Abstract
Transcriptomics (at the level of single cells, tissues and/or whole organisms) underpins many fields of biomedical science, from understanding the basic cellular function in model organisms, to the elucidation of the biological events that govern the development and progression of human diseases, and the exploration of the mechanisms of survival, drug-resistance and virulence of pathogens. Next-generation sequencing (NGS) technologies are contributing to a massive expansion of transcriptomics in all fields and are reducing the cost, time and performance barriers presented by conventional approaches. However, bioinformatic tools for the analysis of the sequence data sets produced by these technologies can be daunting to researchers with limited or no expertise in bioinformatics. Here, we constructed a semi-automated, bioinformatic workflow system, and critically evaluated it for the analysis and annotation of large-scale sequence data sets generated by NGS. We demonstrated its utility for the exploration of differences in the transcriptomes among various stages and both sexes of an economically important parasitic worm (Oesophagostomum dentatum) as well as the prediction and prioritization of essential molecules (including GTPases, protein kinases and phosphatases) as novel drug target candidates. This workflow system provides a practical tool for the assembly, annotation and analysis of NGS data sets, also to researchers with a limited bioinformatic expertise. The custom-written Perl, Python and Unix shell computer scripts used can be readily modified or adapted to suit many different applications. This system is now utilized routinely for the analysis of data sets from pathogens of major socio-economic importance and can, in principle, be applied to transcriptomics data sets from any organism.
INTRODUCTION
Transcriptomics is the molecular science of examining, simultaneously, the transcription of all genes at the level of the cell, tissue and/or whole organism, allowing inferences regarding cellular functions and mechanisms. The ability to measure the transcription of thousands of genes simultaneously has led to major advances in all biomedical fields, from understanding the basic function in model organisms, such as the free-living nematode Caenorhabditis elegans (1–3) or the vinegar fly, Drosophila melanogaster (4–6), to studying molecular events associated with the development and progression of human diseases, including cancer (7–9) and neurodegenerative disorders (10–12), to the exploration of the mechanisms of survival, drug-resistance and virulence/pathogenicity of bacteria (13,14) and other socioeconomically important pathogens, such as parasites (15–20). For more than a decade, transcriptomes have been determined by sequencing expressed sequence tags (ESTs) using the conventional Sanger method (21,22), whereas levels of transcription have been established quantitatively or semi-quantitatively by real-time polymerase chain reaction (PCR) (23) and/or cDNA microarrays (24). The use of these technologies has been accompanied by an increasing demand for analytical tools for the efficient annotation of nucleotide sequence data sets, particularly within the framework of large-scale EST projects (25). With a substantial expansion of EST sequencing has come the development of algorithms for sequence assembly, analysis and annotation, in the form of individual programs (26–28) and integrated pipelines (29,30), some of which have been made available on the worldwide web (29,31,32). However, the cost and time associated with large-scale sequencing using a conventional (Sanger) method and/or the design of customized analytical tools (e.g. cDNA microarray) have driven the search for alternative methods for transcriptomic studies (33).
In the last few years, there has been a massive expansion in the demand for and access to low cost, high-throughput sequencing, attributable mainly to the development of next-generation sequencing (NGS) technologies, which allow massively parallelized sequencing of millions of nucleic acids (33,34). These sequencing platforms, such as 454/Roche (35; http://www.454.com/) and Illumina/Solexa (36; http://www.illumina.com/), have transformed transcriptomics by decreasing the cost, time and performance limitations presented by previous approaches. This situation has resulted in an explosion of the number of EST sequences deposited in databases worldwide, the majority of which is still awaiting detailed functional annotation. However, the high-throughput analysis of such large data sets has necessitated significant advances in computing capacity and performance, and in the availability of bioinformatic tools to distil biologically meaningful information from raw sequence data.
Sequences generated by NGS are significantly shorter (454/Roche: ∼400 bases; Illumina/ABI-SOLiD: ∼60 bases) than those determined by Sanger sequencing (0.8–1 kb), which poses a challenge for assembly. In addition, the data files generated by these technologies are often gigabytes to terabytes (1 × 109 to 1 × 1012 bytes) in size, substantially increasing the demands placed on data transfer and storage, such that many web-based interfaces are not suited for large-scale analyses. The bioinformatic processing of large data sets usually requires access to powerful computers and support from bioinformaticians with significant expertise in a range of programming languages (e.g. Perl and Python). This situation has limited the accessibility of high-throughput sequencing technologies to some (smaller) research groups, and has thus restricted somewhat the ‘democratization’ of large-scale genomic and/or transcriptomic sequencing. Clearly, user-friendly and flexible bioinformatic pipelines are needed to assist researchers from different disciplines and backgrounds in accessing and taking full advantage of the advances heralded by NGS. Increasing the accessibility to high-throughput sequencing will have major benefits in a range of areas, including the investigation of pathogens. The exploration of the transcriptomes of pathogens has major implications in improving our understanding of their development and reproduction, survival in and interactions with the host, virulence, pathogenicity, the diseases that they cause and drug resistance (17–20,37–39), and has the potential to pave the way to novel approaches for treatment, diagnosis and control. In the present study, we (i) constructed a semi-automated, bioinformatic workflow system for the analysis and annotation of large-scale sequence data sets generated by NGS, (ii) demonstrated its utility by profiling differences in the transcriptome of an economically important parasite, Oesophagostomum dentatum (Strongylida), throughout its development, and (iii) indicated the broader applicability of this system to different types of transcriptomic data sets.
METHODS
Sequence data sets
For this study, original cDNA sequence data sets representing four distinct developmental stages of O. dentatum [i.e. third-stage (L3) and fourth-stage (L4) larvae as well as adult female and male worms] were produced and stored as described previously (40). Total RNA (10 µg) from each stage and/or sex was used to construct a normalised cDNA library; each library was sequenced using a Genome Sequencer™ (GS) Titanium FLX (Roche Diagnostics) as described previously (18). FASTA- and associated files, with short-read sequence quality scores of each data set, were extracted from each SFF-file; sequence adaptors were clipped using the ‘sff_extract’ software (http://bioinf.comav.upv.es/sff_extract/index.html).
Bioinformatic components for the construction of the workflow system
Five components (1–5), documented in a series of peer-reviewed, international publications, were selected based on the parameters of general applicability, ease of use, versatility and efficiency. Once constructed, the workflow system was applied to the analysis of the O. dentatum data sets.
Assembly
The Contig Assembly Program (CAP3 v.3; 31) was used to cluster sequences (with quality scores) into contigs and singletons from individual or combined (i.e. pooled) data sets, employing a minimum sequence overlap of 40 nucleotides and an identity threshold of 90%. This program was selected to enable the assembly of relatively long sequences and to remove redundant short-reads (41).
Similarity searching
BLASTn and BLASTx algorithms (42) were used to compare contigs and singletons with sequences available in public databases [i.e. NCBI (www.ncbi.nlm.nih.gov) and EMBL-EBI Parasite Genome Blast Server (www.ebi.ac.uk); April 2010], to identify putative homologues in range of other organisms (cut-off: <1E-05). For nematodes, WormBase (release WS200; www.wormbase.org) was interrogated extensively for relevant information on C. elegans orthologues/homologues, including transcriptomic, proteomic, RNA interference (RNAi) phenotypes and interactomic data.
Prediction and annotation of peptides
The program ESTScan (32) was used to conceptually translate peptides from assembled contigs and singletons. InterProScan (available at http://www.ebi.ac.uk/InterProScan/; 27) and gene ontology (GO; 43) were used to classify peptides (based on their putative function/s). Biological pathways were inferred from C. elegans for each peptide using the KEGG Orthology-Based Annotation System software (KOBAS; 44) and displayed using the iPath tool (http://pathways.embl.de/data_mapping.html; 45).
In silico subtraction
A BLASTn algorithm, employing a stringent cut-off (cut-off: <1E-15; 17), was used to examine differential transcription between data sets by subtraction in silico. Peptides corresponding to transcripts that were unique to a particular data set were assigned parental (i.e. level 1) InterPro terms and compared, using a BLASTp algorithm (cut-off: <1E-15), with peptides inferred from the assembly of sequences from combined data sets. The subtraction approach allows qualitative (not quantitative) differences between or among samples to be established.
Probabilistic functional networking of protein-encoding genes, and drug target prediction
Interaction networks among C. elegans orthologues of differentially transcribed molecules were inferred using an established approach (46). The druggability of C. elegans homologues of molecules unique to a particular O. dentatum data set or common to all data sets was inferred using a published method (18). Briefly, the InterPro domains of predicted proteins were compared with those linked to known, small molecular drugs, which follow the ‘Lipinsky rule of 5′ regarding bioavailability (47,48). GO terms were mapped to Enzyme Commission (EC) numbers, and a list of enzyme-targeting drugs was compiled based on data available in the BRENDA database (www.brenda-enzymes.info; 49,50). The C. elegans orthologues/homologues included in this list were ranked according to the ‘severity’ of non-wild-type RNAi phenotypes (including lethality or sterility of different developmental stages; see www.wormbase.org; release WS200).
RESULTS
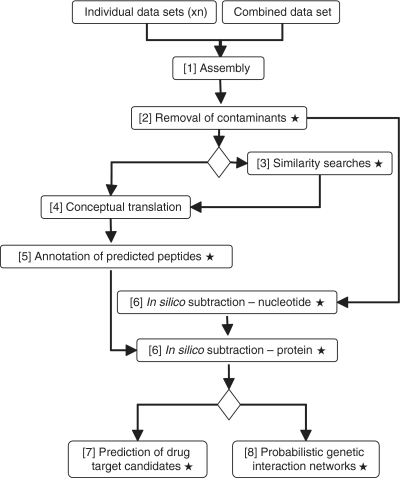
A semi-automated bioinformatic workflow system (Figure 1), incorporating five key bioinformatic components, was constructed and linked using customized Perl, Python and Unix shell computer scripts (listed in Supplementary File S1 and accessible via http://research.vet.unimelb.edu.au/gasserlab/index.html). This system was then assessed for the assembly, analysis and functional annotation of each or all of the four sequence data sets for O. dentatum. The specificity of the in silico subtraction step was verified using independent experimental evidence.
Figure 1.
Bioinformatic analyses of the Oesophagostomum dentatum data sets. Stars indicate analyses performed using custom-written Perl, Python and/or Unix shell computer scripts, accessible via http://research.vet.unimelb.edu.au/gasserlab/index.html. [1] Individual and combined expressed sequence tags (EST) data sets are assembled using CAP3 (compiled Linux 64-bit executable) to generate consensus sequences. [2] Assembled contigs with high similarity (cut-off: <1E-15) to nucleotide sequences of the vertebrate host (Sus scrofa) are eliminated. [3] Database similarity searches (for individual or combined data sets) are carried out using BLASTn and BLASTx (compiled Linux 64-bit executable; 42), embedded in custom-built Unix shell scripts. [4] Sequences (from the individually and combined assembled data sets) are conceptually translated into peptide sequences using ESTScan (compiled Linux 64-bit executable with a Perl wrapper). [5] Domains/motifs within translated peptides are identified via InterProScan (Perl wrapper) and linked to biological pathways in C. elegans using KOBAS (stand-alone Python application; 44). Functional annotation of the predicted peptides is performed by gene ontology (Perl wrapper; 27). [6] The individually assembled data sets are subtracted from one another (in both directions) using a BLASTn algorithm (42) embedded in a custom-built Unix shell script; proteins inferred from subtracted transcripts are assigned parental (i.e. level 1) InterPro terms and subtracted from one another using a BLASTp algorithm, embedded in a custom-built Unix shell script. [7] Potential drug target candidates for each of the individually assembled and/or in silico subtracted data sets are predicted and ranked according to the ‘severity’ of the non-wild-type RNAi phenotypes observed for the corresponding C. elegans orthologues/homologues (custom-built Unix shell scripts). [8] Probabilistic interaction networks among C. elegans orthologues of subtracted molecules are predicted (command lines).
Assembly and detailed annotation and analyses of the O. dentatum data sets
A total of 1 826 367 sequences (244 ± 32 bases; i.e. mean length ± standard deviation) were determined for L3, L4 as well as adult female and male of O. dentatum. Following the clipping of adapter sequences, only sequences of >100 bases (n = 1 800 874; 98.6%) were included in further analyses. The numbers of contigs assembled for each of the four data sets are listed in Table 1. The assembly of the sequences of all four data sets yielded 36 233 contigs (516 ± 316 bases in length) and 452 528 singletons (Table 1); sequences (n = 115) with similarity (cut-off: <1E-15) to potential host molecules were excluded. The L3 data set had the largest number of sequence clusters with orthologues/homologues in C. elegans (n = 32 904; Table 1) and in organisms other than nematodes (n = 14 731; Table 1), whereas the L4 data set included the largest number of clusters with orthologues/homologues in other parasitic nematodes (n = 38 634; Table 1).
Table 1.
Summary of the nucleotide sequence data for the adult female, adult male, and third (L3) and fourth (L4) larval stages of Oesophagostomum dentatum prior to and following in silico subtraction as well as detailed bioinformatic annotation and analyses
| Female | Male | L3 | L4 | Combined | |
|---|---|---|---|---|---|
| Expressed sequence tags (ESTs) | |||||
| Number of unassembled ESTs | 336 131 | 490 645 | 503 566 | 496 025 | 1 826 367 |
| Contigs (average length ± SD) | 23 807 (483 ± 290) | 29 043 (484 ± 289) | 30 176 (465 ± 281) | 26 349 (498 ± 308) | 36 233 (516 ± 316) |
| Singletons | 23 303 (233 ± 50) | 37 248 (243 ± 45) | 49 341 (227 ± 57) | 36 875 (242 ± 40) | 452 528 (244 ± 37) |
| Total | 47 110 | 66 291 | 79 517 | 63 224 | 488 761 |
| Containing an open reading frame (%) | 38 504 (81.7) | 52 787 (80) | 57 818 (73) | 50 533 (80) | 85 395 (17.5) |
| Returning InterProScan results (%) | 20 229 (43) | 26 496 (40) | 27 297 (47.2) | 26 121 (51.7) | 56 940 (66.7) |
| Gene ontology (%) | 9970 (25.9) | 12 386 (23.5) | 12 763 (22.1) | 12 735 (25.2) | 25 216 (30) |
| Number of biological process terms | 17 031 | 19 510 | 19 705 | 19 645 | 19 346 |
| Cellular component | 8864 | 10 091 | 10 926 | 10649 | 11 007 |
| Molecular function | 30 482 | 35 934 | 34 904 | 35 241 | 35 182 |
| With orthologues in C. elegans | 23 485 (50) | 28 643 (43.2) | 32 904 (41.4) | 30 000 (47.4) | |
| Other parasitic nematodes (%) | 17 533 (37.2) | 21 553 (32.5) | 23 748 (29.9) | 38 634 (61) | |
| Other organisms (%) | 12 011 (25.5) | 13 843 (21) | 14 731 (18.5) | 14 332 (22.7) | |
| KOBAS (number of biological pathways predicted) | 256 | 254 | 249 | 255 | |
| In silico subtracted data sets | |||||
| Number of ESTs (contigs + singletons) | 3451 (671 + 2780) | 10 344 (2902 + 7442) | 14 380 (2752 + 11 628) | 7520 (1280 + 6240) | |
| Containing an open reading frame (%) | 2397 (70) | 7117 (69) | 7222 (50.2) | 4789 (63.7) | |
| Predicted peptides | |||||
| Returning InterProScan results (%) | 521 (21.7) | 1179 (16.6) | 1224 (17) | 989 (20.7) | |
| Gene ontology (%) | 376 (15.7) | 840 (11.8) | 760 (10.5) | 652 (13.6) | |
| Number of biological process terms | 314 | 625 | 684 | 527 | |
| Cellular component | 177 | 355 | 412 | 359 | |
| Molecular function | 563 | 1259 | 1073 | 948 | |
| With homologues in C. elegans (%) | 824 (23.9) | 1834 (17.7) | 2252 (15.6) | 1589 (21.1) | |
| Other parasitic nematodes (%) | 558 (16.1) | 1212 (11.7) | 1384 (9.6) | 1052 (14) | |
| Other organisms (%) | 159 (4.6) | 123 (1.2) | 176 (1.2) | 137 (1.8) | |
| KOBAS (number of biological pathways predicted) | 7 | 16 | 18 | 23 |
Of the four assembled data sets, the L3 set included the largest number of sequence clusters with predicted open reading frames (ORFs; n = 57 818; Table 1), of which 27 297 (47.2%) could be annotated functionally using InterPro terms and 12 763 (22.1%) could be assigned GO terms, including 19 705 ‘biological process’, 10 926 ‘cellular component’ and 34 904 ‘molecular function’. The numbers of peptides inferred from sequence clusters in the adult female, adult male and/or L4 data sets, which could be assigned InterPro and/or GO terms, are given in Table 1. In total, 85 395 peptides were predicted for all sequences from all four data sets, representing 17.5% of clusters (Table 1); 56 940 (66.7%) of them could be mapped to known proteins defined by 31 982 different domains, the most represented being ‘SCP-like extracellular’ (IPR014044; 1.2% of the peptides mapping to a conserved protein motif), ‘NAD(P)-binding’ (IPR016040; 1.1%) and ‘proteinase inhibitor I2, Kunitz metazoa’ (IPR002223; 1%) (Table 2). GO annotation allowed 56 940 (66.7%) inferred proteins to be assigned to 19 346 ‘biological process’, 11 007 ‘cellular component’ and 35 182 ‘molecular function’ terms (Table 1). The predominant terms were ‘metabolic process’ (GO:0008152; 10.9%), ‘proteolysis’ (GO:0006508; 7%) and ‘translation’ (GO:0006412; 5.4%) for ‘biological process’; ‘intracellular’ (GO:0005622; 17.5%), ‘membrane’ (GO:0016020; 15.6%) and ‘nucleus’ (GO:0005634; 11.6%) for ‘cellular component’ and ‘ATP binding’ (GO:0005524; 7.5%); ‘catalytic activity’ (GO:0003824; 7%) and ‘binding’ (GO:0005488; 4.6%) for ‘molecular function’ (Table 3). Proteins inferred from the combined assembly were predicted to be involved in 262 different biological pathways, defined by 64 unique KEGG terms, of which ‘peptidases’ (12%), ‘other enzymes’ (8%) and ‘antigen processing and presentation’ (5.5%) were predominant (see Supplementary File S2). A display of biological pathways, defined by KEGG terms, inferred from predicted peptides and mapped to the complement of known pathways in C. elegans, is shown in Supplementary Figure S1.
Table 2.
The 20 most represented (InterPro) protein domains inferred from peptides conceptually translated from individual contigs for Oesophagostomum dentatum [combined assembly of data for adult female, adult male, and the third (L3) and fourth (L4) larval stages] and InterPro protein domains (level 1) assigned to predicted peptides unique to each stage or sex following in silico subtraction
| InterPro description | InterPro code | Number of predicted peptides (%) |
|---|---|---|
| Combined assembly (31 982)a | ||
| SCP-like extracellular | IPR014044 | 377 (1.2) |
| NAD(P)-binding domain | IPR016040 | 365 (1.1) |
| Proteinase inhibitor I2, Kunitz metazoa | IPR002223 | 339 (1) |
| Zinc finger, LIM-type | IPR001781 | 332 (1) |
| WD40 repeat | IPR001680 | 312 (0.9) |
| Ankyrin | IPR002110 | 257 (0.8) |
| EF-HAND 2 | IPR018249 | 247 (0.7) |
| WD40 repeat, subgroup | IPR019781 | 242 (0.7) |
| Allergen V5/Tpx-1 related | IPR001283 | 236 (0.7) |
| Protein kinase-like | IPR011009 | 220 (0.6) |
| RNA recognition motif, RNP-1 | IPR000504 | 216 (0.6) |
| WD40 repeat 2 | IPR019782 | 215 (0.6) |
| Protease inhibitor I4, serpin | IPR000215 | 207 (0.6) |
| Src homology-3 domain | IPR001452 | 201 (0.6) |
| Peptidase C1A, papain C-terminal | IPR000668 | 194 (0.6) |
| C-type lectin | IPR001304 | 183 (0.5) |
| Kelch repeat type 1 | IPR006652 | 183 (0.5) |
| Annexin repeat | IPR018502 | 183 (0.5) |
| Protein kinase, core | IPR000719 | 172 (0.5) |
| EF-HAND 1 | IPR018247 | 168 (0.5) |
| Female (139)a | ||
| Chitin binding protein, peritrophin-A | IPR002557 | 18 (8.6) |
| Basic-leucine zipper (bZIP) transcription factor | IPR004827 | 10 (4.8) |
| DNA primase, small subunit | IPR002755 | 6 (2.9) |
| p53-like transcription factor, DNA-binding | IPR008967 | 5 (2.4) |
| DNA-binding HORMA | IPR003511 | 4 (2) |
| Acyl-CoA dehydrogenase/oxidase | IPR013786 | 3 (1.4) |
| Frizzled-like domain | IPR020067 | 3 (1.4) |
| Lipid transport protein | IPR001747 | 3 (1.4) |
| PreATP-grasp-like fold | IPR016185 | 3 (1.4) |
| UbiA prenyltransferase | IPR000537 | 3 (1.4) |
| Male (243)a | ||
| PapD-like | IPR008962 | 16 (4) |
| Major sperm protein | IPR000535 | 15 (3.7) |
| C-type lectin | IPR018378 | 6 (1.5) |
| Phosphoenolpyruvate carboxykinase | IPR008209 | 6 (1.5) |
| Protein of unknown function DUF236 | IPR004296 | 6 (1.5) |
| Scramblase | IPR005552 | 6 (1.5) |
| ClpX, ATPase regulatory subunit | IPR004487 | 5 (1.3) |
| Galactose oxidase/kelch | IPR011043 | 5 (1.3) |
| Ribosomal protein S2 | IPR001865 | 5 (1.3) |
| Amidinotransferase | IPR003198 | 4 (1) |
| L3 (220)a | ||
| RmlC-like jelly roll fold | IPR014710 | 17 (4.5) |
| Six-bladed beta-propeller, TolB-like | IPR011042 | 10 (2.7) |
| Protein of unknown function DUF590 | IPR007632 | 9 (2.4) |
| 7TM GPCR, serpentine receptor class r (Str), Nematode | IPR019428 | 8 (2.1) |
| Acyltransferase ChoActase/COT/CPT | IPR000542 | 7 (1.9) |
| Putative DNA binding | IPR009061 | 7 (1.9) |
| 7TM GPCR, serpentine receptor class e (Sre), Nematode | IPR004151 | 6 (1.6) |
| Nuclear hormone receptor, ligand-binding, core | IPR000536 | 6 (1.6) |
| Coenzyme A transferase | IPR004165 | 5 (1.3) |
| Ion transport | IPR005821 | 5 (1.3) |
| L4 (249)a | ||
| Peptidase M24, methionine aminopeptidase | IPR001714 | 7 (2.2) |
| FAD-binding, type 2 | IPR016166 | 4 (1.3) |
| Oxysterol-binding protein | IPR000648 | 4 (1.3) |
| Translation protein SH3-like | IPR008991 | 4 (1.3) |
| Tubulin/FtsZ, GTPase domain | IPR003008 | 4 (1.3) |
| 6-phosphogluconate dehydrogenase | IPR008927 | 3 (1) |
| Peptidase C13, legumain | IPR001096 | 3 (1) |
| Aminoacyl-tRNA synthetase | IPR015413 | 3 (1) |
| Adenosylcobalamin biosynthesis, ATP | IPR016030 | 3 (1) |
| Aspartate/other aminotransferase | IPR000796 | 2 (0.6) |
aNumber of unique InterPro domains assigned to predicted peptides in each data set
Table 3.
Functions predicted for proteins encoded in the transcriptome of Oesophagostomum dentatum (combined assembly), based on gene ontology (GO)
| GO description (GO code) | Number of predicted peptides (%) |
|---|---|
| Biological process (19 346)a | |
| Metabolic process (GO:0008152) | 2102 (10.9) |
| Proteolysis (GO:0006508) | 1361 (7) |
| Translation (GO:0006412) | 1033 (5.4) |
| Transport (GO:0006810) | 816 (4.2) |
| Protein amino acid phosphorylation (GO:0006468) | 763 (4) |
| Cellular component (11 007) | |
| Intracellular (GO:0005622) | 1925 (17.5) |
| Membrane (GO:0016020) | 1717 (15.6) |
| Nucleus (GO:0005634) | 1279 (11.6) |
| Integral to membrane (GO:0016021) | 1159 (10.5) |
| Ribosome (GO:0005840) | 736 (6.7) |
| Molecular function (35 182) | |
| ATP binding (GO:0005524) | 2645 (7.5) |
| Catalytic activity (GO:0003824) | 2449 (7) |
| Binding (GO:0005488) | 1622 (4.6) |
| Zinc ion binding (GO:0008270) | 1229 (3.5) |
| Oxidoreductase activity (GO:0016491) | 1226 (3.5) |
| Protein binding (GO:0005515) | 1206 (3.4) |
| Nucleic acid binding (GO:0003676) | 919 (2.6) |
| DNA binding (GO:0003677) | 788 (2.2) |
| Structural constituent of ribosome (GO:0003735) | 755 (2.1) |
| Nucleotide binding (GO:0000166) | 717 (2) |
aTotal number of unique GO terms assigned to predicted peptides.
The parental (=level 2) GO categories were assigned according to (InterPro) domains inferred from proteins with homology to functionally annotated molecules.
Using BLASTn algorithms, subsets of 3451, 10 344, 14 380 and 7520 nucleotide sequences were identified as being uniquely transcribed in adult female, adult male, L3 and L4, respectively (Table 1). The accuracy of the in silico subtraction process was verified using independent evidence from a previous analysis of differential transcription between adult females and males of O. dentatum using a microarray-based approach (51). This verification showed that all 220 female- and 171 male-enriched molecules characterized previously (51; GenBank accession numbers AM157797-AM158083) were contained exclusively within the female and male data sets, respectively, following in silico subtraction (data available upon request). Based on these findings, the specificity of the subtraction process, calculated using the Wilson score (52) at a confidence interval of 95%, ranged from 98% to 100%. Of the 139 parental functional domains assigned to predicted peptides unique to the adult female data set, ‘chitin-binding protein, peritrophin-A’ (IPR002557; 8.6%) and ‘basic-leucine zipper (bZIP) transcription factor’ (IPR004827; 4.8%) were highly represented. Of the 243 protein motifs identified amongst the predicted peptides that were unique to the adult male data set, ‘PapD-like’ (IPR008962; 4%) and ‘major-sperm protein’ (IPR000535; 3.7%) were most represented. For the L3 data set, 220 unique protein motifs were identified, of which ‘RmlC-like jelly roll fold’ (IPR014710; 4.5%) and ‘six-bladed beta-propeller’ (IPR011042; 2.7%) had the highest representation. In contrast, of the 249 protein motifs unique to L4 data set, ‘peptidase M24, methionine aminopeptidase’ (IPR0011714; 2.2%) and ‘FAD-binding’ (IPR016166; 1.3%) were the predominant domains (Table 2). The number of ‘biological process’, ‘cellular component’ and ‘molecular function’ terms assigned to peptides unique to each of the individually assembled data sets is given in Table 1. The KOBAS analysis assigned 7, 16, 18 and 23 KEGG terms to inferred peptides exclusive to the adult female, adult male, L3 and L4 data sets, respectively; of the 23 KEGG terms assigned to L4, 20 could be mapped to known pathways in C. elegans (Supplementary Figure S2).
Probabilistic genetic interaction networking predicted 215 C. elegans orthologues, representing sequence clusters unique to the adult female of O. dentatum, to interact directly with a total of 1729 other genes (range: 1–277), including some (e.g. lin-12, mom-5, glp-1, ppk-1, tbx-2 and rnr-1; Supplementary Figure S3, and Supplementary File S3) that are essential to embryogenesis and reproduction (see www.wormbase.org). The 373 C. elegans orthologues of sequence clusters unique to the adult male of O. dentatum were predicted to interact directly with a total of 1710 other genes (range: 1–117; Supplementary File S3). Amongst them were genes involved in sperm development (i.e. ima-3) and motility (i.e. act-2) (Supplementary Figure S3, and Supplementary File S3; www.wormbase.org). A total number of 387 and 323 C. elegans orthologues of L3- and L4-unique molecules, respectively, were predicted to interact with 790 (range: 1–122; Supplementary File S3) and 1058 (range: 1–59; Supplementary File S3) other genes, respectively, including some involved in embryonic and/or larval viability (i.e. scc-1, tba-4, cct-3, pfd-3 and mcm-4) and larval development (i.e. let-711) (Supplementary Figure S3 and Supplementary File S3; www.wormbase.org).
The 2397 predicted peptides unique to the adult female of O. dentatum had significant homology (cut-off: >1E-05) to 261 C. elegans orthologues/homologues (data not shown), of which 151 were associated with EC numbers linked to ‘druggable’ enzymes and/or InterPro domains (Table 4); of these, 92 were associated with non-wild-type RNAi phenotypes, including adult lethality (n = 3), embryonic and/or larval lethality (n = 44) and/or adult sterility (n = 65). Of the 541 C. elegans homologues of the 7117 predicted peptides unique to the adult male of O. dentatum, 375 were associated with EC numbers linked to ‘druggable’ enzymes and/or InterPro domains (Table 4). Of these, 205 were associated with the RNAi phenotypes ‘embryonic and/or larval lethality’ and 196 to ‘sterility’ (Table 4). Of the 565 unique C. elegans homologues of predicted peptides unique to the L3 of O. dentatum, 344 were associated with EC numbers linked to ‘druggable’ enzymes and/or InterPro domains (Table 4); 121 of these were linked to RNAi phenotypes ‘embryonic and/or larval lethality’ and 165 to ‘sterility’ (Table 4). Amongst the 416 C. elegans homologues of predicted peptides unique to the L4 stage of O. dentatum, 283 could be associated with EC numbers linked to ‘druggable’ enzymes and/or InterPro domains (Table 4). Sixty-three of these homologues were associated with RNAi phenotypes ‘embryonic and/or larval lethality’ and 72 to ‘sterility’ (Table 4). Examples of ‘druggable’ molecules unique to each of the data sets, together with examples of effective BRENDA compounds, are given in Table 4 and Supplementary Figure S4; the complete lists, together with the list of ‘druggable’ molecules common between two or among more data sets, are available from the primary author upon request.
Table 4.
Examples of C. elegans orthologues of contigs unique to each Oesophagostomum dentatum adult female, adult male and the third (L3) and fourth (L4) larval stages, following in silico subtraction, ranked according to the ‘severity’ of the RNAi phenotype/s observed, and for which inferred peptides were associated with ‘druggable’ (InterPro) domains and/or Enzyme Commission (EC) numbers as well as examples of candidate compounds linked to these domains, predicted using the BRENDA database. The number of the C. elegans ortologues predicted to interact with each of the molecules listed is also indicated
| Contig code | C. elegans gene ID | Gene name | RNAi phenotypes | Protein description | Druggable IPR domain (description) | Examples of BRENDA compounds | No. of predicted interacting genes |
|---|---|---|---|---|---|---|---|
| Female (151) | |||||||
| Contig722 | T23G5.1 | rnr-1 | Embryonic lethal, embryonic defects, larval lethal, larval arrest, sterile | Ribonucleotide reductase | IPR000788 (ribonucleotide) | D-phosphoserine | 35 |
| Contig18241 | F44F4.2 | egg-3 | Embryonic lethal, maternal sterile, sterile progeny | Protein tyrosine phosphatase | IPR000242 (protein tyrosine) | 4-nitrophenyl phosphate | – |
| Contig15526 | T21E3.1 | egg-4 | Embryonic lethal, maternal sterile | Protein tyrosine phosphatase | IPR000242 (protein tyrosine) | 4-nitrophenyl phosphate | – |
| Contig10671 | Y110A7A.4 | Embryonic lethal, reduced brood size | Thymidylate synthase | IPR000398 (thymidylate) | 5,10-methylenetetrahydrofolate + deoxyuridine phosphate | 26 | |
| E6SSEER01EX2TA | F17C8.1 | acy-1 | Embryonic defects, larval arrest | Adenylyl cyclase | IPR001054 (denylyl) | 3′,5′-cAMP + diphosphate | 2 |
| Male (375) | |||||||
| Contig12350 | W03A5.1 | Embryonic lethal, embryonic defects | Fibroblast/platelet-derived growth factor receptor and related receptor tyrosine kinase | IPR001254 (serine proteases) | Cleaved azocasein | – | |
| Contig10801 | T04B2.2 | frk-1 | Embryonic lethal, embryonic defects | Protein tyrosine kinase | IPR001245 (tyrosine protein kinase) | ADP + a phosphoprotein | – |
| Contig13376 | T04B2.2 | frk-1 | Embryonic lethal, embryonic defects | Protein tyrosine kinase | IPR001245 (tyrosine protein kinase) | ADP + a phosphoprotein | – |
| Contig10782 | ZK354.6 | Embryonic defects | Casein kinase | IPR001245 (tyrosine protein kinase) | ADP + a phosphoprotein | – | |
| Contig13084 | C25A8.5 | Aldicarb resistant | Protein tyrosine kinase | IPR001254 (serine proteases | Cleaved azocasein | – | |
| L3 (344) | |||||||
| Contig10987 | T04D3.4 | gcy-35 | Embryonic lethal, larval arrest | Adenylate/guanylate kinase | IPR001054 (guanylate cyclase) | 3′,5′-cAMP + diphosphate | 1 |
| Contig17117 | B0240.3 | daf-11 | Embryonic lethal, slow growth | Transmembrane guanylate cyclase | IPR001054 (guanylate cyclase) | 3′,5′-cAMP + diphosphate | 27 |
| Contig10518 | R01E6.1b | odr-1 | Slow growth | Guanylate cyclase | IPR001054 (guanylate cyclase) | 3′,5′-cAMP + diphosphate | – |
| Contig10600 | C24G6.2b | Fibroblast/platelet-derived growth factor receptor and related receptor tyrosine kinase | IPR11009 (protein kinase) | Cleaved azocasein | – | ||
| Contig1406 | R134.2 | gcy-2 | Slow growth | Guanylyl cyclase | IPR001054 (guanylate cyclase) | 3′,5′-cAMP + diphosphate | – |
| Contig11765 | Y46H3A.1 | srt-42 | Extended life span | 7-transmembrane receptor | IPR11009 (protein kinase) | ADP + a phosphoprotein | – |
| L4 (283) | |||||||
| Contig23920 | T05G5.3 | Embryonic lethal, embryonic defects, maternal sterile | Protein kinase PCTAIRE and related kinases | IPR000719 (protein kinase) | ADP + a phosphoprotein | 139 | |
| Contig1501 | K12D12.1 | top-2 | Embryonic lethal, embryonic defects, larval arrest | DNA topoisomerase type II | IPR002205 (DNA girase) | Catenated DNA networks + ADP + phosphate | 39 |
| Contig2892 | C46A5.4 | Protruding vulva | IPR002007 (animal haem peroxidase | 2-Amino-9,10a-dihydro-3H-phenoxazin-3-one | – | ||
| Contig20741 | C46A5.4 | Protruding vulva | IPR002007 (animal haem peroxidase) | 2-Amino-9,10a-dihydro-3H-phenoxazin-3-one | – | ||
| Contig25779 | R11A5.7 | Dumpy | Zinc carboxypeptidase | IPR000834 (Zinc carboxypeptidases) | 4-chlorocinnamic acid + L- β-phenyllactate | 5 |
DISCUSSION
Technical considerations
We demonstrated the utility of an integrated bioinformatic workflow system for the analysis and annotation of large sequence data sets produced by NGS. This system is considered useful for researchers with basic expertise in computer programming but without the means for developing bioinformatic pipelines or purchasing expensive soft- or hardware packages. The system constructed here was appraised according to: (i) computational time required to perform the analyses, (ii) ease of use, (iii) compatibility with different computer operating systems, (iv) ability to focus the analyses on answering relevant biological questions and (v) general applicability.
The majority of the software incorporated in the bioinformatic workflow was derived from existing application tools (e.g. CAP3 = maximum length of 50 kb) available as web-based interfaces, and originally designed for the analysis and annotation of a relatively small number of sequences. These applications were adapted here to face the challenges presented by the need to analyse large sequence data sets in a time-efficient manner. Indeed, the original sequence data sets described herein, which included a total of ∼2 million sequences (244 ± 32 bases), could be analysed and annotated using a 2 CPU Linux computer with 8 processor cores, within ∼2000 computing hours corresponding to ∼240 man-hours (one computing hour = 1 hour of computing time on one processor core). Based on our experience, the same analyses, conducted using web-based interfaces, require several months to complete. However, an advantage of web-based software tools with extensive graphical interfaces is that no knowledge of computing and/or programming is required (29). The process of developing, trouble-shooting, maintaining and updating scripts can be involved and challenging, laborious and time-consuming. On the other hand, the use of a command line (which consists of a series of standardized commands) to execute pre-existing scripts, such as the Perl, Python and Unix shell, which have been written and made available here, overcomes this limitation. Furthermore, although these scripts have been written and optimized using the Linux operational system, the output files (generated in the form of text or tab delimited files) can be readily viewed, analysed and modified in a range of different operating systems, such as Microsoft Windows and Mac OS, thus being broadly applicable.
A key goal for scientists focusing on the analyses of large NGS data sets is to distil, from large amounts of raw data, biologically meaningful information about the organism under investigation. For example, some pathogens, such as parasitic worms, have complex life cycles and thus represent a challenging group of organisms for genomic and transcriptomic studies, because different life stages can express various sets of genes which are involved in development, reproduction, host–parasite interactions and/or disease (17,37–39). Understanding these aspects should have important implications for finding new ways of disrupting biological processes and pathways, and thus could facilitate the prediction and prioritization of new drug and/or vaccine targets. In addition, compared with the free-living nematode C. elegans, there is a paucity of knowledge on the fundamental molecular biology of parasitic worms (17,39,53). However, extensive information is available on the functions of C. elegans genes through the use of gene silencing and/or transgenesis (see www.wormbase.org). This knowledge, together with the results of comparative analyses of genetic data sets, revealed that parasitic nematodes usually share ∼50–70% of genes with C. elegans (54,55), indicating the utility of this free-living nematode as a model to explore molecular aspects of development, survival and reproduction in some parasitic nematodes (18,38,51,56,57).
Biological interpretations from the annotated data set
The bioinformatic workflow system constructed here was utilized to explore differential transcription in O. dentatum. Several reports indicate that this nematode provides a unique model system for studying fundamental aspects of the molecular biology of gastrointestinal strongylid nematodes (58). The in silico subtraction approach identified 139 and 243 protein motifs specific to the adult female and male of O. dentatum, respectively. Most of these molecules could be linked, using KOBAS analyses and genetic interaction networking, to pathways associated with reproductive processes. For instance, a large number of female-specific molecules encoded proteins containing a ‘chitin-binding protein, peritrophin A’ domain (i.e. n = 18; Table 2). This domain was also found to be highly represented amongst the molecules enriched in the female of the pig roundworm, Ascaris suum (59). These proteins are hypothesized to have crucial roles in pathways linked to developmental and reproductive processes, based on the knowledge that the corresponding C. elegans homologues (containing one or more peritrophin-A domains) CPG-1/CEJ-1 and CPG-2 are essential for the synthesis of the eggshell as well as for early embryonic development (60). The production and maturation of oocytes has also been shown, in C. elegans, to be regulated by nematode-specific bipartite signalling molecules, the major-sperm proteins (MSPs) (61,62). Numerous sequences unique to the adult male of O. dentatum represented MSPs (n = 15; c.f. Table 2), in accordance with previous studies of male-enriched data sets of other species of strongylid nematodes, including Trichostrongylus vitrinus (63), Haemonchus contortus (38), as well as the filarioid Brugia malayi (64–66), and A. suum (59). Based on the observation that MSPs from various nematodes, including C. elegans, are characterized by a significant amino acid sequence conservation (i.e. ∼64%) (67), a similar role has been proposed for these proteins in processes linked to the maturation of oocytes in the uterus of female nematodes (61,62).
In addition to molecules unique to adult female and male of O. dentatum, the predicted proteins exclusive to the larval stages of this parasite could be linked, using InterPro and/or GO classification and/or probabilistic genetic interaction networking, to biological pathways associated with larval development and/or interactions with the vertebrate host (see Table 2). For example, a large number of molecules unique to the L4 stage (n = 10) were inferred to represent proteases. In parasitic nematodes, proteases have been proposed to facilitate the survival of the parasite by mediating, for instance, tissue penetration, feeding and/or immune evasion (68–70). Indeed, O. dentatum L4s are known to evoke immunological reactions that result in the encapsulation of the larvae in nodules with aggregations of neutrophils and eosinophils (58,71). In addition, somatic extracts of and supernatants from in vitro maintenance cultures of O. dentatum L4s have been shown to induce the proliferation of porcine mononuclear cells in vitro (72). These observations suggest an active role for L4-specific proteases in the modulation of the host’s immune response, which (as proposed for other biological systems) could consist of: (i) the direct digestion of antibodies (68); (ii) cleavage of cell-surface receptors for cytokines (73) and/or (iii) direct lysis of immune cells (74). In parasitic nematodes, other molecules have been proposed to play immuno-modulatory roles during the invasion of the host, the migration through tissues as well as feeding. Amongst them, proteins containing a ‘sperm-coating protein (SCP)-like extracellular domain’ (InterPro: IPR014044), also called SCP/Tpx-1/Ag5/PR-1/Sc7 (SCP/TAPS; Pfam accession number no. PF00188), were highly represented in the transcriptome of O. dentatum (see Table 2). Members of the SCP/TAPS protein family have been identified in various eukaryotes, including plants, arthropods, snakes, mammals as well as free-living and parasitic helminths (75). These molecules have been studied mainly in the hookworms Ancylostoma caninum and Necator americanus, and are commonly referred as to Ancylostoma secreted proteins (i.e. ASPs; 75). Due to their abundance in the excretory/secretory (ES) products from serum-activated L3s (=aL3s) of A. caninum and to the high levels of mRNAs encoding ASPs in aL3s compared with non-activated, ensheathed L3s (L3s), these molecules have been hypothesized to play a major role in the transition from the free-living to the parasitic stage of this species (39,76). Other ASP homologues have been characterized for the adult stage of hookworms, and suggested to play a role in the initiation, establishment and/or maintenance of the host-parasite relationship (39,77,78). Although a male-biased transcription of ASP homologues had been reported for O. dentatum (51), results from the present study show that the transcription of SCP/TAPS molecules occurs in all developmental stages studied herein. As the sequences analysed were generated from normalized cDNA libraries, the differences in levels of transcription of genes encoding SCP/TAPS throughout the life cycle of O. dentatum could not be inferred. Future work could involve, for instance, the application of the present bioinformatic workflow tool to the analysis of data generated (e.g. by Illumina sequencing) from non-normalized cDNA libraries of O. dentatum, which would allow quantitative rather than qualitative differences in transcription to be determined for genes encoding SCP/TAPS, to assist in the study of the biological function(s) of these molecules (75). The O. dentatum-pig model could also provide a useful means of exploring the biological role/s of these molecules in the development and reproduction of this nematode as well as its interactions with the host. Several features of O. dentatum, including its short life-cycle, its ability to survive and grow in culture in vitro for weeks through several moults, and the possibility of rectally transplanting worms (e.g. from in vitro culture) into the host without the need for surgical intervention (58,79), offer an opportunity to experimentally test hypotheses formulated based on the interpretation of results from bioinformatic analyses. Bioinformatically guided interpretations of NGS data sets are also increasingly playing an important role in the identification of putative drug targets (80), due to the possibility of using predictive algorithms to prioritize and select sets of molecules for experimental studies both in vitro and in vivo (81–83), potentially leading to a significant reduction in the cost associated with drug discovery and development (84). For instance, in the present study, subsets of molecules without known host (pig) homologues were identified and predicted to represent targets for intervention. Amongst them, protein kinases and phosphatases were the most abundantly represented (Table 4). Previously, in O. dentatum, a catalytic subunit of a serine/threonine protein phosphatase (PP1) was characterized (Od-mpp1); gene silencing by RNAi of the corresponding C. elegans homologue resulted in a significant reduction (30–40%) in the numbers of F2-progeny produced (56). Based on these findings, it is tempting to speculate that some pathways, involving phosphatases/kinases, represent key targets for nematocidal drugs.
Concluding remarks
Here, we demonstrated, using a large test data set derived from different stages/sexes of a parasitic worm (O. dentatum), that our bioinformatic workflow system provides a practical tool for the assembly, annotation and analysis of NGS data. The custom-written Perl, Python and Unix shell computer scripts, accessible via the web, can be readily adapted to suit the requirements of researchers conducting transcriptomic studies in their particular discipline. This workflow system is now routinely used by our research group for the analysis of data sets from a range of pathogens of major socio-economic importance and has been applied more broadly to data sets representing other organisms, including mammals. Thus, this integrated system should be a user-friendly and efficient tool for biologists involved in transcriptomic studies in any field on any organism.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Australian Research Council; Australian Academy of Science; the Australian-American Fulbright Commission (to R.B.G.); National Human Genome Research Institute and National Institutes of Health (to M.M.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Staff at WormBase are gratefully acknowledged. The Austrian Ministry for Science and Research approved the animal experimentation (BMWF-68.205/0103-II/10b/2008) and is also acknowledged. C.C. is in receipt of an International Postgraduate Research Scholarship from the Australian Government and a fee-remission scholarship from The University of Melbourne as well as the Clunies Ross (2008) and Sue Newton (2009) awards from the School of Veterinary Science of the same university.
REFERENCES
- 1.McKay SJ, Johnsen R, Khattra J, Asano J, Baillie DL, Chan S, Dube N, Fang L, Goszczynski B, Ha E, et al. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb. Symp. Quant. Biol. 2003;68:159–169. doi: 10.1101/sqb.2003.68.159. [DOI] [PubMed] [Google Scholar]
- 2.Portman DS. Profiling C. elegans gene expression with DNA microarrays. WormBook. 2006;20:1–11. doi: 10.1895/wormbook.1.104.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golden TR, Melov S. Gene expression changes associated with aging in C. elegans. WormBook. 2007;12:1–12. doi: 10.1895/wormbook.1.127.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stathopoulos A, Levine M. Whole-genome expression profiles identify gene batteries in Drosophila. Dev. Cell. 2002;3:464–465. doi: 10.1016/s1534-5807(02)00300-3. [DOI] [PubMed] [Google Scholar]
- 5.Gupta V, Oliver B. Drosophila microarray platforms. Brief. Funct. Genomic Proteomic. 2003;2:97–105. doi: 10.1093/bfgp/2.2.97. [DOI] [PubMed] [Google Scholar]
- 6.Vibranovski MD, Lopes HF, Karr TL, Long M. Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet. 2009;5:e1000731. doi: 10.1371/journal.pgen.1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizuarai S, Irie H, Schmatz DM, Kotani H. Integrated genomic and pharmacological approaches to identify synthetic lethal genes as cancer therapeutic targets. Curr. Mol. Med. 2008;8:774–783. doi: 10.2174/156652408786733676. [DOI] [PubMed] [Google Scholar]
- 8.Ren S, Liu S, Howell P, Jr, Xi Y, Enkemann SA, Ju J, Riker AI. The impact of genomics in understanding human melanoma progression and metastasis. Cancer Control. 2008;15:202–215. doi: 10.1177/107327480801500303. [DOI] [PubMed] [Google Scholar]
- 9.Santos ES, Blaya M, Raez LE. Gene expression profiling and non-small-cell lung cancer: where are we now? Clin. Lung Cancer. 2009;10:168–173. doi: 10.3816/CLC.2009.n.023. [DOI] [PubMed] [Google Scholar]
- 10.Greene JG. Gene expression profiles of brain dopamine neurons and relevance to neuropsychiatric disease. J. Physiol. 2006;575:411–416. doi: 10.1113/jphysiol.2006.112599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mufson EJ, Counts SE, Che S, Ginsberg SD. Neuronal gene expression profiling: uncovering the molecular biology of neurodegenerative disease. Prog. Brain Res. 2006;158:197–222. doi: 10.1016/S0079-6123(06)58010-0. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka F, Niwa J, Ishigaki S, Katsuno M, Waza M, Yamamoto M, Doyu M, Sobue G. Gene expression profiling toward understanding of ALS pathogenesis. Ann. NY Acad. Sci. 2006;1086:1–10. doi: 10.1196/annals.1377.011. [DOI] [PubMed] [Google Scholar]
- 13.Chan VL. Bacterial genomes and infectious diseases. Pediatr. Res. 2003;54:1–7. doi: 10.1203/01.PDR.0000066622.02736.A8. [DOI] [PubMed] [Google Scholar]
- 14.Jackson RW, Giddens SR. Development and application of in vivo expression technology (IVET) for analysing microbial gene expression in complex environments. Infect. Disord. Drug Targets. 2006;6:207–240. doi: 10.2174/187152606778249944. [DOI] [PubMed] [Google Scholar]
- 15.Li BW, Rush AC, Mitreva M, Yin Y, Spiro D, Ghedin E, Weil GJ. Transcriptomes and pathways associated with infectivity, survival and immunogenicity in Brugia malayi L3. BMC Genomics. 2009;10:267. doi: 10.1186/1471-2164-10-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ranganathan S, Menon R, Gasser RB. Advanced in silico analysis of expressed sequence tag (EST) data for parasitic nematodes of major socio-economic importance–fundamental insights toward biotechnological outcomes. Biotechnol. Adv. 2009;27:439–448. doi: 10.1016/j.biotechadv.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Cantacessi C, Campbell BE, Young ND, Jex AR, Hall RS, Presidente PJA, Zawadzki JL, Zhong W, Aleman-Meza B, Loukas A, et al. Differences in transcription between free-living and CO2-activated third-stage larvae of Haemonchus contortus. BMC Genomics. 2010;11:266. doi: 10.1186/1471-2164-11-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cantacessi C, Mitreva M, Jex AR, Young ND, Campbell BE, Hall RS, Doyle MA, Ralph SA, Rabelo EM, Ranganathan S, et al. Massively parallel sequencing and analysis of the Necator americanus transcriptome. PLoS Negl. Trop. Dis. 2010;4:e684. doi: 10.1371/journal.pntd.0000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young ND, Hall RS, Jex AR, Cantacessi C, Gasser RB. Elucidating the transcriptome of Fasciola hepatica - a key to fundamental and biotechnological discoveries for a neglected parasite. Biotechnol. Adv. 2010;28:222–231. doi: 10.1016/j.biotechadv.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Young ND, Campbell BE, Hall RS, Jex AR, Cantacessi C, Laha T, Sohn WM, Sripa B, Loukas A, Brindley PJ, et al. Unlocking the transcriptomes of the carcinogens Clonorchis sinensis and Opisthorchis viverrini. PLoS Negl. Trop. Dis. 2010;4:e719. doi: 10.1371/journal.pntd.0000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977;265:687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- 23.Wang AM, Doyle MV, Mark DF. Quantitation of mRNA by the polymerase chain reaction. Proc. Natl Acad. Sci. USA. 1989;86:9717–9721. doi: 10.1073/pnas.86.24.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat. Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 25.Clifton SW, Mitreva M. Strategies for undertaking expressed sequence tag (EST) projects. Methods Mol. Biol. 2009;533:13–32. doi: 10.1007/978-1-60327-136-3_2. [DOI] [PubMed] [Google Scholar]
- 26.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 27.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, Copley R, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flicek P, Birney E. Sense from sequence reads: methods for alignment and assembly. Nat. Methods. 2009;6:S6–S12. doi: 10.1038/nmeth.1376. [DOI] [PubMed] [Google Scholar]
- 29.Nagaraj SH, Deshpande N, Gasser RB, Ranganathan S. ESTExplorer: an expressed sequence tag (EST) assembly and annotation platform. Nucleic Acids Res. 2007;35:W143–W147. doi: 10.1093/nar/gkm378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagaraj SH, Gasser RB, Nisbet AJ, Ranganathan S. In silico analysis of expressed sequence tags from Trichostrongylus vitrinus (Nematoda): comparison of the automated ESTExplorer workflow platform with conventional database searches. BMC Bioinf. 2008;9:S10. doi: 10.1186/1471-2105-9-S1-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iseli C, Jongeneel CV, Bucher P. ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1999;1:138–148. [PubMed] [Google Scholar]
- 33.Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 34.Metzker ML. Sequencing technologies - the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 35.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moser JM, Freitas T, Arasu P, Gibson G. Gene expression profiles associated with the transition to parasitism in Ancylostoma caninum larvae. Mol. Biochem. Parasitol. 2005;143:39–48. doi: 10.1016/j.molbiopara.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 38.Campbell BE, Nagaraj SH, Hu M, Zhong W, Sternberg PW, Ong EK, Loukas A, Ranganathan S, Beveridge I, McInnes RL, et al. Gender-enriched transcripts in Haemonchus contortus–predicted functions and genetic interactions based on comparative analyses with Caenorhabditis elegans. Int. J. Parasitol. 2008;38:65–83. doi: 10.1016/j.ijpara.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Datu BJ, Gasser RB, Nagaraj SH, Ong EK, O'Donoghue P, McInnes R, Ranganathan S, Loukas A. Transcriptional changes in the hookworm, Ancylostoma caninum, during the transition from a free-living to a parasitic larva. PLoS Negl. Trop. Dis. 2008;2:e130. doi: 10.1371/journal.pntd.0000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joachim A, Ruttkowski B. Cytosolic glutathione S-transferases of Oesophagostomum dentatum. Parasitology. 2008;135:1215–1223. doi: 10.1017/S0031182008004769. [DOI] [PubMed] [Google Scholar]
- 41.Soderlund C, Johnson E, Bomhoff M, Descour A. PAVE: program for assembling and viewing ESTs. BMC Genomics. 2009;10:400. doi: 10.1186/1471-2164-10-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006;34:W720–W724. doi: 10.1093/nar/gkl167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Letunic I, Yamada T, Kanehisa M, Bork P. iPath: interactive exploration of biochemical pathways and networks. Trends Biochem. Sci. 2008;33:101–103. doi: 10.1016/j.tibs.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 46.Zhong W, Sternberg PW. Genome-wide prediction of C. elegans genetic interactions. Science. 2006;311:1481–1484. doi: 10.1126/science.1123287. [DOI] [PubMed] [Google Scholar]
- 47.Lipinski C, Lombardo F, Dominy B, Feeney P. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 48.Hopkins AL, Groom CR. The druggable genome. Nat. Rev. Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 49.Robertson JG. Mechanistic basis of enzyme-targeted drugs. Biochemistry. 2005;44:5561–5571. doi: 10.1021/bi050247e. [DOI] [PubMed] [Google Scholar]
- 50.Chang A, Scheer M, Grote A, Schomburg I, Schomburg D. BRENDA, AMENDA and FRENDA the enzyme information system: new content and tools in 2009. Nucleic Acids Res. 2009;37:D588–D592. doi: 10.1093/nar/gkn820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cottee PA, Nisbet AJ, Abs El-Osta YG, Webster TL, Gasser RB. Construction of gender-enriched cDNA archives for adult Oesophagostomum dentatum by suppressive-subtractive hybridization and a microarray analysis of expressed sequence tags. Parasitology. 2006;132:691–708. doi: 10.1017/S0031182005009728. [DOI] [PubMed] [Google Scholar]
- 52.Wilson EB. Probable inference, the law of succession, and statistical inference. J. Am. Stat. Assoc. 1927;22:209–212. [Google Scholar]
- 53.Nikolaou S, Gasser RB. Prospects for exploring molecular developmental processes in Haemonchus contortus. Int. J. Parasitol. 2006;36:859–868. doi: 10.1016/j.ijpara.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 54.Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, Vierstraete A, Vanfleteren JR, Mackey LY, Dorris M, Frisse LM, et al. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–75. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- 55.Parkinson J, Mitreva M, Whitton C, Thomson M, Daub J, Martin J, Schmid R, Hall N, Barrell B, Waterston RH, et al. A transcriptomic analysis of the phylum Nematoda. Nat. Genet. 2004;36:1259–1267. doi: 10.1038/ng1472. [DOI] [PubMed] [Google Scholar]
- 56.Boag PR, Ren P, Newton SE, Gasser RB. Molecular characterisation of a male-specific serine/threonine phosphatase from Oesophagostomum dentatum (Nematoda: Strongylida), and functional analysis of homologues in Caenorhabditis elegans. Int. J. Parasitol. 2003;33:313–325. doi: 10.1016/s0020-7519(02)00263-1. [DOI] [PubMed] [Google Scholar]
- 57.Hu M, Zhong W, Campbell BE, Sternberg PW, Pellegrino MW, Gasser RB. Elucidating ANTs in worms using genomic and bioinformatic tools–biotechnological prospects? Biotechnol. Adv. 2010;28:49–60. doi: 10.1016/j.biotechadv.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 58.Gasser RB, Cottee P, Nisbet AJ, Ruttkowski B, Ranganathan S, Joachim A. Oesophagostomum dentatum: potential as a model for genomic studies of strongylid nematodes, with biotechnological prospects. Biotechnol. Adv. 2007;25:281–293. doi: 10.1016/j.biotechadv.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 59.Cantacessi C, Zou FC, Hall RS, Zhong W, Jex AR, Campbell BE, Ranganathan S, Sternberg PW, Zhu XQ, Gasser RB. Bioinformatic analysis of abundant, gender-enriched transcripts of adult Ascaris suum (Nematoda) using a semi-automated workflow platform. Mol. Cell. Probes. 2009;23:205–217. doi: 10.1016/j.mcp.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 60.Olson SK, Bishop JR, Yates JR, Oegema K, Esko JD. Identification of novel chondroitin proteoglycans in Caenorhabditis elegans: embryonic cell division depends on CPG-1 and CPG-2. J. Cell. Biol. 2006;173:985–994. doi: 10.1083/jcb.200603003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller MA, Nguyen VQ, Lee MH, Kosinski M, Schedl T, Caprioli RM, Greenstein D. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science. 2001;291:2144–2147. doi: 10.1126/science.1057586. [DOI] [PubMed] [Google Scholar]
- 62.Miller MA, Ruest PJ, Kosinski M, Hanks SK, Greenstein D. An Eph receptor sperm-sensing control mechanism for oocyte meiotic maturation in Caenorhabditis elegans. Genes Dev. 2003;17:187–200. doi: 10.1101/gad.1028303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nisbet AJ, Gasser RB. Profiling of gender-specific gene expression for Trichostrongylus vitrinus (Nematoda: Strongylida) by microarray analysis of expressed sequence tag libraries constructed by suppressive-subtractive hybridisation. Int. J. Parasitol. 2004;34:633–643. doi: 10.1016/j.ijpara.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 64.Li BW, Rush AC, Tan J, Weil GJ. Quantitative analysis of gender-regulated transcripts in the filarial nematode Brugia malayi by real-time RT-PCR. Mol. Biochem. Parasitol. 2004;137:329–337. doi: 10.1016/j.molbiopara.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 65.Li BW, Rush AC, Crosby SD, Warren WC, Williams SA, Mitreva M, Weil GJ. Profiling of gender-regulated gene transcripts in the filarial nematode Brugia malayi by cDNA oligonucleotide array analysis. Mol. Biochem. Parasitol. 2005;143:49–57. doi: 10.1016/j.molbiopara.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 66.Moreno Y, Geary TG. Stage- and gender-specific proteomic analysis of Brugia malayi excretory-secretory products. PLoS Negl. Trop. Dis. 2008;2:e326. doi: 10.1371/journal.pntd.0000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cottee PA, Nisbet AJ, Boag PR, Larsen M, Gasser RB. Characterization of major sperm protein genes and their expression in Oesophagostomum dentatum (Nematoda: Strongylida) Parasitology. 2004;129:479–490. doi: 10.1017/s003118200400561x. [DOI] [PubMed] [Google Scholar]
- 68.Hotez PJ, Prichard DI. Hookworm infection. Sci. Am. 1995;6:42–48. doi: 10.1038/scientificamerican0695-68. [DOI] [PubMed] [Google Scholar]
- 69.Williamson AL, Brindley PJ, Knox DP, Hotez PJ, Loukas A. Digestive proteases of blood-feeding nematodes. Trends Parasitol. 2003;19:417–423. doi: 10.1016/s1471-4922(03)00189-2. [DOI] [PubMed] [Google Scholar]
- 70.Bethony JM, Loukas A, Hotez PJ, Knox DP. Vaccines against blood-feeding nematodes of humans and livestock. Parasitology. 2006;133:S63–S79. doi: 10.1017/S0031182006001818. [DOI] [PubMed] [Google Scholar]
- 71.Stockdale PH. Necrotic enteritis of pigs caused by infection with Oesophagostomum spp. Br. Vet. J. 1970;126:526–530. doi: 10.1016/s0007-1935(17)48138-3. [DOI] [PubMed] [Google Scholar]
- 72.Freigofas R, Leibold W, Daugschies A, Joachim A, Schuberth HJ. Products of fourth-stage larvae of Oesophagostomum dentatum induce proliferation in naïve porcine mononuclear cells. J. Vet. Med. B Infect. Dis. Vet. Public Health. 2001;48:603–611. doi: 10.1046/j.1439-0450.2001.00483.x. [DOI] [PubMed] [Google Scholar]
- 73.Björnberg F, Lantz M, Gullberg U. Metalloproteases and serineproteases are involved in the cleavage of the two tumour necrosis factor (TNF) receptors to soluble forms in the myeloid cell lines U-937 and THP-1. Scand. J. Immunol. 1995;42:418–424. doi: 10.1111/j.1365-3083.1995.tb03675.x. [DOI] [PubMed] [Google Scholar]
- 74.Robinson BW, Venaille TJ, Mendis AH, McAleer R. Allergens as proteases: an Aspergillus fumigatus proteinase directly induces human epithelial cell detachment. J. Allergy Clin. Immunol. 1990;86:726–731. doi: 10.1016/s0091-6749(05)80176-9. [DOI] [PubMed] [Google Scholar]
- 75.Cantacessi C, Campbell BE, Visser A, Geldhof P, Nolan MJ, Nisbet AJ, Matthews JB, Loukas A, Hofmann A, Otranto D, et al. A portrait of the “SCP/TAPS” proteins of eukaryotes – developing a framework for fundamental research and biotechnological outcomes. Biotech. Adv. 2009;27:376–388. doi: 10.1016/j.biotechadv.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 76.Hawdon JM, Jones BF, Hoffman DR, Hotez PJ. Cloning and characterization of Ancylostoma-secreted protein. A novel protein associated with the transition to parasitism by infective hookworm larvae. J. Biol. Chem. 1996;271:6672–6678. doi: 10.1074/jbc.271.12.6672. [DOI] [PubMed] [Google Scholar]
- 77.Zhan B, Liu Y, Badamchian M, Williamson A, Feng J, Loukas A, Hawdon JM, Hotez PJ. Molecular characterisation of the Ancylostoma-secreted protein family from the adult stage of Ancylostoma caninum. Int. J. Parasitol. 2003;33:897–907. doi: 10.1016/s0020-7519(03)00111-5. [DOI] [PubMed] [Google Scholar]
- 78.Mulvenna J, Hamilton B, Nagaraj S, Smyth D, Loukas A, Gorman J. Proteomic analysis of the excretory/secretory component of the blood-feeding stage of the hookworm, Ancylostoma caninum. Mol. Cell Proteomics. 2009;8:109–121. doi: 10.1074/mcp.M800206-MCP200. [DOI] [PubMed] [Google Scholar]
- 79.Joachim A, Ruttkowski B, Daugschies A. Comparative studies on the development of Oesophagostomum dentatum in vitro and in vivo. Parasitol. Res. 2001;87:37–42. doi: 10.1007/s004360000305. [DOI] [PubMed] [Google Scholar]
- 80.Krasky A, Rohwer A, Schroeder J, Selzer PM. A combined bioinformatics and chemoinformatics approach for the development of new antiparasitic drugs. Genomics. 2007;89:36–43. doi: 10.1016/j.ygeno.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 81.Caffrey CR, Rohwer A, Oellien F, Marhöfer RJ, Braschi S, Oliveira G, McKerrow JH, Selzer PM. A comparative chemogenomics strategy to predict potential drug targets in the metazoan pathogen, Schistosoma mansoni. PLoS One. 2009;4:e4413. doi: 10.1371/journal.pone.0004413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Keil M, Marhofer RJ, Rohwer A, Selzer PM, Brickmann J, Korb O, Exner TE. Molecular visualization in the rational drug design process. Front. Biosci. 2009;14:2559–2583. doi: 10.2741/3398. [DOI] [PubMed] [Google Scholar]
- 83.Doyle MA, Gasser RB, Woodcroft BJ, Hall RS, Ralph SA. Drug target prediction and prioritization: using orthology to predict essentiality in parasite genomes. BMC Genomics. 2010;11:222. doi: 10.1186/1471-2164-11-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pong SW, Shiang R. Biopharmaceutical Drug Design and Development. 2010. The use of bioinformatics and chemogenomics in drug discovery. 2nd edn., Humana Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.