Abstract
Ku or DNA ligase 4-independent alternative end joining (alt-EJ) repair of DNA double-strand breaks (DSBs) frequently correlates with increased junctional microhomology. However, alt-EJ also produces junctions without microhomology (apparent blunt joins), and the exact role of microhomology in both alt-EJ and classical non-homologous end joining (NHEJ) remains unclear. To better understand the degree to which alt-EJ depends on annealing at pre-existing microhomologies, we examined inaccurate repair of an I-SceI DSB lacking nearby microhomologies of greater than four nucleotides in Drosophila. Lig4 deficiency affected neither frequency nor length of junctional microhomology, but significantly increased insertion frequency. Many insertions appeared to be templated. Based on sequence analysis of repair junctions, we propose a model of synthesis-dependent microhomology-mediated end joining (SD-MMEJ), in which de novo synthesis by an accurate non-processive DNA polymerase creates microhomology. Repair junctions with apparent blunt joins, junctional microhomologies and short indels (deletion with insertion) are often considered to reflect different repair mechanisms. However, a majority of each type had structures consistent with the predictions of our SD-MMEJ model. This suggests that a single underlying mechanism could be responsible for all three repair product types. Genetic analysis indicates that SD-MMEJ is Ku70, Lig4 and Rad51-independent but impaired in mus308 (POLQ) mutants.
INTRODUCTION
DNA double-strand breaks (DSBs) are a diverse set of molecular lesions, requiring a complex and flexible repertoire of DSB repair processes. DSB repair mechanisms are broadly classified as homologous recombination (HR) or end joining (EJ) (1). Unlike most HR processes, which consult an external template, end-joining ligates the DNA ends without regard to the original sequence and is therefore potentially mutagenic. Inaccurate end-joining repair causes both local sequence alterations and large-scale changes in genome architecture (2).
Eukaryotic cells possess multiple genetically distinct end-joining mechanisms. The best-characterized is commonly referred to as ‘canonical non-homologous end joining’ (C-NHEJ) (3). C-NHEJ requires two conserved ‘core’ complexes: the DNA ligase 4 (Lig4)/XRCC4 and the Ku70/80 (Ku) heterodimers. The Ku complex mediates end synapsis and recruits other repair factors (4,5). Lig4 carries out the final ligation, and may have a role in end alignment (6,7).
Eukaryotic cells also possess ‘alternative’ end-joining mechanisms (alt-EJ), which do not require Ku or Lig4 (8–11). The identity of the alt-EJ machinery is still under investigation (12–17). Severe phenotypes observed in C-NHEJ deficient cells suggest that alt-EJ may be less efficient and less accurate than C-NHEJ (18,19). Nevertheless, some DSBs may be preferentially repaired by alt-EJ (12,20). Recent studies demonstrate unexpectedly efficient alt-EJ in mice (21–24). Additionally, recent results from our lab implicate mus308, the Drosophila ortholog of vertebrate DNA polymerase theta, in alt-EJ (Chan, Yu and McVey, in revision).
Given that the core C-NHEJ components play important roles in break end synapsis, an obvious question is how DSB ends are synapsed in C-NHEJ deficient cells. EJ of DSBs with long GC-rich overhangs is less dependent on C-NHEJ (25,26), suggesting that base-pairing interactions alone can mediate end synapsis. Supporting this hypothesis, end-joining repair junctions from C-NHEJ deficient cells frequently have structures consistent with annealing at direct repeats flanking the DSB (microhomologies). It has therefore been proposed that alt-EJ proceeds via a mechanism called microhomology-mediated end joining (MMEJ) (27). In MMEJ, 5′-3′ resection or unwinding allows base-pairing across the break. This results in deletion of one half of the direct repeat, along with any intervening sequence. MMEJ therefore produces junctions with nucleotides that cannot be unambiguously assigned to either side of the break (junctional microhomologies).
The relationship between alt-EJ and MMEJ is unclear. C-NHEJ deficiency generally increases average frequency and/or length of junctional microhomology. However, microhomologies are found at both C-NHEJ and alt-EJ mediated repair junctions and alt-EJ can produce repair junctions without microhomology (21). Also, non-genetic factors such as sequence context (28) and break end structure (10,29,30) influence microhomology use. It is also unclear why some microhomologies appear to be used preferentially over others (12). MMEJ observed in budding yeast appears to be different from MMEJ observed in higher eukaryotes, both in terms of genetic requirements and microhomology length (13,27). Together, these observations suggest that there may be multiple alt-EJ mechanisms with varying degrees of microhomology dependence.
The fruit fly, Drosophila melanogaster, is one of the few model organisms in which alt-EJ can be readily observed in wild-type individuals. We have previously shown that end-joining repair of P-element-induced DSBs in Drosophila typically proceeds by MMEJ and is unaffected in lig4 mutant flies (20). Conversely, studies using the I-SceI endonuclease to induce DSBs support a role for Lig4 in Drosophila EJ (31,32). This raises the question of whether alt-EJ repair of an I-SceI DSB in flies uses an MMEJ mechanism similar to that observed at P-element DSBs or a different type of alt-EJ.
To address this question, we examined inaccurate repair junctions of a chromosomal I-SceI DSB in C-NHEJ proficient and deficient flies. Annealing at pre-existing microhomologies did not appear to be important for end-joining repair in any genetic background. Sequence analysis of repair junctions suggested a model we term synthesis-dependent MMEJ (SD-MMEJ). We show that end-joining junctions with and without microhomology, as well as junctions with insertions, have structures consistent with predictions made by our SD-MMEJ model. Thus, in this system, a single mechanism accounts for multiple classes of end-joining repair products usually thought to be produced by different mechanisms.
MATERIALS AND METHODS
Fly stocks and crosses
Flies were maintained according to standard methods on cornmeal-agar medium in a 25° incubator on a 12 h light–dark cycle.
The [Iw]7 construct (33) was used in all experiments. The [Iw]7 construct is a P-element integrated on chromosome 2 at 53F8 containing the 18 bp I-SceI site (not normally present in the Drosophila genome) and a mini-white gene 733 bp downstream (Figure 1). Location of the integrated [Iw]7 construct was confirmed by inverse PCR. Alignments are based on our sequencing data using the PE5′ and 3jnB primers (see below).
Figure 1.
The [Iw] construct. The [Iw] construct is a chromosomally integrated P-element containing the 18 bp I-SceI site and a mini-white gene 733 bp downstream of the I-SceI site. I-SceI expression induces a single complementary ended DSB with 4 bp 3′ overhangs. Iterative cycles of cutting and repair continue until inaccurate end-joining repair or gene conversion destroys the I-SceI site. white expression facilitates identification of end-joining repair events. A 4 bp microhomology (CCCT, underlined) is present at the break site. No other ≥4 bp microhomologies are available near the DSB. Primers used to amplify repair products are indicated by arrows.
The I-SceI sources used were either the UIE[I-SceI]2R (34) expression construct on the right arm of chromosome 2, which is under the control of a ubiquitin promoter, or the hsp70[I-SceI]1A construct (33) integrated on chromosome 3, which is under the control of a constitutive hsp70 promoter fragment.
Mutant alleles used in this study were lig4169a (20), spnA057 (35), spnA093 (35), ku707B2.2 (31), ku70ex8 (31), mus3082003 (36) and mus308Sz-29 (37). Transheterozygotes were used whenever possible to avoid second site effects.
Male flies undergoing germline DSB induction were generated by mating females of appropriate genotype harboring the [Iw] construct to males of appropriate genotype harboring an I-SceI expression construct. Germline repair events were recovered in the next generation by appropriate individual matings. Virtually all chromosomes that had been cut had undergone a repair event altering or eliminating the I-SceI cut site. Thus, although an indeterminate number of accurate repair events may have occurred prior to inaccurate repair, accurate repair is not quantified in this assay. Progeny that inherited a chromosome repaired by EJ were identified by having pigmented eyes. Since the [Iw] construct was present on only one chromosome, DSB repair by gene conversion or EJ with a deletion to the right >700 bp results in the loss of white expression. On average, >90% of progeny flies inheriting [Iw] chromosomes from each mating had pigmented eyes or were the results of gene conversion (determined by PCR), indicating that EJ events with large deletions is not a major repair pathway in this system. Only independent events were sequenced.
Repair junction analysis
DNA was extracted from individual flies according to (38). PCR was carried out using the primers PE5′ (GATAGCCGAAGCTTACCGAAGT) and jn3′B (GGACATTGACGCTATCGACCTA), which amplify a 1.3 kb region and are approximately equidistant from the I-SceI site. A ∼400 bp secondary PCR product was used as an internal PCR control. Amplified products were gel purified (GenScript) and sequenced using the PE5′ primer.
Sequence analysis and modeling
Alignments were done with ClustalW software and/or manual inspection. Repeated sequences were identified via the EMBOSS PALINDROME software (39) or by a suffix-tree based repeat finding program. Computer programs written in our lab were used to estimate the frequency of SD-MMEJ consistent junctions by random chance alone. All simulations and calculations used the sequence interval shown in Figure 3. The probability of SD-MMEJ consistent blunt joins occurring by random chance at this DSB was estimated by a Monte Carlo simulation run three separate times on 10 000 randomly generated apparent blunt joins. The probability of SD-MMEJ consistent junctional microhomologies by random chance alone was calculated by enumeration of microhomology junctions. The probability of SD-MMEJ consistent indels with 1–3 bp insertions was estimated by analysis of 110 randomly generated junctions with 1–3 bp insertions. Randomly generated SD-MMEJ consistent repair junctions were used to estimate the expected frequency of SD-MMEJ consistent repair junctions in primer groups I–IV. Statistical analyses were carried out in Excel, SPSS or GraphPad Prism.
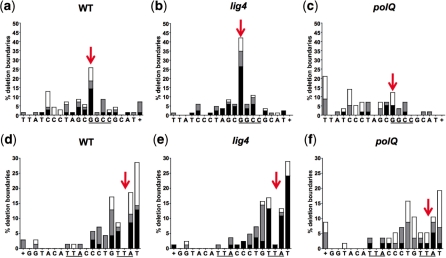
Figure 3.
Combined effects of sequence context and genotype on deletion boundary frequency. Frequency as percentage of total repair products of deletion boundaries associated with net insertions (black), junctional microhomologies (gray) and apparent blunt joins (white) in wild-type, lig4 and polQ genetic backgrounds. ‘Deletion boundary’ is defined as the position (including any junctional microhomologies) where an uninterrupted match between the original sequence and the repair product resumes. X-axis indicates top strand sequence. Deletions extending beyond the sequence shown are represented by a plus sign (+). (a–c) Boundaries of deletions to the right of the bottom strand nick. The underlined sequence (GGCC) is the left half of a GGCC repeat. Arrows show clustering of deletion boundaries at this repeat in wild-type and lig4 but not polQ sequences. (d–f) Deletion boundaries to the left of the top strand nick. The underlined sequence (TTA) is a 3 bp direct repeat. Note different overall shape of the histograms of deletion boundaries to the right and left of the DSB in all genotypes (compare a–c, d–f). Arrows show virtual absence of 2 bp deletions in wild-type and lig4 but not polQ sequences. WT: n = 70; lig4: n = 83; polQ: n = 57.
RESULTS
C-NHEJ deficiency does not alter length or frequency of junctional microhomology
End-joining repair of P-element-induced DSBs in Drosophila often produces repair products with microhomologies similar to those seen in Ku-independent MMEJ in budding yeast. To investigate microhomology use in alt-EJ of a non-P-element DSB in flies, we used transgenic flies with one copy of the [Iw] DSB substrate to examine Ku-independent repair of a complementary ended I-SceI DSB (33).
The [Iw]/ I-SceI system (Figure 1) induces a single complementary ended chromosomal DSB. High levels of I-SceI expression ensure repeated cycles of cutting and repair until inaccurate EJ or gene conversion destroys the I-SceI site.
Unlike P-element DSBs, the [Iw] DSB lacks ≥5 bp microhomologies near the break. In the [Iw] system, MMEJ at ≥5 bp of pre-existing microhomology results in deletion of over 70 bp. This enables a PCR-based assay to rule out use of ≥5 bp microhomologies.
In wild-type flies (n = 90), 87.8% of independently recovered repair events yielded a PCR product inconsistent with use of ≥5 bp microhomologies (data not shown). Such products constituted 82.9% of repair events in ku70 mutants (n = 82), which is not significantly different from wild-type (P = 0.39, Fisher's Exact Test). Similar results were obtained for lig4 mutants. Of the 469 junctions sequenced in this study (Supplemental Tables S3–S8), only one involved annealing at 5 bp microhomology and junctional microhomologies >5 bp were not observed.
In vertebrates, alt-EJ is often characterized by larger deletions and more frequent short junctional microhomologies. Comparison of [Iw] repair junctions from C-NHEJ proficient and deficient flies showed no differences in deletion size (Supplementary Figure S1a), frequency of junctional microhomologies (Figure 2) or length of junctional microhomologies (Supplementary Table S1). Together, these results indicate that alt-EJ of this DSB is not characterized by the increased junctional microhomology typically observed in budding yeast and higher eukaryotes.
Figure 2.
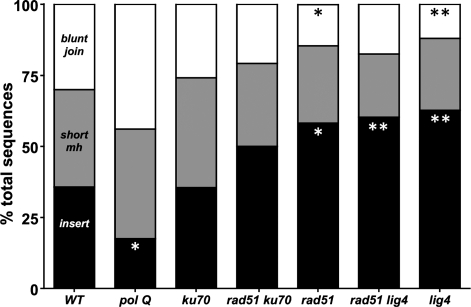
C-NHEJ deficiency increases frequency of net insertions. Relative proportions of end-joining junction types (insertion, microhomology, apparent blunt join) by genotype. The relative proportion of junctions with insertions is significantly decreased relative to wild-type in polQ mutants (white asterisk; P = 0.03, Fisher’s exact test) and significantly increased relative to wild-type (WT) in rad51 (P = 0.02), rad51 lig4 (P < 0.01) and lig4 (P < 0.01) mutants. The relative proportion of apparent blunt joins is significantly decreased relative to WT in rad51 (black asterisk; P = 0.05) and lig4 (P < 0.01) flies (Fisher's Exact Test). The relative proportion of repair products with junctional microhomology is not significantly different from wild-type in any genotype assayed. WT: n = 70; polQ: n = 57; ku70: n = 62; rad51 ku70: n = 48; rad51: n = 55; rad51 lig4: n = 63; lig4: n = 83.
Alt-EJ produces insertions at repair junctions
A large number of [Iw] repair junctions had insertions (Supplementary Figure S1b; Chan, Yu and McVey, in revision). The frequency of insertions was significantly increased relative to wild-type in lig4 mutants and decreased in mus308 mutants, which lack DNA polymerase theta (PolQ) (Figure 2; Chan, Yu and McVey, in revision).
Many inserted sequences appeared to have been templated from sequence near the DSB. In Drosophila, P-element DSBs can be repaired by a hybrid HR/EJ pathway in which some Rad51-dependent repair synthesis takes place before repair is completed by EJ (40). To ask if these insertions were produced by a similar process, we sequenced repair junctions from rad51, rad51 ku70 and rad51 lig4 flies. In all HR-deficient and C-NHEJ/HR double mutant genetic backgrounds, insertion frequency was comparable to or higher than wild-type (Figure 2). Therefore, the insertions result from an alternative end-joining mechanism that requires neither Ku nor Lig4 but is impaired in polQ mutants.
Genotype specific effects of sequence context on repair outcome
To gain insight into the mechanisms responsible for the insertions, we carried out fine-level sequence analysis of the repair junctions. This revealed patterns suggesting that genetic background and sequence composition interact to influence repair outcome. Deletion boundary frequency histograms (Figure 3; Supplementary Figure S2) showed that to the right of the DSB, deletion boundaries cluster at a 4 bp GGCC repeat in wild-type and lig4 but not polQ flies (Figure 3a–c, arrows). The sequence to the left of the DSB lacks a comparable repeat and similar clustering was not observed (Figure 3d–f). However, deletions of 3 bp to the left of the top strand nick (arrow) were virtually absent in wild-type and lig4 flies. This bias was also attenuated in polQ flies. Together, these observations suggest that flanking sequence influences repair outcome and that PolQ contributes to this sequence specific effect.
Templated insertions and deletion boundaries correlate with specific patterns of short repeats in the original sequence
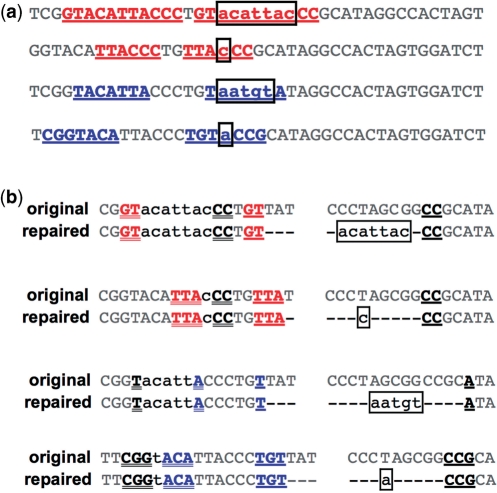
Many inserted sequences formed direct or inverted repeats with nearby sequence unaffected by the repair event, suggesting that the insertions may have been templated. Sequence analysis to identify potential insertion templates revealed that repeated motifs included not only the insertion itself, but also flanking sequence on both sides of the insertion (Figure 4a; Supplementary Figure S3).
Figure 4.
Repair products contain repeated motifs with a consistent pattern. (a) Sequences of representative repair products with insertion/deletion (indel) events. In each case, the repair event contains a direct (red) or inverted (blue) repeat. The repeated motif (underlined) comprises the insertion (boxed, lowercase) and flanking sequence on each side. Repeats are found both in repair products with insertions of sufficient length to be identified as templated and in repair products with apparently random short insertions. Sequences are (top to bottom) Supplementary Table S8, sequence 83; Supplementary Table S5, sequences 1–10; Supplementary Table S8, sequence 125; Supplementary Table S5, sequence 15. Additional similar examples are given in Supplementary Figure S3. (b) Repair events from panel a above, aligned with the original [Iw] sequence. Only top strands are shown. Whitespace indicates the top strand nick. Dashes indicate bases deleted in the repair product. Net insertions are boxed and in lowercase. The apparent insertion template is in lowercase not boxed. Red/blue, underlined: Short direct (red) or inverted (blue) repeat near but not spanning the DSB. Black, underlined: short direct or inverted repeat spanning the DSB.
Alignment of these repair products with the original [Iw] sequence (Figure 4b; Supplementary Figure S3) showed a consistent relationship between two short repeats present in the original sequence, the insertion template and the repair product. The insertion template and the deletion were each bounded by two short repeats. One of the short repeats was situated entirely to one side of the DSB; the other short repeat spanned the DSB.
The SD-MMEJ model
DNA polymerase theta is unique in possessing both an A-family DNA polymerase domain and an SF2 helicase-like domain (37). Evidence that PolQ may be involved in an alt-EJ mechanism that generates templated insertions, together with the patterns that emerged from our sequence analysis, suggested the following model, which we term SD-MMEJ.
In SD-MMEJ, a short direct or inverted repeat (Figures 5a and 6a) primes synthesis of de novo microhomologies at DSB ends. Resection or unwinding exposes short direct or inverted repeats near the DSB (Figures 5b and 6b). These short ‘primer repeats’ mediate formation of transient secondary structures (Figures 5c and 6c), which prime synthesis by a template dependent non-processive DNA polymerase (Figures 5d and 6d). If the newly synthesized 3′ end is complementary to sequence on the other side of the DSB, dissociation or unwinding of the structure (Figures 5e and 6e) allows annealing across the break. The result of this process is creation of a repeated motif that contains the insertion and flanking sequence on both sides (Figures 5g and 6g).
Figure 5.
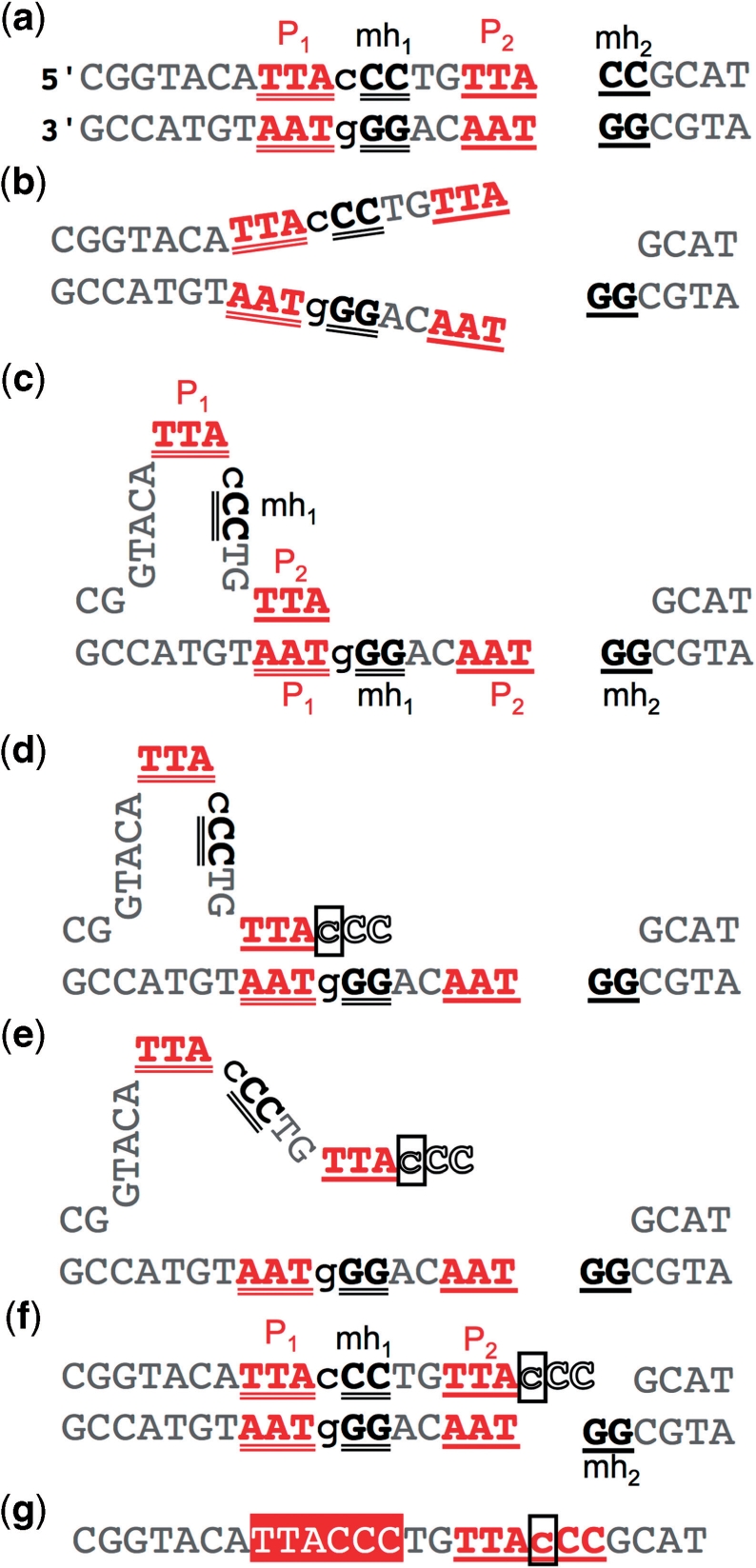
Loop-out SD-MMEJ produces repair products with direct repeats. Example is taken from Figure 4. Both strands are shown. Deleted bases are not shown. (a) Loop-out SD-MMEJ requires two short direct repeats. The ‘primer repeat’ (P1, P2; red) is located completely to one side of the DSB. The ‘microhomology repeat’ (mh1, mh2; black) spans the DSB. Notational convention is that P2 and mh2 are closest to the DSB. (b) The DNA double helix is unwound through P1. (c) P2 on the top strand base-pairs with P1 on the bottom strand. mh1 and P1 on the top strand are looped out. (d) A template-dependent DNA polymerase extends through the end of mh1. Newly synthesized bases are in white type outlined in black. The base that will form the net insertion is boxed and in lowercase. (e) The loop-out structure dissociates or is unwound. (f) P1 and P2 re-anneal as originally. This DSB end now has a single stranded 3′ overhang that is complementary to mh2. (g) The resulting repair product showing the net insertion and the direct repeat spanning the junction created by loop-out SD-MMEJ repair. (Compare with Figure 4b.) The part of the repeat that was present in the original sequence (white text on red) comprises P1, mh1 and the sequence between them. The part of the repeat containing the insertion (underlined, red) comprises P2, mh2 and a copy of the sequence between P1 and mh1. This convention is used throughout the remaining figures/tables.
Figure 6.
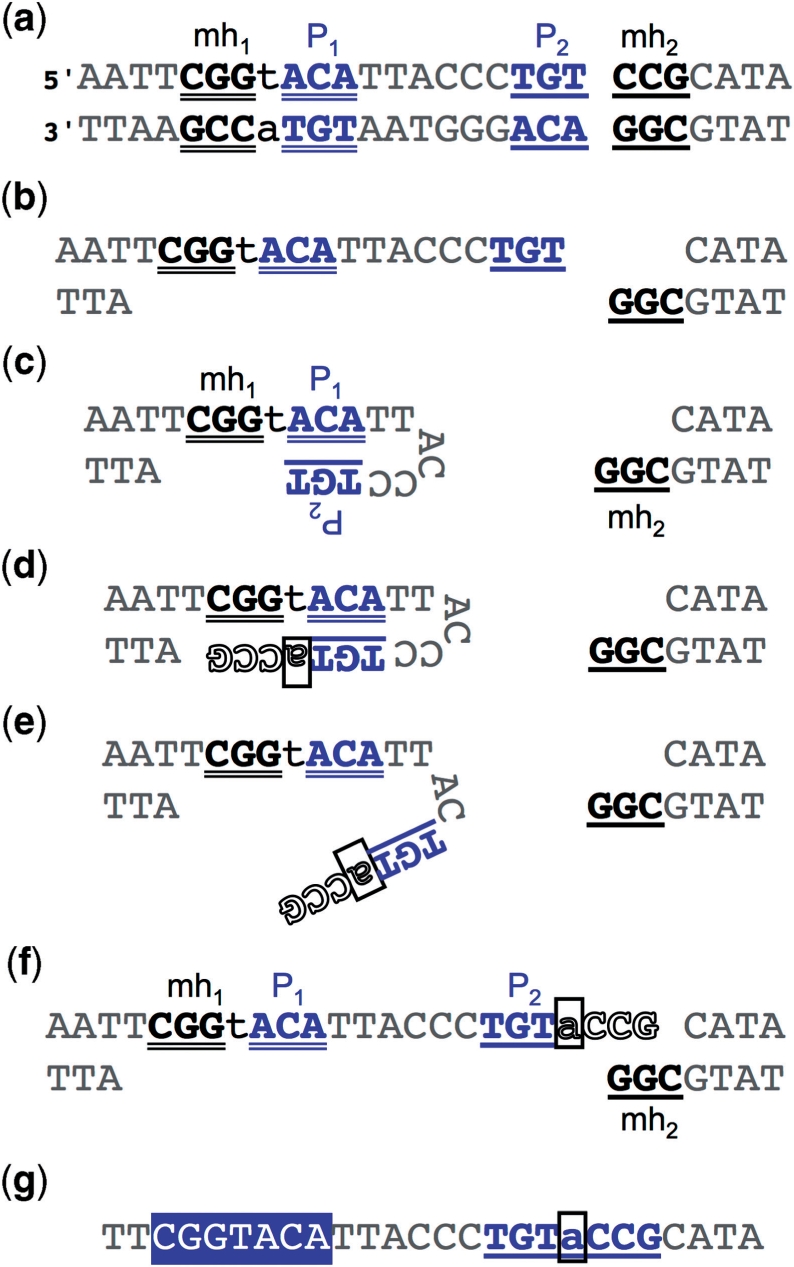
Snap-back SD-MMEJ produces repair products with inverted repeats. Example is taken from Figure 4. Both strands are shown. Deleted bases are not shown. (a) Snap-back SD-MMEJ requires two short direct repeats. The ‘primer repeat’ (P1, P2; blue) is located completely to one side of the DSB. The ‘microhomology repeat’ (mh1, mh2; black) spans the DSB. Notational convention is that P2 and mh2 are closest to the DSB. (b) The DNA double helix is resected or unwound unwound through mh1. In contrast to loop-out SD-MMEJ (compare Figure 5, panels b and c), ssDNA may be generated by either nuclease or helicase activity. (c) P2 base-pairs with P1, forming a hairpin with a 3′-end. (d) A template-dependent DNA polymerase extends through the end of mh1. Newly synthesized bases are in white type outlined in black. The base that will form the net insertion is boxed and in lowercase. (e) The hairpin dissociates or is unwound. (f) P1 and P2 re-anneal as originally. This DSB end now has a single stranded 3′ overhang complementary to mh2. (g) The resulting repair product showing the net insertion and the inverted repeat spanning the junction created by snap-back SD-MMEJ repair. (Compare Figure 4b). The part of the repeat that was present in the original sequence (white text on blue) comprises mh1, P1 and the sequence between them. The part of the repeat containing the insertion (underlined, blue) comprises the reverse complements of P2, mh2 and the reverse complement of the sequence intervening between mh1 and P1. This convention is used throughout the remaining figures/tables.
In addition to the simple templated insertions described in Figure 4, we also recovered a substantial number of complex insertions comprising multiple overlapping copies of nearby sequence. This class of complex insertion is consistent with SD-MMEJ involving multiple rounds of synthesis and dissociation from one or both sides of the DSB (Supplementary Figure S4). Facilitation of complex events by coupled polymerase/helicase activity is consistent with the observation that rare insertions observed in polQ mutants never exceeded 12 bp in length. The variance of the insertion lengths seen in polQ mutants was significantly smaller than wild-type (P = 0.05, Levene's test), indicating that failure to observe longer complex insertions in polQ mutants is unlikely to be an artifact of small sample size (Supplementary Figure S1b).
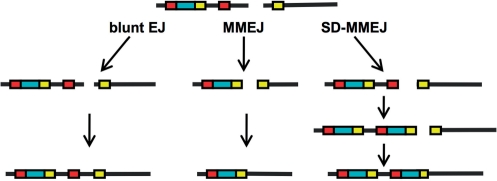
SD-MMEJ produces multiple types of repair products without net insertions
SD-MMEJ as illustrated in Figures 5 and 6 necessarily produces a repair product with a direct or inverted repeat. The repeated motif comprises the net insertion and flanking sequence on both sides of the insertion. The size of the net insertion is equal to the distance between P1 and mh1 (Figures 5a and 6a). If P1 and mh1 are directly adjacent, SD-MMEJ will produce a repair product with no net insertion. SD-MMEJ products without net insertions can either resemble blunt joins or have junctional microhomology (Figure 7). However, SD-MMEJ products without insertions will still have the same repeat structure as SD-MMEJ products with insertions. This criterion can be used to classify repair junctions without net insertions as consistent or not consistent with repair by SD-MMEJ (Supplementary Figures S5 and S6).
Figure 7.
SD-MMEJ repair can produce repair junctions without insertions, which may or may not have microhomology. If P1 and mh1 are directly adjacent, SD-MMEJ will not produce an insertion. To efficiently summarize the proposed SD-MMEJ mechanism, this ‘insertion’ of length zero is indicated by a box containing no sequence. In the case of ambiguous deletion breakpoints (junctional microhomologies), the ambiguous bases are flanked by boxes containing dots. Each panel shows alignment of the original sequence and repair product, the primer and microhomology repeats and the longer repeat created in the repair product. Notational conventions are as in Figures 5 and 6. (a and b) Apparent blunt joins consistent with SD-MMEJ repair. Box with dash indicates the point of ligation (0 bp insertion). (c and d) SD-MMEJ consistent repair products with junctional microhomologies. In both of these cases, SD-MMEJ results in annealing at longer microhomologies than would MMEJ at the microhomologies present in the repair product. a: sequence H, Supplementary Table S7. b: sequence B, Supplementary Table S7. c: sequence O, Supplementary Table S8. d: sequence F, Supplementary Table S8.
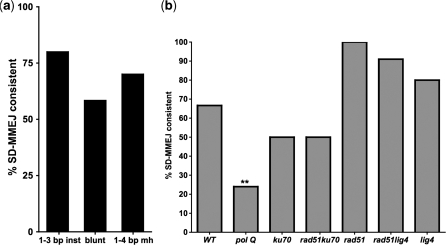
We analyzed all repair product sequences with 1–3 bp insertions, apparent blunt joins and junctional microhomologies to determine if they had repeat motifs consistent with SD-MMEJ within ±20 bp of the junction. A repair product was classified as consistent with repair by SD-MMEJ if the junction, plus at least one base on either side, was contained within half of a direct or inverted repeat of at least 4 bp. By these criteria, the expected frequencies of SD-MMEJ consistent 1–3 bp insertions, apparent blunt joins and microhomologies by random chance alone were estimated to be, respectively, 18, 43 and 54 percent (see ‘Materials and Methods’ section).
The observed frequencies of SD-MMEJ consistent repair junctions with 1–3 bp insertions, apparent blunt joins and junctions with microhomology were, respectively, 80.0, 58.4 and 70.1 percent (Figure 8a). All of these are significantly greater than predicted by random chance (P < 0.01; P < 0.05; P = 0.05; Fisher's Exact Test). Further supporting the hypothesis that repair products with and without net insertions could have been formed by the same mechanism, the proportion by genotype of SD-MMEJ consistent apparent blunt joins followed the same genotype specific patterns as net insertion frequency (Figure 8b; compare Figure 2). Importantly, relative to wild-type, significantly fewer apparent blunt joins and junctional microhomologies recovered from polQ flies were SD-MMEJ consistent (Figure 8b and data not shown).
Figure 8.
A majority of 1–3 bp insertions, apparent blunt joins and microhomologies are SD-MMEJ consistent. (a) 80.0% of all repair products with 1–3 bp insertions (inst), 58.4% of all apparent blunt joins and 70.1% of all junctions with microhomologies are SD-MMEJ consistent [a ≥4 bp direct or inverted repeat that contains the breakpoint(s) is present within ±20 bp of the junction]. For all three junction types, the observed frequency of SD-MMEJ consistent repair products is significantly higher than the estimated frequency expected by random chance for this sequence: 1–3 bp insertions (n = 71), P < 0.01; apparent blunt joins (n = 101), P < 0.05; microhomologies (n = 134) P = 0.05; Fisher's exact test. (b) The proportions by genotype of SD-MMEJ consistent blunt joins follow a pattern similar to the proportions of net insertions by genotype (compare Figure 2). The proportion of polQ blunt joins consistent with the SD-MMEJ model is significantly decreased relative to wild-type (**P < 0.01, Fisher's exact test). This is consistent with a role for PolQ in SD-MMEJ. WT: n = 21; polQ: n = 25; ku70 n = 16; rad51 ku70: n = 10; rad51: n = 8; rad51 lig4: n = 11; lig4: n = 10.
Categorization of repair products according to SD-MMEJ parameters supports a single underlying mechanism
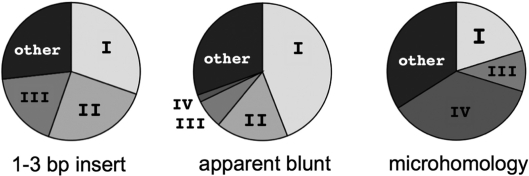
A significant majority of repair junctions from each of three apparently distinct structural classes (short insertions, apparent blunt joins and junctional microhomologies) had structures consistent with repair by SD-MMEJ (Figure 8). If SD-MMEJ is in fact producing all three types of repair junctions, grouping of SD-MMEJ consistent repair products by primer repeat should result in groups of structurally similar repair products comprising more than one of the above junction types. Side by side comparison of SD-MMEJ consistent 1–3 bp insertions, apparent blunt joins and microhomologies with the same SD-MMEJ primer repeat in fact demonstrates such similarities (Tables 1–4).
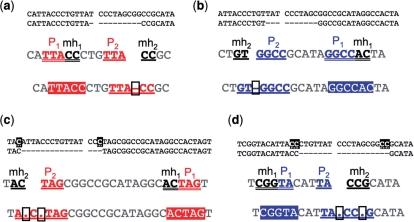
Table 1.
Examples of repair junctions with primer repeat I
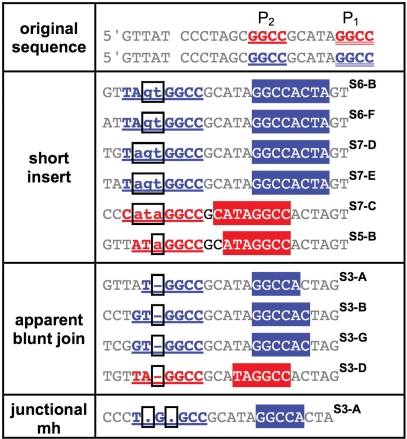 |
Categorizing SD-MMEJ consistent repair junctions by primer repeat reveals structural similarities among multiple repair junction types. Representative examples of SD-MMEJ-consistent sequences with the specified primer repeats (top row) are shown. Superscripts cross-refer Supplementary Tables S3–S8. Primer repeat group I corresponds to SD-MMEJ at a 4 bp direct and inverted repeat. Insertion junctions, apparent blunt joins and junctional microhomologies are all represented. The deletion resulting from SD-MMEJ at this primer repeat corresponds to the cluster of deletion boundaries to the right of the DSB (Figure 3a–c). This primer group contains the most frequently recovered junctional microhomology (Supplementary Table S2, top row).
Table 2.
Examples of repair junctions with primer repeat II
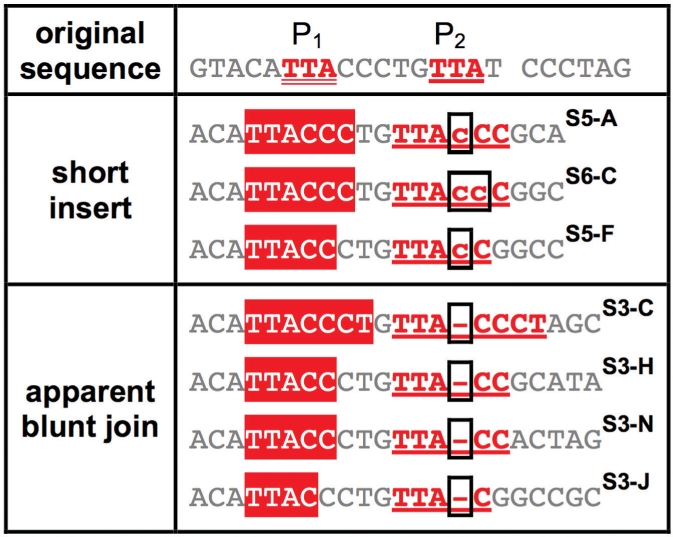 |
Categorizing SD-MMEJ consistent repair junctions by primer repeat reveals structural similarities among multiple repair junction types. Representative examples of SD-MMEJ-consistent sequences with the specified primer repeats (top row) are shown. Superscripts cross-refer Supplementary Tables S3–S8. Primer repeat group II corresponds to loop-out SD-MMEJ at a 3 bp TTA direct repeat to the left of the DSB. Both short inserts and apparent blunt joins are represented. Use of this repeat for SD-MMEJ at this repeat is consistent with the absence of deletion boundaries 2 bp to the left of the top strand nick (Figure 2d–f).
Table 3.
Examples of repair junctions with primer repeat III
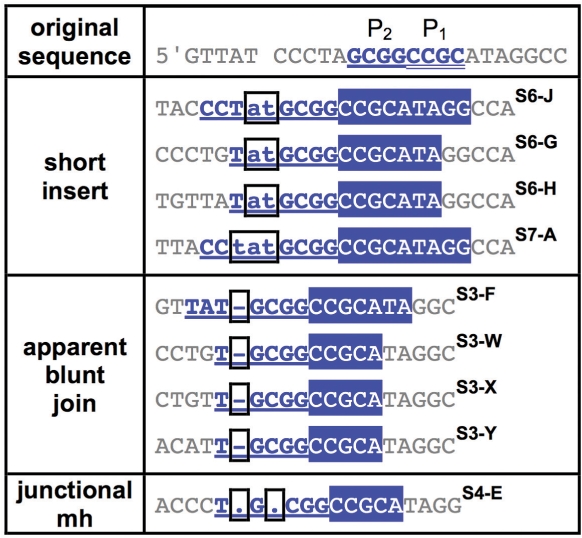 |
Categorizing SD-MMEJ consistent repair junctions by primer repeat reveals structural similarities among multiple repair junction types. Representative examples of SD-MMEJ-consistent sequences with the specified primer repeats (top row) are shown. Superscripts cross-refer Supplementary Tables S3–S8. Primer repeat group III corresponds to SD-MMEJ at a 4 bp inverted repeat to the right of the DSB. Short inserts, apparent blunt joins, and junctional microhomologies are all represented.
Table 4.
Examples of repair junctions with primer repeat IV
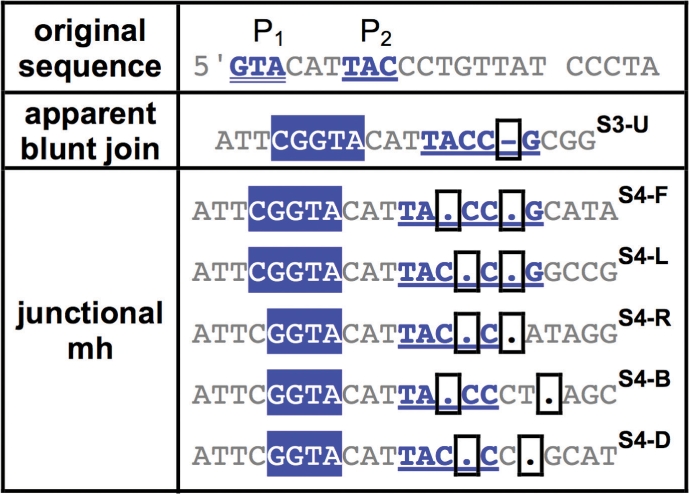 |
Categorizing SD-MMEJ consistent repair junctions by primer repeat reveals structural similarities among multiple repair junction types. Representative examples of SD-MMEJ-consistent sequences with the specified primer repeats (top row) are shown. Superscripts cross-refer Supplementary Tables S3–S8. Primer repeat group IV corresponds to a 3 bp inverted repeat to the left of the DSB. Junctional microhomologies and an apparent blunt join are represented.
Many of the most frequently used primer repeats are the same for multiple junction types. A small set of frequently used primer repeats accounts for a substantial portion of SD-MMEJ consistent repair products (Figure 9). Chi-square contingency table analysis comparing experimentally observed ratios of repair junctions in the five classes in Figure 9 with ratios obtained from randomly generated blunt joins and microhomologies indicates that the observed frequencies of specific SD-MMEJ primer repeats are unlikely due to random chance (P < 0.01 and P = 0.01, respectively). Together, these results suggest that SD-MMEJ may be responsible for a wide variety of superficially different end-joining repair junction structures.
Figure 9.
Most SD-MMEJ consistent repair junctions accounted for by four primer repeats. Quantification of the proportion of SD-MMEJ consistent 1–3 bp insertions, apparent blunt joins and junctional microhomologies with primer repeats I–IV as described in Tables 1–4 or a different primer repeat (other). Chi-square contingency tables used to compare the observed ratios of the five categories with the ratios derived from randomly generated blunt joins and microhomologies showed that, for both SD-MMEJ consistent blunt joins and microhomologies, the observed proportions of primer repeats in repair products was significantly different from random chance (P < 0.01 and P = 0.01, respectively). Ratios of primer repeats expected from random chance was estimated from 200 randomly generated SD-MMEJ consistent blunt joins and directly calculated by enumeration for microhomology junctions. Similar methods did not generate a sufficient number of random SD-MMEJ consistent 1–3 bp insertions to populate a contingency table.
DISCUSSION
In this study, we provide experimental evidence that end-joining junction structures typically considered diagnostic of fundamentally different repair mechanisms can be most parsimoniously explained by a single mechanism, which we term synthesis-dependent MMEJ (SD-MMEJ). Genetic evidence suggests that SD-MMEJ in Drosophila is a form of Ku and Lig4 independent alt-EJ that involves mus308, the Drosophila ortholog of vertebrate DNA polymerase theta (PolQ).
Historically, one of the most difficult to explain aspects of end-joining repair has been the diverse spectrum of repair events produced and the variability of this spectrum both within and between studies. The SD-MMEJ model accounts for many apparently idiosyncratic patterns observed in repair junction sequences. For example, the frequency of particular junctional microhomologies in this study did not correlate with microhomology length or distance from the DSB, but correlated strongly with whether or not the resulting repair product was SD-MMEJ consistent (Supplementary Table S2).
The SD-MMEJ model suggested by our results provides a fresh perspective on the interplay of sequence context and genetic background during end-joining repair. Previous evidence suggests that sequence context can modulate the effect of C-NHEJ deficiency in potentially complex ways. For example, studies comparing different class switch recombination junctions from individual C-NHEJ deficient patients have found increased microhomology at some junctions, but not others (28).
Comparison of our results with reports in the literature confirms that differences in sequence not immediately adjacent to the break can cause breaks with identical end structure to be repaired differently. A previous study using a different I-SceI based system in Drosophila (34) reported a repair product spectrum much closer to that expected for ‘typical’ C-NHEJ. This is consistent with the observation that the sequence features that most strongly influenced repair outcome in our study are outside the I-SceI site.
Similar contrasts are apparent in different studies of I-SceI DSBs in vertebrate-derived systems. Some studies report few insertions (41). Others report sequence capture (42) or extensive rearrangements (43). Templated insertions strikingly similar to those observed in our study have also been found at inaccurate I-SceI repair junctions in the U937 lymphoblastoid cell line (44).
This last example raises the possibility that templated SD-MMEJ may operate in vertebrate systems. Templated insertions have been reported at translocation junctions in lymphomas (45–47) and PolQ expression is increased in several cancers (48). These observations suggest further exploration of the idea that templated SD-MMEJ involving DNA polymerase theta may contribute to genome instability in humans.
Our SD-MMEJ model both contrasts and shares features with previous models of templated mutagenesis. SD-MMEJ is probably most similar to the model proposed by Scaringe et al. to explain templated mutagenesis during lesion bypass (49). Some SD-MMEJ consistent insertions resemble templated nucleotides at T-DNA insertions in plants (50). Longer SD-MMEJ consistent insertions (Supplementary Figure S4) resemble, on a much smaller scale, the chromosomal rearrangements explained by the FoSTes model (51). One substantial difference between SD-MMEJ and several microhomology-mediated mechanisms, including FoSTeS, is that SD-MMEJ does not involve DNA replication. Loop-out SD-MMEJ superficially resembles slippage during DNA replication but is not precipitated by highly repetitive sequences (52).
SD-MMEJ is perhaps most novel in that it does not require spontaneous formation of DNA secondary structures that persist without external stabilization (53,54). Some intermediate structures repeatedly suggested by our sequence analysis, such as snap-back priming between two adjacent nucleotides, are thermodynamically unfavorable. Interestingly, the notion that such structures could be stabilized by protein binding is supported by crystallographic data showing that M. tuberculosis LigD with its ancillary polymerase domain can mediate a hairpin between two adjacent nucleotides (55).
We speculate that in SD-MMEJ, transient base pairing interactions are stabilized by DNA polymerase binding. This addresses the apparently paradoxical observation that some microhomologies that appear to have been created by SD-MMEJ are not longer than pre-existing microhomologies available near the DSB. It seems likely that annealing at a short microhomology together with polymerase binding could provide better end stabilization than base pairing alone. This is supported by recent evidence from budding yeast that a DNA polymerase can have end-bridging activity and can mediate stabilizing interactions via base stacking (56). A role for polymerase-mediated end synapsis in vertebrate MMEJ would explain why some C-NHEJ deficient systems exhibit increased frequency of microhomologies apparently too short to mediate effective end synapsis via base pairing alone. It is also consistent with observation of longer microhomologies in Saccharomyces cerevisiae MMEJ because budding yeast does not have an ortholog of POLQ.
Our results support the hypothesis that alt-EJ may comprise multiple pathways or mechanisms with varying requirements for microhomology (Figure 10; Supplementary Figures S5 and S6). Previous studies have documented differences between Ku and Lig4 independent alt-EJ (41,57,58). Our SD-MMEJ model reveals striking differences between Ku and Lig4 independent alt-EJ. In ku70 mutants, approximately half of the repair junctions without insertions or microhomology did not fit the SD-MMEJ model and are likely true blunt joins. This suggests that in ku70 mutants, other proteins, such as the MRN complex, may tether break ends. However, nearly all of the rare apparent blunt joins recovered from lig4 mutants were SD-MMEJ consistent (Figure 8b). We speculate that this may reflect a need for especially stable break end synapsis to maintain the ends in proximity during recruitment of an alternative DNA ligase.
Figure 10.
Multiple mechanisms for alt-EJ. Alt-EJ can proceed by a variety of mechanisms: direct ligation of non-complementary blunt ends (left), annealing at pre-existing microhomologies (center) or synthesis of de novo microhomologies via SD-MMEJ (right). Deletions during direct ligation of blunt ends are not influenced by repeated sequences. MMEJ at pre-existing microhomologies (yellow) involves deletion of one of the two repeated motifs as well as all sequence intervening between the repeated motifs. SD-MMEJ synthesizes de novo microhomologies via priming at a repeat situated entirely to one side of the DSB (red). Any sequence intervening between the primer and the template for the new microhomology (blue) is copied and inserted at the break site. Identically colored boxes represent direct repeats. See also Supplementary Figures S5 and S6.
In sum, our results contribute to the emerging idea that end-joining repair is best conceptualized not as a set of distinct pathways, but as a flexible network of interacting repair strategies (59,60). Such flexibility could encompass the ability to take advantage of local sequence features to facilitate repair. Thus, characterization of sequence specific effects will likely provide important new insights into the fundamental nature of end-joining repair.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation (MCB 0643253) and the Ellison Medical Foundation (New Scholar in Aging Award to M.M.). Funding for open access charge: Ellison Medical Foundation, National Science Foundation (MCB-0643253).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank D. Kane, S. Mirkin, A. Thorn and members of the McVey Lab for helpful comments, W. Engels and Y. Rong for the generous gift of fly stocks and T. Yu for programming expertise.
REFERENCES
- 1.Pardo B, Gomez-Gonzalez B, Aguilera A. DNA repair in mammalian cells: DNA double-strand break repair: how to fix a broken relationship. Cell Mol. Life Sci. 2009;66:1039–1056. doi: 10.1007/s00018-009-8740-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lieber MR, Gu J, Lu H, Shimazaki N, Tsai AG. Nonhomologous DNA end joining (NHEJ) and chromosomal translocations in humans. Subcell. Biochem. 2010;50:279–296. doi: 10.1007/978-90-481-3471-7_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeFazio LG, Stansel RM, Griffith JD, Chu G. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 2002;21:3192–3200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol. Cell. 2006;22:511–519. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 6.Lee JW, Yannone SM, Chen DJ, Povirk LF. Requirement for XRCC4 and DNA ligase IV in alignment-based gap filling for nonhomologous DNA end joining in vitro. Cancer Res. 2003;63:22–24. [PubMed] [Google Scholar]
- 7.Gu J, Lu H, Tippin B, Shimazaki N, Goodman MF, Lieber MR. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J. 2007;26:1010–1023. doi: 10.1038/sj.emboj.7601559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taccioli GE, Rathbun G, Oltz E, Stamato T, Jeggo PA, Alt FW. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 10.Bogue MA, Wang C, Zhu C, Roth DB. V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal, and hybrid joint formation. Immunity. 1997;7:37–47. doi: 10.1016/s1074-7613(00)80508-7. [DOI] [PubMed] [Google Scholar]
- 11.Liang F, Romanienko PJ, Weaver DT, Jeggo PA, Jasin M. Chromosomal double-strand break repair in Ku80-deficient cells. Proc. Natl Acad. Sci. USA. 1996;93:8929–8933. doi: 10.1073/pnas.93.17.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee K, Lee SE. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics. 2007;176:2003–2014. doi: 10.1534/genetics.107.076539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Windhofer F, Iliakis G. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 15.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 16.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 18.Liang F, Jasin M. Ku80-deficient cells exhibit excess degradation of extrachromosomal DNA. J. Biol. Chem. 1996;271:14405–14411. doi: 10.1074/jbc.271.24.14405. [DOI] [PubMed] [Google Scholar]
- 19.Secretan MB, Scuric Z, Oshima J, Bishop AJ, Howlett NG, Yau D, Schiestl RH. Effect of Ku86 and DNA-PKcs deficiency on non-homologous end-joining and homologous recombination using a transient transfection assay. Mutat. Res. 2004;554:351–364. doi: 10.1016/j.mrfmmm.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 20.McVey M, Radut D, Sekelsky JJ. End-joining repair of double-strand breaks in drosophila melanogaster is largely DNA ligase IV independent. Genetics. 2004;168:2067–2076. doi: 10.1534/genetics.104.033902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 22.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 23.Boboila C, Jankovic M, Yan CT, Wang JH, Wesemann DR, Zhang T, Fazeli A, Feldman L, Nussenzweig A, Nussenzweig M, et al. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc. Natl Acad. Sci. USA. 2010;107:3034–3039. doi: 10.1073/pnas.0915067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boboila C, Yan C, Wesemann DR, Jankovic M, Wang JH, Manis J, Nussenzweig A, Nussenzweig M, Alt FW. Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. J. Exp. Med. 2010;207:417–427. doi: 10.1084/jem.20092449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandoval A, Labhart P. High G/C content of cohesive overhangs renders DNA end joining ku-independent. DNA Repair (Amst) 2004;3:13–21. doi: 10.1016/j.dnarep.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 26.Daley JM, Wilson TE. Rejoining of DNA double-strand breaks as a function of overhang length. Mol. Cell. Biol. 2005;25:896–906. doi: 10.1128/MCB.25.3.896-906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McVey M, Lee SE. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kotnis A, Du L, Liu C, Popov SW, Pan-Hammarstrom Q. Non-homologous end joining in class switch recombination: the beginning of the end. Philos. Trans. Roy. Soc. Lond. B. Biol. Sci. 2009;364:653–665. doi: 10.1098/rstb.2008.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dahm-Daphi J, Hubbe P, Horvath F, El-Awady RA, Bouffard KE, Powell SN, Willers H. Nonhomologous end-joining of site-specific but not of radiation-induced DNA double-strand breaks is reduced in the presence of wild-type p53. Oncogene. 2005;24:1663–1672. doi: 10.1038/sj.onc.1208396. [DOI] [PubMed] [Google Scholar]
- 30.Paull TT, Gellert M. A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc. Natl Acad. Sci. USA. 2000;97:6409–6414. doi: 10.1073/pnas.110144297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson-Schlitz DM, Flores C, Engels WR. Multiple-pathway analysis of double-strand break repair mutations in drosophila. PLoS Genet. 2007;3:e50. doi: 10.1371/journal.pgen.0030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei DS, Rong YS. A genetic screen for DNA double-strand break repair mutations in drosophila. Genetics. 2007;177:63–77. doi: 10.1534/genetics.107.077693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rong YS, Golic KG. The homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in drosophila. Genetics. 2003;165:1831–1842. doi: 10.1093/genetics/165.4.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Preston CR, Flores CC, Engels WR. Differential usage of alternative pathways of double-strand break repair in drosophila. Genetics. 2006;172:1055–1068. doi: 10.1534/genetics.105.050138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staeva-Vieira E, Yoo S, Lehmann R. An essential role of DmRad51/SpnA in DNA repair and meiotic checkpoint control. EMBO J. 2003;22:5863–5874. doi: 10.1093/emboj/cdg564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koundakjian EJ, Cowan DM, Hardy RW, Becker AH. The zuker collection: a resource for the analysis of autosomal gene function in drosophila melanogaster. Genetics. 2004;167:203–206. doi: 10.1534/genetics.167.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris PV, Mazina OM, Leonhardt EA, Case RB, Boyd JB, Burtis KC. Molecular cloning of drosophila mus308, a gene involved in DNA cross-link repair with homology to prokaryotic DNA polymerase I genes. Mol. Cell. Biol. 1996;16:5764–5771. doi: 10.1128/mcb.16.10.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gloor GB, Preston CR, Johnson-Schlitz DM, Nassif NA, Phillis RW, Benz WK, Robertson HM, Engels WR. Type I repressors of P element mobility. Genetics. 1993;135:81–95. doi: 10.1093/genetics/135.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 40.McVey M, Adams M, Staeva-Vieira E, Sekelsky JJ. Evidence for multiple cycles of strand invasion during repair of double-strand gaps in drosophila. Genetics. 2004;167:699–705. doi: 10.1534/genetics.103.025411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guirouilh-Barbat J, Rass E, Plo I, Bertrand P, Lopez BS. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc. Natl Acad. Sci. USA. 2007;104:20902–20907. doi: 10.1073/pnas.0708541104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin Y, Waldman AS. Promiscuous patching of broken chromosomes in mammalian cells with extrachromosomal DNA. Nucleic Acids Res. 2001;29:3975–3981. doi: 10.1093/nar/29.19.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Honma M, Izumi M, Sakuraba M, Tadokoro S, Sakamoto H, Wang W, Yatagai F, Hayashi M. Deletion, rearrangement, and gene conversion; genetic consequences of chromosomal double-strand breaks in human cells. Environ. Mol. Mutagen. 2003;42:288–298. doi: 10.1002/em.10201. [DOI] [PubMed] [Google Scholar]
- 44.Varga T, Aplan PD. Chromosomal aberrations induced by double strand DNA breaks. DNA Repair (Amst) 2005;4:1038–1046. doi: 10.1016/j.dnarep.2005.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jager U, Bocskor S, Le T, Mitterbauer G, Bolz I, Chott A, Kneba M, Mannhalter C, Nadel B. Follicular lymphomas' BCL-2/IgH junctions contain templated nucleotide insertions: novel insights into the mechanism of t(14;18) translocation. Blood. 2000;95:3520–3529. [PubMed] [Google Scholar]
- 46.Welzel N, Le T, Marculescu R, Mitterbauer G, Chott A, Pott C, Kneba M, Du MQ, Kusec R, Drach J, et al. Templated nucleotide addition and immunoglobulin JH-gene utilization in t(11;14) junctions: implications for the mechanism of translocation and the origin of mantle cell lymphoma. Cancer Res. 2001;61:1629–1636. [PubMed] [Google Scholar]
- 47.Murga Penas EM, Callet-Bauchu E, Ye H, Gazzo S, Berger F, Schilling G, Albert-Konetzny N, Vettorazzi E, Salles G, Wlodarska I, et al. The t(14;18)(q32;q21)/IGH-MALT1 translocation in MALT lymphomas contains templated nucleotide insertions and a major breakpoint region similar to follicular and mantle cell lymphoma. Blood. 2009;11:2214–2219. doi: 10.1182/blood-2009-08-236265. [DOI] [PubMed] [Google Scholar]
- 48.Kawamura K, Bahar R, Seimiya M, Chiyo M, Wada A, Okada S, Hatano M, Tokuhisa T, Kimura H, Watanabe S, et al. DNA polymerase theta is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int. J. Cancer. 2004;109:9–16. doi: 10.1002/ijc.11666. [DOI] [PubMed] [Google Scholar]
- 49.Scaringe WA, Li K, Gu D, Gonzalez KD, Chen Z, Hill KA, Sommer SS. Somatic microindels in human cancer: the insertions are highly error-prone and derive from nearby but not adjacent sense and antisense templates. Hum. Mol. Genet. 2008;17:2910–2918. doi: 10.1093/hmg/ddn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gorbunova V, Levy AA. Non-homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucleic Acids Res. 1997;25:4650–4657. doi: 10.1093/nar/25.22.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 52.Feschenko VV, Lovett ST. Slipped misalignment mechanisms of deletion formation: analysis of deletion endpoints. J. Mol. Biol. 1998;276:559–569. doi: 10.1006/jmbi.1997.1566. [DOI] [PubMed] [Google Scholar]
- 53.Ripley LS. Model for the participation of quasi-palindromic DNA sequences in frameshift mutation. Proc. Natl Acad. Sci. USA. 1982;79:4128–4132. doi: 10.1073/pnas.79.13.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greenblatt MS, Grollman AP, Harris CC. Deletions and insertions in the p53 tumor suppressor gene in human cancers: confirmation of the DNA polymerase slippage/misalignment model. Cancer Res. 1996;56:2130–2136. [PubMed] [Google Scholar]
- 55.Brissett NC, Pitcher RS, Juarez R, Picher AJ, Green AJ, Dafforn TR, Fox GC, Blanco L, Doherty AJ. Structure of a NHEJ polymerase-mediated DNA synaptic complex. Science. 2007;318:456–459. doi: 10.1126/science.1145112. [DOI] [PubMed] [Google Scholar]
- 56.Daley JM, Wilson TE. Evidence that base stacking potential in annealed 3′ overhangs determines polymerase utilization in yeast nonhomologous end joining. DNA Repair (Amst) 2008;7:67–76. doi: 10.1016/j.dnarep.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iwabuchi K, Hashimoto M, Matsui T, Kurihara T, Shimizu H, Adachi N, Ishiai M, Yamamoto K, Tauchi H, Takata M, et al. 53BP1 contributes to survival of cells irradiated with X-ray during G1 without Ku70 or artemis. Genes Cells. 2006;11:935–948. doi: 10.1111/j.1365-2443.2006.00989.x. [DOI] [PubMed] [Google Scholar]
- 58.Schulte-Uentrop L, El-Awady RA, Schliecker L, Willers H, Dahm-Daphi J. Distinct roles of XRCC4 and Ku80 in non-homologous end-joining of endonuclease- and ionizing radiation-induced DNA double-strand breaks. Nucleic Acids Res. 2008;36:2561–2569. doi: 10.1093/nar/gkn094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu J, Lieber MR. Mechanistic flexibility as a conserved theme across 3 billion years of nonhomologous DNA end-joining. Genes Dev. 2008;22:411–415. doi: 10.1101/gad.1646608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lieber MR, Lu H, Gu J, Schwarz K. Flexibility in the order of action and in the enzymology of the nuclease, polymerases, and ligase of vertebrate non-homologous DNA end joining: relevance to cancer, aging, and the immune system. Cell Res. 2008;18:125–133. doi: 10.1038/cr.2007.108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.