / E. coli | Specificity | Comments | Acc. No. | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Partially overlapping with 5′-end of ORF | ||||||||||||
| StyR-9 | 145 | > | 11 | 1071490 | No | +/− | +/− | S. typhi,S. para C | FJ746436 | |||
| StyR-15 | 306 | > | 129 | 1070087 | Yes | nd | nd | Sal. sp. | FJ746437 | |||
| StyR-29 | 245 | < | 116 | 1067535 | Yes | +/+ | +/− | FJ746440 | ||||
| StyR-64 | 108 | < | 30 | 1071363 | Yes | nd | nd | FJ746443 | ||||
| StyR-77 | 114 | > | 49 | 1069758 | Yes | nd | nd | FJ746446 | ||||
| StyR-84 | 77 | < | 8 | 1067059 | Yes | nd | nd | FJ746447 | ||||
| StyR-145 | 327 | > | 108 | 1069487 | No | nd | nd | S. typhi,S. para C | FJ746448 | |||
| StyR-228 | 111 | > | 2 | 1070302 | No | −/− | −/− | Sal. sp. | FJ746452 | |||
| StyR-255 | 243 | > | 100 | 1067871 | Yes | −/− | −/− | FJ746453 | ||||
| StyR-282 | 86 | > | 72 | 1068122 | Yes | nd | nd | FJ746457 | ||||
| StyR-296 | 293 | > | 71 | 1069730 | Yes | +/+ | +/+ | FJ746454 | ||||
| StyR-335 | 263 | < | 97 | 1069119 | Yes | −/− | −/− | FJ746455 | ||||
| StyR-364 | 176 | > | 77 | 1067975 | Yes | nd | nd | FJ746456 | ||||
| Partially overlapping with 3′-end of ORF | ||||||||||||
| StyR-6 | 159 | > | 112 | 1068275 | Yes | nd | nd | FJ746435 | ||||
| StyR-26 | 223 | < | 150 | 1067805 | Yes | nd | nd | FJ746438 | ||||
| StyR-27 | 146 | > | 29 | 1071267 | No | +/− | +/− | FJ746439 | ||||
| StyR-51 | 87 | > | 4 | 1067741 | Yes | +/+ | +/+ | FJ746441 | ||||
| StyR-68 | 95 | > | 43 | 1068476 | Yes | nd | nd | FJ746444 | ||||
| StyR-74 | 87 | < | 12 | 1069270 | Yes | nd | nd | ρ-ind. term. | FJ746442 | |||
| StyR-76 | 241 | > | 112 | 1069147 | Yes | nd | nd | FJ746445 | ||||
| StyR-149 | 83 | > | 19 | 1070930 | No | nd | nd | Sal. sp. | FJ746449 | |||
| StyR-165 | 105 | > | 40 | 1069332 | Yes | nd | nd | FJ746450 | ||||
| StyR-192 | 71 | > | 33 | 1071394 | Yes | nd | nd | ρ-ind. term. | FJ746451 | |||
npcRNAs, reference identifications of S. typhi npcRNA candidates; cDNA (nt), length of cDNA clones in nt; Orient., orientation of npcRNA; Overlap, number of nucleotides overlapping the ORF; Gene ID, ID of protein-coding genes; Presence in E. coli, npcRNA is present in the E. coli K12 genome; N. blot S. typhi/E. coli, northern blot was positive (+) or negative (−) for the given transcript; nd, not done; Growth reg. S. typhi /E. coli, npcRNA was (+) or was not (−) growth phase-regulated; Specificity, species specificity; Sal sp, Salmonella species specific; S. typhi, S. typhi specific; Comments, ρ-ind. term., probable ρ-independent terminator detected; Acc. No., accession number of npcRNA sequence in DDBJ/EMBL/GenBank databases.
Northern blot analyses revealed growth stage-specific expression for five of the novel npcRNA candidates in this category (Figure 6). StyR-29 represents another example of an npcRNA candidate that overlaps the area of the 5′-UTR and a portion of the ORF of the rpsT ribosomal protein S20 coding gene. Expression of StyR-29 is a bit complex, as the start of transcription is located further upstream and complementarily to the hypothetical t0046 ORF. StyR-29 expression analysis revealed three distinct transcripts of 77 nt, 140 nt and 245 nt (Figure 6). Our cDNA library contains the longest (245 nt). The 245 nt and 77 nt transcripts were specific to the stationary phase of S. typhi growth. In contrast, the 140-nt RNA fragment was present throughout all the growth stages in both S. typhi and E. coli (Figure 6). Therefore, the three forms of StyR-29 might regulate expression in multiple modes.
npcRNAs from the S. typhi Pathogenicity Islands
The emergence of pathogenic strains of enteric bacteria and their adaptation to new environments is accompanied by the acquisition of foreign DNA segments termed ‘genetic islands’. These islands are arranged as clusters of genes and encode virulence determinants (11,14). Of the 97 novel npcRNA candidates discussed, 10 mapped to SPI regions, suggesting that they might have various unknown virulence-associated functions (Figure 7).
StyR-327 is located in the intergenic region between ssaE and sseA of SPI-2 (Figure 7). Sequences similar to this intergenic region exist in multiple copies in Salmonella and E. coli species (at least six times in Salmonella species). We detected terminal A+T-rich inverted repeat sequences flanking these regions; therefore, the intergenic region harboring StyR-327 might represent a possible miniature inverted repeat transposable element (MITE) in bacterial genomes (73). The S. typhi SPI-3 harbors an additional copy of such an intergenic MITE with sequence similarity to StyR-327 (Figure 7). Northern blot analysis indicated a growth stage-specific expression of StyR-327 (Figure 5B). Bacterial MITE sequences are predominantly found in intergenic regions carrying regulatory elements or even short ORFs fused to pre-existing genes, potentially changing their regulation or function (73). Thus, StyR-327 might be a functional riboregulator within a bacterial MITE sequence.
In addition to three putative npcRNAs in SPI-5 (StyR-24 and StyR-103) and SPI-6 (StyR-341), we identified six npcRNAs candidates in SPI-7 (StyR-9, StyR-101, StyR-137, StyR-143, StyR-161 and StyR-381). StyR-9 overlaps with the translational start codon of the hypothetical protein gene t4341, and northern blot analysis showed growth stage-specific expression (Figure 6). The RNA shares 89% sequence similarity with StyR-10, the latter being conserved in E. coli. StyR-9 may be a paralog of StyR-10, having originated via genomic duplication. Interestingly, the two npcRNA candidates show different expression patterns (Figures 2 and 6) and both form specific RNP complexes with the S. typhi Hfq protein (Supplementary Data and Supplementary Figure S4), further indicating that they might function as riboregulators. StyR-101 and StyR-137 are cis-encoded in the antisense orientation to ORFs t4228 and t4317, respectively, suggesting possible functions in regulating these genes. StyR-161 is restricted to S. typhi and S. paratyphi C and its transcription is growth stage-regulated (Figure 2). StyR-381 is constitutively expressed and restricted to S. typhi and S. dublin (Figures 2 and 7). These observations suggest that npcRNAs are prominently represented in and expressed from pathogenic islands of the bacteria and have potentially different functions related to virulence.
CONCLUSION
We present an experimental survey of the small npcRNA transcriptome in S. typhi that causes typhoid fever. Ninety-seven novel npcRNA candidates were identified and characterized. Northern blot analyses identified 33 npcRNA candidates that were differentially expressed during S. typhi growth. ρ-Independent transcription terminators and/or promoters were predicted for 10 of the putative npcRNAs. Most of the npcRNAs were derived from intergenic (24) or antisense ORF (39) regions. Phylogenetic analysis predicted that 63 of the novel putative npcRNA candidates had homologs in E. coli. Northern blot signals supported the phylogenetic predictions for 19 of these npcRNA homologs, further implicating their functional significance. On the other hand, three npcRNA candidates were specific to S. typhi, while 22 were restricted to Salmonella strains. Some of these candidates are potentially involved in host-specific pathogenicity. Furthermore, we identified 10 SPI-derived npcRNA candidates that might be involved in regulatory mechanisms of virulence, antibiotic resistance pathways or the pathogenic specificity of S. typhi. Eleven npcRNA candidates were of repetitive origin, including possible riboregulators for transposase and toxin/antitoxin related genes. Thirty-nine of the cis-encoded npcRNA candidates were transcribed in the antisense orientation to ORFs of protein-coding genes, further supporting previous observations of widespread sense–antisense regulatory networks in bacteria. We also detected small stable npcRNAs candidates overlapping with the 5′-UTRs of a number of ribosomal protein-coding genes (individual or organized in an operon). This could point to an additional layer of regulation during ribosome biogenesis. In vitro binding assays showed that six of the npcRNA candidates, representing different subgroups, interacted with the S. typhi Hfq protein, further supporting an important role of Hfq in npcRNA regulatory networks.
A particularly interesting case is StyR-3 that shares the promoter region with the ramA gene. We show that RamR, the transcriptional repressor protein of ramA, interacted with both the promoter region of ramA and StyR-3 npcRNA. The RNP complex was formed with an apparent K50 of about 300 nM, similar to that of the observed DNP complex. Moreover, StyR-3 specifically competed with RamR protein from the DNP complex. These findings point to a potential function of StyR-3 npcRNA in regulating ramA gene expression, which is associated with MDR in S. typhi. Our findings set the stage for further experimentation, to confirm the functions of numerous novel npcRNA candidates in the pathogenesis of S. typhi and its interactions with its human host cells.
ACCESSION NUMBER
GenBank: FJ746361–FJ746457.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Nationales Genomforschungsnetz (NGFNII-EP 0313358A and NGFNIII 01GS0808 to J.B. and T.S.R.); Fundamental Research Grant Scheme, Ministry of Higher Education, Malaysia (FRGS 203/CIPPT/6711116 to T.H.T.). Funding for open access charge: NGFNIII 01GS0808.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Ravichandran Manickam for helpful discussions and Marsha Bundman for editorial assistance.
REFERENCES
- 1.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 2.Sledjeski DD, Gupta A, Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 3.Steglich C, Futschik ME, Lindell D, Voss B, Chisholm SW, Hess WR. The challenge of regulation in a minimal photoautotroph: non-coding RNAs in Prochlorococcus. PLoS Genet. 2008;4:e1000173. doi: 10.1371/journal.pgen.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner EG, Simons RW. Antisense RNA control in bacteria, phages, and plasmids. Annu. Rev. Microbiol. 1994;48:713–742. doi: 10.1146/annurev.mi.48.100194.003433. [DOI] [PubMed] [Google Scholar]
- 5.Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawamoto H, Koide Y, Morita T, Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- 7.Wassarman KM. Small RNAs in bacteria: diverse regulators of gene expression in response to environmental changes. Cell. 2002;109:141–144. doi: 10.1016/s0092-8674(02)00717-1. [DOI] [PubMed] [Google Scholar]
- 8.Romby P, Vandenesch F, Wagner EG. The role of RNAs in the regulation of virulence-gene expression. Curr. Opin. Microbiol. 2006;9:229–236. doi: 10.1016/j.mib.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Vogel J, Papenfort K. Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 2006;9:605–611. doi: 10.1016/j.mib.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu. Rev. Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 11.Deng W, Liou SR, Plunkett G., Mayhew GF, Rose DJ, Burland V, Kodoyianni V, Schwartz DC, Blattner FR. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 2003;185:2330–2337. doi: 10.1128/JB.185.7.2330-2337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker S, Dougan G. The genome of Salmonella enterica serovar Typhi. Clin. Infect. Dis. 2007;45(Suppl. 1):S29–33. doi: 10.1086/518143. [DOI] [PubMed] [Google Scholar]
- 13.Pfeiffer V, Sittka A, Tomer R, Tedin K, Brinkmann V, Vogel J. A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol. Microbiol. 2007;66:1174–1191. doi: 10.1111/j.1365-2958.2007.05991.x. [DOI] [PubMed] [Google Scholar]
- 14.Padalon-Brauch G, Hershberg R, Elgrably-Weiss M, Baruch K, Rosenshine I, Margalit H, Altuvia S. Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res. 2008;36:1913–1927. doi: 10.1093/nar/gkn050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luban S, Kihara D. Comparative genomics of small RNAs in bacterial genomes. Omics. 2007;11:58–73. doi: 10.1089/omi.2006.0005. [DOI] [PubMed] [Google Scholar]
- 17.Vogel J. A rough guide to the non-coding RNA world of Salmonella. Mol. Microbiol. 2009;71:1–11. doi: 10.1111/j.1365-2958.2008.06505.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 19.Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 20.Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35:1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Majdalani N, Vanderpool CK, Gottesman S. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol. 2005;40:93–113. doi: 10.1080/10409230590918702. [DOI] [PubMed] [Google Scholar]
- 22.Sharma AK, Payne SM. Induction of expression of hfq by DksA is essential for Shigella flexneri virulence. Mol. Microbiol. 2006;62:469–479. doi: 10.1111/j.1365-2958.2006.05376.x. [DOI] [PubMed] [Google Scholar]
- 23.Perkins TT, Kingsley RA, Fookes MC, Gardner PP, James KD, Yu L, Assefa SA, He M, Croucher NJ, Pickard DJ, et al. A strand-specific RNA-Seq analysis of the transcriptome of the typhoid bacillus Salmonella typhi. PLoS Genet. 2009;5:e1000569. doi: 10.1371/journal.pgen.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawano M, Reynolds AA, Miranda-Rios J, Storz G. Detection of 5′- and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res. 2005;33:1040–1050. doi: 10.1093/nar/gki256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EG. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Selinger DW, Cheung KJ, Mei R, Johansson EM, Richmond CS, Blattner FR, Lockhart DJ, Church GM. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol. 2000;18:1262–1268. doi: 10.1038/82367. [DOI] [PubMed] [Google Scholar]
- 27.Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raabe CA, Sanchez CP, Randau G, Robeck T, Skryabin BV, Chinni SV, Kube M, Reinhardt R, Ng GH, Manickam R, et al. A global view of the nonprotein-coding transcriptome in Plasmodium falciparum. Nucleic Acids Res. 2010;38:608–617. doi: 10.1093/nar/gkp895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khanam T, Rozhdestvensky TS, Bundman M, Galiveti CR, Handel S, Sukonina V, Jordan U, Brosius J, Skryabin BV. Two primate-specific small non-protein-coding RNAs in transgenic mice: neuronal expression, subcellular localization and binding partners. Nucleic Acids Res. 2007;35:529–539. doi: 10.1093/nar/gkl1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mullis KB, Faloona FA. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Meth. Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 31.Rozhdestvensky TS, Tang TH, Tchirkova IV, Brosius J, Bachellerie JP, Huttenhofer A. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res. 2003;31:869–877. doi: 10.1093/nar/gkg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abouzeed YM, Baucheron S, Cloeckaert A. ramR mutations involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 2008;52:2428–2434. doi: 10.1128/AAC.00084-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng J, Cui S, Meng J. Effect of transcriptional activators RamA and SoxS on expression of multidrug efflux pumps AcrAB and AcrEF in fluoroquinolone-resistant Salmonella Typhimurium. J. Antimicrob. Chemother. 2009;63:95–102. doi: 10.1093/jac/dkn448. [DOI] [PubMed] [Google Scholar]
- 34.Benito Y, Kolb FA, Romby P, Lina G, Etienne J, Vandenesch F. Probing the structure of RNAIII, the Staphylococcus aureus agr regulatory RNA, and identification of the RNA domain involved in repression of protein A expression. RNA. 2000;6:668–679. doi: 10.1017/s1355838200992550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carmichael GG. Antisense starts making more sense. Nat. Biotechnol. 2003;21:371–372. doi: 10.1038/nbt0403-371. [DOI] [PubMed] [Google Scholar]
- 36.Ozoline ON, Deev AA. Predicting antisense RNAs in the genomes of Escherichia coli and Salmonella typhimurium using promoter-search algorithm PlatProm. J. Bioinform. Comput. Biol. 2006;4:443–454. doi: 10.1142/s0219720006001916. [DOI] [PubMed] [Google Scholar]
- 37.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 38.Panek J, Bobek J, Mikulik K, Basler M, Vohradsky J. Biocomputational prediction of small non-coding RNAs in Streptomyces. BMC Genomics. 2008;9:217. doi: 10.1186/1471-2164-9-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzalez N, Heeb S, Valverde C, Kay E, Reimmann C, Junier T, Haas D. Genome-wide search reveals a novel GacA-regulated small RNA in Pseudomonas species. BMC Genomics. 2008;9:167. doi: 10.1186/1471-2164-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adhya S, Gottesman M. Promoter occlusion: transcription through a promoter may inhibit its activity. Cell. 1982;29:939–944. doi: 10.1016/0092-8674(82)90456-1. [DOI] [PubMed] [Google Scholar]
- 41.Nasser W, Rochman M, Muskhelishvili G. Transcriptional regulation of fis operon involves a module of multiple coupled promoters. EMBO J. 2002;21:715–724. doi: 10.1093/emboj/21.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 43.Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest AR, Grimmond SM, Schroder K, et al. Tiny RNAs associated with transcription start sites in animals. Nat. Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 44.Hirota K, Miyoshi T, Kugou K, Hoffman CS, Shibata T, Ohta K. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature. 2008;456:130–134. doi: 10.1038/nature07348. [DOI] [PubMed] [Google Scholar]
- 45.Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 46.Barrandon C, Spiluttini B, Bensaude O. Non-coding RNAs regulating the transcriptional machinery. Biol. Cell. 2008;100:83–95. doi: 10.1042/BC20070090. [DOI] [PubMed] [Google Scholar]
- 47.George AM, Hall RM, Stokes HW. Multidrug resistance in Klebsiella pneumoniae: a novel gene, ramA, confers a multidrug resistance phenotype in Escherichia coli. Microbiology. 1995;141 (Pt 8):1909–1920. doi: 10.1099/13500872-141-8-1909. [DOI] [PubMed] [Google Scholar]
- 48.Chollet R, Chevalier J, Bollet C, Pages JM, Davin-Regli A. RamA is an alternate activator of the multidrug resistance cascade in Enterobacter aerogenes. Antimicrob. Agents Chemother. 2004;48:2518–2523. doi: 10.1128/AAC.48.7.2518-2523.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yassien MA, Ewis HE, Lu CD, Abdelal AT. Molecular cloning and characterization of the Salmonella enterica Serovar Paratyphi B rma Gene, which confers multiple drug resistance in Escherichia coli. Antimicrob. Agents Chemother. 2002;46:360–366. doi: 10.1128/AAC.46.2.360-366.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iben JR, Draper DE. Specific interactions of the L10(L12)4 ribosomal protein complex with mRNA, rRNA, and L11. Biochemistry. 2008;47:2721–2731. doi: 10.1021/bi701838y. [DOI] [PubMed] [Google Scholar]
- 51.Rivas E, Eddy SR. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics. 2001;2:8. doi: 10.1186/1471-2105-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 2007;10:102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 53.Brantl S. Antisense-RNA regulation and RNA interference. Biochim. Biophys. Acta. 2002;1575:15–25. doi: 10.1016/s0167-4781(02)00280-4. [DOI] [PubMed] [Google Scholar]
- 54.Duhring U, Axmann IM, Hess WR, Wilde A. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc. Natl Acad. Sci. USA. 2006;103:7054–7058. doi: 10.1073/pnas.0600927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adamski FM, McCaughan KK, Jorgensen F, Kurland CG, Tate WP. The concentration of polypeptide chain release factors 1 and 2 at different growth rates of Escherichia coli. J. Mol. Biol. 1994;238:302–308. doi: 10.1006/jmbi.1994.1293. [DOI] [PubMed] [Google Scholar]
- 56.Craigen WJ, Caskey CT. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986;322:273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- 57.Abaibou H, Pommier J, Benoit S, Giordano G, Mandrand-Berthelot MA. Expression and characterization of the Escherichia coli fdo locus and a possible physiological role for aerobic formate dehydrogenase. J. Bacteriol. 1995;177:7141–7149. doi: 10.1128/jb.177.24.7141-7149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Christman MF, Storz G, Ames BN. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc. Natl Acad. Sci. USA. 1989;86:3484–3488. doi: 10.1073/pnas.86.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bolker M, Kahmann R. The Escherichia coli regulatory protein OxyR discriminates between methylated and unmethylated states of the phage Mu mom promoter. EMBO J. 1989;8:2403–2410. doi: 10.1002/j.1460-2075.1989.tb08370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Warne SR, Varley JM, Boulnois GJ, Norton MG. Identification and characterization of a gene that controls colony morphology and auto-aggregation in Escherichia coli K12. J. Gen. Microbiol. 1990;136:455–462. doi: 10.1099/00221287-136-3-455. [DOI] [PubMed] [Google Scholar]
- 61.Pomposiello PJ, Demple B. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 2001;19:109–114. doi: 10.1016/s0167-7799(00)01542-0. [DOI] [PubMed] [Google Scholar]
- 62.Hiasa H, Marians KJ. Tus prevents overreplication of oriC plasmid DNA. J. Biol. Chem. 1994;269:26959–26968. [PubMed] [Google Scholar]
- 63.Pelletier AJ, Hill TM, Kuempel PL. Termination sites T1 and T2 from the Escherichia coli chromosome inhibit DNA replication in ColE1-derived plasmids. J. Bacteriol. 1989;171:1739–1741. doi: 10.1128/jb.171.3.1739-1741.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hill TM, Tecklenburg ML, Pelletier AJ, Kuempel PL. Tus, the trans-acting gene required for termination of DNA replication in Escherichia coli, encodes a DNA-binding protein. Proc. Natl Acad. Sci. USA. 1989;86:1593–1597. doi: 10.1073/pnas.86.5.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee EH, Kornberg A. Features of replication fork blockage by the Escherichia coli terminus-binding protein. J. Biol. Chem. 1992;267:8778–8784. [PubMed] [Google Scholar]
- 66.Lindahl T, Ljungquist S, Siegert W, Nyberg B, Sperens B. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J. Biol. Chem. 1977;252:3286–3294. [PubMed] [Google Scholar]
- 67.Varshney U, Hutcheon T, van de Sande JH. Sequence analysis, expression, and conservation of Escherichia coli uracil DNA glycosylase and its gene (ung) J. Biol. Chem. 1988;263:7776–7784. [PubMed] [Google Scholar]
- 68.Kawano M, Oshima T, Kasai H, Mori H. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol. Microbiol. 2002;45:333–349. doi: 10.1046/j.1365-2958.2002.03042.x. [DOI] [PubMed] [Google Scholar]
- 69.Pedersen K, Gerdes K. Multiple hok genes on the chromosome of Escherichia coli. Mol. Microbiol. 1999;32:1090–1102. doi: 10.1046/j.1365-2958.1999.01431.x. [DOI] [PubMed] [Google Scholar]
- 70.Brosius J, Dull TJ, Sleeter DD, Noller HF. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 71.Ma C, Simons RW. The IS10 antisense RNA blocks ribosome binding at the transposase translation initiation site. EMBO J. 1990;9:1267–1274. doi: 10.1002/j.1460-2075.1990.tb08235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beuzon CR, Marques S, Casadesus J. Repression of IS200 transposase synthesis by RNA secondary structures. Nucleic Acids Res. 1999;27:3690–3695. doi: 10.1093/nar/27.18.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Delihas N. Small mobile sequences in bacteria display diverse structure/function motifs. Mol. Microbiol. 2008;67:475–481. doi: 10.1111/j.1365-2958.2007.06068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.