Abstract
Metnase is a human protein with methylase (SET) and nuclease domains that is widely expressed, especially in proliferating tissues. Metnase promotes non-homologous end-joining (NHEJ), and knockdown causes mild hypersensitivity to ionizing radiation. Metnase also promotes plasmid and viral DNA integration, and topoisomerase IIα (TopoIIα)-dependent chromosome decatenation. NHEJ factors have been implicated in the replication stress response, and TopoIIα has been proposed to relax positive supercoils in front of replication forks. Here we show that Metnase promotes cell proliferation, but it does not alter cell cycle distributions, or replication fork progression. However, Metnase knockdown sensitizes cells to replication stress and confers a marked defect in restart of stalled replication forks. Metnase promotes resolution of phosphorylated histone H2AX, a marker of DNA double-strand breaks at collapsed forks, and it co-immunoprecipitates with PCNA and RAD9, a member of the PCNA-like RAD9–HUS1–RAD1 intra-S checkpoint complex. Metnase also promotes TopoIIα-mediated relaxation of positively supercoiled DNA. Metnase is not required for RAD51 focus formation after replication stress, but Metnase knockdown cells show increased RAD51 foci in the presence or absence of replication stress. These results establish Metnase as a key factor that promotes restart of stalled replication forks, and implicate Metnase in the repair of collapsed forks.
INTRODUCTION
Cellular systems that maintain genome stability are critical for cancer suppression. The failure to accurately repair DNA damage, including single-strand damage and double-strand breaks, is strongly linked to cancer initiation and progression. DNA damage is caused by intrinsic factors associated with cellular metabolism, such as reactive oxygen species and hydrogen peroxide, and extrinsic factors, such as ionizing radiation, UV radiation and chemotherapeutic agents including reactive chemicals, topoisomerase poisons and hydroxyurea (HU) which depletes nucleotide pools (1,2). Cells are particularly vulnerable to DNA damage during DNA replication because many DNA lesions cause replication forks to stall. Cellular responses to replication stress are extremely important in cancer therapy, as a number of chemotherapeutic drugs target DNA metabolism and cause replication stress, including topoisomerase poisons and HU. Cells respond to stalled forks in several ways. Single-stranded DNA (ssDNA) bound by RPA accumulates at stalled forks and is a major signal for downstream events including fork repair and checkpoint activation. The replisome at stalled forks is stabilized by proteins that function in DNA repair and the DNA damage checkpoint response, including RPA, ATR-ATRIP, ATM, BLM and INO80 (3–6); the action of these proteins may preserve the fork structure while the damage is repaired, allowing replication to resume. Alternatively, error-prone translesion synthesis (TLS) polymerases may be recruited to monoubiquitinated proliferating cell nuclear antigen (PCNA), allowing lesion bypass in a damage tolerance pathway (7,8). Type I and II topoisomerases play key roles in normal DNA replication. Topoisomerase I (type I) is thought to play a major role in relaxing positive supercoils produced in front of replication forks during duplex DNA unwinding by the replicative helicase. Topoisomerase IIα (TopoIIα), a type II enzyme, has roles in chromosome condensation and decatenation, is also present in the replisome, and is proposed to relax positive supercoils ahead of replication forks (9–11). Although it is known that topoisomerase poisons cause replication stress, specific roles for topoisomerases in response to replication stress have not been defined.
If stalled forks are not restarted in a timely manner, they may be converted to unusual DNA structures and collapse creating a one-ended double-strand break or ‘double-strand end’ (DSE). Certain types of damage, such as single-strand breaks, may cause direct fork collapse to DSEs. As with double-strand breaks, the checkpoint kinases ATM and ATR are recruited to DSEs and activated, leading to histone H2AX phosphorylation (γ-H2AX) in the vicinity of DSEs (12). This chromatin modification is important for fork repair and checkpoint activation, and once collapsed forks are repaired, γ-H2AX is replaced by unmodified H2AX (13–15). Homologous recombination (HR), involving RAD51-mediated strand invasion, plays a major role in restarting stalled and collapsed forks (5). NHEJ factors also play a role in cell survival after replication stress (16).
Replication stress activates the intra-S checkpoint (5). ssDNA–RPA at stalled forks is bound by ATRIP leading to activation of its obligate binding partner ATR. ATR activation depends on RAD17 (plus Rfc2-5) loading of the RAD9–HUS1–RAD1 complex (9-1-1; a scaffold and processivity factor structurally related to PCNA) through a RAD9–RPA interaction. RAD9 recruits TopBP1, an essential factor for ATR activation. ATR phosphorylates RAD17, which recruits Claspin to be phosphorylated by ATR, and phosphorylated RAD17-Claspin promotes ATR phosphorylation/activation of Chk1 kinase which phosphorylates proteins that stabilize the stalled fork and prevent late origin firing.
Metnase is a human protein that interacts with DNA ligase IV, TopoIIα, Pso4 and NBS1, and promotes NHEJ, DNA integration and TopoIIα-dependent chromosome decatenation (17–20). Metnase has SET (protein methylase) and nuclease domains. It methylates histone H3 at lysines 4 and 36, which are associated with ‘open’ chromatin and may increase accessibility of repair factors to damaged DNA. Metnase knockdown confers mild sensitivity to ionizing radiation (17). Because Metnase functions in NHEJ and regulates TopoIIα activity, we investigated whether it plays a role in replication or replication fork restart after stress. We show here that Metnase promotes cell proliferation, and cell survival after replication stress caused by HU, the topoisomerase I poison camptothecin (CPT), and UV-B. Metnase does not influence replication fork progression, but it strongly influences restart of stalled forks. Additionally, it co-immunoprecipates with PCNA and RAD9. We further show that Metnase promotes resolution of HU-induced γ-H2AX foci, and enhances TopoIIα-dependent relaxation of positively supercoiled DNA. These results establish Metnase as a key regulatory factor in the human replication stress response.
MATERIALS AND METHODS
Cell lines, RNAi-suppression of Metnase and expression of V5-tagged Metnase
Cell lines were cultured in D-MEM with 10% (v/v) fetal bovine serum supplemented with 100 U/ml penicillin and 100 µg/ml streptomycin, or 1× antimycotic/antibiotic (Invitrogen, Carlsbad, CA). Metnase was overexpressed in HEK-293 and HEK-293T cells as described (19). V5-tagged Metnase expression was confirmed by western blot with a monoclonal antibody against the V5 tag (Invitrogen). Metnase was down-regulated by transfecting cells with a pRNA/U6-Metnase RNAi vector and selecting in growth medium with 150–200 µg/ml hygromycin B (Sigma-Aldrich, St. Louis, MO), or with an Metnase shRNA vector (pRS-shMetnase). Control cells were transfected with empty pRNA/U6 or pRS-shGFP vectors. Metnase expression was measured by RT–PCR and by western blots using antibodies to native Metnase as described (17). As noted below, ‘stable’ knockdown of Metnase causes growth defects, and cell lines typically revert or cease to grow after ∼2 months. This necessitates frequent reconstruction of Metnase knockdown cell lines (designated shMet-11, 47, etc.). For each new construction, Metnase knockdown was confirmed by western blot and RT-PCR.
Cell proliferation and replication stress sensitivity assays
Cell proliferation was analyzed in triplicate in treated or mock-treated populations incubated in fully supplemented media at 37°C, 5% CO2, and at indicated times cells were harvested and counted with a Coulter counter. Cell sensitivity to CPT and HU was determined by seeding 1000 cells per 10 cm (diameter) dish in drug-free medium (to determine plating efficiency, PE), and 100 000 cells per dish in medium with CPT or HU, incubating for indicated times, then cells were rinsed with PBS, fresh growth medium was added, and cells were incubated for 12–14 days before colonies were stained with 0.1% crystal violet in methanol and counted. For UV sensitivity, cells were seeded and incubated for 24 h as above, rinsed with PBS, exposed to UVB in a biological safety cabinet equipped with a Phillips UVB fluorescent bulb, then fresh growth medium was added and cells were incubated and colonies scored. UV doses were determined by using a UVX dosimeter (UVP, Upland, CA). PE was calculated as the number of colonies divided by the number of cells plated without drug or UVB treatment. Percent survival was calculated as the number of colonies formed with drug or UVB treatment divided by the number of cells plated times the PE.
Cell cycle distributions and cell death
Cell cycle distributions were measured by fixing cells with 70% ethanol and staining with 0.2 mg/ml propidium iodide in a fresh solution containing 1% (v/v) Triton X-100 and 2 U of DNAse-free RNAse (all from Sigma-Aldrich) for 15 min at 37°C or 30 min at room temperature. Samples were analyzed using a FACScan or a FACScalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ). The percentages of cells in G1, S or G2/M phases were calculated by dividing the number of cells in each cell cycle stage by the total number of PI positive cells after normalizing to controls that were not stained with PI and converting values to percentages. Apoptosis and cell death were analyzed by flow-cytometric measurement of annexin-V expression and propidium iodide incorporation by using the Annexin V-FITC Apoptosis Detection Kit I (BD Pharmingen, San Diego, CA). All flow cytometry data were analyzed with FlowJo software (Ashland, OR, http://www.flowjo.com/).
DNA replication by BrdU incorporation and DNA fiber analysis
Log phase cells, or cells treated with 5 mM HU for 3 or 18 h were washed with PBS and released into fully supplemented D-MEM containing 10 μM BrdU. Aliquots were removed at indicated times, cells were fixed, permeabilized, and stained using the FITC BrdU Flow Kit (BD Pharmingen), and analyzed by flow cytometry as above. DNA fibers were analyzed as described (21). Briefly, cells were grown in 6-well tissue culture dishes, 20 μM IdU was added to growth medium, mixed and incubated for 10 min at 37°C. Media was removed and cells were washed with PBS, followed by a 100 μM thymidine wash. Then, cells were either treated with HU or mock treated, medium was replaced with fresh medium containing 20 μM CldU and cells were incubated for 20 min at 37°C. Cells were harvested, resuspended in PBS, 2500 cells were transferred to a positively charged microscope slide (Superfrost/Plus, Daigger, Vernon Hills, IL), lysed with 6 μl of 0.5% SDS, 200 mM Tris–HCl, pH 7.4, 50 mM EDTA and incubated at room temperature for ∼5 min. Slides were tilted to allow DNA to spread via gravity, covered with aluminum foil, air-dried for 8 min, fixed for 5 min with 3:1 methanol:acetic acid (prepared fresh), air dried for 8 min, and stored in 70% ethanol at 4°C overnight. Slides were deproteinized in 2.5 N HCl at 37°C for 1 h, blocked with 5% BSA and labeled sequentially for 1 h each with mouse anti-BrdU antibody (BD Biosciences, San Jose, CA), secondary goat anti-mouse Alexa 568 (Invitrogen), rat anti-BrdU (Accurate Chemical, Westbury, NY) and secondary donkey anti-rat Alexa 488 (Invitrogen). Slides were mounted in PermaFluor aqueous, self-sealing mounting medium (Thermoscientific, Waltham, MA). DNA fibers were visualized using an LSM 510 confocal microscope (Zeiss, Thornwood, NY) optimized for each Alexa dye. Images were analyzed using Zeiss LSM Image Browser software.
Analysis of γ-H2AX and RAD51 foci
Cells were treated with 10 mM HU for indicated times in fully supplemented D-MEM, released into fully supplemented D-MEM for indicated times, harvested, cytospun, fixed, and processed for immunofluorescence microscopy with γ-H2AX or RAD51 primary antibodies and appropriate secondary antibodies as described previously (22,23). Images were obtained with a Zeiss 510 inverted microscope or a Radiance 2100 inverted confocal microscope (BioRad, Hercules, CA) fitted with appropriate filter sets for DAPI and FITC/Alexa fluors. Images were optimized consistently with Photoshop or ImageJ software (http://rsb.info.nih.gov/ij/).
Protein immunoprecipitation
Protein samples were pre-treated with 4 U of DNaseI, incubated at 37°C for 10 min, immunoprecipitated using 0.5–5 mg of protein and antibodies to V5, PCNA (Abcam, Cambridge, MA), RAD9 (Abcam), or TopoIIα (TopoGEN, Port Orange, FL), samples were incubated overnight at 4°C, then 25 μl of A/G (1:1) agarose beads (Invitrogen) were added and incubated for 1 h at 4°C, centrifuged at 300 × g for 2 min at 4°C. Supernatants were removed and beads were washed four times with M-PER buffer (Thermoscientific). Beads were centrifuged at 300 × g for 2 min at 4°C, boiled for 10 min, and centrifuged at 300 × g for 2 min at 4°C. The supernatants were transferred to new tubes, samples were boiled for 10 min, separated by SDS–PAGE, and analyzed by western blotting.
Relaxation of positive supercoiled DNA
Positively supercoiled DNA was prepared as described (9). Positive supercoil relaxation reactions were performed in a total volume of 20 μl in 10 mM Tris–HCl, pH 7.9, 175 mM KCl, 0.1 mM EDTA, 5 mM MgCl2, 2.5% glycerol, 0.5 mM ATP (USB Co., Cleveland, OH), 2 U TopoIIα, 180 ng Metnase (when noted) and 0.3 μg DNA. Aliquots were removed at indicated times and reactions were stopped with 4 μl of 0.77% SDS, 77 mM EDTA. Products were separated on 1% agarose gels and densitometry was performed using Image J software. Background values were subtracted from signals, resulting values were normalized to signals at initial time points, and plotted as function of time in two independent experiments.
RESULTS
Metnase promotes cell proliferation
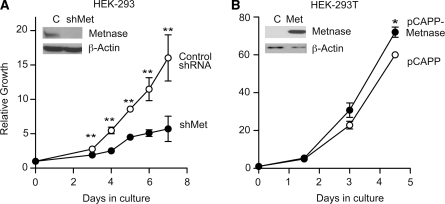
Metnase is expressed in a wide variety of human tissues (17) and in all human cell lines tested, except those transformed by T-antigen such as HEK-293T cells (unpublished results). Overexpression of Metnase in HEK-293T increases cell proliferation (19). HEK-293 cells express Metnase, but not T-antigen, and stable shRNA knockdown of Metnase in HEK-293 cells significantly reduced cell proliferation rate compared to control cells (Figure 1A). We have shown previously and confirmed in this study that Metnase overexpression in HEK-293T increases cell proliferation (Figure 1B). Moreover, cells stably transfected with Metnase shRNA vectors either cease to proliferate after 2–3 months or revert to normal. These results indicate that Metnase promotes proliferation of human cells, and suggest that Metnase is very important for growth of human cells that do not express T antigen.
Figure 1.
Metnase promotes cell proliferation. (A) Cell growth was monitored in HEK-293 cells transfected with shGFP (control) or shMetnase vectors. (B) Cell growth was monitored in HEK-293-T cells, which do not normally express Metnase, transfected with the pCAPP-Metnase expression vector or empty pCAPP. Plotted are averages (±SD) of two to three determinations per time-point. *P < 0.05, **P < 0.01. Metnase expression is shown in representative western blots with β-actin loading control (insets). Cell growth was measured by harvesting cells at indicated times and counting cells with a Coulter counter.
Metnase promotes cell survival and DNA replication after replication stress
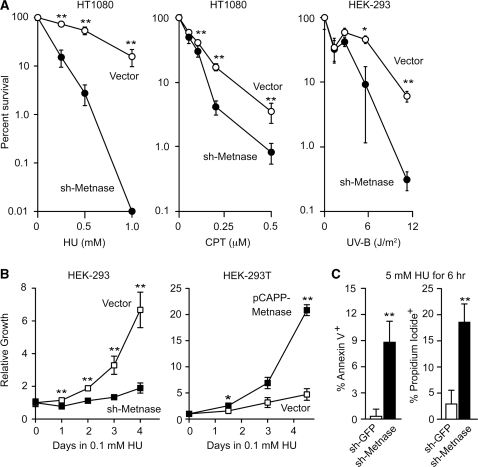
The effect of Metnase on cell proliferation, coupled with its DNA repair properties and functional interaction with TopoIIα (17,19), suggested that Metnase may have a role in replication and/or rescuing cells from replication stress at sites of spontaneous DNA damage. We therefore tested whether Metnase regulates sensitivity to replication stress induced by HU, CPT and UV-B (Figure 2A). Metnase knockdown sensitized HT1080 cells to 1 mM HU by more than 1000-fold (P = 0.01), and to 0.2–0.5 µM CPT by nearly 10-fold (P ≤ 0.011) (all statistical analyses in this study were performed by using t-tests). Metnase knockdown sensitized HEK-293 cells to a UV-B dose of 11.2 J/m2 by nearly 20-fold (P = 0.007). When cultured in a low concentration of HU (0.1 mM), HEK-293 cells proliferated at a slow rate, but Metnase knockdown cells showed almost no proliferative capacity; this effect specifically reflects the Metnase defect since Metnase overexpression in HEK-293T significantly enhanced proliferation under these conditions (Figure 2B). The hypersensitivity of Metnase knockdown cells to replication stress reflects, at least in part, enhanced cell death via apoptosis, as shown by the nearly 30-fold increase in the apoptosis marker annexin V, and >6-fold increase in inviable cells (unable to exclude propidium iodide) (both P < 0.005). The marked sensitivity of Metnase knockdown cells to replication stress contrasts with their mild sensitivity to ionizing radiation (17).
Figure 2.
Metnase promotes cell survival after DNA replication stress. (A) Average percent cell survival (± SD) after HU, CPT or UV-B treatments measured as relative plating efficiency for HT1080 or HEK-293 cells stably transfected with control or shRNA-Metnase vectors. Data are from two to three independent experiments per condition; *P = 0.0127, **P ≤ 0.01. (B) Average growth rates (±SD) of control HEK-293 and sh-Metnase knockdown cells, and control HEK-293T or Metnase overexpression cells in medium containing 0.1 mM HU; data are from two to three independent experiments per cell line. (C) HEK-293 control or Metnase knockdown cells were treated with 5 mM HU for 6 h and the percentages of cells expressing annexin V or incorporating propidium iodide were determined by flow cytometry. Values are averages (±SD) from three independent experiments.
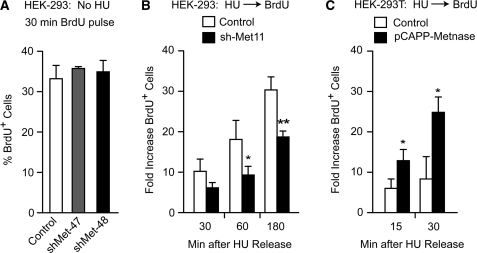
To investigate the mechanism by which Metnase promotes cell proliferation and resistance to replication stress, we tested whether Metnase expression level influenced DNA replication by measuring BrdU incorporation and cell cycle distributions by flow cytometry, in unstressed cells and after release from replication stress. In log phase (untreated) HEK-293 cells, Metnase knockdown had no effect on BrdU incorporation during a 30 min incubation (Figure 3A). However, when HEK-293 Metnase knockdown cells were pretreated with 5 mM HU for 3 h and then released into BrdU medium, BrdU incorporation was significantly reduced (∼2-fold, Figure 3B). The opposite effect was seen with Metnase overexpression in HEK-293T cells treated with HU for 18 h as BrdU incorporation was significantly increased (Figure 3C). Although neither over- nor underexpression of Metnase significantly affected cell cycle distributions of unstressed cells (Supplementary Figure S1A), when treated with 5 mM HU for 18 h and released, HEK-293T cells overexpressing Metnase entered S-phase more rapidly than control cells (seen 1 h after release from HU), and entered G2 phase more rapidly (seen 7 h after release from HU) (Supplementary Figure S1B). Somewhat stronger effects were seen when Metnase was overexpressed in HEK-293 cells (Supplementary Figure S1C); this may reflect the fact that HEK-293T cells show robust proliferation even though they do not express Metnase. When HEK-293 Metnase knockdown cells were treated with 5 mM HU for 18 h and released, the opposite effect was seen. In two independent knockdown cell lines, there were marked accumulations of S phase cells 10 and 18 h after release from HU (Supplementary Figure S1D), indicating that Metnase knockdown prolongs S phase after replication stress. These results support the idea that Metnase promotes DNA replication in cells recovering from replication stress.
Figure 3.
Metnase promotes DNA replication after release from replication stress. (A) Log phase HEK-293 cells expressing normal or low levels of Metnase were incubated with 10 µM BrdU for 30 min and average percentages (±SD) of BrdU+ cells are shown for two determinations per strain. (B) HEK-293 control and Metnase knockdown cells were treated with 5 mM HU for 3 h and released into medium with 10 µM BrdU. Average fold increases (±SD) in the percentage of BrdU+ cells relative to untreated cells (no HU, no BrdU) are plotted for three independent experiments per cell line. *P = 0.042, **P = 0.0047. (C) BrdU incorporation after HU release from HEK-293T control and Metnase overexpression cells as in panel B, except cells were treated with HU for 18 h; *P ≤ 0.03.
Metnase promotes replication fork restart
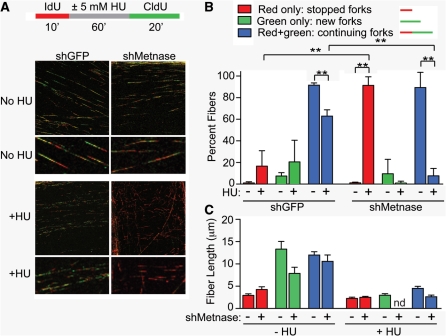
To gain a better understanding of the role of Metnase in replication and the replication stress response, we analyzed replication fork restart, new origin firing, and replication speed by using DNA fiber analysis. Log phase HEK-293 cells stably transfected with vectors expressing shRNA targeting Metnase, or GFP as control, were labeled with IdU for 10 min, then incubated with or without 5 mM HU for 1 h, briefly washed with thymidine and then incubated with CldU for 20 min. Cells were lysed on glass slides and DNA fibers were stretched by gravity, fixed, IdU was stained red and CldU was stained green, and DNA fibers were quantified using confocal-microscopy (Figure 4A and B). In untreated control cells, ∼90% of fibers showed adjacent red–green signals indicative of continuing forks. Typically, we did not observe gaps between red and green signals on individual fibers in controls that were not treated with HU, perhaps because replication is slowed by media changes, which included a thymidine wash. Approximately 10% of fibers had only green signals indicating forks that initiated after IdU was removed (‘new forks’). When control cells were treated with HU, continuing forks (those that stalled and restarted) were moderately reduced to ∼65% (P = 0.0014), new forks that initiated after HU treatment showed a slight but not statistically significant increase to ∼20%, and ∼15% of forks stopped and failed to restart. The pattern observed with untreated Metnase knockdown cells was similar to untreated wild-type cells, with predominantly continuing forks and a small percentage of new forks. Strikingly, when Metnase knockdown cells were treated with HU, the percentage of stopped forks greatly increased (to ∼90%) and there was a corresponding large decrease in the percentage of continuing forks (both P ≤ 0.0008). New forks were extremely rare in HU treated Metnase knockdown cells, however, new forks are also rare in untreated cells, and the decrease with HU treatment was not statistically significant (P = 0.3). These results provide direct evidence that Metnase plays a critical role in restarting stalled replication forks, and further suggest that Metnase may regulate new origin firing when cells experience replication stress.
Figure 4.
Metnase promotes replication fork restart. (A) IdU and CldU labeling scheme is shown above representative confocal microscope images of DNA fibers, with IdU stained red and CldU stained green. (B) Quantification of fiber types. At least 150 fibers were scored per treatment, per cell line for each of three experiments; **P ≤ 0.0014. (C) Fiber lengths were measured by using LSM 510 Image Browser software. Plotted are averages (±SD) of triplicate experiments in which 150–500 fibers were scored per treatment, per experiment. nd, none detected.
To determine whether Metnase regulates the speed of replication, we measured average fiber lengths. As expected, red fibers were shorter than green since cells were treated with IdU (red) for 10 min and CldU (green) for 20 min. Fibers were longer in unstressed cells than after HU treatment (Figure 4C). However, Metnase had no effect on fiber lengths in either HU treated or untreated cultures. We conclude that Metnase regulates the efficiency of replication fork restart, and possibly initiation after replication stress, but it has no effect on the speed of ongoing forks.
Metnase promotes resolution of γ-H2AX induced by replication stress
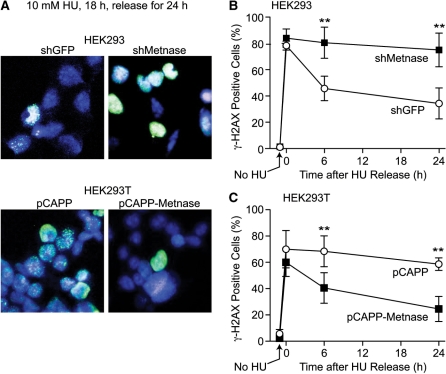
Replication stress causes fork collapse to DSEs marked by phosphorylation of histone H2AX to γ-H2AX. Elimination of the γ-H2AX signal over time reflects DSE/fork repair. Metnase and classical NHEJ proteins promote survival after replication stress and influence replication fork restart (21,24–26) (this study), and Metnase promotes NHEJ and interacts with the key NHEJ protein DNA LigIV (17,18). We therefore tested whether Metnase influences resolution of HU-induced γ-H2AX by treating cells with 10 mM HU for 18 h, then releasing into normal growth medium and examining γ-H2AX by immunofluorescence microscopy. Because HEK-293 and HEK-293T cells used in these experiments adhere poorly, cells were cytospun prior to fixation and immunocytochemical staining. For this reason, individual γ-H2AX foci are not always detectable. Instead γ-H2AX signals typically appear as diffuse nuclear staining and cells were scored as either γ-H2AX positive or negative (representative images are shown in Figure 5A). Consistent with the enhanced sensitivity of Metnase knockdown cells to HU, γ-H2AX persisted longer in the knockdown cells, with significant differences from controls at both 6 and 24 h after release from HU (Figure 5B, P < 0.0001). Similarly, overexpression of Metnase in HEK-293T cells accelerated the resolution of γ-H2AX signals (Figure 5C, P ≤ 0.0055). Note that in all four cell lines, similar percentages of cells were γ-H2AX positive at the end of the 18 h HU treatment. These results indicate that Metnase promotes resolution of γ-H2AX after cells are released from replication stress, but Metnase does not prevent fork collapse to DSEs over the course of this relatively long HU treatment.
Figure 5.
Metnase promotes resolution of replication stress-induced γ-H2AX. (A) Representative confocal microscope images of HEK-293 and HEK-293T cells over- or under-expressing Metnase were treated with 10 mM HU for 18 h and released into growth medium for 24 h. Aliquots of cells were removed at indicated times, cytospun, stained with DAPI (blue) and antibodies to γH2AX (green) and imaged by confocal microscopy. (B, C) Percentage of γ-H2AX positive cells among total DAPI stained cells. An average of >190 cells were counted per slide, 10 slides per experiment. Values are averages (±SD); **P ≤ 0.0055.
Metnase is not required for replication stress-induced RAD51 focus formation
HR is thought to play a major role in resolving stalled and collapsed replication forks. We showed previously that Metnase does not affect HR repair of chromosomal DSBs induced by I-SceI nuclease (17). To determine whether Metnase influences fork restart/repair by affecting RAD51 filament formation we examined RAD51 foci in HEK-293 cells stably transfected with control (shGFP) or Metnase knockdown (shMet) vectors, and treated with 10 mM HU for 4 h or mock treated (Supplementary Figure S2). As expected, control cells showed increased RAD51 foci after HU, reflected as statistically significant (3–5-fold) increases in the percentage of cells with at least one RAD51 focus, or >5 foci. Interestingly, two independent Metnase knockdown cell lines showed high percentages of foci-positive cells (and cells with >5 foci) in the absence of HU, but foci were not further increased by HU treatment. Although the percentage of RAD51 foci-positive cells increased with HU in control cells, and was high in HU-treated and untreated Metnase knockdown cells, the average number of RAD51 foci per cell among cells with foci was not affected by HU treatment or Metnase expression level (Supplementary Figure S2C). These results indicate that Metnase is not required for RAD51 focus formation after replication stress, that Metnase suppresses RAD51 focus formation in unstressed cells, and that cells produce a limited number of RAD51 foci when experiencing replication stress from endogenous DNA damage, HU, or Metnase knockdown (see ‘Discussion’ section).
Metnase co-immunoprecipitates with PCNA and RAD9
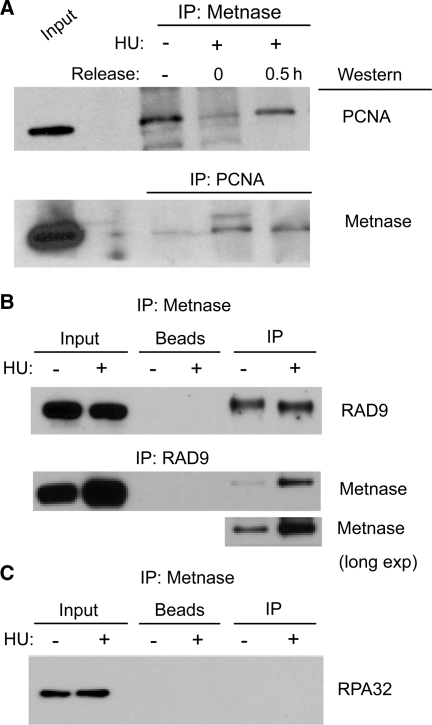
Because Metnase is involved in the replication stress response, we explored its interactions with proteins at the replication fork. PCNA is a key scaffold protein that mediates binding of numerous proteins in the replisome and promotes replication processivity (7). Metnase co-immunoprecipitated with PCNA, and vice versa, in unstressed cells and after treatment with HU (Figure 6A). PCNA-interacting proteins share a conserved binding motif, the PIP box. Metnase has a highly conserved PIP box (Supplementary Table S1) suggesting that it directly interacts with PCNA. Interestingly, Metnase also co-immunoprecipitated with RAD9, a member of the 9-1-1 complex that is structurally and functionally related to PCNA, and that is recruited to stalled and/or collapsed replication forks (Figure 6B). Although this interaction appeared stronger when RAD9 was immunoprecipitated from HU treated cells, a similar enhancement was not seen with HU treatment when Metnase was immunoprecipitated. Metnase did not co-immunoprecipitate with the 32 kDa subunit of RPA (Figure 6C), indicating that Metnase is present within the replisome, but is not closely associated with ssDNA at stalled forks. These results indicate that Metnase is closely associated with replication stress factors that control TLS, fork processing via HR mechanisms, and checkpoint signaling.
Figure 6.
Metnase interacts with PCNA and RAD9, but not RPA32. (A) Reciprocal co-immunoprecipitation of V5-tagged Metnase and native PCNA from cells treated with 5 mM HU for 18 h, tested immediately or 30 min after release from HU, or untreated. Input represents 0.5% of immunoprecipitation. Results are representative of at least three independent experiments. (B and C) Co-immunoprecipitation of V5-tagged Metnase with native RAD9 and native RPA as in panel A, except HU treated cells were only tested immediately after treatment.
Metnase interacts with TopoIIα and promotes TopoIIα-dependent relaxation of positively supercoiled plasmid DNA
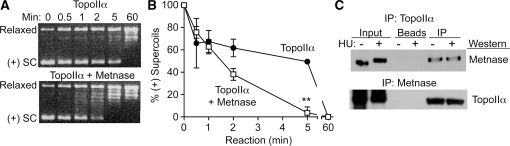
Metnase interacts with TopoIIα and promotes TopoIIα-dependent chromosome decatenation (19). TopoIIα is present in the replisome (11) and has been implicated in DNA replication through relaxation of positive supercoils that accumulate ahead of replication forks (9). We found that Metnase significantly enhanced TopoIIα-dependent relaxation of positive supercoils during a 5 min time course, but Metnase was not required to achieve full relaxation within an hour (Figure 7A and B). To gain insight into whether Metnase functions in the replication stress response through its interaction with TopoIIα, we tested whether the interaction between Metnase and TopoIIα was affected by replication stress. HEK-293 cells were treated with 5 mM HU for 3 h, and cell extracts were prepared and analyzed by co-immunoprecipitation of Metnase and TopoIIα. As shown in Figure 7C, Metnase and TopoIIα show a robust interaction regardless of which protein was immunoprecipitated, but this interaction was not affected by HU treatment. These results suggest that Metnase interaction with TopoIIα may promote TopoIIα processing of DNA structures in front of replication forks.
Figure 7.
Metnase interacts with TopoIIα and stimulates relaxation of positive supercoils. (A) Predominantly positively-supercoiled plasmid DNA samples were treated with TopoIIα (2 U) with or without Metnase (180 ng) for indicated times, and topological forms were detected on ethidium bromide stained agarose gels. (B) Gel images were scanned and the percentage of positively-supercoiled DNA remaining at each time point was quantitated. Values are averages (±SD) of two determinations per condition, normalized to 100% at t = 0; **P = 0.007. (C) Co-immunoprecipitation of V5-tagged Metnase and native TopoIIα; data presented as in Figure 6B.
DISCUSSION
Although Metnase appeared very late in evolution, in anthropoid primates (27), it influences several important aspects of DNA metabolism including NHEJ, DNA integration, chromosome translocation and chromosome decatenation (17–20,28). Through interaction with TopoIIα, it regulates cellular resistance to common chemotherapeutics (22,29). The present study establishes another important role for Metnase in the replication stress response. Given how late Metnase appeared in evolution, it is not surprising that it does not influence replication fork progression. Instead, Metnase functions during replication stress since Metnase affected BrdU incorporation, S phase progression and fork restart by DNA fiber analysis only when cells were subjected to replication stress (Figures 3 and 4, Supplementary Figure S1). Metnase knockdown conferred a marked defect in fork restart during a 20 min period after a brief (1 h) HU treatment (Figure 4). This result indicates that Metnase plays a key role in restarting stalled forks, because the brief HU treatment causes mainly fork stalling. Also, when forks collapse, restart is largely dependent on HR, an inherently slow process that involves RAD51 replacement of RPA on ssDNA, and strand invasion of sister chromatids by RAD51 filaments (5). When cells were subjected to longer periods of replication stress, Metnase promoted resolution of γ-H2AX (Figure 5), which marks collapsed forks. This indicates that Metnase also promotes repair of collapsed forks. Another late-evolving protein that functions in replication fork restart is PARP-1. PARP-1 is not found in yeast, but is present in higher eukaryotes. PARP-1 recruits the ancient DNA repair endonuclease MRE11 to stalled forks, which is proposed to process structures at stalled forks, leading to RPA recruitment and eventual restart via HR (30).
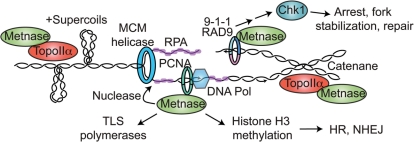
Metnase might promote replication fork restart/repair in a variety of ways, as illustrated in Figure 8. Metnase promotes NHEJ (17) and other factors involved in NHEJ are known to promote cell survival after replication stress (16,25). NHEJ factors might promote rejoining of DSEs at collapsed forks, but it seems that this type of repair would be highly inaccurate (and genome destabilizing) since each collapsed fork produces only a single broken end. It is possible that NHEJ factors promote fork restart indirectly through interactions with HR factors (23). When replication forks stall, the initial cellular response is to stabilize the replisome to prevent fork collapse. Metnase does not appear to play a role in fork stabilization over an extended period of replication stress, because altering Metnase levels had no effect on the percentage of cells with collapsed forks (γ-H2AX positive) after an 18 h HU treatment (Figure 5). Although it is clear that Metnase promotes resolution of γ-H2AX, further studies are required to determine whether this reflects enhanced repair of collapsed forks via NHEJ, HR, or other mechanisms such as fork rescue from adjacent replicons. It is also possible that the Metnase nuclease plays a role in fork processing, as with Mus81 (31).
Figure 8.
Potential roles of Metnase in the replication stress response. See text for details.
Metnase is not required for HR stimulated by I-SceI nuclease-induced DSBs (17) nor for RAD51 focus formation in response to replication stress (Supplementary Figure S2). Thus, Metnase does not affect RAD51 filament formation, which involves RAD51 replacement of RPA on ssDNA; this is consistent with the lack of interaction between Metnase and RPA (Figure 6C). Metnase knockdown cells show high frequencies of RAD51 foci, but foci do not increase upon treatment with HU. This suggests that Metnase knockdown itself is a form of replication stress, consistent with the slow growth phenotype (Figure 1A), and that RAD51 focus formation is somehow limited, for example, because of RAD51 availability, or because multiple forks are sequestered in a limited number of ‘repair centers’ (32,33). The repair center concept can also account for the similar number of RAD51 foci per cell with foci in HU treated or untreated wild-type and Metnase knockdown cells (Supplementary Figure S2C).
Metnase could promote fork restart through its interactions with the replisome factors PCNA and RAD9. Although it is not yet known whether Metnase interacts directly with these proteins, the fact that the Metnase SET domain has a conserved PIP box is highly suggestive of direct interactions with these related proteins. Regardless, our results clearly place Metnase in the vicinity of stalled replication forks. The Metnase SET domain encodes a protein methylase, and Metnase is known to methylate histone H3 and itself (17,19). Metnase could regulate PCNA and/or RAD9 function by methylating these proteins. PCNA regulates TLS through direct interactions with TLS polymerases (7), thus Metnase may enhance fork restart after UV by enhancing TLS at UV lesions.
RAD9 has well-established roles in the intra-S checkpoint response (5). Metnase could promote fork restart by influencing checkpoint activation; it may also affect downstream checkpoint-dependent processes such as inhibition of origin firing. We are currently investigating the effects of Metnase on checkpoint factors downstream of RAD9, including Chk1. Metnase is not required for the p53/Chk2 arm of the DNA damage checkpoint response since replication stress-induced cell death in Metnase knockdown cells shows a robust apoptotic response (Figure 2C). Metnase may have a more general role in fork restart through chromatin modification in the vicinity of stalled and collapsed forks. It is noteworthy that Metnase methylates histone H3 lysines 4 and 36, which are specifically associated with ‘open’ chromatin (17). Thus, Metnase could promote fork restart by enhancing access of repair factors to stalled and collapsed forks.
Finally, Metnase could influence fork restart through its direct interaction with TopoIIα, another factor within the mammalian replisome (11). TopoIIα is proposed to relax positive supercoils that form ahead of replication forks (9). When replication forks stall, the MCM helicase complex can continue to unwind duplex DNA, uncoupled from the replicative polymerases, producing excess ssDNA that is bound by RPA and triggering the intra-S checkpoint (5,34). Continued DNA unwinding by MCM will also increase positive supercoiling that may drive unusual DNA structures at stalled forks (34). By enhancing TopoIIα-dependent relaxation of positive supercoils, Metnase could promote a favorable topological state that results in timely fork restart, particularly when unusual structures form, such as ‘chicken feet’, since the resolution of such structures is probably dependent on the topology of the stalled fork. Local topology could also influence restart of collapsed forks since HR-mediated invasion of broken ends into sister chromatids requires unwinding of the sister duplex. Note that the models described above are not mutually exclusive: Metnase may have different roles depending on the specific structures at stalled or collapsed replication forks.
Prior studies have established that Metnase is highly expressed in actively proliferating tissues (17). We have recently shown that Metnase is frequently overexpressed in leukemia and breast cancer cell lines, and importantly, down-regulating Metnase greatly enhances tumor cell sensitivity to common chemotherapeutics including epididophylotoxins and anthracyclines (22,29). The current study establishes Metnase as a critical factor in the replication stress response. Metnase is therefore an excellent target for therapeutic strategies that block DNA synthesis, or that exploit defects of tumor cells in replication fork restart (35,36), and it may thus prove to be an important target in the treatment of a wide variety of tumor types.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health [R01 CA100862 and R01 GM084020 to J.A.N., R01 HL093606 to R.H., R01 GM033944 to N.O., F31 CA132628 to L.P.D.H., and A.C.G. was supported by T32 CA09582]. Images in this article were generated in the University of New Mexico Cancer Center Fluorescence Microscopy Facility which received support from the National Institutes of Health [P30 CA118100]; National Science Foundation [MCB9982161]; National Center for Research Resources [S10 RR14668, P20 RR11830, S10 RR19287, S10 RR016918]; University of New Mexico Health Sciences Center, and the University of New Mexico Cancer Center. Flow cytometry data was generated in the Flow Cytometry Shared Resource Center supported by the University of New Mexico Health Sciences Center and the University of New Mexico Cancer Center. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Miret Aladjem and Amanda Wraith Kijas for assistance in establishing the DNA fiber assay.
REFERENCES
- 1.Hanks S, Coleman K, Reid S, Plaja A, Firth H, FitzPatrick D, Kidd A, Méhes K, Nash R, Robin N, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat. Genet. 2004;36:1159–1161. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 2.Burma S, Chen BP, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair. 2006;5:1042–1048. doi: 10.1016/j.dnarep.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 3.Zou Y, Liu Y, Wu X, Shell SM. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J. Cell. Physiol. 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimada K, Oma Y, Schleker T, Kugou K, Ohta K, Harata M, Gasser SM. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr. Biol. 2008;18:566–575. doi: 10.1016/j.cub.2008.03.049. [DOI] [PubMed] [Google Scholar]
- 5.Budzowska M, Kanaar R. Mechanisms of dealing with DNA damage-induced replication problems. Cell Biochem. Biophys. 2009;53:17–31. doi: 10.1007/s12013-008-9039-y. [DOI] [PubMed] [Google Scholar]
- 6.Davies SL, North PS, Hickson ID. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat. Struct. Mol. Biol. 2007;14:677–679. doi: 10.1038/nsmb1267. [DOI] [PubMed] [Google Scholar]
- 7.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Niimi A, Brown S, Sabbioneda S, Kannouche PL, Scott A, Yasui A, Green CM, Lehmann AR. Regulation of proliferating cell nuclear antigen ubiquitination in mammalian cells. Proc. Natl Acad. Sci. USA. 2008;105:16125–16130. doi: 10.1073/pnas.0802727105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McClendon AK, Rodriguez AC, Osheroff N. Human topoisomerase IIalpha rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J. Biol. Chem. 2005;280:39337–39345. doi: 10.1074/jbc.M503320200. [DOI] [PubMed] [Google Scholar]
- 10.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 11.Jiang HY, Hickey RJ, Abdel-Aziz W, Tom TD, Wills PW, Liu J, Malkas LH. Human cell DNA replication is mediated by a discrete multiprotein complex. J. Cell. Biochem. 2002;85:762–774. doi: 10.1002/jcb.10182. [DOI] [PubMed] [Google Scholar]
- 12.Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 2001;276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- 13.Chanoux RA, Yin B, Urtishak KA, Asare A, Bassing CH, Brown EJ. ATR and H2AX cooperate in maintaining genome stability under replication stress. J. Biol. Chem. 2008;284:5994–6003. doi: 10.1074/jbc.M806739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Downey M, Durocher D. γH2AX as a checkpoint maintenance signal. Cell Cycle. 2006;5:1376–1381. doi: 10.4161/cc.5.13.2899. [DOI] [PubMed] [Google Scholar]
- 15.Keogh MC, Kim JA, Downey M, Fillingham J, Chowdhury D, Harrison JC, Onishi M, Datta N, Galicia S, Emili A, et al. A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- 16.Lundin C, Erixon K, Arnaudeau C, Schultz N, Jenssen D, Meuth M, Helleday T. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol. Cell. Biol. 2002;22:5869–5878. doi: 10.1128/MCB.22.16.5869-5878.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SH, Oshige M, Durant ST, Rasila KK, Williamson EA, Ramsey H, Kwan L, Nickoloff JA, Hromas R. The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair. Proc. Natl Acad. Sci. USA. 2005;102:18075–18080. doi: 10.1073/pnas.0503676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hromas R, Wray J, Lee SH, Martinez L, Farrington J, Corwin LK, Ramsey H, Nickoloff JA, Williamson EA. The human set and transposase domain protein Metnase interacts with DNA Ligase IV and enhances the efficiency and accuracy of non-homologous end-joining. DNA Repair. 2008;7:1927–1937. doi: 10.1016/j.dnarep.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williamson EA, Rasila KK, Corwin LK, Wray J, Beck BD, Severns V, Mobarak C, Lee SH, Nickoloff JA, Hromas R. The SET and transposase domain protein Metnase enhances chromosome decatenation: regulation by automethylation. Nucleic Acids Res. 2008;36:5822–5831. doi: 10.1093/nar/gkn560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beck BD, Park SJ, Lee YJ, Roman Y, Hromas RA, Lee SH. Human PSO4 is a Metnase (SETMAR) binding partner that regulates Metnase function in DNA repair. J. Biol. Chem. 2008;283:9023–9030. doi: 10.1074/jbc.M800150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimura T, Torres MJ, Martin MM, Rao VA, Pommier Y, Katsura M, Miyagawa K, Aladjem MI. Bloom's syndrome helicase and Mus81 are required to induce transient double-strand DNA breaks in response to DNA replication stress. J. Mol. Biol. 2008;375:1152–1164. doi: 10.1016/j.jmb.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wray J, Williamson EA, Fnu S, Lee S.-H, Libby E, Willman CL, Nickoloff JA, Hromas R. Metnase mediates chromosome decatenation in acute leukemia cells. Blood. 2009;114:1852–1858. doi: 10.1182/blood-2008-08-175760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shrivastav M, Miller CA, De Haro LP, Durant ST, Chen BP, Chen DJ, Nickoloff JA. DNA-PKcs and ATM co-regulate DNA double-strand break repair. DNA Repair. 2009;8:920–929. doi: 10.1016/j.dnarep.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnaudeau C, Tenorio Miranda E, Jenssen D, Helleday T. Inhibition of DNA synthesis is a potent mechanism by which cytostatic drugs induce homologous recombination in mammalian cells. Mutat. Res. 2000;461:221–228. doi: 10.1016/s0921-8777(00)00052-5. [DOI] [PubMed] [Google Scholar]
- 25.Arnaudeau C, Lundin C, Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 2001;307:1235–1245. doi: 10.1006/jmbi.2001.4564. [DOI] [PubMed] [Google Scholar]
- 26.Shao RG, Cao CX, Zhang H, Kohn KW, Wold MS, Pommier Y. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 1999;18:1397–1406. doi: 10.1093/emboj/18.5.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cordaux R, Udit S, Batzer MA, Feschotte C. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc. Natl Acad. Sci. USA. 2006;103:8101–8106. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wray J, Williamson EA, Farrington J, Chester S, Kwan L, Weinstock D, Jasin M, Lee S.-H, Nickoloff JA, Hromas R. The transposase domain protein Metnase/SETMAR suppresses chromosomal translocations. Cancer Genet. Cytogenet. 2010 doi: 10.1016/j.cancergencyto.2010.04.011. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wray J, Williamson EA, Royce M, Shaheen M, Beck BD, Lee SH, Nickoloff JA, Hromas R. Metnase mediates resistance to topoisomerase II inhibitors in breast cancer cells. PLoS ONE. 2009;4:e5323. doi: 10.1371/journal.pone.0005323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28:2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 2007;14:1096–1104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- 32.Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl Acad. Sci. USA. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wray J, Liu J, Nickoloff JA, Shen Z. Distinct RAD51 associations with RAD52 and BCCIP in response to DNA damage and replication stress. Cancer Res. 2008;68:2699–2707. doi: 10.1158/0008-5472.CAN-07-6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forsburg SL. The MCM helicase: linking checkpoints to the replication fork. Biochem. Soc. Trans. 2008;36:114–119. doi: 10.1042/BST0360114. [DOI] [PubMed] [Google Scholar]
- 35.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 36.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.