Abstract
Single-stranded DNA (ssDNA)-specific exonucleases (ssExos) are expected to be involved in a variety of DNA repair pathways corresponding to their cleavage polarities; however, the relationship between the cleavage polarity and the respective DNA repair pathways is only partially understood. To understand the cellular function of ssExos in DNA repair better, genes encoding ssExos were disrupted in Thermus thermophilus HB8 that seems to have only a single set of 5′–3′ and 3′–5′ ssExos unlike other model organisms. Disruption of the tthb178 gene, which was expected to encode a 3′–5′ ssExo, resulted in significant increase in the sensitivity to H2O2 and frequency of the spontaneous mutation rate, but scarcely affected the sensitivity to ultraviolet (UV) irradiation. In contrast, disruption of the recJ gene, which encodes a 5′–3′ ssExo, showed little effect on the sensitivity to H2O2, but caused increased sensitivity to UV irradiation. In vitro characterization revealed that TTHB178 possessed 3′–5′ ssExo activity that degraded ssDNAs containing deaminated and methylated bases, but not those containing oxidized bases or abasic sites. Consequently, we concluded that TTHB178 is a novel 3′–5′ ssExo that functions in various DNA repair systems in cooperation with or independently of RecJ. We named TTHB178 as T. thermophilus exonuclease I.
INTRODUCTION
Single-stranded DNA (ssDNA)- and double-stranded DNA (dsDNA)-specific exonucleases are essential for DNA replication, repair and recombination. Functional defects of exonucleases are known to have a profound impact on human diseases, such as Aicardi–Goutieres syndrome, familial chilblain lupus and ataxia telangiectasia-like disorder (1–3). The ssDNA-specific exonucleases (ssExos) are categorized by their cleavage polarity; from 3′ to 5′ (3′–5′) and from 5′ to 3′ (5′–3′). A variety of DNA repair pathways require ssExos to process the intermediate DNA structures generated during the reactions (4–6). As the diverse intermediate DNA structures are yielded depending on the repair pathway, living cells are considered to require several kinds of ssExos with different polarities. For example, a 5′–3′ ssExo is required for the early stage of double-strand break (DSB) repair. DSB is a potentially lethal lesion that is spontaneously generated in normal cells and also generated by external factors including ultraviolet (UV)-C (100–280 nm) (7). Bacterial DSB repair mainly occurs through homologous recombination (8). In this mechanism, an ssExo with 5′–3′ polarity processes the termini of a dsDNA to a 3′-overhanging structure, generating an entry point for downstream enzymes.
However, the type of ssExo polarity required for other DNA repair pathways is unclear. First, in DNA mismatch repair (MMR), which corrects a mismatched base generated during DNA replication, an ssExo is required for the excision of the ssDNA region containing the mismatched base. Most organisms, except Escherichia coli and its closely related species, are thought to adopt the system in which the MutS/MutL complex (or its counterpart) nicks the 3′- and 5′-sides of the mismatched base (9–15). The ssExo polarity responsible for the removal of the error-containing ssDNA region in bacteria is unknown. Second, it also remains to be established how cells process intermediates in the repair of deaminated bases, such as xanthine, hypoxanthine and uracil. Endonuclease V is known to hydrolyse the phosphodiester bond at the 3′-side of the deaminated lesion (16,17); however, the downstream reaction is unclear. ssExos might be involved in the downstream reaction. Third, the role of ssExos is completely uncertain in the repair of other damages, such as oxidized bases, methylated bases or abasic sites in DNA.
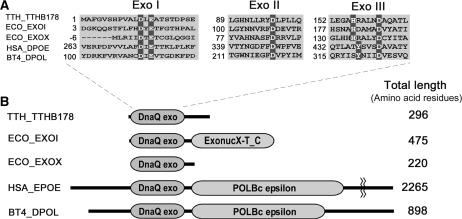
Thermus thermophilus HB8 has a small genome size, ∼2.2 Mb, and an extremely high optimum growth temperature, 75°C (18). Proteins from this eubacterial strain are extremely stable and suitable for in vitro characterization. Therefore, we selected T. thermophilus HB8 for systematic study of the structures and functions of all proteins from a single organism (18). We have already investigated many DNA repair enzymes from the strain (9,19–21), including a 5′–3′ ssExo, RecJ. It seemed that T. thermophilus HB8 possesses a 3′–5′ ssExo, TTHB178, in addition to RecJ. We found the DnaQ exonuclease motif in the N-terminal region of TTHB178 (Figure 1) that had been annotated as a functionally unknown protein. Most of the exonuclease domains of 3′–5′ ssExos are categorized into the DnaQ superfamily (22,23). The 3′–5′ exonuclease domains of the DnaQ superfamily share three conserved exonuclease motifs (Exo I–III) containing negatively charged amino acid residues (24), which coordinate two divalent metal ions to catalyse the phosphodiester bond cleavage (25,26). Recent reports have described the expression of the tthb178 gene under the control of a transcriptional regulator, cyclic AMP-receptor protein (CRP), in T. thermophilus HB8 (27), implying that tthb178 plays a certain kind of biological role in the cell.
Figure 1.
Amino acid sequence alignments of TTHB178 and the proteins belonging to the DnaQ superfamily. (A) Exonuclease motifs I, II and III of TTHB178 and exonucleases belonging to the DnaQ superfamily. The numbers to the left of the motifs indicate the distances from the protein N-termini. The predicted active site residues are highlighted in dark grey. (B) Schematic diagrams of TTHB178 and the other DnaQ superfamily proteins. DnaQ exo, ExonucX-T_C and POLBc epsilon mean the exonuclease domain of the DnaQ superfamily, the SH3-like and helical domains of E. coli EXOI and the DNA polymerase domain of type-B family DNA polymerases, respectively. TTH_TTHB178, T. thermophilus HB8 TTHB178; ECO_EXOI, E. coli ExoI; ECO_EXOX, E. coli ExoX; HSA_DPOE, Homo sapiens DNA polymerase ε; BT4_DPOL, bacteriophage T4 DNA polymerase.
As there seems to be no other candidate for the ssExos in T. thermophilus HB8 except for the proofreading domains of DNA polymerases, this organism is expected to have a single set of 3′–5′ ssExo (TTHB178) and 5′–3′ ssExo (RecJ). In majority of the organisms, including E. coli, yeast and humans, redundancy of the same polarity of ssExos makes it difficult to clarify the relationship between their cleavage polarities and their cellular functions (Supplementary Table S1). In contrast, T. thermophilus HB8 is suitable for investigating the difference in cellular functions between 3′–5′ and 5′–3′ ssExos. Although we cannot exclude the possibility that T. thermophilus HB8 has additional ssExo with sequences different from known enzymes, gene disruption of an ssExo is expected to affect the phenotypes of the disruption mutants directly.
In this study, we investigated the phenotypes of the disruption mutants of the tthb178 and recJ genes under DNA-damaging conditions. We also prepared the tthb178 gene product and examined its biochemical activity against various types of DNA in vitro. The results suggest that TTHB178 is a 3′–5′ ssExo that functions in several DNA repair pathways not only cooperatively with but also independently of RecJ.
MATERIALS AND METHODS
Transcription analysis of tthb178
Thermus thermophilus HB8 cells were cultured overnight in TT broth (0.8% polypeptone, 0.4% yeast extract, 0.2% NaCl, 0.4-mM MgCl2 and 0.4-mM CaCl2; pH 7.2) at 70°C, diluted 100-fold in fresh TT broth and the diluted culture was incubated at 70°C. At each time point, cells were harvested by centrifugation at 2300g for 10 min at 4°C and stored at −20°C. Purification of mRNA was carried out by using an RNeasy mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol (28). The cDNA was synthesized by reverse transcription–PCR using forward primer 5′-ACCTCTACGCCTTCCTCCTC-3′ and reverse primer 5′-CTCCTTGATTCTCTGGGCGG-3′. The amplified fragment was 332 bp.
Disruptions of tthb178 and recJ
The gene null mutants of T. thermophilus HB8 were constructed by using a previously reported procedure (29). The plasmids for gene disruption were derivatives of the pGEM-T Easy vector (Promega Co., Madison, WI, USA), constructed by inserting the thermostable kanamycin-resistance gene, HTK (30), flanked by ∼500-bp upstream and downstream sequences of the tthb178 and recJ genes (Supplementary Figure S1A). The 500-bp DNA fragments from upstream and downstream of the tthb178 gene were amplified by PCR using primer sets 5′-ACTCGGGCGGCACGATC-3′ and 5′-ATATGGTACCCGCCGTCAACGGGTACCG-3′, and 5′-ATATCTGCAGCATGTTGGTTACGCTGCA-3′ and 5′-GCGCCGCCTCCACCACCT-3′, respectively (the underlining indicates KpnI and PstI sites, respectively). The 500-bp DNA fragments from upstream and downstream of the recJ gene were also amplified by PCR using primer sets 5′-CGGGACCCTCTTGGGCCT-3′ and 5′-ATATGGTACCCGCCGTCAACGGGTACCA-3′, and 5′-ATATCTGCAGCATGTTGGTTACGCTGCA-3′ and 5′-CGGGGGCCTCCACCACCC-3′, respectively (the underlining indicates KpnI and PstI sites, respectively). The amplified fragments were digested with KpnI and PstI, respectively, to obtain fragments I and II. The HTK gene was also amplified by PCR from plasmid pUC18/HTK (30) by using 5′-ATATGGTACCCGTTGACGGCGGGATATG-3′ and 5′-ATATCTGCAGCGTAACCAACATGATTAA-3′ as primers (the underlining indicates KpnI and PstI sites, respectively). The amplified fragment was then treated with KpnI and PstI to obtain fragment III. Fragments I, II and III were ligated into the pGEM-T Easy vector. For double gene knockout, the thermostable hygromycin-B-resistance gene was used for disrupting the recJ gene (Supplementary Figure S1B). The hygromycin-B-resistance gene was amplified by PCR from plasmid pHG206 (31) by using 5′-GAATTCGAGCTCGGTACCCG-3′ and 5′-ATATCTGCAGGAATTCGAGGTCGCTACCCG-3′ as primers (the underlining indicates KpnI and PstI sites, respectively). The amplified fragment was then digested with KpnI and PstI to obtain fragment IV. Fragments I and II from the recJ gene and fragment IV were ligated into pGEM-T Easy vector. The plasmid was transformed into T. thermophilus HB8 cells as previously described (29). Disruptions of the tthb178 and recJ genes were confirmed by PCR amplification using the isolated genomic DNAs as templates (Supplementary Figure S2). The absence of the mRNA transcribed from tthb178 and recJ was also confirmed by reverse transcription–PCR (Supplementary Figure S3).
Estimation of spontaneous mutation rates
The spontaneous mutation rate of T. thermophilus HB8 was estimated based on the frequency of streptomycin-resistant strains measured by means of the modified Luria–Delbrück fluctuation test (32). Cultured T. thermophilus HB8 wild-type (WT) and disruptants in the mid-exponential growth phase (A660 = 1.0–1.5) were appropriately diluted in TT broth and spread on TT agar plates with or without 50 μg/ml streptomycin. The numbers of colonies formed, colony-forming units (CFUs), were counted after incubation at 70°C for 24 h. The surviving fractions were expressed as the average obtained from at least three independent experiments. The spontaneous mutation rates (%) were calculated according to the formula mutation rate (%) = M/N × 100, where M is the counted CFUs on the TT plates containing 50 μg/ml streptomycin and N is the mean of the CFUs on the TT plates without streptomycin.
Examination of the sensitivities to UV irradiation and H2O2 addition
Thermus thermophilus HB8 cells in the mid-exponential growth phase were spread on TT agar plates and irradiated with 254-nm UV light at the dose rate of 1.9 J m−2 s−1 for 40 s. The CFUs were counted after incubation at 70°C for 24 h and the surviving fractions were expressed as the average obtained from at least three independent experiments.
The sensitivity to H2O2 addition was measured as follows. Thermus thermophilus HB8 cells in the mid-exponential growth phase were mixed with equal volumes of 0-, 10-, 20- and 100-mM H2O2. The cells were further incubated at 70°C for 20 min and spread on TT agar plates. The CFUs were counted after incubation at 70°C for 24 h and the surviving fractions were expressed as the average obtained from at least three independent experiments.
Overexpression and purification of TTHB178
Escherichia coli Rosetta(DE3) (Novagen, Madison, WI, USA) was transformed with pET-11a/tthb178 (RIKEN BioResource Center, Tsukuba, Japan) and the transformed cells were cultured in L-broth containing 50 μg/ml ampicillin. When the cell density reached 1 × 108 cells/ml, isopropyl-1-thio-β-d-galactopyranoside was added to the culture to induce tthb178 gene expression. The cells were further cultured for 6 h and harvested by centrifugation at 9000g under 4°C and stored at −20°C until use.
All the steps for TTHB178 purification except the heat treatment were performed at 4°C. The frozen cells were suspended in 50-mM Tris–HCl and 5-mM EDTA (pH 8.0; buffer A) and disrupted by sonication. The cell lysate was treated at 60°C for 10 min and the supernatant was recovered after centrifugation at 34 000g for 1 h. (NH4)2SO4 was gradually added to the solution to a final concentration of 1.5 M. The solution was applied to a Toyopearl Ether-650M column (bed volume of 20 ml; Tosoh Corp., Tokyo, Japan) equilibrated with 50-mM Tris–HCl, 1.5-M (NH4)2SO4, 100-mM KCl and 5-mM EDTA (pH 8.0; buffer B). The column was washed with buffer B and then eluted with a linear gradient of 1.5–0-M (NH4)2SO4 in 50-mM Tris–HCl, 100-mM KCl and 5-mM EDTA (pH 8.0). SDS–PAGE revealed that the target protein was eluted at 0.7–0.6-M (NH4)2SO4. The fractions containing TTHB178 were dialysed twice against 5 l of buffer A. The dialysed solution was diluted to 50 ml with buffer A and loaded onto a Toyopearl SuperQ-650M column (bed volume of 20 ml; Tosoh Corp.) equilibrated with buffer A. The column was substantially washed with buffer A and the proteins were eluted with a linear gradient of 0–1 M KCl in buffer A. The target protein was eluted at 100–150-mM KCl. The fractions containing the target protein were dialysed twice against 5 l of 10-mM potassium phosphate and 5-mM EDTA (pH 7.4; buffer C). The dialysed solution was diluted to 50 ml with buffer C and loaded onto a hydroxyapatite column, BioScale CHT5-I (bed volume of 20 ml; Bio-Rad Laboratories Inc., Hercules, CA, USA) equilibrated with buffer C. The flow-through fraction was collected. The proteins were eluted with a linear gradient of 10–500 mM K3PO4 (pH 7.4) and 5-mM EDTA. Purified TTHB178 was concentrated to 10 mg/ml in 20-mM Tris–HCl, 100-mM KCl and 60% glycerol (pH 8.0) and stored at −20°C. Peptide mass fingerprinting (33) confirmed that the purified protein was TTHB178.
Size-exclusion chromatography
Size-exclusion chromatography was performed 25°C by using a Superdex 75 HR column (1 × 30 cm; GE Healthcare Biosciences) in an ÄKTA system (GE Healthcare Biosciences). The 100 μl of purified TTHB178 (0.75 mg/ml) was loaded onto the column and eluted at a flow rate of 0.5 ml/min with 20-mM Tris–HCl and 100-mM KCl (pH 8.0). The elution profile was monitored by recording the absorbance at 280 nm. The column was calibrated using apoferritin (443 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), thyroglobulin (66.9 kDa) and cytochrome c (12.4 kDa).
Dynamic light scattering experiment
The 2.0 mg/ml of TTHB178 was prepared in 20-mM Tris–HCl and 100-mM KCl, pH 7.5, and was passed through 0.02 μm Whatman Anodisc 13 Supported Membrane Filter. The 12 μl of the protein solution was loaded into a quartz cuvette and then analysed by dynamic light scattering instrument, DynaPro MSXTC/12/F with a gallium–arsenite diode laser, DynaPro-99-E-50 (Protein Solutions Inc., Charlottesville, VA, USA) at 20°C. The data were analysed using the Dynamics version 6.3.18 (Proteins Solutions Inc., Charlottesville, VA, USA). The sample was analysed a minimum of 10 times and the resulting data were analysed to estimate apparent molecular weight assuming a globular protein in an aqueous solution. The hydrodynamic radius (Rh) value was calculated with the Stokes–Einstein equation (Equation 1) using the obtained translational diffusion coefficient (DT):
| (1) |
where kB is the Boltzmann constant, T the absolute temperature, η the solvent viscosity and Rh the hydrodynamic radius. Molecular mass of the protein in the solution was estimated from Rh using an empirical curve of known proteins (Equation 2).
| (2) |
Mass analysis by using Fourier transform ion cyclotron mass spectrometer
The products of the exonuclease reaction were analysed by Fourier transform ion cyclotron mass spectrometer (FT-ICR MS) with electrospray ionization. In brief, 21-mer ssDNA (21f) was reacted with 3-μM TTHB178 in 20-mM Hepes–KOH, 100-mM KCl and 5-mM MgCl2, pH 7.5, at 37°C for 0, 1, 5 and 10 min, respectively. Each reactant was mixed with ion-pairing agent, butyl dimethyl ammonium carbonate, pH 8.0 to a final 25-mM concentration. The mixture was loaded onto a self-made reverse-phase column using C18 Empore disk (3M Co., St Paul, MN, USA), after equilibration with 25-mM butyl dimethyl ammonium carbonate. The column was further washed with 5% acetonitrile containing 25-mM butyl dimethyl ammonium carbonate and then the products were eluted with 50% acetonitrile. Basic additives, piperidine (pH 9) and imidazole (pH 8) for mass analysis of nucleic acids under negative mode were added to each eluent to a final concentration of 25 mM, respectively. The resulting solution was subjected to an APEX IV, FT-ICR MS shielded with 9.4 T magnet (Brüker Daltonics Inc., MA, USA), by electrospray ionization under 2 μl/min flow rate as describe in a previous report (34).
Exonuclease assays
Single-stranded oligonucleotides were synthesized (BEX Co., Tokyo, Japan) and their 5′-termini were radiolabelled with [γ-32P]ATP using T4 polynucleotide kinase (Takara Bio, Shiga, Japan) at 37°C for 1 h. The substrates with 3′-overhanging, Y and gapped flap structures were yielded by hybridizing 50sf with 40sr, 50sf with 28sr and 50sf with 28sr and 21sr (Table 2), respectively. In the case of 3′-end labelling, an oligonucleotide (Table 1; 21r; 5′-GGGTGTTGCTTTAGTTGTCAT-3′) was radiolabelled with [α-32P]cordycepin-5′-triphosphate (PerkinElmer Life & Analytical Sciences, Boston, MA, USA) by using terminal deoxynucleotidyl transferase (Promega Co., Fitchburg, WI, USA). The radiolabelled substrates were incubated with 3-μM TTHB178 in 20-mM Hepes–KOH, 100-mM KCl and 5-mM MgCl2 (pH 7.5). The total reaction volume was 10 μl. The reaction temperatures and times were as indicated in the figure legends. The reactions were stopped by the addition of an equal volume of phenol, CHCl3 and isoamyl alcohol (25:24:1) as well as 1 μl of 100-mM EDTA. The mixture was centrifuged at 15 000g at 4°C for 10 min and the aqueous phase was mixed with an equal volume of sample buffer (5-mM EDTA, 80% deionized formamide, 10-mM NaOH, 0.1% bromophenol blue and 0.1% xylene cyanol). The samples were analysed by electrophoresis through denaturing 25% polyacrylamide gels with 1× TBE buffer (89-mM Tris–borate and 2-mM EDTA), and the gels were dried and placed in contact with an imaging plate. The substrates and products were detected and analysed with a BAS2500 image analyzer (Fuji Photo Film, Tokyo, Japan). In the kinetic analysis, 10-nM radiolabelled substrates were mixed with 0.1, 1, 5, 10, 50, 100, 300, 500 or 1000-μM of non-labelled substrates and then reacted with 100-nM TTHB178 for 30 min at 37°C or 5 min at 60°C, respectively. The values of the initial rates were calculated based on the amount of undegraded substrates according to a previously described procedure (21,35,36). The kcat and KM values were determined by fitting the data to the Michaelis–Menten equation using Igor 4.03 (WaveMetrics, Lake Oswego, OR, USA).
Table 2.
Steady-state kinetic parameters of the TTHB178 exonuclease activity for ssDNAs
| Length (mer) | Temperature (°C) | kcat (s−1) | KM (μM) | kcat/KM (s−1 M−1) |
|---|---|---|---|---|
| 10 | 37 | 0.71 ± 0.23 | 280 ± 83 | 2.6 × 103 ± 0.19 |
| 60 | 7.5 ± 0.023 | 480 ± 16 | 16 × 103 ± 0.49 | |
| 21 | 37 | 0.40 ± 0.15 | 350 ± 31 | 1.1 × 103 ± 0.49 |
| 60 | 5.3 ± 2.10 | 440 ± 120 | 12 × 103 ± 3.70 | |
| 28 | 37 | 0.23 ± 0.12 | 260 ± 105 | 0.83 × 103 ± 0.20 |
| 60 | 1.9 ± 0.48 | 290 ± 58 | 7.0 × 103 ± 2.40 | |
| 40 | 37 | 0.17 ± 0.016 | 240 ± 45 | 0.72 × 103 ± 0.066 |
| 60 | 2.3 ± 0.10 | 320 ± 47 | 7.5 × 103 ± 1.40 | |
| 50 | 37 | 0.19 ± 0.046 | 130 ± 52 | 1.7 × 103 ± 0.58 |
| 60 | 1.0 ± 0.41 | 160 ± 39 | 6.4 × 103 ± 0.87 |
Table 1.
Sequences of oligonucleotides used in this study
| Name | Length | Sequence |
|---|---|---|
| 10f | 10-mer | GGCCAGGTGG |
| 21f | 21-mer | ATGACAACTAAAGCAACACCC |
| 21r | 21-mer | GGGTGTTGCTTTAGTTGTCAT |
| 21rna | 21-mer | ATGACAACTAAAGCAACACCC |
| 21sr | 21-mer | CCTAGCGGCTGCCACCTGGCC |
| 28sr | 28-mer | GGGCACCATGCGGGCGGCCAAAATGCCC |
| 40sr | 40-mer | CGGGCGGCCTCCCCTCCACCCTAGCGGCTGCCACCTGGCC |
| 50sf | 50-mer | GGCCAGGTGGCAGCCGCTAGGGTGGAGGGGAGGCCGCCCGCATGGTGCCC |
| O6m | 18-mer | GCCCGGCCAmGCTGCAGTT |
| O4m | 18-mer | GCCCGGCCAmTCTGCAGTT |
| 8OG | 21-mer | TACTGTTGAoxoGTTGGTTGTGGG |
| rAP21 | 21-mer | TACTGTTGArAPTTGGTTGTGGG |
| AP21 | 21-mer | TACTGTTGAAPGTTGGTTGTGGG |
| Ura21 | 21-mer | TACTGTTGAUraTTGGTTGTGGG |
| Hyp30 | 30-mer | GCTCGTAGAGCGGTCHypTAGTCAAGATACCG |
| Xan30 | 30-mer | GCTCGTAGAGCGGTCXanTAGTCAAGATACCG |
mG, O6-methylguanine; mT, O4-methylthymine; oxoG, 8-oxoguanine; rAP, reduced abasic site; AP, abasic site; Ura, uracil; Hyp, hypoxanthine; Xan, xanthine.
RESULTS
Sequence comparison between TTHB178 and DnaQ superfamily exonucleases
The tthb178 gene encoded a protein that comprised 296 amino acid residues and whose N-terminal region showed significant sequence similarity to DnaQ superfamily exonucleases such as E. coli ExoI and ExoX (37,38) and the proofreading domains of the DNA polymerases. These regions contained exonuclease motifs I–III that included conserved Asp, Glu and His residues (Figure 1A). The DnaQ superfamily exonucleases had a wide variety of protein lengths and showed sequence diversities except for the exonuclease motifs (Figure 1B). Among them, the length of TTHB178 was comparable to that of E. coli ExoX; however, their C-terminal regions showed no detectable sequence similarity.
Expression of tthb178 in T. thermophilus HB8 cells
Unlike recJ, tthb178 has been annotated as a hypothetical protein, and there was no evidence for the in vivo expression of tthb178 before the recent report on the CRP-dependent expression of tthb178 in cells (27). We first performed time course transcription analysis of the tthb178 gene in T. thermophilus HB8 cells by using reverse transcription–PCR. Transcription of tthb178 was detected from the mid-log phase to the late stationary phase (Supplementary Figure S4). The gene was more actively transcribed in the late stationary phase (no. 6 in the figure) than in the mid-log phase (no. 3) or stationary phase (nos. 4 and 5). The result strongly suggested that tthb178 is not a pseudo-gene and is required for a cellular function in T. thermophilus HB8.
Phenotypes of tthb178 and recJ disruptants
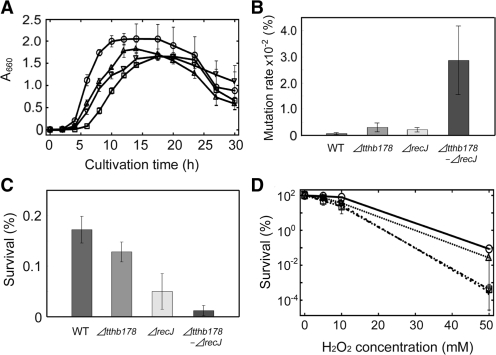
To investigate the cellular functions of ssExos in vivo, we generated a tthb178 disruptant (Δtthb178), recJ disruptant (ΔrecJ) and tthb178–recJ double disruptant (Δtthb178–ΔrecJ). All the disruptants grew in rich medium, indicating that these two genes were not essential under the condition examined. However, all of them exhibited a relatively long lag time prior to the exponential growth compared with the WT (Figure 2A). These disruptants also showed lower maximum cell density than the WT during the stationary phase (Figure 2A). Furthermore, the disruptants aggregated in the log phase whereas the WT did not (data not shown). Elongated cells of the disruptants were observed in the late log, stationary and death phases, unlike in the case of the WT (data not shown). These results suggested that T. thermophilus HB8 requires TTHB178 as well as RecJ for optimal growth under conditions without external DNA-damaging stress.
Figure 2.
Effects of the disruptions of the tthb178 and recJ genes on the growth of T. thermophilus HB8. (A) Growth curves of the WT (circles), ΔrecJ (triangles), Δtthb178 (inverted triangles) and the double disruptant (squares). Growth was monitored by measuring the absorbance at 660 nm. (B) Spontaneous mutation rates of each strain to the streptomycin-resistant strain. (C) Sensitivity to 254-nm UV-C irradiation. The survival ratios are shown as a bar graph. (D) Sensitivity to H2O2. The survival ratios were plotted against the H2O2 concentration. The symbols are the same as in (A). In all panels, the data represent the averages of at least three independent experiments and each bar indicates the SD.
To examine the possible involvement of TTHB178 and RecJ in DNA repair processes in T. thermophilus HB8, we first measured the spontaneous mutation rate of the disruptants to a streptomycin-resistant strain (39). The spontaneous mutation rates of Δtthb178 and ΔrecJ were approximately four-fold and three-fold higher than that of the WT, respectively (Figure 2B). Interestingly, Δtthb178–ΔrecJ showed a significantly higher rate than the single-disruption cells (Figure 2B). The streptomycin-resistant strains obtained here must have mutations within the rRNA gene (39,40) and such spontaneous mutagenesis can be accelerated by defects in several DNA repair systems such as MMR. Therefore, the observed increase in mutation frequency suggested that both TTHB178 and RecJ are involved in DNA repair.
We then examined the growth phenotypes of these disruptants under DNA-damaging conditions. The disruption of tthb178 did not affect the sensitivity to UV-C irradiation at 254 nm (Figure 2C). On the other hand, ΔrecJ exhibited three-fold higher sensitivity to UV-C than the WT and Δtthb178 (Figure 2C). The major damages caused by UV-C irradiation are cyclobutane pyrimidine dimers, pyrimidine-pyrimidone (6-4) photoproducts and DSBs (41–45). The observed increase in the sensitivity indicated that RecJ is intimately involved in the repair of these lesions whereas TTHB178 is not. Nevertheless, it should be noted that the survival ratio of UV-C-irradiated Δtthb178–ΔrecJ was lower than that of ΔrecJ (Fig. 2C). This result raised the possibility that TTHB178 also participates in the repair pathway for those lesions in the cell strain lacking the recJ gene product.
In contrast, the disruption of tthb178 caused drastic increase in the sensitivity to H2O2, whereas the disruption of recJ did not (Figure 2D). In addition, Δtthb178–ΔrecJ exhibited a similar survival ratio to Δtthb178. Reactive oxygen species generated from H2O2 are responsible for the oxidation and deamination of bases, which result in transversion and transition mutations, respectively (46,47). Therefore, the increased sensitivity of Δtthb178 to H2O2 suggested that TTHB178 is involved in the repair pathway for such damaged bases. Thus, our in vivo experiments indicated that the tthb178 and recJ genes are required for several DNA repair pathways in T. thermophilus HB8.
Exonuclease activity of TTHB178
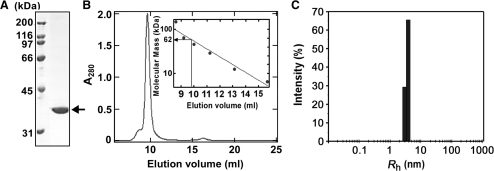
In order to characterize TTHB178 biochemically, we overexpressed TTHB178 in E. coli and purified it to homogeneity (Figure 3A). Size-exclusion chromatography was performed to examine the self-association ability of TTHB178. The result showed that TTHB178 was eluted with a single peak corresponding to an apparent molecular mass of 62 kDa (Figure 3B). As the molecular mass of TTHB178 was calculated to be 33 kDa according to its amino acid sequence, this result implied that TTHB178 exists in a dimeric state in solution. The observed shoulder of the main peak might represent the slight tendency of TTHB178 to form a larger complex in the solution. Dynamic light scattering experiment was also performed to evaluate the dimerization ability of TTHB178. The measurement gave an Rh value of 3.6 nm, suggesting that the molecular mass of the particle in the TTHB178 solution is about 66 kDa (Figure 3C). Thus, the result of dynamic light scattering experiment also supports the dimerization of TTHB178.
Figure 3.
Preparation of recombinant TTHB178. (A) Recombinant TTHB178 was purified as described in the ‘Materials and Methods’ section and then subjected to SDS–PAGE. The 3 μg of protein was loaded on the gel. The calculated molecular mass of TTHB178 is 33 kDa. The arrow indicates the band of TTHB178. (B) Size-exclusion chromatography. TTHB178 (0.75 mg/ml) was loaded onto a Superdex 75 HR column. The apparent molecular mass of the main peak was estimated to be ∼62 kDa, from the calibration curve shown in the inset. Apoferritin (443 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), thyroglobulin (66.9 kDa) and cytochrome c (12.4 kDa) were used as molecular size markers. (C) Dynamic light scattering measurement. Rh was calculated on the basis of the observed DT as described in ‘Materials and Methods’ section.
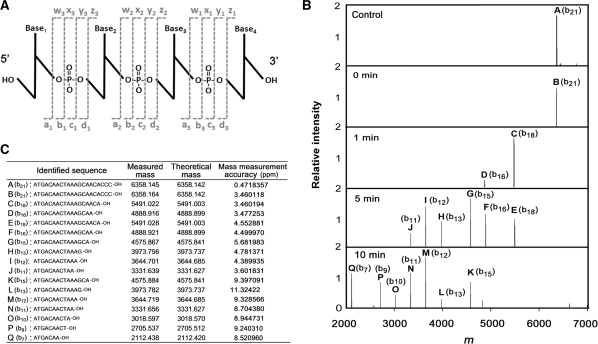
To test the prediction that TTHB178 has 3′–5′ exonuclease activity, the activity was examined using ssDNA as a substrate. The ssDNA that reacted with TTHB178 was subjected to FT-ICR MS analysis. FT-ICR MS is a powerful tool for the characterization of nuclease activity as it achieves precise and simultaneous identification of the length, nucleotide content, and nature of the 5′-termini and 3′-termini of all products (Figure 4A). As the reaction time increased, the shorter products became obvious and all of them were the b-series ions that lack the 3′-terminal region of the substrate DNA (Figure 4B and C). This result strongly indicated that TTHB178 possesses exonuclease activity that degrades ssDNA from the 3′-end to the 5′-end and that TTHB178 hydrolyses a phosphodiester bond at the 3′-side of the phosphate.
Figure 4.
Analysis of the exonuclease activity by using FT-ICR MS. (A) The nomenclature scheme used for oligonucletide ions (72). The four possible cleavages are indicated by the lower case letters a, b, c and d for ions containing the 5′-OH group and w, x, y and z for ions containing the 3′-OH group. The numerical subscripts indicate the number of bases from the respective termini. FT-ICR MS can achieve the simultaneous identification of these ions. (B) The deconvoluted mass spectra of the product ssDNAs of TTHB178. The 21-mer ssDNA (21f) was reacted with 3-μM TTHB178 at 37°C. The product ssDNAs were purified as described in the ‘Materials and Methods’ section and then analysed by using FT-ICR MS. The reaction time is shown in the panel. (C) The measured and theoretical masses of each peak (named A to Q) are listed and the corresponding sequences are also shown as the ‘identified sequence’. The measured mass coincided with the theoretical mass ∼10-ppm mass measurement accuracy.
The 3′–5′ exonuclease activity of TTHB178 was also confirmed by electrophoretic analyses using 5′-end-labelled ssDNAs as substrates. As shown in Figure 5A, TTHB178-digested products exhibited a ladder pattern of DNA fragments on the gel, which suggested that TTHB178 degrades ssDNA from the 3′-end to the 5′-end. We confirmed that the elution profile of the observed exonuclease activity from Superdex 75 HR column was exactly matched with that of TTHB178 (Supplementary Figure S5), indicating that the observed activity is derived from TTHB178.
Figure 5.
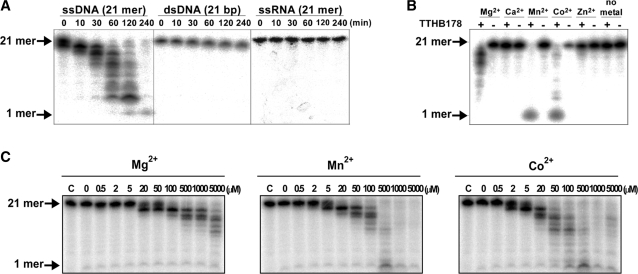
Analyses of TTHB178 exonuclease activity by using 5′ radiolabelled substrates. (A) Substrate specificity of TTHB178 exonuclease activity. TTHB178 (3 μM) was incubated with 10-nM 5′-end-labelled 21-mer ssDNA (21f), 21-bp dsDNA (21f + 21r) or 21-mer ssRNA (21rna) in the presence of 5-mM Mg2+ at 37°C. The reaction time is shown at the top of the panels. (B) Dependence of the exonuclease activity on divalent metal ions. The 10-nM 5′-end-labelled ssDNA (21f) was incubated with 3-μM TTHB178 at 37°C for 2 h. The reaction mixture contained 5 mM of the respective divalent metal ions. (C) Dependence of the exonuclease activity on the concentrations of Mg2+, Mn2+ and Co2+. The 10-nM 5′-end-labelled ssDNA (21f) was incubated with 3-μM TTHB178 at 37°C for 2 h in the presence of various concentrations of divalent cations. The concentrations of the divalent metal ions are indicated at the top of the panels.
The results also showed that TTHB178 specifically cleaves an ssDNA (Figure 5A). No activity against ssRNA and dsDNA was observed in spite of the prolonged reaction time (up to 4 h). It was also shown that the activity required divalent cations such as Mg2+, Mn2+ or Co2+ (Figure 5B and C). Our previous study showed that the intracellular concentrations of Mn (0.16 mM) and Co ions (not detected) are significantly lower than that of Mg ions (35 mM) in T. thermophilus HB8 cells (48). The concentrations of Mn2+ and Co2+ were thought to be insufficient for the activation of TTHB178 exonuclease activity in vivo. Therefore, the assays for exonuclease activity were carried out in the presence of Mg2+.
The steady-state kinetic parameters of the TTHB178 exonuclease activity were determined (Table 2) based on the reduction rate of undegraded substrates (21,35,36). The reaction temperature did not affect the KM values of the respective substrates, but the kcat values at 60°C were higher than those at 37°C. As the substrates became shorter, the KM values became higher, showing the preferential binding of TTHB178 to longer ssDNAs. On the other hand, the kcat values of longer ssDNAs were lower than those of the shorter ones. Therefore, the kcat/KM values, the index of the efficiency of an enzyme, were not affected by the lengths of the substrates. The kcat/KM values were 4- to 10-fold higher at 60°C than at 37°C, which indicated that the observed digestion of ssDNA was certainly performed by a protein from a thermophile (i.e. T. thermophilus HB8) and not by a contaminated protein from a host cell.
DNA structure and lesion specificity of the exonuclease activity
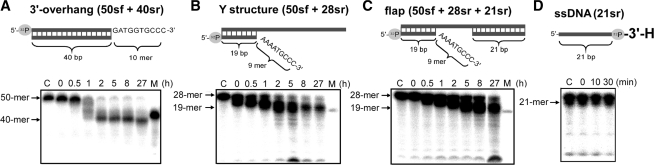
We further tested the specificity of the TTHB178 exonuclease activity to the structure and lesion of substrate DNA. The 3′-overhanging, Y and gapped flap structures were used as substrates. These are possible intermediate DNA structures generated during the processes of several DNA repair pathways (49). The results indicated that TTHB178 can digest the ssDNA regions of the three substrates (Figure 6A–C). The Y and gapped flap structures mimicked the intermediate structures, which were generated by the unwinding of nicked dsDNAs by a DNA helicase. Furthermore, TTHB178 hardly degraded the ssDNA whose 3′-terminus was radiolabelled with [α-32P]cordycepin-5′-monosphate (Figure 6D). The cordycepin-labelled ssDNA had 3′-H instead of 3′-OH at its 3′-terminus. The 3′-OH group of the substrate would be essential for the TTHB178 activity.
Figure 6.
Exonuclease activity of TTHB178 against various DNA structures. (A–C) The 3′-overhanging (50sf + 40sr) (A), Y structure (50sf + 20sr) (B) and gapped flap structure (50sf + 21sr + 28sr) (C) DNAs were reacted with 3-μM TTHB178 for various reaction periods. The reaction time is indicated at the top of the panels. Assays for the Y structure and gapped flap structure were carried out at 20°C to stabilize the short dsDNA region of the substrates. As TTHB178 showed relatively weak activity at 20°C compared with that at 37°C or 60°C, the assays were performed for a prolonged reaction time. The assay for 3′-overhanging DNA was carried out at 37°C. ‘C’ means the substrate incubated without TTHB178 for 27 h. (D) Activity for an ssDNA with a 3′-H terminus. The substrate 21-mer ssDNA (21r) was 3′-end-labelled with [α-32P]cordycepin-5′-triphosphate and reacted with 3-μM TTHB178 at 37°C. The reaction time is indicated at the top of the panel. ‘C’ means the substrate incubated without TTHB178 for 30 min. In all the panels, the digested products were analysed by electrophoresis through denaturing 8 and 25% polyacrylamide gels. ‘M’ means the 40- (in A) and 19-mer (in B and C) of marker DNAs. The bands seen near the bottom of the gels in B and C might be experimental artefacts because they were not observed reproducibly.
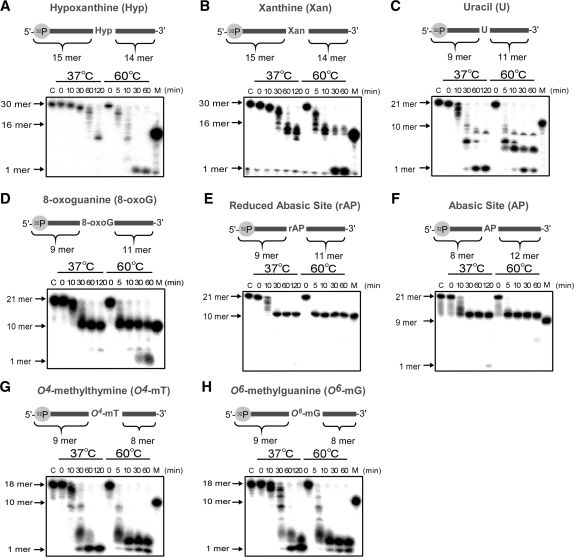
We also examined the exonuclease activity for ssDNA substrates containing various kinds of damaged bases. As a result, TTHB178 degraded ssDNAs containing hypoxanthine, xanthine, uracil, O4-methylguanine and O6-methylthymine (Figure 7A–D). At 37°C, the degradation stopped at the positions of hypoxanthine and xanthine in the respective substrates, but the degradation proceeded beyond these non-canonical bases at 60°C. In contrast, only slight activity was observed even at 60°C when the substrate contained 8-oxoguanine or an abasic site (Figure 7E and F). Based on these results, it is considered that TTHB178 can be involved in the excision step of the DNA repair pathways for deaminated and methylated bases.
Figure 7.
Excision assay for ssDNAs containing various kinds of damaged bases. The 5′-end-labelled ssDNA containing a damaged base was reacted with 3-μM TTHB178 at 37 or 60°C. The respective substrates contained hypoxanthine (A), xanthine (B), uracil (C), 8-oxoguanine (D), a reduced abasic site (E), an abasic site (F), O4-methylthymine (G) and O6-methylguanine (H). In all the panels, ‘C’ means the substrate incubated at 60°C for 60 min without TTHB178. ‘M’ means the 16- (in A and B), 10- (in C, D, E, G and H) and 9-mer (in F) marker DNAs. The reaction time is shown at the top of the panels.
DISCUSSION
Our study showed that TTHB178 possesses 3′–5′ exonuclease activity. This result supports the prediction that TTHB178 is a member of the DnaQ superfamily exonucleases. Detailed biochemical analyses revealed that TTHB178 has several characteristic features that are similar to or different from those of E. coli ExoI and ExoX. First, gel-filtration chromatography suggested that TTHB178 can form a homodimer in solution (Figure 3) unlike ExoI and ExoX. It is reported that human DnaQ superfamily ssExos, TREX1 and TREX2, also can exist as a dimer (50,51). The dimerization ability of TREX2 has been discussed in a model of allosteric effect to ensure the cooperative binding to substrate DNA (52). It will be interesting to elucidate the significance of dimerization in DNA-binding activity of TTHB178. Second, TTHB178 specifically degraded ssDNA, but not dsDNA and ssRNA (Figure 5A). Such a strict specificity is similar to that of E. coli ExoI and ExoX (37,38,53). Finally, TTHB178 did not degrade an abasic site-containing ssDNA (Figure 7E and F), indicating that TTHB178 does not possess DNA deoxyribophosphodiesterase activity as exhibited by E. coli ExoI (54–56). From these results, we concluded that TTHB178 is a novel ssDNA-specific 3′–5′ exonuclease and named this protein as T. thermophilus exonuclease I (Tth ExoI).
Tth ExoI exhibited relatively high KM value compared with other ssExos. It may be reasonable to suppress the non-specific ssDNA-binding activity of an ssExo until it is required. For example, ssExo activity of Neisseria meningitidis exonuclease VII is regulated by the interaction between the large and small subunits in response to environmental signals (57). The ssDNA-binding activity of Tth ExoI might be enhanced by the interaction with other proteins such as single-stranded DNA-binding protein that has been reported to interact with Haemophilus influenzae RecJ (35), E. coli RecJ (58) and E. coli ExoI (59).
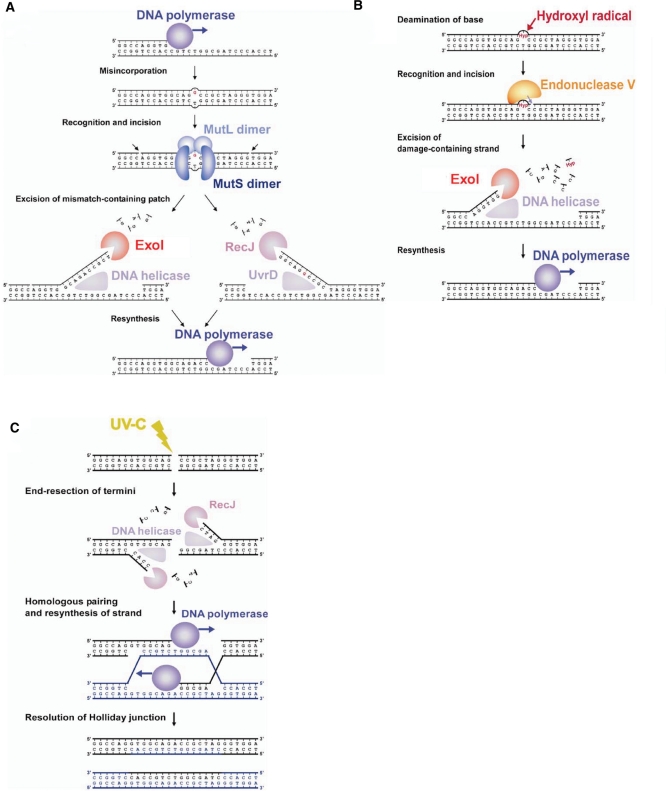
The most striking result in this study is that Δtthb178 showed a phenotype associated with defects in DNA repair systems. Δtthb178 and ΔrecJ showed a higher spontaneous mutation rate than the WT (Figure 2B and D). DNA mismatches generated by errors of DNA replication are the major source of spontaneous mutation (49). MMR is responsible for the repair of such mismatches and deficiency of MMR genes, such as mutS or mutL, results in a considerable increase in the frequency of spontaneous mutation (10,15). In majority of the organisms, the required exonuclease polarity for MMR is unknown, although exonuclease activity should be necessary for the removal of the mismatch-containing patch generated by the MutS/MutL complex. The spontaneous mutation rate of Δtthb178–ΔrecJ was much higher than that of Δtthb178 or ΔrecJ (Figure 2B). The severe effect of the double disruption can be interpreted as follows: Tth ExoI and RecJ function in two parallel pathways during MMR. Consequently, we propose a model for MMR in T. thermophilus HB8. In this model, Tth ExoI and RecJ act as ssExos with opposite polarities, which excise the mismatch-containing DNA patch after the unwinding of dsDNA by DNA helicases (Figure 8A). The results of our in vitro experiments support the ability of Tth ExoI to cleave the liberated ssDNA regions in unwound dsDNAs (Figure 6B and C).
Figure 8.
Proposed models of the DNA repair pathways in T. thermophilus HB8. (A) The model of MMR. A DNA mismatch is generated by misincorporation of a base during DNA replication. The MutS/MutL complex recognises the mismatch and nicks the 3′- and 5′-sides of the incorrect base to create a DNA patch for removal. DNA helicases, such as UvrD, and RecJ or ExoI excise the error-containing patch. DNA polymerase fills the gap to complete the repair. (B) The model of the repair pathway for deaminated bases. Reactive oxygen species, such as hydroxyl radicals, attack the base to yield deaminated bases. Endonuclease V recognises a deaminated base and hydrolyses the second phosphodiester bond of the 3′-side of the bases. A DNA helicase unwinds the chain, and then, ExoI digests the lesion-containing ssDNA. (C) The model of DSB repair. UV-C irradiation causes DSBs in DNA. RecJ processes the termini to the 3′-overhanging structure in cooperation with a DNA helicase. The homologous pairing and re-synthesis of the DNA strand yield Holliday junctions. The resolution of Holliday junctions completes the repair.
Intriguingly, the disruption of tthb178 resulted in a great increase in the sensitivity to H2O2, whereas that of recJ did not (Figure 2D). In addition, Δtthb178–ΔrecJ showed similar H2O2 sensitivity as Δtthb178. H2O2-induced oxidative stress is known to cause base oxidation and base deamination (47). These results raise the possibility that Tth ExoI is involved in repair pathways for such damaged bases whereas RecJ is not.
It is well-known that oxidized base such as 8-oxoguanine or deaminated base such as uracil is repaired through base excision repair pathway. In this repair system, the N-glycosidic bonds of the damaged bases are incised by specific DNA glycosylases (60–62) to yield mainly an abasic site, a 5′-deoxyribose-5-phosphate residue. However, the exonuclease assay in this study clearly showed that Tth ExoI cannot excise the abasic site-containing ssDNA. A previous study also showed that RecJ has no 5′-deoxyribose-5-phosphatase activity (54). Thus, it is suggested that Tth ExoI and RecJ do not play a crucial role in the base excision repair pathway in T. thermophilus HB8.
The deaminated bases are also repaired by the alternative repair pathway that has not been completely revealed even in E. coli. In this repair pathway, endonuclease V is reported to nick the 3′-side of the lesion such as uracil, xanthine and hypoxanthine (46,63,64). Thermus thermophilus HB8 also possesses a gene encoding endonuclease V (TTHA1347). Our biochemical study showed that Tth ExoI cleaves the uracil-, xanthine-, hypoxanthine-containing ssDNA. On the basis of our results and those of the previous studies, we propose a model of a repair pathway for those deaminated bases (Figure 8B). In this model, Tth ExoI starts the excision from the entry point introduced by endonuclease V. As endonuclease V nicks the 3′-side of the lesion, the 3′–5′ ssExo but not the 5′–3′ ssExo is expected to be required for removing the lesion (16,17). This model is in good agreement with our result that the recJ disruption did not affect the sensitivity to H2O2. In addition, as shown in Supplementary Figure S4, tthb178 was actively transcribed especially in the late stationary phase. This result also implies the involvement of tthb178 in the repair, because cells in the late stationary phase are thought to be under serious oxidative stress, which can yield deaminated bases.
Our in vivo experiments also indicate that RecJ but not Tth ExoI plays a significant role in the repair of DNA damages caused by UV-C irradiation. UV-C irradiation mainly induces pyrimidine dimers, (6-4) photoproducts and DSBs (44). As pyrimidine dimers and (6-4) photoproducts are repaired mainly by nucleotide excision repair (NER) (49), our results indicate the possible involvement of RecJ in NER. However, it is believed that ssExos do not play a critical role in the NER pathway (65). Instead, RecJ is suggested to be involved in the recovery of replication forks at blocking DNA lesions caused by UV irradiation (66). Our result also can be interpreted in the context of the rescue of arrested replication forks after UV-C irradiation. It should be mentioned that the double disruption of tthb178 and recJ caused the more severe sensitivity to UV-C irradiation than the single disruption of recJ. It can be speculated that Tth ExoI and RecJ participate in separate pathways for the repair of UV-induced damage, and the Tth ExoI-dependent pathway might be stimulated when the RecJ-dependent one is inactive.
Unlike pyrimidine dimers or (6–4) photoproducts, DSBs directly result in cell death; therefore, we cannot exclude the possibility that the increase in the UV sensitivity of ΔrecJ suggests the involvement of RecJ in DSB repair. DSB repair, in bacteria, is performed mainly by homologous recombination. In general, there are two pathways in bacterial homologous recombination: the RecBCD and RecF pathways (67,68). The RecBCD complex and another 5′–3′ ssExo process the termini of dsDNAs to generate the 3′-overhanging structure in the RecBCD and RecF pathways, respectively. As T. thermophilus HB8 lacks the genes encoding RecBCD, the RecF pathway is expected to be dominant in this bacterium, and 5′–3′ exonuclease activity of RecJ may be required for the end-resection step in this pathway (Figure 8C).
In conclusion, the results of our in vivo and in vitro experiments suggest that 3′–5′ ssExo Tth ExoI participates in the excision step of MMR and the repair of deaminated bases, while 5′–3′ ssExo RecJ is involved in the excision step of MMR and the repair of UV-C-induced damages. The need for Tth ExoI and RecJ in other repair pathways such as the repair of methylated bases remains to be investigated. It is also to be examined whether Tth ExoI plays other roles in addition to DNA repair. Interestingly, it was recently reported that phage infection induces the expression of tthb178 in a CRP-dependent manner in vivo (69). Upon phage infection, T. thermophilus CRP up-regulates the transcription of not only tthb178 but also a variety of clustered regularly interspaced short palindromic repeat (CRISPR)-associated genes, so-called cas genes (27), which have been implicated as the components of a host defense system against invading foreign replicons (70,71). The 3′–5′ exonuclease activity of Tth ExoI may also be utilized for the bacterial host defense system.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Ministry of Education, Culture, Sports, Science and Technology of Japan, Grant-in-Aid for Scientific Research (grant numbers 20570131 to R.M., 19770083 to N.N. and 20870042 to K.F.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Akeo Shinkai and Yashihiro Agari for their valuable discussion on this study. They also thank Dr Yoshinori Koyama for providing pHG305 plasmid and Naoko Aoki for her excellent help in the construction of the plasmid for gene disruption.
REFERENCES
- 1.Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat. Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 2.Lee-Kirsch MA, Gong M, Chowdhury D, Senenko L, Engel K, Lee YA, de Silva U, Bailey SL, Witte T, Vyse TJ, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat. Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 3.Fukuda T, Sumiyoshi T, Takahashi M, Kataoka T, Asahara T, Inui H, Watatani M, Yasutomi M, Kamada N, Miyagawa K. Alterations of the double-strand break repair gene MRE11 in cancer. Cancer Res. 2001;61:23–26. [PubMed] [Google Scholar]
- 4.Viswanathan M, Burdett V, Baitinger C, Modrich P, Lovett ST. Redundant exonuclease involvement in Escherichia coli methyl-directed mismatch repair. J. Biol. Chem. 2001;276:31053–31058. doi: 10.1074/jbc.M105481200. [DOI] [PubMed] [Google Scholar]
- 5.Lombardo M-J, Aponyi I, Ray MP, Sandigursky M, Franklin WA, Rosenberg SM. xni-deficient Escherichia coli are proficient for recombination and multiple pathways of repair. DNA Repair. 2003;2:1175–1183. doi: 10.1016/s1568-7864(03)00135-6. [DOI] [PubMed] [Google Scholar]
- 6.Burdett V, Baitinger C, Viswanathan M, Lovett ST, Modrich P. In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc. Natl Acad. Sci. USA. 2001;98:6765–6770. doi: 10.1073/pnas.121183298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenstein BS, Ducore JM. Induction of DNA strand breaks in normal human fibroblasts exposed to monochromatic ultraviolet and visible wavelengths in the 240-546 nm range. Photochem. Photobiol. 1983;38:51–55. doi: 10.1111/j.1751-1097.1983.tb08365.x. [DOI] [PubMed] [Google Scholar]
- 8.Cromie GA, Connelly JC, Leach D.RF. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol. Cell. 2001;8:1163–1174. doi: 10.1016/s1097-2765(01)00419-1. [DOI] [PubMed] [Google Scholar]
- 9.Tachiki H, Kato R, Masui R, Hasegawa K, Itakura H, Fukuyama K, Kuramitsu S. Domain organization and functional analysis of Thermus thermophilus MutS protein. Nucleic Acids Res. 1998;26:4153–4159. doi: 10.1093/nar/26.18.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunkel TA, Erie DA. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 11.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLα in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 12.Kadyrov FA, Holmes SF, Arana ME, Lukianova OA, O’Donnell M, Kunkel TA, Modrich P. Saccharomyces cerevisiae MutLα is a mismatch repair endonuclease. J. Biol. Chem. 2007;282:37181–37190. doi: 10.1074/jbc.M707617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Modrich P. Mechanisms in eukaryotic mismatch repair. J. Biol. Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 15.Fukui K, Nishida M, Nakagawa N, Masui R, Kuramitsu S. Bound nucleotide controls the endonuclease activity of mismatch repair enzyme MutL. J. Biol. Chem. 2008;283:12136–12145. doi: 10.1074/jbc.M800110200. [DOI] [PubMed] [Google Scholar]
- 16.Dalhus B, Arvai AS, Rosnes I, Olsen OE, Backe PH, Alseth I, Gao H, Cao W, Tainer JA, Bjoras M. Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair. Nat. Struct. Mol. Biol. 2009;16:138–143. doi: 10.1038/nsmb.1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moe A, Ringvoll J, Nordstrand LM, Eide L, Bjoras M, Seeberg E, Rognes T, Klungland A. Incision at hypoxanthine residues in DNA by a mammalian homologue of the Escherichia coli antimutator enzyme endonuclease V. Nucleic Acids Res. 2003;31:3893–3900. doi: 10.1093/nar/gkg472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yokoyama S, Hirota H, Kigawa T, Yabuki T, Shirouzu M, Terada T, Ito Y, Matsuo Y, Kuroda Y, Nishimura Y, et al. Structural genomics projects in Japan. Nat. Struct. Biol. 2000;7:943–945. doi: 10.1038/80712. [DOI] [PubMed] [Google Scholar]
- 19.Fukui K, Masui R, Kuramitsu S. Thermus thermophilus MutS2, a MutS paralogue, possesses an endonuclease activity promoted by MutL. J. Biochem. 2004;135:375–384. doi: 10.1093/jb/mvh045. [DOI] [PubMed] [Google Scholar]
- 20.Morita R, Nakagawa N, Kuramitsu S, Masui R. An O6-methylguanine-DNA methyltransferase-like protein from Thermus thermophilus interacts with a nucleotide excision repair protein. J. Biochem. 2008;144:267–277. doi: 10.1093/jb/mvn065. [DOI] [PubMed] [Google Scholar]
- 21.Yamagata A, Masui R, Kakuta Y, Kuramitsu S, Fukuyama K. Overexpression, purification and characterization of RecJ protein from Thermus thermophilus HB8 and its core domain. Nucleic Acids Res. 2001;29:4617–4624. doi: 10.1093/nar/29.22.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Zhu L, Deutscher MP. Oligoribonuclease is encoded by a highly conserved gene in the 3′-5′ exonuclease superfamily. J. Bacteriol. 1998;180:2779–2781. doi: 10.1128/jb.180.10.2779-2781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moser MJ, Holley WR, Chatterjee A, Mian IS. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 1997;25:5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernad A, Blanco L, Lazaro J, Martin G, Salas M. A conserved 3′–>5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989;59:219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- 25.Breyer WA, Matthews BW. Structure of Escherichia coli exonuclease I suggests how processivity is achieved. Nat. Struct. Mol. Biol. 2000;7:1125–1128. doi: 10.1038/81978. [DOI] [PubMed] [Google Scholar]
- 26.Busam R. Structure of Escherichia coli exonuclease I in complex with thymidine 5′-monophosphate. Acta Crystallogr. D Biol. Crystallogr. 2008;64:206–210. doi: 10.1107/S090744490706012X. [DOI] [PubMed] [Google Scholar]
- 27.Shinkai A, Kira S, Nakagawa N, Kashihara A, Kuramitsu S, Yokoyama S. Transcription activation mediated by a cyclic AMP receptor protein from Thermus thermophilus HB8. J. Bacteriol. 2007;189:3891–3901. doi: 10.1128/JB.01739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishii T, Sootome H, Shan L, Yamashita K. Validation of universal conditions for duplex quantitative reverse transcription polymerase chain reaction assays. Anal. Biochem. 2007;362:201–212. doi: 10.1016/j.ab.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 29.Hashimoto Y, Yano T, Kuramitsu S, Kagamiyama H. Disruption of Thermus thermophilus genes by homologous recombination using a thermostable kanamycin-resistant marker. FEBS Lett. 2001;506:231–234. doi: 10.1016/s0014-5793(01)02926-x. [DOI] [PubMed] [Google Scholar]
- 30.Hoseki J, Yano T, Koyama Y, Kuramitsu S, Kagamiyama H. Directed evolution of thermostable kanamycin-resistance gene: a convenient selection marker for Thermus thermophilus. J. Biochem. 1999;126:951–956. doi: 10.1093/oxfordjournals.jbchem.a022539. [DOI] [PubMed] [Google Scholar]
- 31.Ooga T, Ohashi Y, Kuramitsu S, Koyama Y, Tomita M, Soga T, Masui R. Degradation of ppGpp by nudix pyrophosphatase modulates the transition of growth phase in the bacterium Thermus thermophilus. J. Biol. Chem. 2009;284:15549–15556. doi: 10.1074/jbc.M900582200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luria SE, Delbruck M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salzano AM, D’Ambrosio C, Scaloni A. Mass spectrometric characterization of proteins modified by nitric oxide-derived species. Meth. Enzymol. 2008;440:3–15. doi: 10.1016/S0076-6879(07)00801-4. [DOI] [PubMed] [Google Scholar]
- 34.Fukui K, Takahata Y, Nakagawa N, Kuramitsu S, Masui R. Analysis of a nuclease activity of catalytic domain of Thermus thermophilus MutS2 by high-accuracy mass spectrometry. Nucleic Acids Res. 2007;35:e100. doi: 10.1093/nar/gkm575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma R, Rao DN. Orchestration of Haemophilus influenzae RecJ exonuclease by interaction with single-stranded DNA-binding protein. J. Mol. Biol. 2009;385:1375–1396. doi: 10.1016/j.jmb.2008.11.041. [DOI] [PubMed] [Google Scholar]
- 36.Perrino FW, de Silva U, Harvey S, Pryor E.E., Jr, Cole DW, Hollis T. Cooperative DNA binding and communication across the dimer interface in the TREX2 3′ –> 5′-exonuclease. J. Biol. Chem. 2008;283:21441–21452. doi: 10.1074/jbc.M803629200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viswanathan M, Lovett ST. Exonuclease X of Escherichia coli. A novel 3′-5′ DNase and DnaQ superfamily member involved in DNA repair. J. Biol. Chem. 1999;274:30094–30100. doi: 10.1074/jbc.274.42.30094. [DOI] [PubMed] [Google Scholar]
- 38.Lehman IR, Nussbaum AL. The deoxyribonucleases of Escherichia coli. J. Biol. Chem. 1964;239:2628–2636. [PubMed] [Google Scholar]
- 39.Melancon P, Lemieux C, Brakier-Gingras L. A mutation in the 530 loop of Escherichia coli 16S ribosomal RNA causes resistance to streptomycin. Nucleic Acids Res. 1988;16:9631–9639. doi: 10.1093/nar/16.20.9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moazed D, Noller HF. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- 41.Bradley MO. Double-strand breaks in DNA caused by repair of damage due to ultraviolet light. J. Supramol. Struct. Cell. Biochem. 1981;16:337–343. doi: 10.1002/jsscb.1981.380160404. [DOI] [PubMed] [Google Scholar]
- 42.Franklin WA, Haseltine WA. The role of the (6-4) photoproduct in ultraviolet light-induced transition mutations in E. coli. Mutat. Res. 1986;165:1–7. doi: 10.1016/0167-8817(86)90002-7. [DOI] [PubMed] [Google Scholar]
- 43.Weinfeld M, Liuzzi M, Paterson MC. Enzymatic analysis of isomeric trithymidylates containing ultraviolet light-induced cyclobutane pyrimidine dimers. II. Phosphorylation by phage T4 polynucleotide kinase. J. Biol. Chem. 1989;264:6364–6370. [PubMed] [Google Scholar]
- 44.Bourre F, Benoit A, Sarasin A. Respective roles of pyrimidine dimer and pyrimidine (6-4) pyrimidone photoproducts in UV mutagenesis of simian virus 40 DNA in mammalian cells. J. Virol. 1989;63:4520–4524. doi: 10.1128/jvi.63.11.4520-4524.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonura T, Smith KC. Enzymatic production of deoxyribonucleic acid double-strand breaks after ultraviolet irradiation of Escherichia coli K-12. J. Bacteriol. 1975;121:511–517. doi: 10.1128/jb.121.2.511-517.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss B. Evidence for mutagenesis by nitric oxide during nitrate metabolism in Escherichia coli. J. Bacteriol. 2006;188:829–833. doi: 10.1128/JB.188.3.829-833.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akagawa M, Suyama K. Oxidative deamination by hydrogen peroxide in the presence of metals. Free Radic. Res. 2002;36:13–21. doi: 10.1080/10715760210167. [DOI] [PubMed] [Google Scholar]
- 48.Kondo N, Nishikubo T, Wakamatsu T, Ishikawa H, Nakagawa N, Kuramitsu S, Masui R. Insights into different dependency of dNTP triphosphohydrolase on metal ion species from intracellular ion concentrations in Thermus thermophilus. Extremophiles. 2008;12:217–223. doi: 10.1007/s00792-007-0118-6. [DOI] [PubMed] [Google Scholar]
- 49.Friedberg EC, Walker GC, Siede W. DNA Repair and Mutagenesis. Washington, DC: American Society for Microbiology; 1995. [Google Scholar]
- 50.Perrino FW, Harvey S, McMillin S, Hollis T. The human TREX2 3′ -> 5′-exonuclease structure suggests a mechanism for efficient nonprocessive DNA catalysis. J. Biol. Chem. 2005;280:15212–15218. doi: 10.1074/jbc.M500108200. [DOI] [PubMed] [Google Scholar]
- 51.de Silva U, Choudhury S, Bailey SL, Harvey S, Perrino FW, Hollis T. The crystal structure of TREX1 explains the 3′ nucleotide specificity and reveals a polyproline II helix for protein partnering. J. Biol. Chem. 2007;282:10537–10543. doi: 10.1074/jbc.M700039200. [DOI] [PubMed] [Google Scholar]
- 52.de Silva U, Perrino FW, Hollis T. DNA binding induces active site conformational change in the human TREX2 3′-exonuclease. Nucleic Acids Res. 2009;37:2411–2417. doi: 10.1093/nar/gkp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lehman IR. The deoxyribonucleases of Escherichia coli. J. Biol. Chem. 1960;235:1479–1487. [PubMed] [Google Scholar]
- 54.Piersen CE, McCullough AK, Lloyd RS. AP lyases and dRPases: commonality of mechanism. Mutat. Res. 2000;459:43–53. doi: 10.1016/s0921-8777(99)00054-3. [DOI] [PubMed] [Google Scholar]
- 55.Sandigursky M, Franklin WA. Escherichia coli single-stranded DNA binding protein stimulates the DNA deoxyribophosphodiesterase activity of exonuclease I. Nucleic Acids Res. 1994;22:247–250. doi: 10.1093/nar/22.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sandigursky M, Franklin WA. DNA deoxyribophosphodiesterase of Escherichia coli is associated with exonuclease I. Nucleic Acids Res. 1992;20:4699–4703. doi: 10.1093/nar/20.18.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morelle S, Carbonnelle E, Matic I, Nassif X. Contact with host cells induces a DNA repair system in pathogenic Neisseriae. Mol. Microbiol. 2005;55:853–861. doi: 10.1111/j.1365-2958.2004.04426.x. [DOI] [PubMed] [Google Scholar]
- 58.Han ES, Cooper DL, Persky NS, Sutera V.A., Jr, Whitaker RD, Montello ML, Lovett ST. RecJ exonuclease: substrates, products and interaction with SSB. Nucleic Acids Res. 2006;34:1084–1091. doi: 10.1093/nar/gkj503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sandigursky M, Mendez F, Bases RE, Matsumoto T, Franklin WA. Protein-protein interactions between the Escherichia coli single-stranded DNA-binding protein and exonuclease I. Radiat. Res. 1996;145:619–623. [PubMed] [Google Scholar]
- 60.Back JH, Park JH, Chung JH, Kim D.S.HL, Han YS. A distinct TthMutY bifunctional glycosylase that hydrolyzes not only adenine but also thymine opposite 8-oxoguanine in the hyperthermophilic bacterium, Thermus thermophilus. DNA Repair. 2006;5:894–903. doi: 10.1016/j.dnarep.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 61.Seeberg E, Eide L, Bjoras M. The base excision repair pathway. Trends Biochem. Sci. 1995;20:391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- 62.Michaels ML, Tchou J, Grollman AP, Miller JH. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry. 2002;31:10964–10968. doi: 10.1021/bi00160a004. [DOI] [PubMed] [Google Scholar]
- 63.Weiss B. Endonuclease V of Escherichia coli prevents mutations from nitrosative deamination during nitrate/nitrite respiration. Mutat. Res. 2001;461:301–309. doi: 10.1016/s0921-8777(00)00062-8. [DOI] [PubMed] [Google Scholar]
- 64.Moe A, Ringvoll J, Nordstrand LM, Eide L, Bjoras M, Seeberg E, Rognes T, Klungland A. Incision at hypoxanthine residues in DNA by a mammalian homologue of the Escherichia coli antimutator enzyme endonuclease V. Nucleic Acids Res. 2003;31:3893–3900. doi: 10.1093/nar/gkg472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 66.Courcelle J, Hanawalt PC. Participation of recombination proteins in rescue of arrested replication forks in UV-irradiated Escherichia coli need not involve recombination. Proc. Natl Acad. Sci. USA. 2001;98:8196–8202. doi: 10.1073/pnas.121008898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dillingham MS, Spies M, Kowalczykowski SC. RecBCD enzyme is a bipolar DNA helicase. Nature. 2003;423:893–897. doi: 10.1038/nature01673. [DOI] [PubMed] [Google Scholar]
- 68.Ivancic-Bace I, Salaj-Smic E, Brcic-Kostic K. Effects of recJ, recQ, and recFOR mutations on recombination in nuclease-deficient recB recD double mutants of Escherichia coli. J. Bacteriol. 2005;187:1350–1356. doi: 10.1128/JB.187.4.1350-1356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Agari Y, Sakamoto K, Tamakoshi M, Oshima T, Kuramitsu S, Shinkai A. Transcription profile of Thermus thermophilus CRISPR systems after phage infection. J. Mol. Biol. 2010;395:270–281. doi: 10.1016/j.jmb.2009.10.057. [DOI] [PubMed] [Google Scholar]
- 70.Sorek R, Kunin V, Hugenholtz P. CRISPR–a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 71.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McLuckey SA, Van Berkel GJ, Glish GL. Tandem mass spectrometry of small, multiply charged oligonucleotides. J. Am. Soc. Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.