Abstract
Five Sib antitoxin RNAs, members of a family of cis-encoded small regulatory RNAs (sRNAs) in Escherichia coli, repress their target mRNAs, which encode Ibs toxins. This target repression occurs only between cognate sRNA–mRNA pairs with an exception of ibsA. We performed co-transformation assays to assess the ability of SibC derivatives to repress ibsC expression, thereby revealing the regions of SibC that are essential for ibsC mRNA recognition. SibC has two target recognition domains, TRD1 and TRD2, which function independently. The target site for TRD1 is located within the ORF of ibsC, whereas the target site for TRD2 is located in the translational initiation region. The TRD1 sequence is sufficient to repress ibsC expression. In contrast, TRD2 requires a specific structure in addition to the recognition sequence. An in vitro structural probing analysis showed that the initial interactions at these two recognition sites allowed base-pairing to progress into the flanking sequences. Displacement of the TRD1 and TRD2 domains of SibC by the corresponding domains of SibD changed the target specificity of SibC from ibsC to ibsD, suggesting that these two elements modulate the cognate target recognition of each Sib RNA by discriminating among non-cognate ibs mRNAs.
INTRODUCTION
Over the past decade, researchers have paid increasing attention to the functions of small regulatory RNAs (sRNAs) as gene-expression regulators in bacteria (1–5). Bacterial sRNAs are typically 50–400 nt in length and found in intergenic regions. Approximately 100 sRNAs have been found in Escherichia coli, and recent systematic searches have indicated that hundreds of sRNAs exist (6–12). Functional studies of sRNAs have shown that most are encoded in trans and that they regulate translation and mRNA stability by base-pairing with their target mRNAs, usually with the help of the Hfq protein (13–17). Alternatively, a small number of sRNAs [e.g. CsrB (18,19) and 6S RNA (20)] regulate gene expression by directly binding to their target proteins. In addition, another class of sRNAs encoded on the strands opposite their targets has been found in various bacterial genomes. Unlike trans-encoded sRNAs, these cis-encoded sRNAs share extensive complementary sequences with their targets through overlapping transcribed regions. A variety of regulation mechanisms of cis-encoded antisense sRNAs have been studied and most of these sRNAs have been well characterized in mobile genetic elements such as plasmids, transposons and phages (21,22). These sRNAs include: RNAI–RNAII of ColE1 and CopA–CopT of R1, which regulate plasmid copy number; RNA-IN/RNA-OUT-controlling IS10 transposition; and the Hok/Sok toxin/antitoxin pair of plasmid R1. A key component of antisense regulation is the recognition of target RNAs by cis-encoded sRNAs. Initial base-pairing is required to form a transient intermediate known as a ‘kissing complex’, which can then form a complete duplex (23,24).
Recent studies have also discovered chromosomal cis-encoded antisense sRNAs and identified their biological functions (25–31). With a few exceptions (26–28), most chromosomal cis-encoded RNAs inhibit the synthesis of toxins whose overexpression leads to cell death or growth stasis (25,29–31). Chromosomal antitoxin RNAs are encoded on the opposite strand of overlapping or adjacent toxin genes and promote the translational repression or degradation of toxin mRNAs (32,33). At least six toxin–antitoxin pairs have been identified in the E. coli K-12 genome (21,32,33). Interestingly, some of these pairs are present in multiple copies. The E. coli K-12 chromosome contains five genes that are homologous to the plasmid-derived Hok/Sok genes (34). In addition, four Ldr/Rdl and five Ibs/Sib pairs are present, each with slight sequence variations (25,31). On the other hand, three toxin–antitoxin pairs (i.e. ShoB/OhsC, SymE/SymR and TisB/IstR-1; note that OhsC and IstR-1 are not cis-encoded) are present in single copies in the E. coli K-12 genome (25,30,35).
The chromosomal toxin/antitoxin systems except for ShoB/OhsC and TisB/IstR-1 are regulated by cis-encoded sRNA interactions, but the detailed mechanism remains unclear (32,33). The simplest mechanism would involve the binding of antitoxin RNA to the translation initiation region (TIR) of toxin mRNA, thereby inhibiting the translation or degrading mRNA. SymR represses the synthesis of SymE by base-pairing to the TIR of symE mRNA, leading to symE mRNA degradation (30). Alternatively, the ldrD translational reporter fusion may be repressed by the portion of the RdlD sequence that does not overlap with the TIR of ldrD mRNA (31).
Five Sib antitoxin RNAs (i.e. SibA, SibB, SibC, SibD and SibE) are transcribed from the repetitive Sib sequences differently located in the E. coli K-12 genome (25). These five antitoxin RNAs are approximately 140 nt in length and share high sequence homology. The opposite strand of each sib gene encodes a toxic Ibs protein that consists of 18–19 highly hydrophobic amino-acid residues. Increased expression of Ibs proteins affects the cell envelope and is lethal to the cell. The toxicity of Ibs proteins can be repressed by the constitutive expression of Sib antitoxin RNAs (25).
Interactions between Sib-ibs mRNAs are unique among cis-encoding sRNA regulatory systems in that the sRNA sequence spans the entire functional mRNA sequence, including the TIR and ORF. Therefore, Sib RNAs have the potential for highly extensive base-pairing with their targets. However, the extent of the RNA–RNA interaction required for regulation, and how this interaction occurs, remain unclear. Furthermore, except for the SibA/ibsA pair, Sib RNAs seem to recognize only their cognate target mRNAs by discriminating among the remaining non-cognate ibs mRNAs, even though the Sib RNAs share extremely high sequence identity (25). As to the SibA/ibsA pair, ibsA mRNA appears to be recognized by non-cognate Sib RNAs (25).
In this study, we sought to identify the essential elements and sequences of SibC RNA required for the recognition of ibsC mRNA and then attempted to determine why SibC RNA is highly selective in its recognition of ibsC mRNAs. We found two target recognition domains of SibC, TRD1 and TRD2, which function independently. The sequence of TRD1 is sufficient to repress ibsC expression, whereas TRD2 has a specific structural requirement. The target recognition sequences of TRD1 and TRD2 are always positioned in highly variable regions of the five Sib RNAs. The repression of ibsD by SibC derivatives containing the TRD1 or TRD2 of SibD indicates that these elements are used to discriminate cognate ibs mRNAs from other non-cognate target RNAs.
MATERIALS AND METHODS
Bacterial strains, plasmids and oligonucleotides
Escherichia coli K-12 strain JM109 was used for the construction of plasmids and strain MG1655 was used as the wild-type control. Strains DY330 and MG1655 were used for the construction of knockout strains. Plasmids pBAD/Myc-His B (Invitrogen), pACYC184, pUC19 and pKK232-8 (Amersham Pharmacia) were used as cloning vectors. Plasmid pSS6 (36) was used for construction of an IPTG-inducible RNA expression vector. The oligonucleotides used in this study are listed in Table 1.
Table 1.
Oligonucleotide sequences
| Name | Sequences (5′→3′)a | Use |
|---|---|---|
| SibCKO1 | CCTGATTGACATCGTTGATTCTTTGACCTAATTTAGTGAG TTATGGACAGCAAGCGAA | ΔsibC/ibsC strain |
| SibCKO2 | GATTTACGATGGCAGGGCAGCATGGGGCTGTAAGCTCAGA AGAACTCGTCAAGAAG | ΔsibC/ibsC strain |
| SibDKO1 | GATTGTGTTGTTACTCGTAAGTTTCGCAGCTATGGACAGC AAGCGAACCG | ΔsibD/ibsD strain |
| SibDKO2 | GCCAGGGATATGTGTTAATGAAACCTTCTTTCAGAAGAAC TCGTCAAGAAG | ΔsibD/ibsD strain |
| MscIIC+1 | GTTTGGCCATATGGGGATTTACGATGGCAGGG | pBAD-ibsC |
| IC+ERI | CCGGAATTCAAGGGTAAGGGAGGATTGCTCC | pBAD-ibsC |
| MscIID+1 | GTTTGGCCATAGCGCTCATCAACAATCG | pBAD-ibsD |
| ID+ERI | CCGGAATTCACAAGGGTGAGGGAGG | pBAD-ibsD |
| tacO-F | CGGATCCATAAATGTGAGCGGATAACATTGACATTGTGAGCGG | pAKA |
| tacO-R | CCCAAGCTTGTCGACGACGTCATTATATTGTTATCCGCTCACAATGTC | pAKA |
| AvaIpHM3919 | CCCTCGGGCAGATAAAGAAGACAGTC | pAKA |
| AatIIC-F | GCGACGTCAAGGGTAAGGGAGGATTG | pAKA-SibC |
| Hnd3C-R | CCCAAGCTTGGCTGTAACGGTAAAG | pAKA-SibC |
| AatIID-F | GCGACGTCACAAGGGTGAGGGAG | pAKA-SibD |
| Hnd3D-R | CCCAAGCTTACGGGAAAGCCCCTCCCGA | pAKA-SibD |
| AatIISC+30 | GCGACGTCAGACTGACTGTTAATAAGC | pAKA-SibC(30–141) |
| AatIISC+42 | GCGACGTCAATAAGCGCTGAAACTTATG | pAKA-SibC(42–141) |
| AatIISC+46 | GCGACGTCAGCGCTGAAACTTATGAG | pAKA-SibC(46–141) pAKA-SibC(46–68) pAKA-SibC(46–78) |
| AatIISC+54 | GCGACGTCAACTTATGAGTAACAGTAC | pAKA-SibC(54–141) |
| AatIISC+62 | GCGACGTCAGTAACAGTACAATCAGTATG | pAKA-SibC(62–141) |
| AatIISC+89 | GCGACGTCAGTCGCATCATAACCCTTC | pAKA-SibC(89–141) |
| T51Gf | CTGACTGTTAATAAGCGCGGAAACTTATGAGTAACAG | pAKA-SibC(U51G) |
| T51Gr | CTGTTACTCATAAGTTTCCGCGCTTATTAACAGTCAG | pAKA-SibC(U51G) |
| C87D-R | TGATGAAGCTCGTCATCATACTGATTGTAC | pAKA-SibC87D |
| C87D-F | AACAGTACAATCAGTATGATGACGAGCTTC | pAKA-SibC87D |
| D71C-R | GATGCGACTTGTCATCATACTGATTGTG | pAKA-SibD71C |
| D71C-F | CAACACAATCAGTATGATGACAAGTCGC | pAKA-SibD71C |
| C26D-R | TTAACAGCCAATCGGGAGGAGCAATCCTC | pAKA-SibC26D |
| C26D-F | GCTCCTCCCGATTGGCTGTTAATAAGCTG | pAKA-SibC26D |
| D27C-R | GTCAGTCTCAGAGAGGGGGGAGAAATC | pAKA-SibD27C |
| D27C-F | CTCCCCCCTCTCTGAGACTGACTGTTAA | pAKA-SibD27C |
| D94C-R | AAGGGTTATGATGAAG | pAKA-SibD94C |
| D94C-F | CATCATAACCCTTCTC | pAKA-SibD94C |
| D114C-R | GCTTACAGAAGGAAAGGGTTATGATGAAG | pAKA-SibD114C |
| D114C-F | CCTTTCCTTCTGTAAGCCCTCGCTTCGGTG | pAKA-SibD114C |
| C48D-R | GTTTCGCAGCTTATTAACAGTCAGTCTC | pAKA-SibC48D |
| C48D-F | GTTAATAAGCTGCGAAACTTACGAG | pAKA-SibC48D |
| 1-78-R | CGTTTTATTTCTGATTGTACTGTTACTC | pAKA-SibC(1–78) |
| 1-78-F | GT ACAATCAGAAATAAAACGAAAGGCTC | pAKA-SibC(1–78) |
| 46-68Fu-F | CTGAAACTTATGAGTAACAAAATAAAACGAAAGG | pAKA-SibC(46–68) |
| AatII8-77rB | GCGACGTCAAGGGTAAAGTATGATGACAAG | pAKA-SibC(1–8::77–141) |
| AatII8-99f | GCGACGTCAAGGGTAATAACCCTTCTCC | pAKA-SibC(1–8::99–141) |
| AatII8-90f | GCGACGTCAAGGGTAAGTCGCATCATAAC | pAKA-SibC(1–8::90–141) |
| AatII8-77(M) | GCGACGTCAAGGGTAAAGTAACTACACAAG TCGGTAGTTAACCCTTCTCCTTC | pAKA-SibC(1–8::77–141M3) |
| HndIII8-77 (M) | CCAAGCTTGGCTGTAACGGTAAACGCCAGC CCGAAGACTGGCGTTGAAGGAGAAGG | pAKA-SibC(1–8::77–141M3) |
| AatII8-29rB | GCGACGTCAGGGAGGATTGCTCCTCCCC TGAAATAAAACGAAAGGC | pAKA-SibC(8–29) |
| AatII77-99rB | GCGACGTCAGTATGATGACAAGTCGCAT CATAAATAAAACGAAAGGC | pAKA-SibC(77–99) |
| ERIpHM459 | CCGGAATTCCAAAAGAGTTTGTAGAAACGC | pAKA pAKA-SibC(1–78) pAKA-SibC(46–68) pAKA-SibC(46–78) pAKA-SibC(8–29) pAKA-SibC(77–99) |
| ERI-T7-ibsC | CGGAATTCTTAATACGACTCACTATAGGCAGCATGGGGCTG | Template for in vitro transcription of ibsC |
| ibsC+159 | AAGGGTAAGGGAGGATTGCTCC | Template for in vitro transcription of ibsC |
| ERI-T7-ibsD | CGGAATTCTTAATACGACTCACTATAGGTCAGCAGGGGGCTG | Template for in vitro transcription of ibsD |
| ibsD+163 | AAGGGTGAGGGAGGATTTCTCC | Template for in vitro transcription of ibsD |
| BHI-SP6-SibC | CGGGATCCATTTAGGTGACACTATAGAAGGGTAAGGGAGGATTGC | pUC-SC |
| SibCSmaIERI | CGGAATTCCCGGGTAAAGCCCTCACCGAAG | pUC-SC |
| BHI-SP6-SibD | CGGGATCCATTTAGGTGACACTATAGACAAGGGTGAGGGAGGAT | pUC-SD |
| SibDSmaIERI | CGGAATTCCCGGGAAAGCCCCTCCC | pUC-SD |
| ICBHIibsC(U) | CCGGATCCTGCTGTACTAATTCCCCTTC | ibsC-CAT fusion plasmid |
| ICHd3ibsC(D) | CCCAAGCTTCTCCTTCAAGCCCTCG | ibsC-CAT fusion plasmid |
| ICBHIibsD(U) | CCGGATCCTAAACCCTCTTCCAGG | ibsD-CAT fusion plasmid |
| ICHd3ibsD(D) | CCCAAGCTTTCCTTCTGTAAGGCC | ibsD-CAT fusion plasmid |
| C1d-R | CATAAGTTTCGCAGCTTATTAACAGTCAG | pAKA-SibC1d |
| C1d-F | GTTAATAAGCTGCGAAACTTATGAGTAAC | pAKA-SibC1d |
| C2d-R | CTTACAGAAGGAAAGGGTTATGATGC | pAKA-SibC2d |
| C2d-R | CCTTTCCTTCTGTAAGCCCTCGC | pAKA-SibC2d |
| 5S+90 | GAGACCCCACACTACCATCGG | Northern |
| cat1 | ACGGTGGTATATCCAGTGAT | primer extension |
| cat2 | ATCTCGTCGAAGCTCGGCGG | primer extension |
| C48-50DHr | GATGAAGCTCGTCATCATACTGATTGTAC | Northern |
| NPSC+141 | GGTAAAGCCCTCACCGAAGCGAGGGCTTG | Northern |
| NPSC46-68 | GTTACTCATAAGTTTCAGCGCT | Northern |
| NPSC77-99 | ATGATGCGACTTGTCATCATACT | Northern |
| NPSC8-29 | CAGGGGAGGAGCAATCCTCCCT | Northern |
aRestriction sites are underlined.
Construction of E. coli strains with mutations in the ibsC/sibC or ibsD/sibD loci
We performed PCR to construct linear DNA inserts (i.e. ibsC::kan or ibsD::kan), in which a kanamycin resistant gene was substituted for the ibs ORF and flanked by the 5′ and 3′ UTRs of ibs. These inserts were then used to generate the ibsC/sibC knockout (ΔibsC/sibC) and ibsD/sibD knockout (ΔibsD/sibD) strains from the wild-type strain MG1655, as previously described (37,38). Each construct was confirmed by sequence analysis of the PCR-amplified knockout regions.
Plasmid construction
Plasmid pAKA, an RNA expression vector and derivative of pACYC184, was constructed. The tac promoter of pSS6 was modified by substituting a lac operator sequence (5′-TCCCTATCAGTGATAGAGA-3′) for the original promoter sequence at two different sites (i.e. immediately upstream of the −35 element as well as between the −10 and −35 elements) and by inserting the AatII/HindIII restriction sites immediately downstream of the −10 element to allow for transcription of a cloned gene beginning at the same 5′ end as that of endogenous RNA. We then performed PCR with primers tacO-F and tacO-R to modify the tac promoter. The PCR product was cloned into the BamHI/HindIII sites of pSS6 to generate pHM1. The modified promoter transcription unit, including the rrnB terminator and the lacI gene of pSS6, were PCR-amplified and cloned into the AvaI/EcoRI sites of the HindIII-inactivated pACYC184 to generate the pAKA vector. The sibC- and sibD-coding sequences were amplified from E. coli MG1655 genomic DNA by PCR and cloned into the AatII/HindIII sites of the pAKA vector, thereby generating pAKA-SibC and pAKA-SibD, respectively. The 5′ truncated SibC derivatives were constructed by amplifying the corresponding SibC coding regions. Substituted derivatives were generated by introducing mutations into pAKA-SibC using the Stratagene QuickChange mutagenesis kit. Chimeric derivatives of SibC/D were constructed according to the SOEing PCR method (39). Briefly, the first PCR reactions were performed using primer pairs AatIIC-F/CD-R and DC-F/Hnd3D-R. The resulting PCR products were mixed and further amplified via a second PCR reaction using the AatIIC-F/Hnd3D-R primer pair to generate the chimeric 5′SibC/3′SibD coding sequences. The 5′SibD/3′SibC coding sequences were also generated using the SOEing PCR method, with primer pair AatIID-F/DC-R and CD-F/Hnd3C-R used for the first reaction and primer pair AatIID-F/Hnd3C-R used for the second reaction. The chimeric DNA fragments were cloned into the AatII/HindIII sites of pAKA. We generated sibC coding sequences containing a 3′ deletion and linked them directly to the rrnB terminator using the SOEing method. The 3′ deleted sibC sequences were cloned into the AatII/EcoRI sites of pAKA. DNA fragments containing the additional 5′ deletion as well as the 3′ deletion were amplified from the 5′ deletion points by PCR using the corresponding 3′ deletion derivatives as templates. If necessary, the sequences of + 1 to + 8 were also added to the 5′ deletion derivatives using the SOEing method. The sequences encoding Sib(1–8::77–141M3), SibC1d, SibC2d and SibC12d were also PCR-amplified using the SOEing method and cloned into the pAKA vector.
The ibsC and ibsD expression plasmids were constructed by amplifying nt −21 to +159 of the ibsC sequence and nt −21 to +165 of the ibsD sequence, relative to the respective +1 transcription start sites. The amplified fragments were then cloned into the MscI/EcoRI sites of pBK vector, a derivative of pBAD/Myc-His B (Invitrogen), where an additional MscI site was introduced to facilitate cloning.
We constructed the ibsC-CAT and ibsD-CAT fusion plasmids to determine the transcription start sites of ibsC and ibsD. Promoter-containing DNA fragments, including the 232 (for ibsC) and 236 (for ibsD) bases upstream of position −8 relative to the translation initiation codon, were obtained by amplifying genomic DNA using primer pairs ICBHIibsC(U)/ICHd3ibsC(D) and ICBHIibsD(U)/ICHd3ibsD(D), respectively. The resulting PCR products were cloned into the BamHI/HindIII sites of pKK232-8 to generate the transcriptional fusion plasmids.
To assess the in vitro transcription of SibC and SibD, the T7 promoter-containing forward primers and SmaI site-containing reverse primers were used to amplify the sibC and sibD structural genes. The amplified DNA fragments were cloned into pUC19 to generate pUC-SC and pUC-SD.
Co-transformation assay
Co-transformation efficiency was assessed by transforming 25 µl of competent cells with 5 ng each of an ibs expression plasmid and a Sib expression plasmid. Cells were then resuspended in a final volume of 300 µl of LB media and placed at 37°C with shaking for 1 h. Cells were incubated on LB plate containing 10 µg/ml of tetracycline, 50 µg/ml of ampicillin and 1 mM of IPTG for 16 h at 37°C. Colony forming ability was then examined.
Preparation of total cellular RNA
Cells were grown overnight in LB broth containing ampicillin (50 μg/ml) and if necessary, tetracycline (10 μg/ml). After performing a 1:200 dilution of the overnight culture in fresh LB, cells were grown at 37°C to an OD600 of 0.5. A final concentration of 1 mM IPTG was added to the cell culture, followed by another incubation period. Total cellular RNAs were extracted from the culture as previously described (37).
Northern blot analysis
In preparation for northern blot analysis, RNA samples (20 µg) isolated from MG1655 cells containing an ibs expression plasmid and a Sib expression plasmid were electrophoresed on a 5% polyacrylamide gel containing 7 M urea and electrotransferred to a Hybond N+ membrane (Amersham Biosciences). The membranes were then hybridized with oligonucleotide probe C48-50DHr for SibC, chimeric SibC/D, and SibD. The C48-50DHr could hybridize with both SibC and SibD. The 5′-deleted SibC derivatives were probed with NPSC+141. The 3′-deleted SibC derivatives were probed with the corresponding Sib-antisense oligonucleotides (NPSC46-78, NPSC77-99 and NPSC8-29). 5S RNA was probed with oligonucleotide 5S+90. Oligonucleotides were labeled at the 5′ end with [γ-32P] ATP and T4 polynucleotide kinase (Takara) and used for hybridization.
Primer extension analysis
Two primers cat1 (5′-ACGGTGGTATATCCAGTGAT) and cat2 (5′-ATCTCGTCGAAGCTCGGCGG) were labeled with [γ-32P] ATP at the 5′ end using T4 polynucleotide kinase. The pimers were then used to analyze ibsC-CAT and ibsD-CAT fusion transcripts. Total cellular RNA was isolated from cells carrying the ibsC-CAT or ibsD-CAT fusion plasmid. The two different primers were separately used for primer extension analysis, as previously described (37).
In vitro transcription
SibC and SibD RNAs were prepared by in vitro transcription using SP6 RNA polymerase (Promega), with SmaI-digested pUC-SC and pUC-SD as templates. ibsC and ibsD mRNA transcripts were generated using T7 polymerase and PCR products that had been amplified using primer pair ERI-T7-ibsC/ibsC+159 and ERI-T7-ibsD/ibsD+163, respectively. The transcripts each contained an extra G sequence at their 5′ end because of the SP6 or T7 promoter sequence.
In vitro RNA-binding assay
In preparation for binding assays, 5′ end-labeled SibC or ibsC RNA (10 nM) was incubated with unlabeled ibsC or SibC (10, 25 or 100 nM) in 10 µl TMN buffer [100 mM Tris–acetate, pH 7.6, 500 mM NaOAc, 25 mM Mg(OAc)2] at 37°C for 15 min. The reaction was stopped by dilution with an equal volume of non-denaturing loading buffer (0.025% xylene cyanol, 2% glycerol, 1× TBE). The reactions were then analyzed on 5% (v/v) polyacrylamide gels.
Chemical and enzymatic structure probing
SibC and ibsC mRNAs were 5′ end-labeled with [γ-32P] ATP using T4 polynucleotide kinase (Takara). The labeled RNA molecules were renatured by heating for 1 min at 95°C, chilling on ice for 3 min and slowly returning to room temperature. The 5′-end-labeled RNAs were pre-incubated with 1 µg yeast tRNA in 5×TMN buffer for 15 min at 37°C. Unlabeled ibsC mRNA or SibC RNA was then added to the reaction, yielding a total reaction volume of 10 µl. The reactions were then incubated at 37°C for 15 min. Subsequently, 1 µl of RNase V1 (0.01 U, Ambion), RNase T1 (0.75 U, Ambion), Pb(II) (25 mM, Sigma Aldrich) or RNase III (1 U, New England Biolabs) was added and the reactions was incubated at room temperature for various time intervals. The RNase III reaction was performed in additional 1 mM DTT. The reaction was stopped by adding 2 µl of 0.5 M EDTA. The RNAs were precipitated and resuspended in 10 µl gel loading buffer II (Ambion). Samples were denatured at 95°C for 3 min and immediately loaded on a 5% (v/v) polyacrylamide-9 M urea sequencing gel. RNase T1 ladders and OH ladders were obtained according to the Ambion manufacturers’ specifications.
The binding rate assay to evaluate RNase-cleavage protection was performed as previously described (40). Briefly, 5′ end-labeled ibsC mRNA (10 nM) was incubated with unlabeled SibC (50 nM) in a total reaction volume of 10 µl in TMN buffer at 37°C for several time points. Samples were immediately cleaved by RNase V1 (0.01U) for 15 min and the cleavage reaction was stopped by ethanol precipitation. The products were analyzed on a 5% polyacrylamide gel as described above.
RESULTS
Co-transformation assays for the evaluation of ibs repression by Sib RNA derivatives
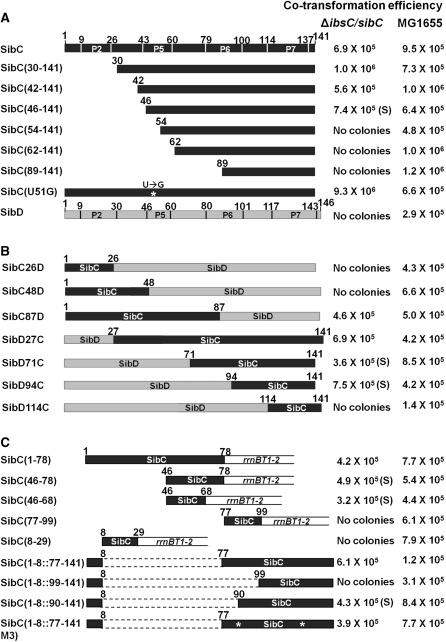
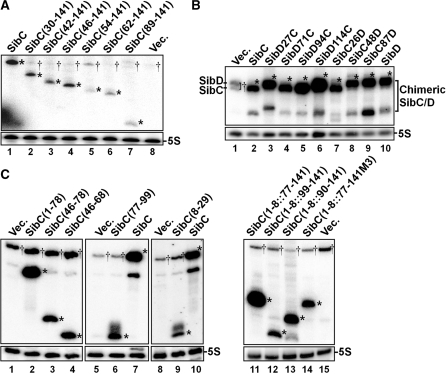
We evaluated the ability of Sib RNA derivatives to repress ibs expression by taking advantage of the fact that Ibs toxicity is repressed by the overexpression of Sib RNA (25). We expressed ibs and Sib RNA from an arabinose-controlled promoter and an IPTG-controlled promoter, respectively, harbored on the plasmid in vivo. Wild-type MG1655 cells were transformed with an ibsC expression plasmid and the transformed cells grew in the absence of arabinose. However, the ΔibsC/sibC strain could not be transformed with the ibsC expression plasmid, indicating that the basal level of ibs expressed by the plasmid is sufficient to induce cell toxicity in the absence of SibC expression and that this cell toxicity can be repressed by endogenously expressed SibC. Indeed, the ibsC expression plasmid could be introduced into the knockout cells by co-transformation with the IPTG-induced SibC expression plasmid (Figure 1). Co-transformants were not generated with a SibD RNA expression plasmid, suggesting that IbsC toxicity is repressed by cognate SibC RNA, rather than by non-cognate SibD, consistent with previous reports (25). Thus, we performed co-transformation assays to evaluate ibsC repression by SibC RNA derivatives. The colony-forming ability of ΔibsC/sibC cells was assessed with co-transformation efficiency in the presence of 1 mM IPTG after cells were transformed with both the RNA and ibsC expression plasmids. We also performed northern blot analyses to confirm the overexpression of Sib RNA derivatives. We found that all derivatives are expressed at higher levels than endogenous SibC (Figure 2).
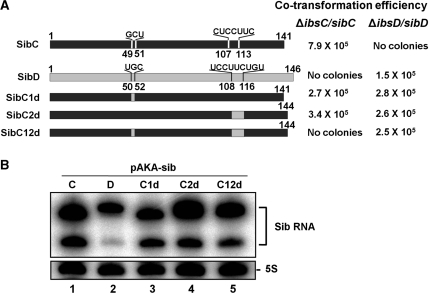
Figure 1.
Co-transformation assays were performed using various derivatives of SibC. The schematically drawn sequences were cloned into pAKA to generate, SibC, SibC derivatives and SibD in vivo. Co-transformation efficiency was assessed in cells co-transformed with 5 ng each of an ibs expression plasmid and a Sib RNA expression plasmid by counting the number of transformants. The data are representative of at least three separate experiments. The error was <20%. (A) SibC derivatives truncated at the 5′ end. (B) SibC/D chimeric derivatives. (C) 3′ Deletion derivatives and minimal motif-carrying derivatives. ‘S’ in parentheses indicates formation of small colonies. Stem-loop regions shown in Figure 3 are indicated in the schematic structure of SibC and SibD.
Figure 2.
Expression of Sib derivatives. MG1655 cells containing the pAKA-Sib plasmids, as shown in Figure 1, were treated with IPTG (final concentration of 1 mM). Total cellular RNAs were prepared from these cells and subjected to northern blot analysis. (A) SibC derivatives truncated at the 5′ end were probed with NPSC+141. (B) SibC/D chimeric derivatives were probed with C48–50DHr. (C) 3′ Deletion derivatives and minimal motif-carrying derivatives were probed with NPSC46-68 (lanes 1–4), NPSC77-99 (lanes 5–7) and NPSC8-29 (lanes 8–10). Vec., vector DNA. Plasmid-borne Sib derivatives and endogenous Sib RNA species are marked with asterisks and daggers, respectively. When Sib RNAs were highly expressed, their truncated species lacking about 30 nt at the 3′ end were also observed.
5′ Deletion analysis of SibC RNA
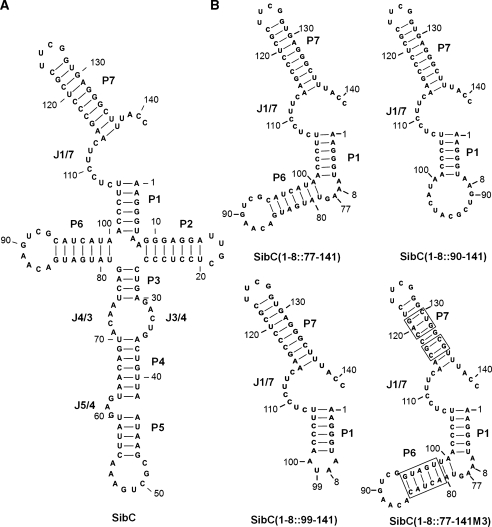
To identify the sequences of SibC RNA that affect ibsC repression, we constructed plasmids expressing SibC derivatives that were sequentially truncated from the 5′ end (Figure 1A). These truncated derivatives were used in co-transformation assays. A 29-nt deletion on the 5′ end [i.e. SibC(30–141)] did not alter colony-forming ability of ΔibsC/sibC strains, compared with the wild-type strain. However, further deletion to +61 [i.e. SibC(62–141)] and +89 [i.e. SibC(90–141)] eliminated colony formation. These results indicate that nt +1 to +29 of SibC RNA are not required for ibsC mRNA recognition, while the nucleotides between +30 and +61 are essential for ibsC mRNA recognition. To further delineate the sequences that are essential for ibsC repression, we constructed three additional deletion derivatives, SibC(42–141), SibC(46–141) and SibC(54–141) (Figure 1A). SibC(42–141) and SibC(46–141) yielded colonies, but SibC(54–141) did not. Hence, we narrowed the essential region to the nucleotides between +46 and +53 of SibC. It is noteworthy that SibC(46–141) yielded smaller colonies than did the other colony-forming plasmids, suggesting that SibC(46–141) is a less efficient repressor. In a secondary structure model of SibC, the sequence between +46 and +53 lies on the P5 loop (Figure 3). However, a U-to-G modification in this loop [i.e. SibC(U51G)] did not affect colony formation (Figure 1A).
Figure 3.
Possible secondary structures of SibC (A) and its derivatives (B). The structures were predicted using the CLC workbench program, version 4.2. The stem regions of SibC are assigned as P1 through P7. The single-stranded regions connected between two stems are denoted by J(preceding stem number)/(next stem number); e.g. J1/2 is the junction region between stems 1 and 2.
SibC/D chimeras
It is possible that the 5′ truncated SibC derivatives were non-functional because the RNA structure was significantly altered by the deletion. Another possibility is that there are multiple recognition sites, which function independently. Because SibD did not repress ibsC expression in the co-transformation assays, even though they share 82% sequence similarity and have a conserved secondary structure, we constructed SibC/SibD chimeric derivatives in which the 5′ or 3′ regions of SibC were replaced with the corresponding regions of SibD (Figure 1B). First, we examined 5′-SibC/3′-SibD chimeras (e.g. SibC26D and SibC48D) and the SibC87D construct where the numbers indicate the 3′ fusion points of SibC. The SibC26D and SibC48D constructs did not repress ibsC expression, whereas SibC87D did. These data indicate that the +1 to +48 sequence of SibC is not involved in target recognition and that the +49 to +87 sequences is sufficient for recognition. This result is consistent with our 5′ deletion analysis showing that the sequence between +46 and +53 is essential for target recognition. We then examined 5′-SibD/3′-SibC chimeras (i.e. SibD27C, SibD71C, SibD94C and SibD114C, where numbers indicate the 5′ fusion points of SibC). As expected, SibD27C repressed ibsC expression. Surprisingly, SibD71C and SibD94C also showed ibsC repression, although their colony sizes were small. This conflicts with the result of our 5′ deletion analysis, which showed that 5′ 53-nt deletion mutants lost the ability to repress transcription. The SibD-extended version of SibD114C did not exhibit ibsC repression. Therefore, it is likely that the +95 to +113 sequence of SibC is included in another independent element capable of repressing ibsC expression.
More deletion analyses
We performed further deletion analyses, including 3′ deletions (Figure 1C). Because the 3′ deletion could disrupt or remove the sibC termination hairpin, constructs were designed so that the deletion points were fused to the rrnB terminator hairpin and transcription would terminate at the terminator. As expected from our observations of SibC/D chimeric derivatives, ibsC expression was repressed by SibC(1–78), which harbored a deletion from the 3′ end to nucleotide +79. Further deletion derivatives SibC(46–78) and SibC(46–68) also repressed ibsC expression, although they yielded small colonies. Therefore, the 23 nt between +46 and +68 are sufficient for ibsC repression. This region covers the P5 loop and includes the essential recognition sequence of +46 to +53, which was demonstrated by 5′ deletion analysis. SibC(8–29) and SibC(77–99), which contained loops P2 and P6, respectively, did not exhibit ibsC repression, indicating that the loop sequence of P5 specifically promotes ibsC repression (Figure 3).
We then sought to determine why a construct harboring a 61-nucleotide deletion on the 5′ end lost the ability to repress ibsC, while the replacement of 93 nt at the 5′ end of SibC with the corresponding SibD sequence in a SibC/D chimeric construct did not alter repression. The replaced SibD sequence in SibC/D chimeric constructs might provide structural platforms that are essential for the function of the remaining SibC sequence. As nt +95 to +113 of SibC are involved in ibsC repression (Figure 1B), we focused on an RNA structure that includes this sequence. Because this structure may contain the P1 stem, we added nt +1 to +8 to the ineffective 5′ deletion derivatives to create the P1 stem. SibC(1–8::77–141) repressed ibsC (Figure 1C), indicating that the P1 stem generated by the addition of the +1 to +8 sequence is required for repression by the +77 to +141 sequence. The SibC(1–8::99–141) construct, which lacked the P6 stem and loop, did not repress ibsC (Figures 1C and 3). To determine if the lack of repression by SibC(1–8::99–141) was due to a sequence or structure requirement, we generated a SibC(1–8::90–141) construct that included more sequence but lacked the P6 structure. SibC(1–8::90–141) repressed ibsC, albeit yielding small colonies (Figure 1C), suggesting that the P6 structure is not crucial but the +90 to +98 nt are required for ibsC repression. Further mutational analysis was performed to define the minimal sequence required for complementary base-pairing in the P1-containing structure. We introduced mutations into stems P6 and P7 of SibC(1–8::77–141) by exchanging the complementary stem sequences so that the mutant constructs would retain the same secondary structure (Figure 3B). SibC(1–8::77–141M3), which contained the 17 nt from +99 to +115, repressed ibsC (Figure 1C), indicating that this contiguous sequence is sufficient for the specific recognition of ibsC mRNA. However, this result is inconsistent with our finding that SibC(1–8::99–141), which carries this 17-nt sequence, was incapable of repression. The reasons for this discrepancy remain unclear, although it is likely that the 17-nt sequence may require an additional sequence element, such as the P6 loop in SibC(1–8::77–141M3), the +90 to +98 sequence in SibC(1–8::90–141), the +94 to +98 sequence in SibD94C. Together, our data indicate that SibC RNA possesses at least two target recognition domains that function independently. These domains are encoded by nt +46 to +68 and by nt +99 to +115, which we have named target recognition domains 1 and 2 (TRD1 and TRD2), respectively. TRD1 consists of the P5 loop, whereas TRD2 contains J1/7 and is active only when bound to a specific structure that contains at least the P1 stem (Figure 3).
Interactions between SibC and ibsC mRNA
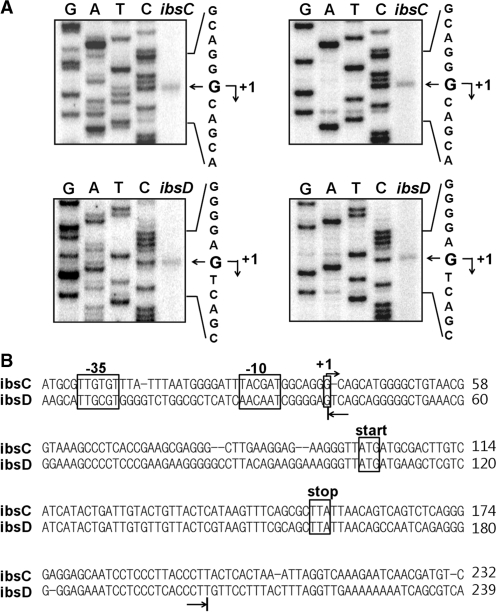
To analyze the interactions between SibC and ibsC mRNA, we performed structure-probing experiments. The 5′ and 3′ ends of Sib RNAs were well defined, but those of the ibs mRNAs were not. Previous northern-blot analysis showed that the ibsC mRNA was approximately 160-nt long (25); thus, we attempted to determine the 5′ ends of the ibsC and ibsD mRNAs by primer extension analysis and to predict the location of the 3′ end by measuring its distance from the 5′ end. The putative promoter regions were cloned into pKK232-8, a promoterless CAT expression vector, and the 5′ ends of the ibs-CAT transcriptional fusion mRNA were determined (Figure 4A). The upstream regions of the ibsC and ibsD genes contained conserved promoter elements and the 5′ ends corresponded with the putative transcription initiation sites (Figure 4B). From these data, we synthesized ibsC and ibsD mRNAs of 159 and 163 nt, respectively, by in vitro transcription.
Figure 4.
Identification of the ibsC and ibsD transcriptional start sites. Total cellular RNAs were prepared from cells containing the ibsC-CAT or ibsD-CAT fusion plasmid, then subjected to primer extension analysis. (A) Extension products from two different primers cat1 (left) and cat2 (right) were electrophoresed on a 5% polyacrylamide sequencing gel containing 7 M urea. The DNA sequencing ladders were prepared using the same primers and ibsC-CAT or ibsD-CAT fusion plasmid DNA as the template. (B) Promoter elements are shown in the ibsC or ibsD sequence. Transcription start sites determined by primer extensions analysis are indicated by arrows. The 5′ and 3′ ends of in vitro-transcribed ibsC and ibsD RNA used for the in vitro binding assay and structural mapping in this study are indicated by the arrows below the sequences.
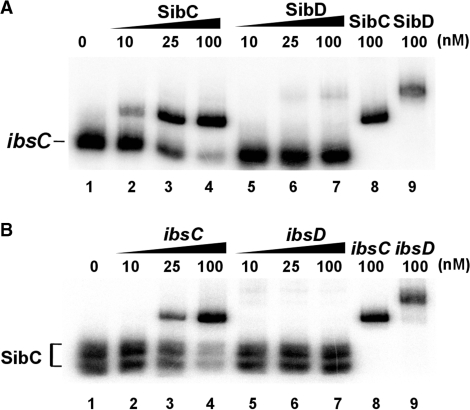
Electrophoretic mobility shift assays showed that SibC specifically binds to ibsC mRNA to form a SibC–ibsC complex at 37°C, whereas non-cognate SibD failed to recognize ibsC mRNA (Figure 5). When SibD and ibsC mRNA were denatured at 95°C and then annealed, the SibD–ibsC complex formed (Figure 5, lane 9). This result indicates that SibD can recognize ibsC mRNA via base-pairing through the denaturation and annealing process, but not naturally.
Figure 5.
Binding assay of SibC RNA with ibsC mRNA. (A) 32P-labeled SibC RNA (10 nM) was incubated at 37°C for 15 min with increasing amounts of unlabeled ibsC mRNA or ibsD mRNA (10 nM to 100 nM). The binding mixtures were divided; one group of samples (lanes 1–7) was loaded directly on the 5% native gel, whereas the other group of samples (lanes 8 and 9) was heated 95°C for 3 min and slowly cooled to room temperature prior to loading on the 5% native gel. (B) Reciprocal interactions between the 32P-labeled ibsC mRNA and the unlabeled SibC RNA were assessed as described in (A).
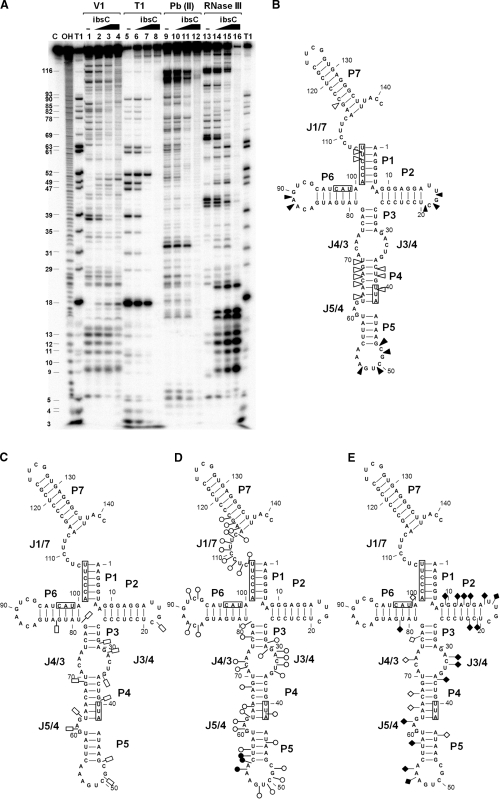
The interactions between SibC and ibsC were analyzed by RNA structure mapping. The 5′ end-labeled SibC RNA was probed with RNase T1, lead(II), RNase V1 and RNase III in the presence of increasing amounts of ibsC mRNA (Figure 6 and Supplementary Figures S1–S4). The effects of ibsC mRNA on each cleavage pattern were observed over most of the SibC sequence, indicating extensive base-pairing of the SibC–ibsC mRNA complex. However, this RNA–RNA complex appears to not form a complete duplex because the cleavage patterns differed from those of a SibC–ibsC duplex generated by denaturation and annealing. In the presence of ibsC mRNA, fewer RNase T1 cleavages were observed within the region spanning G18 to G90. Prominent reductions of RNase T1 cleavages were observed at G47 and G49 within the P5 stem and loop (Figure 6A, lanes 6 and 7; Figure 6C). Binding of ibsC mRNA extensively reduced lead(II) cleavages of SibC RNA (Figure 6D). The reduced regions were extended from G29 to C116. Fewer lead(II) cleavages were observed in the region spanning C106 to C116; however, RNase T1 cleavages were not altered because this region contains no G nucleotides (Figure 6A, lanes 10 and 11; Figure 6C). Two regions were predominantly protected: the G48 to G50 residues of stem P5 and the C107 to C113 residues of J1/7. Interestingly, A54 became more susceptible to lead(II) cleavage upon binding to ibsC mRNA, suggesting a marked structural rearrangement of the P5 loop after binding. RNase V1 cleavage patterns revealed an increase in cleavage in three loop regions of stems P2, P5 and P6. Therefore, these three regions are likely to interact with ibsC mRNA. TRD1 and TRD2 are located in P5 and J1/7; hence, the interaction of stems P2 and P6 with ibsC mRNA should be non-productive. RNase V1-sensitive sites near the J7/1 region became resistant to the enzyme and this resistance was also found in the complete duplex (Figure 6A, lanes 3 and 4; Figure 6B). These findings indicate that interactions in this region promote a marked structural change that might result in longer duplex formation. RNase III, a double-strand-specific nuclease that cleaves full duplexes of about 20 or more base-pairs (41), almost completely digested the fully base-paired SibC–ibsC mRNA duplex by generating only short fragments, while the enzyme partially digested the SibC–ibsC mRNA complex with generating more cleavage products (Figure 6A, lane 15 and 16). These results suggest that the SibC–ibsC mRNA complex with unpaired regions is a poorer substrate for RNase III than the complete duplex. The RNase III treatment of the complete duplex generated fragments of 9–21 and 21–34 nt from the labeled 5′ ends of SibC and ibsC mRNA, respectively (Figures 6 and 7). These cleavages are quite consistent with the observation that the cleavage site of RNase III is 10–14 bp away from 3′ ends of RNA duplex (42), considering that the complete SibC–ibsC mRNA duplex has a 5′ overhang of 18 nt in the ibsC mRNA sequence and that the RNase III cleavage of this duplex could result in that much longer fragments from the labeled 5′ ends of ibsC mRNA (Figure 7) than SibC in the duplex (Figure 6). As for the SibC–ibsC mRNA complex, RNase III-cleavage patterns markedly changed as the concentration of target RNA increased, suggesting that interactions between SibC and ibsC promote overall structural changes in RNA.
Figure 6.
Interaction of SibC with ibsC mRNA. (A) 32P-labeled SibC RNA (10 nM) was partially digested with RNase V1, RNase T1, lead(II) and RNase III in the absence (lanes 1, 5, 9 and 13) and presence of increasing amounts of unlabeled ibsC mRNA (25 nM final concentration: lanes 2, 6, 10 and 14; 100 nM final concentration: lanes 3, 7, 11 and 15). The fully paired SibC–ibsC mRNA complex was generated by heating and annealing with 100 nM ibsC. The complex was then used for digestion (lanes 4, 8, 12 and 16). Untreated SibC and alkaline ladders are shown in lanes C and OH, respectively. Lane T1 corresponds to the RNase T1 ladders of denatured SibC RNA. The positions of the cleaved G residues are shown on the left side of the gel. The altered sites of RNase V1 (B), RNase T1 (C), lead(II) (D) and RNase III (E) cleavage/protection in the presence of ibsC mRNA are shown in the secondary structure of SibC. The complementary sequences corresponding to the S/D region, start codon and stop codon of ibsC mRNA are indicated by boxes. Increasing and decreasing cleavages are indicated by solid and open symbols, respectively.
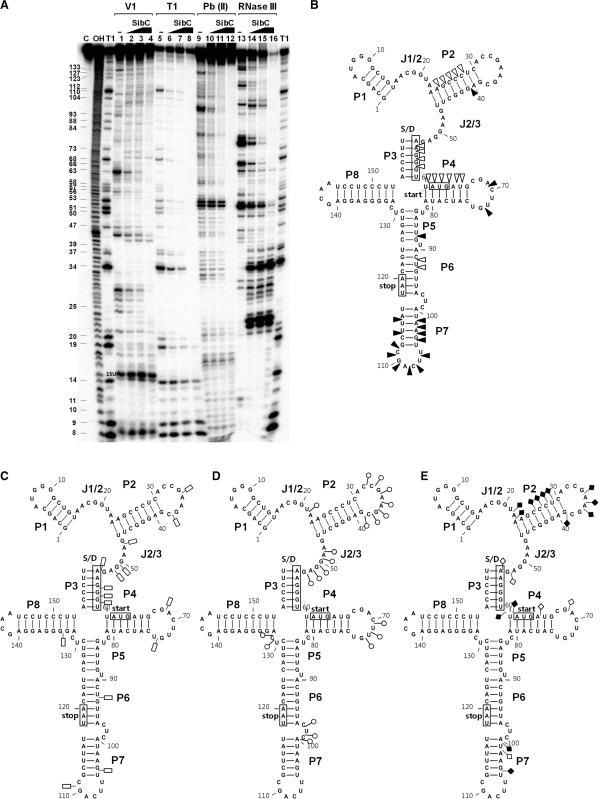
Figure 7.
Interaction of ibsC mRNA with SibC. (A) A reciprocal enzymatic and chemical footprint experiment was performed by labeling the 5′ end of ibsC mRNA, as described in Figure 6. The RNase V1 (B), RNase T1 (C), lead(II) (D) and RNase III (E) protected or cleaved residues are shown in the secondary structure model of ibsC. The S/D sequence, start codon and stop codon are indicated by boxes. Increasing and decreasing cleavages are indicated by solid and open symbols, respectively.
We performed a reciprocal probing experiment using 5′-labeled ibsC mRNA (Figure 7 and Supplementary Figures S5–S8). The addition of SibC changed the cleavage patterns of several regions within ibsC mRNA. The effects of SibC RNA were observed throughout the ibsC mRNA sequence. RNaseT1 and lead(II) probing data showed that cleavages were reduced over most of the ibsC mRNA sequence. RNase T1 cleavages at G50, G51 and G53 of J2/3 were completely blocked, likely due to base-pairing with C107, C109 and C110 of TRD2 of SibC. However, this region in the absence of SibC was cleaved by lead(II) but not by RNase V1, indicating that this region forms a special structure that undergoes dynamic changes in response to interactions with SibC. The three loop regions (i.e. loops P2, P4 and P7) became sensitive to RNaseV1 cleavage (Figure 7A, lanes 2 and 3: Figure 7B). The P7 loop of ibsC mRNA is complementary to the P5 loop of SibC, which includes the TRD1. The P7 loop contains a YUNR motif, which was previously reported to be involved in the initial RNA–RNA recognition (43). This YUNR motif might also play a crucial role for the initial contact between ibsC and SibC. The ibsC mRNA exhibited distinct RNase III-cleavage patterns in response to SibC binding, similar to the SibC probing data.
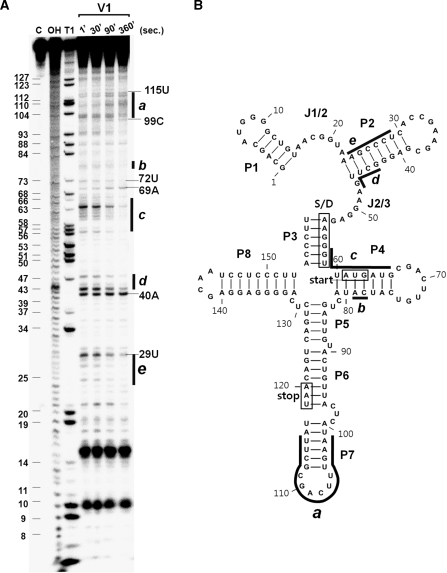
Our structure probing data showed that SibC–ibsC mRNA interaction occurred at multiple sites. We performed a time-course experiment to determine whether TRD1 and TRD2 are the initial interaction sites from which RNA–RNA base-pairing commences. The 5′ end-labeled ibsC was probed with RNase V1 at various time points during incubation with unlabeled SibC (Figure 8 and Supplementary Figure S9). Cleavages at the P7 stem-loop region that could be recognized by TRD1 gradually increased with the reaction time. RNase V1-resistance of the J2/3 region, which is complementary to TRD2, was not observed in the presence of SibC because this region was already resistant to RNase V1 without SibC; however, cleavage decreased in the flanking stems (i.e. P2 and P3), suggesting that the stems are rearranged to form a longer duplex. In contrast, cleavages at residues A69 and U72 of the P4 loop were visible immediately but further changes were not observed over time. This may explain why interactions between the P6 loop of SibC and the complementary P4 loop of ibsC were non-productive in our co-transformation assays. These results indicate that two independent recognition elements (i.e. TRD1 and TRD2) are initial contact sites that allow base-pairing to proceed into nearby sequences, thereby promoting longer RNA–RNA duplex formation. When the interaction is followed by pseudo-first-order kinetics, densitometric analysis of cleavage bands in the P7 loop and P2 stem of ibsC yields binding rate constants of ∼5 × 105 and ∼2 × 105 M−1s−1, respectively. These values are similar to previously reported information about the stable antisense/sense RNA complex (∼106 M−1s−1) (21).
Figure 8.
Time course of the interactions between SibC and ibsC RNA. (A) 32P-labeled ibsC RNA (10 nM) was incubated with 50 nM of unlabeled SibC RNA at 37°C for 1, 30, 90 and 360 s. Samples were withdrawn from each time point and digested with RNase V1 for 15 min. The reaction was stopped by ethanol precipitation. Lanes C and OH correspond to untreated ibsC RNA and alkaline hydrolysis ladders, respectively. Lane T1 corresponds to G ladders generated by RNase T1 under denaturing conditions. The regions showing significant changes in the intensities of bands are denoted as a to e. (B) The regions showing significant changes during the time course of incubation are also shown in the secondary structure model of ibsC mRNA.
Specificity between SibC and ibsC mRNA
We have identified two target recognition domains of SibC: TRD1 and TRD2. Because Sib RNAs do not cross-react with non-cognate ibs mRNAs with the exception of the ibsA/SibA pair, we tested whether these two elements influence target specificity. We compared the sequences of five pairs of Sib RNA species. The TRD1 and TRD2 domains are positioned within highly variable regions of the five Sib RNAs. Furthermore, our results showed that these two elements exhibit high sequence variation and can severely disrupt the contiguity of a sequence when the Sib RNAs occur in any combination of pairs (Supplementary Figure S10), suggesting that these elements play a role in target discrimination. Thus, we replaced one or both of the elements in SibC with those of SibD. The changes from G49CU51 to UGC and from C107UCCUUC113 to UCCUUCUGU allowed SibC to base-pair with ibsD through mutated TRD1 and TRD2 elements, respectively. Mutated TRD1 and TRD2 elements can form contiguous 22 and 25-bp helices, respectively, with ibsD mRNA. A change in either one of the two TRD elements of SibC represses ibsC (Figure 9). However, a change in both elements eliminated the repressive activity of SibC, even though the mutant SibC still contained approximately 100 nt that were complementary to the ibsC mRNA. These data provide further evidence that SibC has two elements for ibsC recognition and that these elements function independently. The Sib derivatives containing either one of the two elements were able to repress ibsD (Figure 9), suggesting that through these elements Sib RNAs can discriminate cognate ibs mRNAs from non-cognate target RNAs.
Figure 9.
Co-transformation assays with SibC derivatives harboring mutations at the TRDs. (A) Co-transformation efficiency was assessed in cells transformed with both an ibs expression plasmid and an Sib RNA expression plasmid, as described in Figure 1. The TRD1 and TRD2 domains of SibC1d and SibC2d were substituted with those of SibD, respectively. SibC12d possesses the TRD1 and TRD2 domains of SibD. (B) To assess the expression of SibC derivatives, MG1655 cells containing pAKA-Sib plasmids were treated with IPTG (final concentrations of 1 mM). Total cellular RNAs were prepared from these cells and subjected to northern blot analysis.
DISCUSSION
In this study, we identified the essential elements and sequences of SibC RNA that are required for the recognition of target ibsC mRNA. SibC RNA has two target recognition domains, TRD1 and TRD2. Either of these domains can repress ibsC expression; therefore, the domains function independently. The target recognition domains TRD1 and TRD2 span the regions of +46 to +68 containing the P5 loop and +99 to +115 including the J1/7 single-stranded region, respectively. TRD1 is sufficient to repress ibsC expression. However, TRD2 appears to require the formation of the P1 stem because TRD2 is functional only in the presence of the sequence of +1 to +8 that could form the P1 stem with the +101 to +106 sequence of TRD2. Since the TRD2 sequence is followed by the P7 transcription terminator stem, the +107 to +114 sequence becomes positioned in the J1/7 region. This positioning of TRD2 in the SibC sequence might be needed to maintain its recognition sequence as a single-stranded segment. In addition to this single-strandedness, TRD2 seems to require another interaction through the P6 loop, because SibC(1–8::99–141), a derivative consisting of the P1 stem and loop, J1/7, and the P7 stem-loop, cannot repress ibsC expression unless it holds an additional GUCG sequence of the P6 loop as in the active SibC(1–8::90–141) and SibC(1–8::77–141M3) derivatives. It is noteworthy; however, that the spatial locations of SibC target sites within ibsC mRNA may be important for the SibC–ibsC mRNA interaction. In case of truncated SibC derivatives, their RNA structures might differ from that of the intact SibC and there is a possibility for generating a new target recognition domain in the truncated derivatives. Nonetheless, they recognized ibsC mRNA only when TRD1 existed. Therefore, the target sequence of TRD1 within ibsC mRNA could be located in an accessible region. The result that this target sequence in the P7 stem-loop of ibsC mRNA is hypersensitive to lead(II) (Figure 7) supports this possibility.
Our in vitro probing analysis revealed that initial interactions by TRD1 and TRD2 promote further base-pairing into the flanking sequences. Therefore, these initial contacts serve as productive target recognition signals. The final SibC–ibsC mRNA complex formed by progressive base-pairings does not seem to be a fully paired duplex because the mapping patterns for enzymes and lead(II) differed from those of the RNA duplex formed through the denaturation and annealing process, suggesting that the partial base-pairing, but not complete duplex formation, occurs in vivo and that the limited number of base-pairing of SibC with ibsC mRNA is enough for the ibsC repression. The sufficiency of the partial base-pairing for the action of cis-encoded sRNA was also previously observed (21). However, the partial base-pairing should be a productive one, just as that through TRD1 or TRD2 of SibC for recognizing ibsC mRNA in this study. The binding rate constants of each TRD domain of SibC for the partial base-pairing were comparable to previously known stable antisense/sense RNA complex, suggesting SibC–ibsC interaction occur as rapidly as other antisense–sense interactions (21).
Repression of ibsC by SibC results in the degradation of ibsC mRNA (25). The target site of TRD1 is located within the ORF of ibsC, whereas that of TRD2 is in the TIR. The function of TRD1 may be similar to the down-regulation of ompD by MicC whose binding to the target sequence within ompD ORF can promote mRNA degradation without inhibition of translation initiation in Salmonella typhimurium (44). The two independent TRDs could be used to assure full repression of ibsC expression until it is needed by the cells. This tight regulation may be essential to protect cells from the highly deleterious effect of the IbsC toxin protein.
There are five ibs/Sib genes. Each Sib RNA recognizes a distinct cognate ibsC mRNA, despite the fact that Sib RNAs and ibs mRNAs are highly homologous each other. Therefore, the TRD1 and TRD2 domains should be involved in the discrimination of non-cognate ibs mRNAs. It is noteworthy that TRD1 and TRD2 are located in highly variable regions in members of the Sib RNA family (Supplementary Figure S10). Even when two Sib RNAs are compared, these regions are always variable. This high sequence variability is required for the discrimination of non-cognate ibs mRNA by Sib RNA. Indeed, we found that the SibC mutants, whose TRD1 or TRD2 sequences were mutated to match those of SibD, repressed ibsD expression. Therefore, it is possible that the Ibs/Sib toxin/antitoxin system was duplicated and evolved to discriminate by assigning TRD1 and TRD2 to variable regions. It has been reported that SibA-deficient cells are viable although these cells express ibsA, which contrast with other Sib RNA-deficient cells that become lethal if cognate ibs mRNAs are expressed (25). Our co-transformation assays also showed that expression of ibsA in ΔibsA/sibA cells had no effect on cell survival (K.Han, unpublished results). These results suggest that other Sib RNA than SibA can recognize ibsA mRNA. Interestingly, the corresponding regions to the +90 to +113 of SibC region are the same between SibA and SibE. This region contains not only the TRD2 sequence but also the GCUU sequence (corresponding to GUCG of SibC) of the P6 loop. Therefore, one may argue that SibE could cross-recognize ibsA mRNA. However, it remains to be further demonstrated because SibA cannot repress ibsE (25).
It remains unclear why expression of the Ibs toxin family is regulated only by their own cognate SibC RNAs. It is possible that expression of each ibs gene is differentially regulated so that distinct Ibs proteins are synthesized under different conditions. The upstream regions of the ibsC and ibsD promoters have putative binding sites for different transcription factors (unpublished results). Alternatively, as the sequences of five Ibs toxin proteins are homologous but not identical, these proteins may have their own specific functions. Similar hydrophobic toxin peptides such as ShoB are believed to play different functions compared with SibC (25). If this is the case, expression of each Ibs protein may need to be regulated separately.
Cis-encoded sRNA may differ from trans-encoded sRNA because it has the potential to form a relatively long contiguous duplex with its target RNA. Although the Sib/ibs pair is operated in cis in the cell, we assessed the ability of SibC derivatives to repress ibsC expression in trans by expressing SibC and ibsC mRNA on two different plasmids. This trans operation might require much more Sib RNA than the naturally occurring cis operations, and cis-regulation could be more effective than trans-regulation owing to the greater local concentrations of interacting RNA molecules. Nonetheless, our findings show that ibsD expression can be repressed by a SibC mutant that is capable of forming a contiguous 22-nt duplex specific to ibsD. Therefore, it seems likely that the mechanism of cis-encoded sRNA does not significantly differ from that of trans-encoded sRNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Korea Research Foundation and the Korean Government (MOEHRD, Basic Research Promotion Fund) (KRF 2008-313-C00542); 21C Frontier Microbial Genomics and Application Center Program (MG08-0201-2-0). Funding for open access charge: 21C Frontier Microbial Genomics and Application Center Program (MG08-0201-2-0).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- 2.Majdalani N, Vanderpool CK, Gottesman S. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol. 2005;40:93–113. doi: 10.1080/10409230590918702. [DOI] [PubMed] [Google Scholar]
- 3.Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu. Rev. Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- 4.Wagner EG. Kill the messenger: bacterial antisense RNA promotes mRNA decay. Nat. Struct. Mol. Biol. 2009;16:804–806. doi: 10.1038/nsmb0809-804. [DOI] [PubMed] [Google Scholar]
- 5.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 7.Chen S, Lesnik EA, Hall TA, Sampath R, Griffey RH, Ecker DJ, Blyn LB. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. Biosystems. 2002;65:157–177. doi: 10.1016/s0303-2647(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 8.Hershberg R, Altuvia S, Margalit H. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res. 2003;31:1813–1820. doi: 10.1093/nar/gkg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rivas E, Klein RJ, Jones TA, Eddy SR. Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr. Biol. 2001;11:1369–1373. doi: 10.1016/s0960-9822(01)00401-8. [DOI] [PubMed] [Google Scholar]
- 10.Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EG. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Zhang Z, Ling L, Shi B, Chen R. Conservation analysis of small RNA genes in Escherichia coli. Bioinformatics. 2004;20:599–603. doi: 10.1093/bioinformatics/btg457. [DOI] [PubMed] [Google Scholar]
- 13.Afonyushkin T, Vecerek B, Moll I, Blasi U, Kaberdin VR. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 2005;33:1678–1689. doi: 10.1093/nar/gki313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Antal M, Bordeau V, Douchin V, Felden B. A small bacterial RNA regulates a putative ABC transporter. J. Biol. Chem. 2005;280:7901–7908. doi: 10.1074/jbc.M413071200. [DOI] [PubMed] [Google Scholar]
- 16.Rasmussen AA, Eriksen M, Gilany K, Udesen C, Franch T, Petersen C, Valentin-Hansen P. Regulation of ompA mRNA stability: the role of a small regulatory RNA in growth phase-dependent control. Mol. Microbiol. 2005;58:1421–1429. doi: 10.1111/j.1365-2958.2005.04911.x. [DOI] [PubMed] [Google Scholar]
- 17.Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 18.Liu MY, Gui G, Wei B, Preston JF, 3rd, Oakford L, Yuksel U, Giedroc DP, Romeo T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 1997;272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- 19.Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, Morozov I, Baker CS, Georgellis D, Babitzke P, Romeo T. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol. Microbiol. 2003;48:657–670. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- 20.Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 21.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 2007;10:102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 22.Wagner EG, Simons RW. Antisense RNA control in bacteria, phages and plasmids. Annu. Rev. Microbiol. 1994;48:713–742. doi: 10.1146/annurev.mi.48.100194.003433. [DOI] [PubMed] [Google Scholar]
- 23.Kolb FA, Engdahl HM, Slagter-Jager JG, Ehresmann B, Ehresmann C, Westhof E, Wagner EG, Romby P. Progression of a loop-loop complex to a four-way junction is crucial for the activity of a regulatory antisense RNA. EMBO J. 2000;19:5905–5915. doi: 10.1093/emboj/19.21.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagner EG, Brantl S. Kissing and RNA stability in antisense control of plasmid replication. Trends. Biochem. Sci. 1998;23:451–454. doi: 10.1016/s0968-0004(98)01322-x. [DOI] [PubMed] [Google Scholar]
- 25.Fozo EM, Kawano M, Fontaine F, Kaya Y, Mendieta KS, Jones KL, Ocampo A, Rudd KE, Storz G. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 2008;70:1076–1093. doi: 10.1111/j.1365-2958.2008.06394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duhring U, Axmann IM, Hess WR, Wilde A. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc. Natl Acad. Sci. USA. 2006;103:7054–7058. doi: 10.1073/pnas.0600927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stork M, Di Lorenzo M, Welch TJ, Crosa JH. Transcription termination within the iron transport-biosynthesis operon of Vibrio anguillarum requires an antisense RNA. J. Bacteriol. 2007;189:3479–3488. doi: 10.1128/JB.00619-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silvaggi JM, Perkins JB, Losick R. Small untranslated RNA antitoxin in Bacillus subtilis. J. Bacteriol. 2005;187:6641–6650. doi: 10.1128/JB.187.19.6641-6650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawano M, Aravind L, Storz G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 2007;64:738–754. doi: 10.1111/j.1365-2958.2007.05688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawano M, Oshima T, Kasai H, Mori H. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol. Microbiol. 2002;45:333–349. doi: 10.1046/j.1365-2958.2002.03042.x. [DOI] [PubMed] [Google Scholar]
- 32.Fozo EM, Hemm MR, Storz G. Small toxic proteins and the antisense RNAs that repress them. Microbiol. Mol. Biol. Rev. 2008;72:579–589. doi: 10.1128/MMBR.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerdes K, Wagner EG. RNA antitoxins. Curr. Opin. Microbiol. 2007;10:117–124. doi: 10.1016/j.mib.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 34.Pedersen K, Gerdes K. Multiple hok genes on the chromosome of Escherichia coli. Mol. Microbiol. 1999;32:1090–1102. doi: 10.1046/j.1365-2958.1999.01431.x. [DOI] [PubMed] [Google Scholar]
- 35.Vogel J, Argaman L, Wagner EG, Altuvia S. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 2004;14:2271–2276. doi: 10.1016/j.cub.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 36.Ko JH, Han K, Kim Y, Sim S, Kim KS, Lee SJ, Cho B, Lee K, Lee Y. Dual function of RNase E for control of M1 RNA biosynthesis in Escherichia coli. Biochemistry. 2008;47:762–770. doi: 10.1021/bi701528j. [DOI] [PubMed] [Google Scholar]
- 37.Kim KS, Lee Y. Regulation of 6S RNA biogenesis by switching utilization of both sigma factors and endoribonucleases. Nucleic Acids Res. 2004;32:6057–6068. doi: 10.1093/nar/gkh939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho SN, Hunt HD, Horton RM, Pullen JK, and Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 40.Greenfield TJ, Franch T, Gerdes K, Weaver KE. Antisense RNA regulation of the par post-segregational killing system: structural analysis and mechanism of binding of the antisense RNA, RNAII and its target, RNAI. Mol. Microbiol. 2001;42:527–537. doi: 10.1046/j.1365-2958.2001.02663.x. [DOI] [PubMed] [Google Scholar]
- 41.Conrad C, Rauhut R. Ribonuclease III: new sense from nuisance. Int. J. Biochem. Cell Biol. 2002;34:116–129. doi: 10.1016/s1357-2725(01)00112-1. [DOI] [PubMed] [Google Scholar]
- 42.Krinke L, Wulff DL. The cleavage specificity of RNase III. Nucleic Acids Res. 1990;18:4809–4815. doi: 10.1093/nar/18.16.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franch T, Petersen M, Wagner EG, Jacobsen JP, Gerdes K. Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J. Mol. Biol. 1999;294:1115–1125. doi: 10.1006/jmbi.1999.3306. [DOI] [PubMed] [Google Scholar]
- 44.Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat. Struct. Mol. Biol. 2009;16:840–846. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.