Abstract
Vasopressin and aldosterone play key roles in the fine adjustment of sodium and water re-absorption in the nephron. The molecular target of this regulation is the epithelial sodium channel (ENaC) consisting of α-, β- and γ-subunits. We investigated mRNA-specific post-transcriptional mechanisms in hormone-dependent expression of ENaC subunits in mouse kidney cortical collecting duct cells. Transcription experiments and polysome gradient analysis demonstrate that both hormones act on transcription and translation. RNA-binding proteins (RBPs) and mRNA sequence motifs involved in translational control of γ-ENaC synthesis were studied. γ-ENaC–mRNA 3′-UTR contains an AU-rich element (ARE), which was shown by RNA affinity chromatography to interact with AU-rich element binding proteins (ARE-BP) like HuR, AUF1 and TTP. Some RBPs co-localized with γ-ENaC mRNA in polysomes in a hormone-dependent manner. Reporter gene co-expression experiments with luciferase γ-ENaC 3′-UTR constructs and ARE-BP expression plasmids demonstrate the importance of RNA–protein interaction for the up-regulation of γ-ENaC synthesis. We document that aldosterone and the V2 receptor agonist dDAVP act on synthesis of α- and γ-ENaC subunits mediated by RBPs as effectors of translation but not by mRNA stabilization. Immunoprecipitation and UV-crosslinking analysis of γ-ENaC–mRNA/HuR complexes document the significance of γ-ENaC–mRNA–3′-UTR/HuR interaction for hormonal control of ENaC synthesis.
INTRODUCTION
The epithelial sodium channel (ENaC) consists of the three subunits α, β and γ and is found in the apical membrane of salt-absorbing epithelia of tissues like kidney, colon or lung, where it constitutes the rate-limiting step in Na+ and water absorption. The number of functional ENaC channels at the cell surface depends on distinct molecular processes like transcription of the ENaC genes, synthesis of protein subunits, storage in vesicles, vesicle trafficking and channel assembly in the membrane and finally on removal of channels from the surface by endocytosis and recycling or degradation by the ubiquitine/proteasome system (for reviews see 1–3). Synthesis of ENaC subunits is tightly controlled by hormones responsible for water and salt homoeostasis like aldosterone, vasopressin or glucocorticoids. The synthesis of α-, β- and γ-subunits is not controlled in a coordinated manner and differs markedly in a tissue- and development-specific way. In most cases, there is a selective change in the levels of one or two subunits, but rarely in all three (4). For instance, in the rat kidney, aldosterone administration leads to an increase in α-ENaC and not in β- and γ-ENaC expression (5). In rat colon, aldosterone infusion leads to an increase in β- and γ-ENaC, but not in α-ENaC expression (6). An increase in circulating vasopressin levels results in a selective increase in the abundance of only β- and γ-subunits in the renal collecting duct (7,8) that was also observed in cultured mouse cortical collecting duct (CCD) cells (9). In lung development, the expression of ENaC subunits changes differently under the influence of O2 and glucocorticoids during the transition from foetal to the post-natal stage (10,11).
Basically, hormonal regulation of ENaC expression is exerted by transcriptional control (9,12–16). In addition, several studies suggest that mRNA related post-transcriptional processes affecting mRNA stability or translational efficiency may play an important role in the synthesis of ENaC subunits (5,17). All studies agree that both the intracellular and the cell surface pools of ENaC subunits turn over rapidly with a half-life in the order of 1–3 hr (18). Thus, controlled ongoing peptide synthesis is a requirement to keep ENaC in a functional state.
Work on translational control of ENaC expression concentrated so far mainly on the role of the 5′-untranslated region (5′-UTR) of the α-subunit (10,17,19,20). In contrast to β- and γ-mRNA the α-mRNA contains an unusually long GC-rich 5′-UTR of ∼550/750 nt (rat/human). This type of 5′-UTR is frequently a target for translational control (21). In rat/mouse and humans different transcripts exist with varying lengths of the UTR. The human 5′-UTR, moreover, is prone to alternative splicing, which leads to α-chains with varying N-termini. In vitro translation experiments and polysome gradient analysis revealed that β- and γ-ENaC mRNAs are much more efficiently translated than α-mRNA (17). That correlates with an opposite feature of the α-mRNA that is more abundant than β- or γ-mRNA. Physiological significance of 5′-UTR mediated α−ENaC translation control was demonstrated by the action of oxygen and glucocorticoids in lung development (19). Mechanistically, it was shown that this type of control affects the 5′-cap binding complex and involves phosphorylation of translation initiation factor eIF4F by mTOR kinase (20).
mRNA-specific translational control is not only mediated by 5′- but also by 3′-UTRs (22–25). In both 5′- and 3′-UTRs cis-elements are found, which interact with RNA-binding proteins (RBPs) as trans-acting factors (26,27). In addition to translational efficiency metabolic stability of mRNA is another relevant feature in post-transcriptional regulation. mRNA stabilization/destabilization is frequently associated with determinants residing in the 3′-UTR. The motifs most intensively studied are AU-rich elements (AREs), which function by interaction with ARE-binding proteins (ARE-BPs). ARE-BPs can mediate mRNA destabilization like AU-element binding factor 1 (AUF1, synonym hnRNP D) or tristetraprolin (TTP) and mRNA stabilization by factors like HuR (synonym ELAV1) (27–29). Some RBPs like HuR, hnRNP-A1 or nucleolin have also been shown to be involved in control of translational efficiency (29–31). Examining 3′-UTRs of ENaC mRNAs it is striking that rat, mouse or human γ-ENaC mRNAs contain conserved AU-rich sections that resemble type III AREs (30).
Here, we investigated post-transcriptional mechanisms that may operate in regulation of ENaC expression. We show that up-regulation of α- and γ-ENaC by aldosterone and vasopressin is not only caused by transcription but also by activation of translation, and not by mRNA stabilization. Mechanistically, the interaction of RBPs like HuR, TTP or FMRP with an ARE in the 3′-UTR is responsible for the activation of γ-ENaC mRNA translation.
MATERIAL AND METHODS
Cell culture
Mouse cortical collecting duct cells (mCCDcl1) were obtained from the laboratory of B. C. Rossier (Department of Pharmacology and Toxicology, Faculty of Biology and Medicine, University of Lausanne, Lausanne, Switzerland). This highly differentiated cell line originates from spontaneous immortalized primary cells of micro-dissected CCDs from mouse kidneys and was described previously (32). The cells were grown on 60 cm2 tissue culture dishes at 37°C and 5% CO2 in a 1:1 mixture of Dulbecco’s modified Eagle’s medium and Ham’s F12 medium (DMEM/Ham’s F-12, Biochrom AG, Berlin, Germany) supplemented with 2% foetal calf serum (FCS, Biochrom AG, Berlin, Germany), 50 U/ml penicillin, 50 µg/ml streptomycin (Biochrom AG, Berlin, Germany), 1 mM HEPES-buffer (Biochrom AG, Berlin, Germany), 2 mg/ml glucose monohydrate (Sigma), 1 × Insulin-Transferrin-Selenium-G supplement (Invitrogen-Gibco), 10 ng/ml epidermal growth factor (Sigma), 50 nM dexamethasone (Sigma) and 1 nM 3,3′,5-triiodo-l-thyronin (Sigma). Subsequently this medium is referred to as ‘growth medium’).
For stimulation experiments cells were seeded on 60 cm2 tissue culture dishes (TPP AG, Switzerland) and incubated with growth medium until cells were grown confluent and began with the formation of domes. Then cells were grown for 24 h in DMEM/Ham’s F12 (Biochrom KG, Berlin, Germany) supplemented with 2 mg/ml Glucose monohydrate (Sigma) and 1 mM HEPES buffer (Biochrom AG, Berlin, Germany). This medium is subsequently referred to as ‘stimulation medium’. For stimulation experiments, cells were incubated for further 24 h with stimulation medium supplemented with either 300 nM aldosterone (32) (Sigma), 10 nM dDAVP (33) (Sigma) or 0.1% Ethanol (Carl Roth) as control.
RNA/protein isolation
For RNA and protein isolation, cells were washed with ice-cold Dulbecco’s phosphate-buffered saline (DPBS). Total RNA was isolated using RNA-Bee reagent (Biozol Diagnostica Vertrieb GmbH) according to the manufacturer’s protocol. RNA preparation was followed by DNase digestion with DNaseI (Fermentas) according to the manufacturer’s protocol, followed by a second RNA isolation using RNA-Bee reagent (Biozol Diagnostica Vertrieb GmbH) according to the manufacturer’s protocol to eliminate all traces of DNase.
For preparation of protein extracts (10 000g supernatants, S10), cell pellets were mixed with 2 × volumes of lysis buffer (10 mM Tris, pH 7.5, 140 mM NaCl, 1 mM EDTA, 25% glycerol, 0.1% SDS, 0.5% Nonidet P-40, 1mM dithiothreitol (DTT), 1 × Complete protease inhibitor mixture; Roche Diagnostics), homogenized with a Polytron-PT300 blender (Kinematica AG, Switzerland) and incubated on ice for 20 min for lysis. The homogenate was centrifuged (10 min, 4°C, 10 000g) and the supernatant is designated S10. Protein extracts were aliquoted, frozen in liquid nitrogen, and stored at −80°C.
RT–PCR
First-strand cDNA synthesis was performed with SuperScript® II reverse transcriptase (Invitrogen) and 3 µg of total RNA using oligo(dT) primers. mRNA levels were quantified by RT–PCR and normalized to 18 S/28 S rRNA signal in stimulation experiments with aldosterone or dDAVP. Therefore, 800 ng of total RNA from the RNA used for the RT reaction were run on an ethidiumbromide agarose gel. This kind of normalization was verified by analysing the same samples by northern blotting. The normalization to stained 18 S/28 S rRNA was chosen because of the induction of the normally used housekeeping genes β-actin and GAPDH under aldosterone and dDAVP stimulation. RNA was used for RT reaction without reverse transcriptase (−RT control) and was subjected to PCR to verify that there was no contamination with genomic DNA. There was no contamination in any case that caused a detectable PCR product, therefore no-RT control is only shown in Figure 10. PCR conditions were used as follows: 5 min at 95°C, cycles were 1 min at 95°C, 30 s of annealing, 1 min at 72°C, final elongation was for 10 min at 72°C. The primers were as follows: α-ENaC-forward 5′-GAGAGGAGAGTGCTCCTCTC, α-ENaC-reverse 5′-GATGGAACAAGCATTTATTGAG; β-ENaC-forward 5′-TAGATCCCCACCCCCACC, β-ENaC-reverse 5′-CCAGTGTTTTCTCTCTTTATTTTC; γ-ENaC-forward 5′-CTCGTCTTCTCTTTCZACAC, γ-ENaC-reverse 5′-GCAGAATAGCTCATGTTG; β-actin-forward 5′-CCGCCCTAGGCACCAGGGTG, β-actin-reverse 5′-GGCTGGGGTGTTGAAGGTCTCAAA. PCR products were separated on 1.5% agarose gels containing 0.5 µg/ml ethidiumbromide.
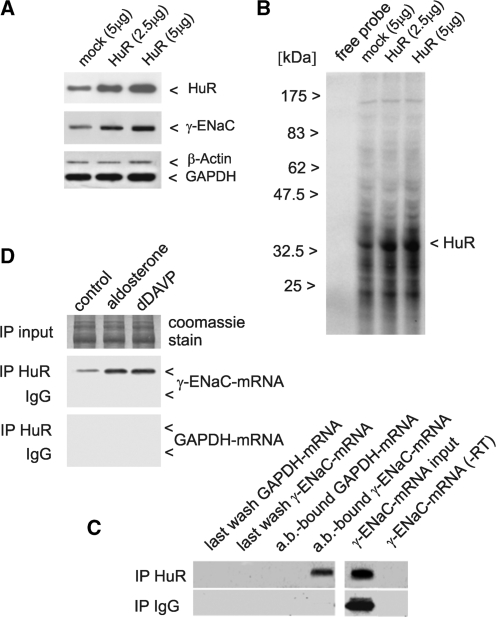
Figure 10.
HuR binds to γ-ENaC 3′-UTR in vitro and to γ-ENaC mRNA in vivo and mediates stimulation of γ-ENaC synthesis in a hormone-dependent way. mCCD cells were transfected for 24 h with 5 µg of empty vector (mock), 2.5 µg or 5 µg of HuR expression vector. Cells were harvested and HuR over-expression and influence on endogenous γ-ENaC protein was determined by western blotting in cytosolic extracts (20 µg) using specific antibodies. Detection of β-actin and GAPDH served as loading controls (A). The same cytosolic extracts were subjected to UV-crosslinking assay using 32P-UTP labelled in vitro transcripts of γ-ENaC mRNA 3′-UTR and showed an increased binding capacity for a 36 kDa protein (HuR) (B). HuR-bound mRNA was co-precipitated from cytosolic extracts of mCCD cells with a HuR-specific antibody, or alternatively the same amount of IgG as a negative control. RNA was isolated from last wash fraction or from antibody-bound (ab-bound) protein-G-sepharose. Isolated RNA was analysed by RT–PCR using primers for detection of γ-ENaC mRNA or GAPDH mRNA as negative control. Input levels of γ-ENaC mRNA were assessed by RT–PCR, furthermore a PCR without RT product (−RT) served as negative control (C). The same immunoprecipitation procedure was repeated with cells pre-treated with aldosterone or dDAVP (Figure 1) to characterize HuR-association of γ-ENaC mRNA under conditions of hormonal stimulation. RT–PCR with γ-ENaC mRNA specific primers demonstrated an increase in the fraction of HuR-bound γ-ENaC mRNA in the presence of both hormones. Equal Input levels of cytosolic extracts were verified by SDS–PAGE and Coomassie staining. Negative controls with IgG for the immunoprecipitation (ip) and detection of GAPDH mRNA in the ab-bound protein-G-sepharose show the specificity of the ip reaction (D). Shown are representative figures of three independent experiments each.
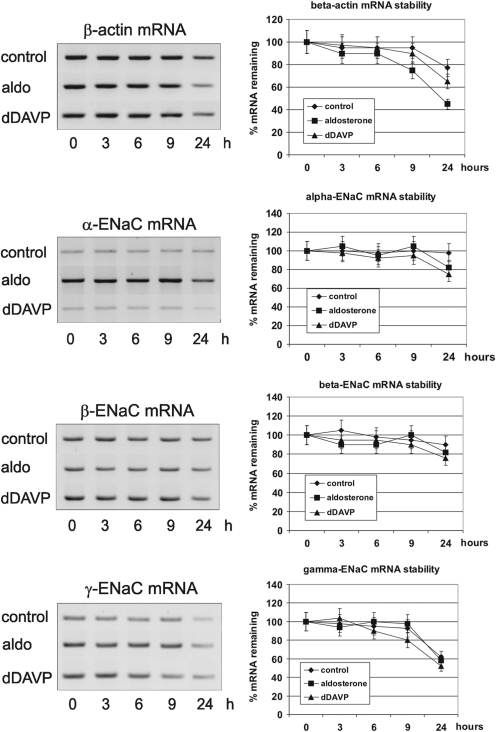
mRNA stability
To test mRNA stability, mCCDcl1 cells were stimulated with aldosterone or dDAVP as described above for 24 h, followed by addition of actinomycin D (MoBiTech, Goettingen, Germany) to a final concentration of 10 µg/ml, for inhibition of transcription. After 0, 3, 6, 9 and 24 h, cells were washed twice with ice-cold DPBS and were then directly harvested with RNA-Bee reagent (Biozol Diagnostica Vertrieb GmbH). RNA was prepared according to the manufacturer’s protocol and estimation of mRNA concentration was performed by RT–PCR as described above. To compare the RNA decay under control conditions with aldosterone and dDAVP stimulation, mRNA levels at the starting point (0 h) were referred to as 100%.
Plasmid constructs
Partial sequences of rat γ-ENaC mRNA (GenBank accession no. NM017046.1) representing the γ-ENaC 5′-UTR (98 nt) and 3′-UTR (933 nt) were amplified by PCR, cloned and transformed using the TOPO® TA Cloning Dual Promoter Kit (Invitrogen). Positive clones were verified by sequencing and used for in vitro transcription. For the generation of luciferase reporter constructs, the pGL3-promotor vector (Promega) was modified as follows: The vector-specific 5′- and 3′-UTRs of luciferase mRNA were replaced by rat γ-ENaC mRNA UTRs or deletion variants. The UTRs and deletion variants of the 3′-UTR were amplified by PCR from subcloned pCR®II-TOPO constructs and restriction sites were added by primer extension. The 5′-UTR of γ-ENaC mRNA was cloned using the pGL3p vector-specific HindIII and NcoI restriction sites and the 3′-UTR (including the poly-A signal) using the XbaI and SalI restriction sites. Generation of construct ‘pGL3p-γENaC3′-UTRdelAU’ was performed by deletion of a unique Psi I fragment of the γ-ENaC 3′-UTR (nt 2869–2958) and religation. Generation of the construct ‘pGL3p-AU-element’ containing the central part of the ARE motif (nt 2865–2916) was done by PCR. The processed vectors were confirmed by sequencing. The resulting vector constructs expressed a constitutively transcribed luciferase transcript with or without the specific γ-ENaC UTRs.
Transfection and luciferase assays
mCCDcl1 cells were grown to ∼70% confluence in 96-well plates (µClear Platte 96K, Greiner BIO-ONE GmbH, Frickenhausen, Germany) and transiently co-transfected with the firefly luciferase pGL3-promoter vector (Promega) or the transformed variants containing the γ-ENaC mRNA UTRs or deletion variants and the ‘Renilla’ luciferase phRL-TK vector (Promega). A ratio (DNA: transfection reagent) of 1:3 was used with the TransFectin™ Lipid Reagent (Bio-Rad) according to the manufacturer’s protocol. Transfection of mCCDcl1 cells with empty pGL3-promoter vector and with the corresponding empty expression-vector for co-transfection experiments served as controls. Co-transfection with the ‘Renilla’ luciferase reporter plasmid was performed for normalization of transfection efficiencies.
For luciferase assays under aldosterone or dDAVP treatment, cells were set to stimulation medium 24 h after seeding and transfected 30 h post-seeding. After seeding 48 h, cells were stimulated by addition of stimulation medium supplemented with either 300 nM aldosterone (Sigma), 10 nM dDAVP (Sigma) or 0.1% ethanol (Carl Roth) as control.
For co-expression experiments with RBPs, the following expression vectors and the corresponding empty vectors were used: pCMV-SPORT6 (empty vector, Invitrogen), pCMV-SPORT6-HuR, pCMV-SPORT6-AUF1, pCMV-SPORT6-TTP, pSG5 (empty vector, Stratagene), pSG5-hnRNP-A1, pEGFP-C1 (BD Biosciences Clontech), pEGFP-FMRP. The luciferase activities were measured with a luminometer (Labsystems Luminoscan RS, Helsinki, Finland) programmed with individual software (Luminoscan RII, Ralf Mrowka) 24 h after transfection as described (34).
Preparation of polysomes, mRNPs and RNA
Polysomes were obtained from S10 protein extracts by centrifugation for 2 h at 100 000g, 4°C in a Beckman SW-41 rotor. The post-polysomal mRNP fraction was sedimented from the S100 supernatant by additional centrifugation for 3 h at 300 000g, 4°C. Polysomal and mRNP pellets were dissolved in TKM-buffer (50 mM Tris, 25 mM KCl, 5 mM MgCl2). RNA isolations from polysomes and RNPs were performed by standard phenol–chloroform extraction.
Sucrose gradient centrifugation
Cytosolic extracts (S10) of mCCDcl1 cells were layered onto 11 ml of a linear 17–51% sucrose gradient (31) (0.5–1.5 M sucrose, 20 mM Tris pH 7.4, 150 mM KCl, 5 mM MgCl2, 1 mM DTT) and centrifuged for 2 h at 36 000 r.p.m. using a Beckman SW-41 rotor. Following sedimentation, the gradient was fractionated from the bottom to the top using a peristaltic pump. The ribosomal profile was determined continuously by measuring absorbance at 254 nm using a 2138 UVICORD-S UV monitor (LKB Bromma). Sucrose gradients were divided into 12 subfractions each, starting with fraction 1 (bottom) to 12 (top). For protein isolation trichloroacetic acid (TCA) was supplemented to a 10% final concentration. Precipitated proteins were sedimented, washed three times with acetone and dissolved in 100 µl buffer (25 mM Tris, 1% SDS). RNA was isolated using the EaZy Nucleic Acid (isolation) RNA Total Kit (VWR International) according to the manufacturer’s protocol.
Western blot analysis
Of proteins 20–40 µg, for analysis of sucrose gradient fractions 5 µl of each fraction, were subjected to 10% SDS–PAGE and transferred to Roti®-PVDF (Roth) by wet electroblotting using a Bio-Rad Mini Trans-Blot transfer cell (Bio-Rad). Membranes were blocked by incubation for 1 h in 1 × TBS-T (20 mM Tris base, 137 mM NaCl, 0.1% Tween-20) containing 5% non-fat dried milk (Carl Roth), followed by incubation with first antibody for either 1 h at RT or overnight at 4°C in 1% milk (1 × TBS, 0,1% Tween-20). Membranes were washed 3 × for 10 min with 1 × TBS, 0.1% Tween-20 and incubated with horseradish peroxidase-conjugated (HRP-conjugated) secondary antibody for 1 h. Then membranes were washed three times for 10 min each with 1 × TBS, 0.1% Tween-20 and chemo-luminescence was detected by using ECL plus Western Blotting Detection System (Amersham Pharmacia Biotech). Membranes were exposed to films (Hyperfilm ECL, GE Healthcare) for 1–5 min. For re-probing, membranes were stripped 5–10 min with 0.2 M NaOH prior to blocking. The following first antibodies were used for detection: anti-αENaC (E4652, Sigma) 1:3000, anti-βENaC (sc21013, Santa Cruz) 1:1000, anti-γENaC (sc22245 santa cruz) 1:500, anti-nucleolin (ab22758, Abcam) 1:2000, anti-HuR (# 07-468, upstate) 1:1000, anti-AUF1 (# 07-260, upstate) 1:3000, anti hnRNP-A2/B1 (BM4520, Acris) 1:500, anti-hnRNP-E1 (BioGenes GmbH) 1:12000, anti-annexin (ab41803, Abcam) 1:1000, anti-ZFP36 (TTP) (ARP34385_P050, AVIA) 1:1000, anti-FMRP (sc28739, Santa Cruz) 1:1000, anti-hnRNP-A1 (sc10032, Santa Cruz) 1:200, anti-β-Actin (MAB1501R, Chemicon) 1:4000, anti-GAPDH (BM429, Acris) 1:6000. As secondary antibodies following HRP-conjugated antibodies were used: goat anti-rabbit (sc2030) 1:30 000, goat anti-mouse (sc2031) 1:100 000, donkey anti-rabbit (sc2317) 1:10 000, donkey anti-goat (sc2033) 1:10 000) and anti-chicken IgY (G135A, Promega) 1:30 000, bovine anti-goat (sc2350) 1:30 000. Secondary antibodies were purchased from Santa Cruz apart anti-chicken IgY. Following antibody detection western blot membranes were stained with Coomassie blue and protein bands were used for normalization of western blot signals for all western blots of cytosolic extracts under hormone treatment with aldosterone or dDAVP. This kind of normalization to stained protein bands was chosen because of the induction of the normally used housekeeping genes β-actin and GAPDH under aldosterone and dDAVP stimulation.
RNA affinity chromatography
For the isolation of mRNA-binding proteins, in vitro transcripts representing the 3′-UTR of γ-ENaC mRNA were generated in the presence of biotinylated CTP (Invitrogen). Cytosolic extracts of mCCD cells (40 mg protein) were incubated with 2.5 µg in vitro labelled transcript for 45 min at room temperature. RNP (ribonucleoprotein) complexes were isolated using 300 µl of streptavidin–agarose/sample (Sigma). Samples without the addition of biotinylated transcripts served as negative control. The agarose beads were centrifuged for 2 min at 5000g and washed five times (10 mM Tris, pH 7.4, 150 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT). The last two washing supernatants were used as control. The RNP complexes were eluted using two times high salt buffer 1 (5 mMTris, pH 7.4, 1 M KCl, 0.75 mM MgCl2) and two times high salt buffer 2 (5 mMTris, pH 7.4, 3 M KCl, 0.75 mM MgCl2). After combination, proteins were precipitated, dissolved and subjected to SDS–PAGE. After Coomassie staining, protein signals representing specific RNA-binding factors were excised. Tryptic digestion of proteins was carried out using ZipPlates (Millipore) without reduction or alkylation. Tryptic fragments were analysed by Reflex IV matrix assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometer (Bruker-Daltonics). Mass spectra were analysed using Mascot software 2.0 with automatic search in NCBI non-redundant databases. Search parameters allowed for one miscleavage and oxidation of methionine. Criteria for positive identification of proteins with MS were set according to the scoring algorithm.
UV-crosslinking assay
In vitro transcripts representing the 3′-UTR of rat γ-ENaC mRNA were radioactively labelled using [α-32P]uridine-5′-triphosphate (800 Ci/mmol, MP Biomedicals GmbH, Heidelberg, Germany). In vitro transcripts were purified over BD Chroma Spin™-100 (DEPC) columns (BD Bioscience). [α-32P]NTP (1–2 ng) labelled in vitro transcript (corresponding to 300 000 c.p.m.) was incubated with 35 μg of cytosolic protein extract for 30 min at room temperature in 10 mM Hepes, pH 7.2, 3 mM MgCl2, 5% glycerol, 1 mM DTT, 150 mM KCl and 2 U/μl RNaseOUT (Invitrogen Life Technologies) in the presence of rabbit rRNA (0.5 μg/μl) as competitor. The samples were exposed to UV light (255 nm, 1.6 J, UV-Stratalinker) on ice, followed by RNase treatment with RNase-A (30 μg/ml final concentration) and RNase-T1 (750 U/ml final concentration) for 1 h at 37°C and subjected to 12% SDS–PAGE and subsequent autoradiography using the Phospho-Imager-System (Fujifilm FLA-3000).
RNA-pulldown assay
Immunoprecipitation of HuR protein-bound mRNA was performed using a protocol from Doller et al. (42) with slight modification. Briefly, mCCD cells were stimulated as described, followed by lysis in a buffer containing 10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.1% Nonidet P-40, 50 mM NaF, 10 mM Na3VO4, 10 mM sodium pyrophosphate, 50 mM disodium glycerol phosphate and 100 U/ml RNasin. Subsequently, cell lysates were immunoprecipitated with 2 μg of either a monoclonal anti-HuR antibody (Santa Cruz) or with the same amount of mouse IgG (Santa Cruz) as negative control overnight at 4°C. To normalize for equal input of RNA before subsequent purification steps, the same amount of extract was subjected to total RNA isolation as described. Subsequently, protein G-Sepharose CL-4B beads (Amersham-Biosciences) were added and incubated for another 2 h. After sedimentation of the beads by centrifugation for 5 min at 3000g, beads were washed twice with low salt buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.2% Triton X-100, 2 mM EDTA, 2 mM EGTA and 0.1% SDS) and twice with high salt buffer (50 mM Tris–HCl, pH 7.5, 500 mM NaCl, 0.2% Triton X-100, 2 mM EDTA, 2 mM EGTA and 0.1% SDS) before total RNA was isolated by using RNA-Bee reagent (Biozol Diagnostica Vertrieb GmbH) as described.
Densitometrical analysis, statistics
Densitometrical analysis of bands from ethidiumbromide stained agarose gels and autoradiographic western blot signals was performed by using Scion Image software (Scion Corp.). If not indicated otherwise, values are presented as mean ± SD. Students’ paired t-test was performed to reveal statistical significances. P-values ≤0.05 were considered significant.
RESULTS
Expression of ENaC subunits in mCCD cells under the action of aldosterone and vasopressin
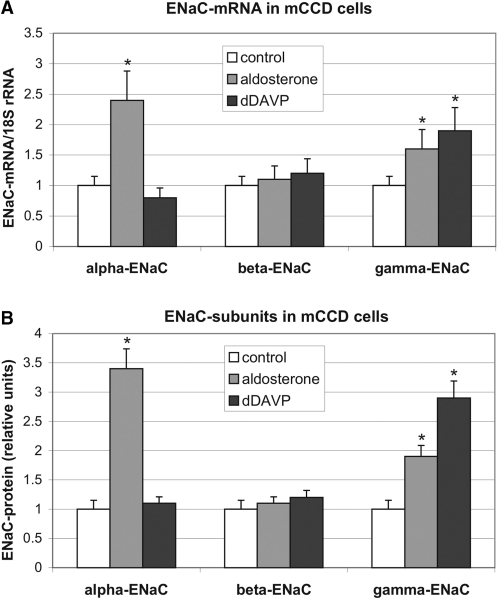
To study post-transcriptional effects on the synthesis of ENaC subunits mouse cortical collecting duct cells (mCCDcl1) expressing functional ENaC channels were validated to respond to aldosterone and the vasopressin agonist dDAVP at the mRNA and protein level. As shown in Figure 1A (mRNA) and B (protein), after 24 h, a marked induction of the α-subunit was observed by aldosterone (2.4-fold at the mRNA level, 3.4-fold at the protein level), but not by dDAVP (0.8 × mRNA, 1.1 × protein). In contrast, the β-subunit did not significantly respond to aldosterone or to dDAVP (1.1/1.2 × mRNA, 1.1/1.2 × protein). The γ-subunit, however, was induced by both, by aldosterone (1.6 × mRNA, 1.9 × protein) and to a somewhat higher extent by dDAVP (1.9 × mRNA, 2.9 × protein).
Figure 1.
Influence of aldosterone and dDAVP on the expression of ENaC in mCCD cells. mCCD cells were cultivated under control conditions (0.1% ethanol) or in the presence of aldosterone (300 nM) or vasopressin agonist dDAVP (10 nM) for 24 h. (A) Total RNA was extracted and relative levels of α-, β- and γ-ENaC mRNAs were quantified by RT–PCR. −RT controls showed no signals (data not shown). Values were normalized to 18 S rRNA as described in ‘Materials and Methods’ section. Data represent mean ± SD (n = 5). Controls were referred to as 1.0. *P ≤ 0.05 compared with control. (B) In cytosolic extracts (S10) α-, β- and γ-ENaC protein was determined by western blotting and normalized to Coomassie-stained western blot membranes as described in ‘Materials and Methods’ section. Signals were quantified by scanning and densitometrical analysis. Data represent mean ± SD (n = 3). Controls were referred to as 1.0. *P ≤ 0.05 compared to control.
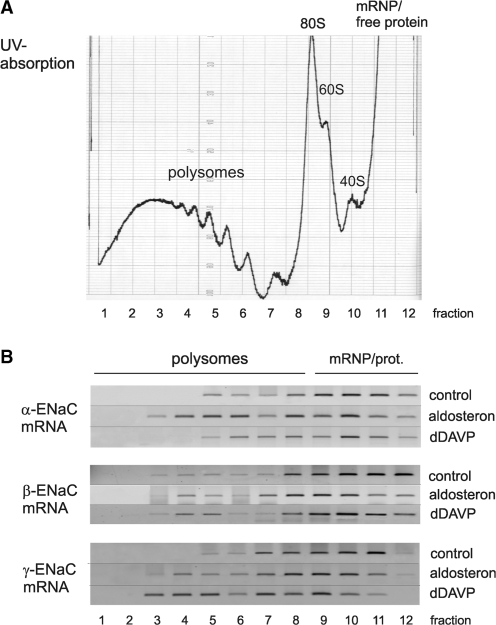
Distribution of ENaC mRNAs between polysomes and translational inactive post-polysomal mRNPs
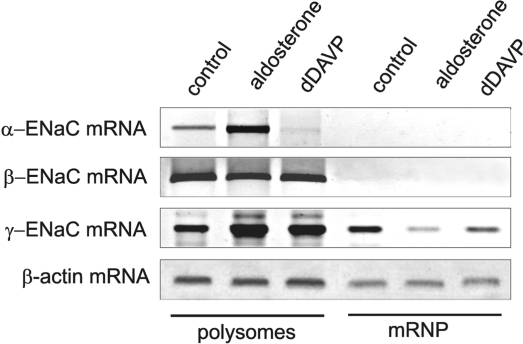
In the cytoplasm of cells, mRNAs reside in two main functional compartments: in a translational active form in polysomes and in a mainly translational inactive state in post-polysomal messenger ribonucleoprotein (mRNP) complexes. To analyse the phenomenon how synthesis of ENaC subunits is regulated at the mRNA level by post-transcriptional events, we isolated polysomes and post-polysomal mRNPs of the cytoplasm of mCCD cells after cultivation in the presence of aldosterone or dDAVP by ultracentrifugation (polysomes = 100 000g pellet of S10, mRNPs = 300 000g pellet of S100) and quantified ENaC mRNAs by RT–PCR. α-,β-and γ-ENaC mRNAs behaved quite differently (Figure 2). In normally cultivated, as well as in aldosterone or dDAVP-treated cells, α- and β-mRNA were completely associated with ribosomes and only minor traces were found in the free mRNP fraction. In opposite, under control conditions, only ∼60% of the γ-mRNA was associated with polysomes, but 40% was found in the translational inactive free mRNP pool. This figure changed considerably after aldosterone or dDAVP treatment. By aldosterone, the majority of the translational inactive γ-ENaC mRNA shifted into the polysomal fraction and only traces remained in the mRNP pool. Similarly, also dDAVP administration lead to such a shift, but the effect was not as pronounced as with aldosterone, ∼10% of the mRNA remained non-ribosomal associated in the free mRNP fraction.
Figure 2.
Distribution of ENaC mRNAs between polysomes and translational inactive post-polysomal mRNPs. mCCD cells were cultivated under control conditions (0.1% ethanol), in the presence of aldosterone (300 nM) or dDAVP (10 nM) for 24 h. Polysomes (100 000g pellet of S10) and post-polysomal mRNP particles (300 000g pellet of S100) were prepared from cytosolic extracts (S10) by ultracentrifugation, and α-, β- and γ-ENaC mRNAs were analysed by RT–PCR. β-Actin mRNA served as a control. (−RT) controls showed no signals (data not shown). A representative figure of three independent experiments is shown.
γ-ENaC mRNA 3′-UTR mediates hormonal stimulation of reporter gene expression
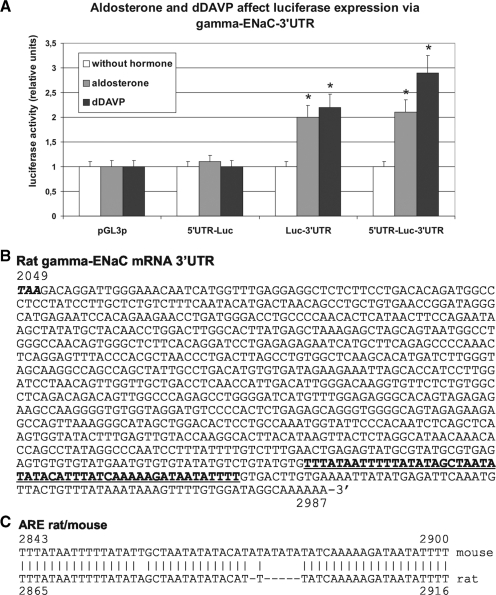
Experiments described in Figure 2 demonstrate that especially γ-ENaC mRNA is characterized by a prominent change in its intracellular localization (shift from post-polysomal RNP to polysomal compartment) and its functional state, presumably by translational activation, in response to hormone treatment. Therefore the focus was set on post-transcriptional control of γ-ENaC expression. To substantiate the link between γ-ENaC UTR and hormonal stimulation of ENaC synthesis, luciferase reporter gene transfection experiments were set up in mCCD cells, under conditions, where luciferase activity was dependent on γ-ENaC mRNA 5′- or 3′-UTR, or their combination (Figure 3A). It is obvious that only γ-ENaC 3′-UTR was able to mediate translational activation in aldosterone or dDAVP-treated cells, not however γ-ENaC 5′-UTR. Compared to non-hormonal stimulated cells, 3′-UTR enhanced luciferase expression in aldosterone-treated cells 2.0-fold and in dDAVP-treated cells 2.3-fold. Although the 5′+3′-UTR combination was even slightly more effective (aldosterone: 2.1-fold, dDAVP: 2.9-fold), the 5′-UTR itself had no significant influence.
Figure 3.
Effect of aldosterone and dDAVP on expression of luciferase γ-ENaC UTR constructs in mCCD cells. (A) mCCD cells were transfected with the original pGL3p-promoter (pGL3p) vector or chimeric variants, where original luciferase mRNA 5′- and/or 3′-UTRs were substituted by rat γ-ENaC 5′- and/or 3′-UTR. Eighteen-hour post-transfection cells were incubated for further 24 h under control conditions (without hormone; 0.1% ethanol) or with aldosterone (300 nM) or dDAVP (10 nM). UTR-dependent luciferase activity was measured 24 h after stimulation. Transfection efficiency was normalized to expression of co-transfected ‘Renilla’ luciferase and relative values were related to pGL3p and control (0.1% ethanol). Data represent mean ± SD (n = 6). *P ≤ 0.05 compared with control. (B) cDNA sequence of the entire 3′-UTR of rat γ-ENaC mRNA (Scnn1g, genbank NM017046) starting with the stop-codon (bold italics) is shown (nt 2049–2987). The AU-rich region deleted in luciferase chimeric constructs used for ARE-BP co-expression experiments is underlined and printed in bold. (C) Alignment of cDNA sequence of γ-ENaC mRNA 3′-UTR ARE of rat (NM017046) and mouse (NM011326).
Characterization of ENaC mRNA stability under the influence of aldosterone and vasopressin
One basic mechanism by which gene expression is modulated post-transcriptionally consists of alteration of metabolic mRNA stability. This is often mediated by specific RBPs interacting with signals in the 3′-UTR of the mRNA (26–28). The 3′-UTR of γ-ENaC mRNA contains an AU-rich sequence (Type III ARE, Figure 3B) in the 3′-terminal region, which is highly conserved between rat and mouse (Figure 3C). Such sequences are known to mediate stabilization/destabilization or translational control by interaction with ARE-BPs. Also the α- and β-mRNA contain relatively long 3′-UTR sequences and may be prone to a similar type of control. Therefore, we analysed the stability of all three ENaC mRNAs in mCCD cells after inhibition of transcription by actinomycin D in dependence of aldosterone and dDAVP action. mRNA levels were observed during 24 h and quantified by RT–PCR (Figure 4). In no case, the hormones lead to an increase in mRNA stability. The results showed furthermore that both hormones did not contribute to the increase in α- or in γ-ENaC mRNA concentration. In contrast, we observed a slight, but not significant, mRNA labilization after actinomycin D blockade. Using α-amanitin instead of actinomycin D as transcription inhibitor and performing mRNA quantification by northern blotting lead to similar results (not shown). Taken together, the increase in α- and γ-mRNA concentration by aldosterone and dDAVP is not generated by mRNA stabilization, but seems to be caused by a pure transcriptional mechanism.
Figure 4.
Stability of ENaC mRNAs in mCCD cells treated with aldosterone and dDAVP. Cells were cultivated for 24 h in the presence of 0.1% ethanol (control), 300 nM aldosterone or 10 nM dDAVP, then transcription was stopped by addition of actinomycin D (10 µM final concentration) and cells were further cultivated for the times indicated. Total RNA was prepared at each time point and relative levels of α-, β- and γ-ENaC mRNA were quantified by RT–PCR including β-actin mRNA as an independent control mRNA. Values were normalized to 18S/28S rRNA. −RT controls showed no signals (data not shown). mRNA levels at time point 0 h of each condition (control, aldosterone, dDAVP) was referred to as 100% mRNA remaining for each transcript. Relative mRNA values of later time points were calculated referring to the corresponding (same culture condition) mRNA value at time point 0 h. Data represent mean ± SD (n = 3).
Aldosterone and dDAVP are effectors of translational efficiency of ENaC mRNAs
A second mechanism, by which post-transcriptional control operates, is to facilitate initiation of translation, which is characterized by enhanced ribosome recruitment of mRNAs leading subsequently to an enhanced translational efficiency. The resulting increase in the average polysome size for a specific mRNA can be characterized by sedimentation of polysomes through sucrose gradients. As shown in Figure 5, we performed polysome gradient analysis with cytosolic lysates (S10) of hormone and non-hormone treated mCCD cells. The lysates were run over 17–51% sucrose gradients (Figure 5A), divided in 12 fractions, and α-, β- and γ-ENaC-specific polysomes were detected by RT–PCR (Figure 5B). The results can be summarized as follows: α-ENaC mRNA was similarly present in intermediate-size polysomes under control conditions as in dDAVP-treated cells, but was more efficiently translated in aldosterone-treated cells. The translation properties of β-ENaC mRNA was neither changed by dDAVP nor by aldosterone. The ribosome recruitment of γ-ENaC mRNA, however, was enhanced by both hormones, visible as a shift of the mRNA signal in the polysomal density profile by two gradient fractions in direction to heavier polysomes. To check polysome integrity, parallel gradients were run after EDTA treatment. As expected, EDTA application lead to a complete dissociation of the polysome complex, detectable as disappearance of the polysome signal in the UV-trace and a shift of mRNA signals to the top fractions 9–12 of the gradient (not shown). In conclusion, translational activation contributes to the induction of α- and γ-chains by aldosterone and V2 receptor stimulation.
Figure 5.
Sucrose polysome gradient analysis: aldosterone and dDAVP increase translational efficiency of α- and γ-ENaC mRNA. (A) Equal amounts of cytosolic extracts of control (0.1% ethanol), aldosterone (300 nM) or dDAVP (10 nM) stimulated mCCD cells (S10) were separated by ultracentrifugation (160 000g, 2 h, Beckman SW 41 rotor) through 17–51% sucrose gradients and divided into 12 fractions. Continuous RNA monitoring (OD254) of a representative gradient fractionation from the bottom (51% sucrose) to the top (17% sucrose) is shown. (B) Total RNA was prepared and relative levels of α-, β- and γ-ENaC mRNAs were monitored by RT–PCR. −RT controls showed no signals (data not shown). Polysome bound mRNAs sediment in fractions 2–8, free mRNPs and cytosolic proteins in fractions 9–12. A representative figure of three independent experiments is shown.
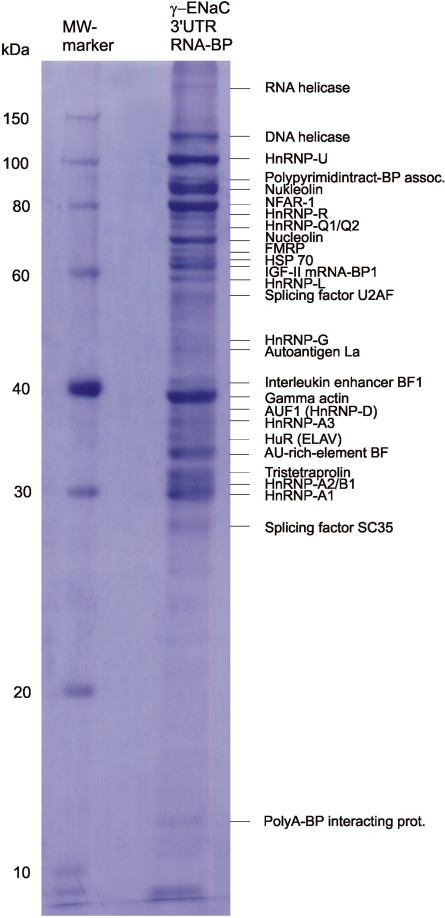
Characterization of γ-ENaC mRNA-binding RBPs by RNA affinity chromatography
Important players in post-transcriptional gene regulation are RBPs. In particular, we were interested in the nature of potential regulative RBPs and how far their interaction to motifs in γ-ENaC 3′-UTR may be changed in response to hormone action. For the general characterization of γ-ENaC 3′-UTR-BPs, we employed RNA affinity chromatography using γ-3′-UTR as an affinity matrix and mCCD cell cytoplasm as a source for RBPs. The subsequent identification of candidate proteins was performed by matrix assisted laser MALDI-TOF mass spectrometry (MS) of affinity purified and finally electrophoretically separated RBPs. Typically, a pattern of ∼20–40 Coomassie stainable polypeptides was obtained. In the example shown, 27 most prominent bands were subjected to the mass spectroscopic identification procedure (Figure 6). All identified proteins out of two (DNA helicase and γ-actin) belonged to the group of RBPs. As discussed above, in γ-3′-UTR an ARE is situated in the 3′-terminal part (Figure 3B). Interestingly, among the identified RBPs, several candidates were found that are discussed as ARE- or general U-tract binding proteins (annexin II, nucleolin, FMRP, AUF1, HuR, TTP and hnRNP-A1).
Figure 6.
Identification of cytosolic proteins bound to γ-ENaC 3′-UTR by RNA affinity chromatography and MALDI-TOF-MS. RBPs were purified by affinity chromatography using biotinylated in vitro transcripts, which represent rat γ-ENaC 3′-UTR, and cytosolic extracts (S100) of mCCD cells. Affinity-purified RBPs were separated by SDS–PAGE and Coomassie-stained bands were subjected to MALDI-TOF-MS for protein identification. A representative figure of three independent experiments is shown.
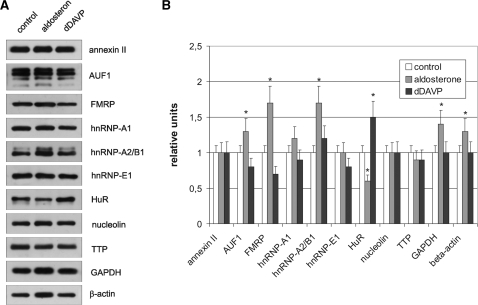
To detect if hormone treatment leads to a significant change in expression of U-tract binding proteins, we quantified the candidate ARE-BPs including hnRNP E1 (which was not bound by γ-3′-UTR) and hnRNP A2/B1 (a close relative to hnRNP A1), as well as non-RBPs GAPDH and β-actin as controls by western blotting (Figure 7). As a result, significant, but no vast differences in the immunological detectable amounts of some analysed proteins were found. In fact, no differences existed within the limits of the methods with annexin II, hnRNP-A1, hnRNP-E1, nucleolin and TTP. The most significant alterations were brought about by aldosterone: an induction was observed with AUF1 (by 30%), FMRP (by 80%) and hnRNP-A2/B1 (by 70%). The only repression was found with HuR (by 40%). Remarkably, also the non-RBPs GAPDH and β-actin used as controls were induced by aldosterone by ∼40 and 50%. dDAVP treatment only lead to a slight induction of two proteins, namely hnRNP A2/B1 by 20% and HuR by 50%. On the other hand it evoked a marginal repression by 20–30% of AUF1, FMRP and hnRNP-E1. The antibody against AUF1 detected four described iso-forms with molecular weights between 37 and 45 kDa (26). There was, however, no significant change in the abundance of the different iso-proteins to each other in the course of hormone treatment.
Figure 7.
Induction of RBPs in mCCD cells by aldosterone and dDAVP. mCCD cells were stimulated with aldosterone or dDAVP as described in the legend of Figure 1. Thirty micrograms of cytosolic extracts (S10) were analysed for expression of RBPs annexin II, AUF1, FMRP, hnRNP-A1, hnRNP-A2/B1, hnRNP-E1, HuR, nucleolin, TTP and the controls β-actin and GAPDH by western blotting using specific antibodies. (A) Representative western blots of five independent experiments of the nine RBPs and the two controls are shown. (B) Autoradiographs were quantified by scanning and densitometrical analysis. Data represent mean ± SD (n = 5). *P ≤ 0.05 compared to control.
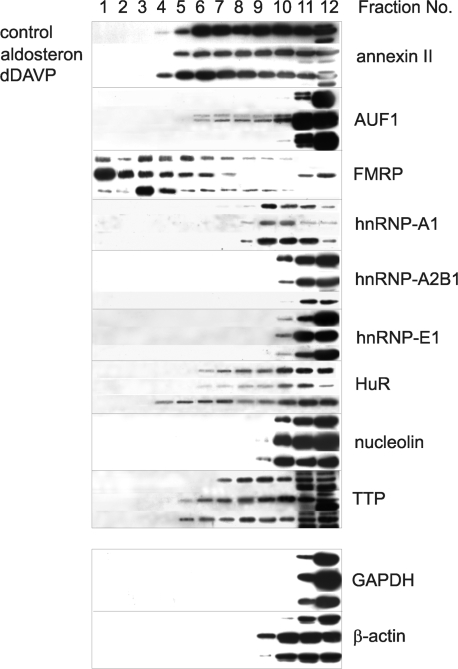
Selected RBPs change their association to polysomal complexes in response to hormone action
As demonstrated above, under hormonal influence γ-ENaC mRNA was efficiently recruited from translational inactive RNPs into polysomes, where protein synthesis occurs. So we asked, if this shift could be accomplished by a re-arrangement of mRNA-bound proteins. For this purpose, we separated the cytoplasm of hormone and non-hormone-treated cells over sucrose density gradients as described in Figure 5, divided the gradient into 12 fractions and analysed the same selection of RBPs as shown in Figure 7 with respect to their polysome association by western blotting. The results are summarized in Figure 8. The gradient can be divided into two main zones: the region of polysomes (bottom region, fractions 2–8) and the region of cytoplasmic proteins including free mRNPs (top region, fractions 9–12). The following proteins were found only in free non-polysomal associated form (fractions 9–12): hnRNP-A2/B1, hnRNP-E1, nucleolin and the control proteins GAPDH and β-actin. By contrast, annexin II, AUF1, FMRP, hnRNP-A1, HuR and TTP were more or less associated with polysomes, which were occupied by ribosomes to a different degree (fractions 2–8). Interestingly, polysomal association of certain ARE-BPs changed in response to hormone action. The most striking alterations were provoked by aldosterone. AUF1, which was not present in polysomes under control conditions, became bound, and this applied only to the two higher molecular weight iso-formes of ∼42 and 45 kDa. Furthermore, FMRP and TTP were increasingly present in larger polysomes of higher translational efficiency. After dDAVP treatment a shift to larger polysomes, i.e. activation of translational efficiency was only observed with polysomes associated with HuR and TTP.
Figure 8.
Aldosterone and dDAVP affect binding of RBPs to polysomes. Polysomes of hormone-treated mCCD cells were separated over sucrose gradients and fractionated as described in the legend of Figure 4. Proteins of each fraction were concentrated by TCA-precipitation redissolved in a volume of 100 µl buffer (25 mM Tris, 1% SDS). Proteins of all fractions (5 µl) of sucrose gradients were analysed for RBPs annexin II, AUF1, FMRP, hnRNP-A1, hnRNP-A2/B1, hnRNP-E1, HuR, nucleolin, TTP and the cytosolic proteins GAPDH and β-actin by western blotting using specific antibodies. A representative figure of three independent experiments is shown.
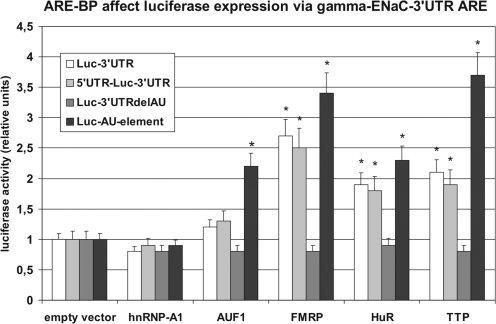
Cell transfection co-expression experiments document significance of RBPs in γ-ENaC expression
To gain insight into the functional role of RBPs in γ-ENaC post-transcriptional control, luciferase reporter gene co-expression experiments were performed using plasmids, containing luciferase coding and complete γ-ENaC 5′- and/or 3′-UTR sequences. This transfection was applied in combination with plasmids expressing the nine selected RBPs described above (Figure 8). The results are shown in Figure 9. An over-expression of all nine RBPs had only marginal effects on luciferase expression when using the basic pGL3p vector (luciferase coding, no γ-ENaC UTRs, 0.8–1.2-fold alteration, not shown). A distinct alteration of luciferase activity however was measurable, when RBP plasmids were co-transfected together with a luciferase construct containing the γ-ENaC 3′-UTR. Remarkably, only the ARE-BPs FMRP, HuR and TTP stimulated reporter gene expression to a significant extent (2.7-, 1.9- and 2.2-fold). AUF1 showed only a slight, non-significant effect (1.2-fold stimulation). All stimulatory effects disappeared completely after deletion of the AU-rich region located in the 3′-terminal part of γ-ENaC 3′-UTR (plasmid Luc-3′-UTRdelAU, see also Figure 3). A very similar result was obtained with constructs containing the authentic 5′+3′-UTR combination of γ-ENaC mRNA. Using this construct, again FMRP, HuR and TTP stimulated luciferase expression significantly (2.2–3.0-fold) and AUF1 slightly (1.3-fold). All other RBPs had only non-significant effects (i.e. hnRNP-A1, -A2/B1 and -E1, annexin II, and nucleolin; only the data for hnRNP-A1 are shown). To prove that the stimulatory effect was due to the ARE itself, the central 52 nt AU-rich sequence of the 3′-UTR (see Figure 3) was tested in the same way. As the results show (Figure 9), the isolated ARE had an even still higher capability to mediate translational activation by ARE-BPs than the complete 3′-UTR (AUF1 2.2-fold, FMRP 3.4-fold, HuR 2.3-fold and TTP 3.7-fold).
Figure 9.
Influence of over-expressed ARE-BPs on the expression of chimeric luciferase plasmids containing γ-ENaC UTRs. mCCD cells were transfected with pGL3p vector or chimeric variants, where original luciferase mRNA UTRs were substituted by rat γ-ENaC 3′-, 5′- and 3′-UTR or the 3′-UTR deletion variants 3′-UTRdelAU (deletion of base 2869–2958) or AU-element (base 2865–2916) of γ-ENaC mRNA. Furthermore, cells were co-transfected with expression vectors encoding for proteins of the RBPs hnRNP-A1, AUF1, FMRP, HuR or TTP. As control cells were co-transfected with empty expression vector. UTR-dependent luciferase activity was measured 24 h post-transfection. Transfection efficiency was normalized to expression of co-transfected ‘Renilla’ luciferase and relative values were normalized to the influence of empty vector control on pGL3p constructs. Data represent mean ± SD (n = 6). *P ≤ 0.05 compared to empty vector.
Aldosterone and dDAVP up-regulate γ-ENaC expression by enhanced binding of HuR to γ-ENaC mRNA 3′-UTR
Polysome association behaviour of γ-ENaC mRNA indicated that aldosterone and dDAVP caused translational activation of γ-ENaC synthesis (Figure 5). Furthermore, in vitro co-expression experiments using luciferase/γ-ENaC 3′-UTR constructs and RBP plasmids proved that RBPs FMRP, HuR and TTP were able to activate reporter gene expression in an ARE-dependent manner (Figure 8). However, this did not prove subsequently the in vivo existence of γ-ENaC mRNA specific mRNP complexes and the functional connection between hormone action and stimulation of interaction of RBPs with γ-ENaC mRNA. For this reason we applied a HuR-specific antibody for immunoprecipitation of HuR associated polysomes of mCCD cells and analysed the role of HuR in hormonal stimulation. HuR served as an example for the analysis of one crucial RBP involved in post-trancriptional control of γ-ENaC expression (Figure 10A–D).
Western blots demonstrate that an over-expression of HuR by a factor of ∼2.0–4.0 over the endogenous level lead to a concentration dependent increase in γ-ENaC protein in the cytoplasm of ∼2.5–3.5-fold (Figure 10A). The result shows that an increase in HuR was sufficient to up-regulate γ-ENaC synthesis. To prove that this HuR dependent up-regulation of γ-ENaC was accomplished by an improved binding of HuR to γ-ENaC 3′-UTR the same cytosolic extract was used in UV-crosslinking analysis. For this purpose 32P-UTP labelled γ-ENaC 3′-UTR in vitro transcripts were incubated with cytoplasm containing gradually over-expressed HuR and analysed for binding. Clearly, a 3–4-fold intensification of a signal migrating at the position of authentic HuR (36 kDa) can be seen (Figure 10B). The supposed ARE binding sequence is composed of 93% of the nucleotides U+A and 7% of G+C (Figure 3). If the labelling of the transcript was repeated with CTP or GTP instead of UTP no signal at the 36 kDa position was visible, i.e. no label transfer occurred (not shown). This demonstrates that the RNA–protein interaction took place predominantly via the nucleotide ‘U’, which is characteristic for an ARE.
An important strategy to verify the in vivo existence of γ-ENaC–mRNA/HuR complexes is to establish a co-precipitation pull-down protocol using a HuR specific antibody and subsequent isolation of the immune complexes by protein-G-sepharose. HuR antibody (ab) bound γ-ENaC mRNA was quantified by RT–PCR. Use of IgG instead of specific antibody, as well as quantification of unspecific binding of GAPDH mRNA served as controls. To assure an equal input, the reaction was normalized by determination of γ-ENaC mRNA prior to the immune reaction, furthermore PCR controls were run omitting the RT reaction. Figure 10C describes the results. It is obvious that the HuR antibody specifically precipitated γ-ENaC–mRNA/HuR, but not GAPDH–mRNA/HuR complexes.
This assay was finally applied to the analysis of mCCD cells, in which γ-ENaC synthesis was stimulated by aldosterone or dDAVP before quantification of immune complexes (Figure 10D). As controls served again the use of unspecific IgG instead of HuR antibody and determination of unspecific GAPDH–mRNA/HuR binding. An equal protein input in the immune reactions was guaranteed by electrophoresis and Coomassie blue staining of an aliquot. It is evident that the increase in γ-ENaC expression by aldosterone or dDAVP action is reflected at the level of mRNP complexes by an increase in the amount of HuR-bound γ-ENaC mRNA (aldosterone: 2.5-fold, dDAVP 3.0-fold).
Taken together, immunoprecipitation of γ-ENaC–mRNA/HuR complexes and characterization of the specific RNA/protein interaction by UV-crosslinking document the significance of γ-ENaC–mRNA–3′-UTR/HuR interaction for hormonal control of ENaC synthesis.
DISCUSSION
We provide several lines of evidence that post-transcriptional mechanisms operate in hormonal control of expression of the genes coding for the subunits of epithelial sodium channel ENaC. This applies in particular for the α- and γ-subunit. We show that the induction of α- and γ-ENaC subunits in mouse kidney CCD cells by aldosterone and dDAVP is due to transcription and additionally to activation of mRNA-specific translation, not however to mRNA stabilization (Figures 4 and 5). The increase in ENaC-subunit protein is partially caused by translational stimulation and is not solely a result of augmented translation due to risen mRNA concentration. The well documented potent induction of the α-subunit by aldosterone (Figure 1) is accompanied by a significant recruitment of the mRNA by ribosomes (i.e. translational activation) as demonstrated by polysome gradient analysis. A similar effect of translational activation was seen with γ-ENaC mRNA responding to both hormones aldosterone and vasopressin (dDAVP) (Figure 5). As demonstrated (Figure 2), a substantial part of γ-ENaC mRNA is stored in an untranslated form in the post-polysomal mRNP compartment until it is activated by hormone action. The mechanisms, however, by which α- and γ-mRNA were mobilized, seem to differ. Without hormonal stimulation, γ-mRNA persisted to a considerable degree at ∼40% in the translational inactive mRNP compartment (Figure 2). This post-polysomal mRNP bound mRNA resource was only occupied by ribosomes to form translational active polysomes during hormonal stimulation. This is in contrast to the behaviour of the α-mRNA. With and without hormone treatment the mRNA was equally complete associated with polysomes. Thus, the activation process was not elicited by recruiting the mRNA from a translational inactive mRNP pool, but it must have proceeded at the polysomal complex itself. This could for instance result from an enhanced cap-dependent initiation caused by phosphorylation of translation initiation factor eIF4F (20). The effect of vasopressin on β-ENaC expression depends considerably on the model and conditions used. In long-term rat infusion experiments, the β-subunit is induced like the γ-subunit (7). In cultured rat CCD cells only short-term dDAVP application (3 h) lead to induction of the β-subunit. After a period of 24 h, however, β-ENaC protein returned to initial levels (9). This corresponds to our experiment achieved with mouse CCD cells (see Figure 1).
A certain discrepancy can be observed if one compares mRNP associated ENaC mRNAs analysed by direct particle precipitation (Figure 2) or by sucrose gradient analysis (Figure 5). In the upper fractions of the sucrose gradient (mRNP fraction), ENaC mRNA signals are found that are not present in the 300 000g precipates, which also represent an mRNP fraction. We think this is due to technical reasons. Whereas precipitation of polysomes and mRNPs occurs under RNase protection in highly concentrated solutions, the sucrose gradients are run without RNAse protection and the RNA becomes much more diluted. This could result in a minor RNA degradation by contaminating RNAses, which shifts some mRNA fragments to the upper fractions. This problem can unfortunately not be overcome by adding RNase inhibitor to the sucrose solution, because during the run it separates from the polysomal complexes and remains on top of the gradient.
Important mediators in mRNA-specific post-transcriptional control are RBPs interacting with UTRs. γ-ENaC mRNA turned out to be the most interesting subunit, because it showed under hormone action a conspicuous shift in intracellular localization connected with a remarkable translational activation. γ-ENaC mRNA contains only a short 5′-UTR of 69 nt (mouse), respectively, 98 nt (rat). The 3′-UTR is much longer (mouse 931 nt, rat 938 nt) and represents a promising target to mediate post-transcriptional control. Mammalian γ-ENaC mRNAs contain a well conserved AU-rich region in the 3′-terminal part of the 3′-UTR (mouse: 58 nt, A+U content 90%, rat: 52 nt, A+U content 89%, see Figure 5B) that often serves as a target for the binding of RBPs involved in stability or translational control (30). Therefore, we focused our interest on RBP candidates interacting with γ-3′-UTR using RNA affinity chromatography. RNA affinity chromatography is not a very specific, but a helpful, method to narrow down potentially regulative RBP candidates, because it has the advantage to allow direct protein identification, when combined with MALDI-TOF-MS (35). Typically, we identified ∼20–40 cytosolic polypeptides by this type of experiment. This is, of course, a certain over-estimation of the number of proteins, which are expected to interact specifically with an mRNA (or UTR, respectively). However, one has to keep in mind, that also low affinity binders and proteins are captured, which do not interact directly with the RNA but keep contact by protein–protein interaction. An argument for the reliability of the method is the fact that, among ∼30 identified RBPs, only two were found of other specificity (Figure 6), i.e. DNA-helicase and γ-actin, which both have a certain capability to bind also to RNA. Interestingly, several RBPs interacting with γ-3′-UTR belonged to the group of ARE-BP. Therefore, we investigated to which extent the identified potential ARE-BPs AUF1, HuR, FMRP, TTP, annexin II, nucleolin and hnRNP proteins A1 or A2/B1 could be involved in hormone-mediated γ-ENaC translational control. We approached this problem by testing the following two hypotheses. First, we asked if ARE-BPs were constitutive components of actively translating polysomes and if hormones were able to change their qualitative association pattern. The result showed that only some RBPs were associated to polysomes and that only AUF1, HuR, FMRP and TTP, which belonged to this group, sedimented with heavier, translational more active, polysomes under the action of aldosterone or dDAVP (Figure 8). Moreover, aldosterone and dDAVP behaved differently: the association of AUF1 and FMRP was altered only by aldosterone whereas that of HuR was only influenced by dDAVP and that of TTP by both hormones.
The other goal was to test the functional significance of RBPs in γ-ENaC expression. Therefore, luciferase reporter genes carrying γ-ENaC UTR sequences were co-expressed together with RBP expression plasmids. As shown in Figure 9, three of the four selected ARE-BPs stimulated luciferase expression between 2.2- and 3.0-fold. This was not dependent if the chimeric plasmid contained also additionally the authentic γ-ENaC 5′-UTR (Figure 9A). A simple mechanistic explanation for the impact of aldosterone and vasopressin on RBP-dependent γ-ENaC translation would result from an induction/repression of RBPs and hence from an altered cytoplasmic availability. To test this hypothesis, we quantified nine RBPs, including AUF1, HuR, FMRP and TTP, by western blotting in the cytoplasm of hormone treated CCD cells (Figure 7). Especially in the group of ARE-BPs, there was a noticeable correlation between RBP concentration and influence on translational efficiency (polysome size). For instance, the induction of AUF1 and FMRP by aldosterone by ∼30% and 70%, respectively, corresponded with behaviour of these proteins to be associated in favour of polysomes of larger size (Figure 8). Likewise, the induction of HuR by dDAVP by ∼50% was found to result in a very similar effect. The HuR concentration, on the other side, dropped down slightly under the influence of aldosterone. From ARE-BP over-expression experiments similar conclusions could be drawn (Figure 9). Three co-transfected ARE-BPs stimulated luciferase expression using chimeric luciferase/γ-ENaC 3′-UTR plasmids (HuR, FMRP and TTP). AUF1, a factor discussed in mRNA destabilization and translational inhibition, had only marginal effects in co-expression experiments (Figure 9), although it was increasingly present in aldosterone-stimulated polysomes (Figure 8). The behaviour of TTP, on the other side, did not fit easily into this scheme. Both, aldosterone and dDAVP did not alter the TTP concentration in the cytoplasm, except for a slight, non-significant repression by ∼5–10%. Nevertheless, under hormone action, TTP was associated with translational more active polysomes. This, however, could also result from a change of its binding properties. TTP is a target of the p38-MAPK/MK2 kinase cascade and alters its RNA affinity by phosphorylation (36). Which kinases were activated by the aldosterone and vasopressin signalling pathways remains to be investigated. A first step could be to determine the phosphorylation status of TTP in the context of aldosterone and vasopressin action using antibodies recognizing phosphorylated TTP. In the course of γ-3′-UTR affinity chromatography another ARE RNA-binding factor was identified by MALDI-TOF analysis (Figure 6, gi 14043072). This factor, however, was not investigated in detail, because no antibody or expression plasmid was available. In summary, from RNA-binding studies and reporter gene co-expression experiments it can be concluded that at least FMRP, HuR and TTP contributed to hormone mediated up-regulation of γ-ENaC synthesis by 3′-UTR dependent translational control.
Although polysome gradient analysis unambiguously demonstrated that mechanisms of post-transcriptional control operate in hormonal control of γ-ENaC expression, and in vitro experiments (co-expression of luciferase-γ-ENaC-3′-UTR and RBP plasmids) suggested that an interaction of γ-ENaC 3′-UTR with certain ARE-BPs may be involved in this mechanism, only immunoprecipitation of specific RNP complexes of ARE-BPs with γ-ENaC mRNA is able to prove this interaction in vivo. Following this strategy, we succeeded with HuR as an example demonstrating its crucial role in hormonal stimulation of γ-ENaC expression in vivo (Figure 10). The following facts supported our hypothesis: (i) over-expression of HuR induced γ-ENaC synthesis (10A), (ii) over-expressed HuR associated to γ-ENaC-3′-UTR-ARE in a concentration dependent manner (10B), (iii) a HuR-antibody is able to pull-down γ-ENaC mRNP/polysomes (10C) and (iv) γ-ENaC induction by aldosterone or dDAVP is connected to augmented formation of γ-ENaC–mRNA/HuR complexes proved by immunoprecipitation (10D). A plausible mechanistic explanation for the improved binding of HuR could be based on an increase in HuR concentration by the hormonal stimulus. The mechanism, however, seems to differ between the two hormones, although the final effect, i.e. the induction of γ-ENaC chains, is very similar. As Figures 8 and 9 demonstrate, dDAVP indeed induced HuR, and also its association to more active polysomes was visible. Aldosterone, on the opposite, slightly decreased HuR concentration and the polysome association was not changed compared to the control. However, one has to keep in mind that the polysome sedimentation in sucrose gradients reflects only the binding behaviour of HuR to polysomal complexes of all mRNAs (Figure 8) and not exclusively to specific γ-ENaC mRNA containing polysomes. But it is tempted to speculate that the concentration of γ-ENaC-specific polysomes may have been elevated in contrast to the overall trend. And indeed, pull-down experiments showed that the concentration of HuR containing γ-ENaC polysomes increased (Figure 10D) and determination of specific γ-ENaC mRNA in sucrose gradients demonstrated that both hormones provoked a favoured occupation of γ-ENaC mRNA with ribosomes (Figure 5). Recently, a target motif for HuR binding has been re-evaluated systematically. It constitutes a 17–20-base long motif rich in uracils that does not necessarily contain the pentamer AUUUA and rarely C and G. This motif was found in almost all mRNAs previously reported to be HuR targets and was preferentially located in 3′-UTRs (37). This characteristics fits perfectly to the γ-ENaC-3′-UTR ARE (Figure 3).
HuR has been shown to modulate translation by binding to mRNA 3′-UTRs of numerous polypeptides. This includes examples like tumour suppressor p53 (38), prothymosin-α (39), cytochrome c (40), CAT-1 (cationic amino acid transporter) (41), cyclooxygenase-2 (42), hypoxia inducible factor (HIFα (43) or MKP-1 (mitogen-activated protein kinase phosphatase-1) (44). The detailed mechanism of HuR/3′-UTR-mediated translational control, however, is poorly understood. As shown for γ-ENaC mRNA, in all cases analysed so far HuR promoted recruiting of mRNAs to polysomes. In the case of prothymosin-α UV-stress was associated with HuR mediated mobilization of target mRNA to the cytoplasm and translational activation, demonstrated by shift to heavier polysomes (39). In post-transcriptional control of CAT-1 activation HuR triggered a favoured release of CAT-1 mRNA from cytoplasmic processing bodies and a relief from miRNA directed translational repression (41). MPK-1 translational activation by oxidative stress was also mediated by HuR, which increased the association of MPK-1 mRNA with the translational machinery (44). Another scenario applies to mRNAs to which HuR binds at the 5′-UTR and acts as a translational inhibitor. In IGF-IR (insulin like growth factor receptor) translation control HuR acts at the 5′-UTR as a translational repressor in a cap and internal ribosomal entry sites (IRES) dependent way (45). Also in the case of p27 (CKI, cyclin-dependent kinase inhibitor) HuR interacts with an IRES element thereby inhibiting translation (46).
The regulation of cytoplasmic action of HuR depends on its phosphorylation, which promotes a shuttle from the nucleus to the cytoplasm and its binding activity (42,44,47). Accordingly, two main principles operate in HuR control, first alterations in HuR trafficking and secondly changes in the HuR binding affinity to the target mRNA in the cytoplasm, in our case γ-ENaC mRNA. Despite the diversity of the mechanisms which operate in intracellular transport and phosphorylation they are regulated by some principal signalling pathways. In the context of HuR phosphorylation the following protein kinases have been described: mitogen-activated protein kinases (MAPK), AMP activated kinase (AMPK) and members of the large family of proteinkinase C (PKC) (28). In the present stage of this study, however, it cannot be differentiated if alterations in the cytoplasmic HuR concentration of mCCD cells in the cause of aldosterone or dDAVP action were due to de novo synthesis, alteration of protein stability or to an altered export from the nucleus. Possibly all three principles are involved. Proteomic studies with inner medullary collecting duct (cells) or CCD cells demonstrated that aldosterone and vasopressin signalling cascades are able to alter expression profiles of a large variety of quite different proteins (48), some of them also belonging to the group of RBPs we studied. But also kinases or phosphatases involved in HuR phosphorylation/dephosphorylation may be affected. Factors extensively analysed in ENaC regulation by aldosterone and vasopressin like SGK1 kinase or Nedd4 participate in gene activation and protein degradation (3). How far these and other factors are involved in synthesis, activation and degradation of HuR and the other candidate RBPs influencing γ-ENaC mRNA translation like FMRP and TTP remains to be determined.
FUNDING
The Deutsche Forschungsgemeinschaft (TH459/5-1) to B.J.T and B.N. Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We acknowledge the excellent technical assistance of Jeannette Werner, Regine Stöbe and Ulrike Neumann. Moreover, we are indebted to Grit Nebrich of the group of Joachim Klose (Human Genetics, Charité, Berlin) who performed the MALDI-TOF-MS analysis.
REFERENCES
- 1.Alvarez de la Rosa DA, Canessa CM, Fyfe GK, Zhang P. Structure and regulation of amiloride-sensitive sodium channels. Ann. Rev. Physiol. 2000;62:573–594. doi: 10.1146/annurev.physiol.62.1.573. [DOI] [PubMed] [Google Scholar]
- 2.Rossier BC, Pradervand S, Schild L, Hummler E. Epithelial sodium channel and the control of sodium balance: interaction between genetic and invironmental factors. Ann. Rev. Physiol. 2002;64:877–897. doi: 10.1146/annurev.physiol.64.082101.143243. [DOI] [PubMed] [Google Scholar]
- 3.Snyder PM. The epithelial Na+ Channel: Cell Surface Insertion and Retrieval in Na+ Homeostasis and Hypertension. Endocr. Rev. 2002;23:258–275. doi: 10.1210/edrv.23.2.0458. [DOI] [PubMed] [Google Scholar]
- 4.Weisz OA, Johnson JP. Noncoordinate regulation of ENaC: paradigm lost? Am. J. Physiol. Renal Physiol. 2003;285:F833–F842. doi: 10.1152/ajprenal.00088.2003. [DOI] [PubMed] [Google Scholar]
- 5.Masilamani S, Kim GH, Mitchell C, Wade JB, Knepper MA. Aldosterone-mediated regulation of ENaC α, β, and γ subunit proteins in rat kidney. J. Clin. Invest. 1999;104:R19–R23. doi: 10.1172/JCI7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Escoubet B, Coureau C, Bonvalet JP, Farman N. Non-coordinated regulation of epithelial Na channel and Na pump subunit mRNAs in kidney and colon by aldosterone. Am. J. Physiol. Cell Physiol. 1997;272:C1482–C1491. doi: 10.1152/ajpcell.1997.272.5.C1482. [DOI] [PubMed] [Google Scholar]
- 7.Ecelbarger C, Kim GH, Terris J, Masilamani S, Mitchell C, Reyes I, Verbalis JG, Knepper MA. Vasopressin-mediated regulation of epithelial sodium channel abundance in rat kidney. Am. J. Physiol. Renal Physiol. 2000;279:F46–F53. doi: 10.1152/ajprenal.2000.279.1.F46. [DOI] [PubMed] [Google Scholar]
- 8.Nicco C, Wittner M, DiStefano A, Jounier S, Bankir L, Bouby N. Chronic exposure to vasopressin upregulated ENaC and sodium transport in the rat renal collecting duct and lung. Hypertension. 2001;38:1143–1149. doi: 10.1161/hy1001.092641. [DOI] [PubMed] [Google Scholar]
- 9.Djelidi S, Fay M, Cluzeau F, Escoubet B, Eugene E, Capurro C, Bonvalet JP, Farman N, Blot-Chabaut M. Transcriptional regulation of sodium transport by vasopressin in renal cells. J. Biol. Chem. 1997;272:32919–31924. doi: 10.1074/jbc.272.52.32919. [DOI] [PubMed] [Google Scholar]
- 10.Otulakowski G, Freywald T, Wen Y, O'Brodovich H. Translational activation and repression by distinct elements within the 5′-UTR of ENaC α-subunit mRNA. Am. J. Lung Cell. Mol. Physiol. 2001;281:L1219–L1231. doi: 10.1152/ajplung.2001.281.5.L1219. [DOI] [PubMed] [Google Scholar]
- 11.Richard K, Ramminger SJ, Inglis SK, Olver RE, Land SC, Wilson SM. O2 can raise fetal pneumocyte Na+ conductance without affecting ENaC mRNA abundance. Biochem. Biophys. Res. Comm. 2003;305:671–676. doi: 10.1016/s0006-291x(03)00832-5. [DOI] [PubMed] [Google Scholar]
- 12.Otulakowski G, Rafii B, Bremner HR, O'Brodovich H. Structure and hormone responsiveness of the gene encoding the α-subunit of the rat amiloride-sensitive epithelial sodium channel. Am. J. Respir. Cell. Mol. Biol. 1999;20:1028–1040. doi: 10.1165/ajrcmb.20.5.3382. [DOI] [PubMed] [Google Scholar]
- 13.Auerbach SD, Loftus RW, Itani OA, Thomas CP. Human amiloride-sensitive epithelial Na+ channel γ subunit promoter: functional analysis and identification of a polypurine-polypyrimide tract with the potential for triplex DNA formation. Biochem. J. 2000;347:105–114. [PMC free article] [PubMed] [Google Scholar]
- 14.Kohler S, Pradervand S, Verdumo C, Merillat AM, Bens M, Vanderwalle A, Beermann F, Hummler E. Analysis of the mouse Scnn1a promoter in cortical collecting duct cells and in transgenic mice. Biochim. Biophys. Acta. 2001;1519:106–110. doi: 10.1016/s0167-4781(01)00228-7. [DOI] [PubMed] [Google Scholar]
- 15.Bremner HR, Freywald T, O'Brodovich HM, Otulakowski G. Promoter analysis of the gene encoding the β-subunit of the rat amiloride-sensitive epithelial sodium channel. Am. J. Physiol. Lung Cell. Mol. Physiol. 2002;282:L124–L134. doi: 10.1152/ajplung.2002.282.1.L124. [DOI] [PubMed] [Google Scholar]
- 16.Verrey F, Fakitsas P, Adam G, Staub O. Early transcriptional control of ENaC (de)ubiquitinilation by aldosterone. Kidney Int. 2008;73:691–696. doi: 10.1038/sj.ki.5002737. [DOI] [PubMed] [Google Scholar]
- 17.Otulakowski G, Rafii B, O'Brodovich H. Differential translational efficiency of ENaC subunits during lung development. Am. J. Respir. Cell. Mol. Biol. 2004;30:862–870. doi: 10.1165/rcmb.2003-0381OC. [DOI] [PubMed] [Google Scholar]
- 18.Rotin D, Kanelis V, Schild L. Trafficking and cell surface stability of ENaC. Am. J. Physiol. Renal Physiol. 2001;281:F391–F399. doi: 10.1152/ajprenal.2001.281.3.F391. [DOI] [PubMed] [Google Scholar]
- 19.Otulakowski G, Rafii B, Harris M, O'Brodovich H. Oxygen and glucocorticoids modulate αENaC mRNA translation in fetal distal lung epithelium. Am. J. Respir. Cell. Mol. Biol. 2006;34:204–212. doi: 10.1165/rcmb.2005-0273OC. [DOI] [PubMed] [Google Scholar]
- 20.Otulakowski G, Duan W, Gandhi S, O’Brodovich H. Steroid and oxygen effects on eIF4F complex, mTOR, and ENaC translation in fetal lung epithelia. Am. J. Respir. Cell. Mol. Biol. 2007;37:457–466. doi: 10.1165/rcmb.2007-0055OC. [DOI] [PubMed] [Google Scholar]
- 21.Pesole G, Mignone F, Gissi C, Grillo G, Liciulli F, Liuni S. Structural and functional features of eukaryotic mRNA untranslated regions. Gene. 2001;276:73–81. doi: 10.1016/s0378-1119(01)00674-6. [DOI] [PubMed] [Google Scholar]
- 22.Thiele BJ, Berger M, Huth A, Reimann I, Schwarz K, Thiele H. Tissue-specific regulation of alternative rabbit 15-lipoxygenase mRNAs differing in their 3′-untranslated regions. Nucleic Acids Res. 1999;27:1828–1836. doi: 10.1093/nar/27.8.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 24.Kuersten S, Goodwin EB. The power of the 3'UTR: Translational control and development. Nature Rev. Genetics. 2003;4:626–636. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- 25.de Moor CH, Meijer H, Lissenden S. Mechanisms of translational control by the 3′UTR in development and differentiation. Seminars Cell. Dev. Biol. 2005;16:49–58. doi: 10.1016/j.semcdb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 26.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the message they carry. Nature Rev. Mol. Cell. Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 27.Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol. Chem. 2008;389:243–255. doi: 10.1515/BC.2008.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eberhardt W, Doller A, Akool El-S, Pfeilschifter J. Modulation of mRNA stability as a novel therapeutic approach. Pharmacol. Therap. 2007;114:56–73. doi: 10.1016/j.pharmthera.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 29.Lopez de Silanes I, Quesada MP, Esteller M. Aberreant regulation of messenger RNA 3′-untranslated region in human cancer. Cell. Oncol. 2007;29:1–17. doi: 10.1155/2007/586139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fähling M, Mrowka R, Steege A, Persson PB, Thiele BJ. hnRNP-A2/B1 modulate collagen prolyl 4-hydroxylase alpha (I) mRNA-stability. J. Biol. Chem. 2006;281:9279–9286. doi: 10.1074/jbc.M510925200. [DOI] [PubMed] [Google Scholar]
- 32.Gaeggeler HP, Gonzalez-Rodriguez E, Jaeger NF, Loffing-Cueni D, Norregaard R, Loffing J, Horisberger JD, Rossier BC. Mineralocorticoid versus glucocorticoid receptor occupancy mediating aldosterone-stimulated sodium transport in a novel renal cell line. J. Am. Soc. Nephrol. 2005;16:878–891. doi: 10.1681/ASN.2004121110. [DOI] [PubMed] [Google Scholar]
- 33.Boulkroun S, Ruffieux-Daidié D, Vitagliano JJ, Poirot O, Charles RP, Lagnaz D, Firsov D, Kellenberger S, Staub O. Vasopressin-inducible ubiquitin-specific protease 10 increases ENaC cell surface expression by deubiquitylating and stabilizing sorting nexin 3. Am. J. Physiol. Renal Physiol. 2008;295:F889–F900. doi: 10.1152/ajprenal.00001.2008. [DOI] [PubMed] [Google Scholar]
- 34.Mrowka R, Steege A, Kaps C, Herzel H, Thiele BJ, Persson PB, Blüthgen N. Dissecting the action of an evolutionary conserved non-coding region on renin promoter activity. Nucleic Acids Res. 2007;35:5120–5129. doi: 10.1093/nar/gkm535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skalweit A, Doller A, Huth A, Kähne T, Persson PB, Thiele BJ. Posttranscriptional control of renin synthesis: Identification of proteins interacting with renin mRNA 3′-untranslated region. Circ. Res. 2003;92:419–427. doi: 10.1161/01.RES.0000059300.67152.4E. [DOI] [PubMed] [Google Scholar]
- 36.Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby W.FC, Blackwell TK, Anderson P. MK2-induced tristetraprolin: 14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl Acad. Sci. USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mazan-Mamczarz K, Galban S, Lopez de Silanes I, Martingale JL, Atasoy U, Keene JD, Gorospe M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl Acad. Sci. USA. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lal A, Kawai T, Yang X, Mazan-Mamczarz K, Gorospe M. Antiapoptotic function of RNA-binding protein HuR effected through prothymosin alpha. EMBO J. 2005;24:1852–1862. doi: 10.1038/sj.emboj.7600661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawai T, Lal A, Yang X, Galban S, Mazan-Mamczarz K, Gorospe M. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol Cell Biol. 2006;26:3295–3307. doi: 10.1128/MCB.26.8.3295-3307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;16:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 42.Doller A, Huwiler A, Müller R, Radeke HH, Pfeilschifter J, Eberhardt W. Protein kinase Cα-dependent phosphorylation of the mRNA-stabilizing factor HuR: Implications for posttranscriptional regulation of cyclooxygenase-2. Mol. Biol. Cell. 2007;18:2137–2148. doi: 10.1091/mbc.E06-09-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galbán S, Kuwano Y, Pullmann R., Jr, Martindale JL, Kim HH, Lal A, Abdelmohsen K, Yang X, Dang Y, et al. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol. Cell. Biol. 2008;28:93–107. doi: 10.1128/MCB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuwano Y, Kim HH, Abdelmohsen K, Pullmann R, Martindale JL, Yang X, Gorospe M. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol. Cell. Biol. 2008;28:4562–4575. doi: 10.1128/MCB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meng Z, King PH, Nabors LB, Jackson NL, Chen CY, Emanuel PD, Blume SW. The ELAV RNA-stability factor HuR binds the 5′-untranslated region of the human IGF-IR transcript and differentially represses cap-dependent and IRES-mediated translation. Nucleic Acids Res. 2005;33:2962–2079. doi: 10.1093/nar/gki603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kullmann M, Göpfert U, Siewe B, Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Gen. Dev. 2002;16:3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doller A, Pfeilschifter J, Eberhardt W. Signalling pathways regulating nucleo-cytoplasmic shuttling of the mRNA-binding protein HuR. Cell. Signal. 2008;20:2165–2173. doi: 10.1016/j.cellsig.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 48.Pisitkun T, Bieniek J, Tchapyjnikov D, Wang G, Wu WW, Shen RF, Knepper MA. High-throughput identification of IMCD proteins using LC-MS/MS. Physiol. Genomics. 2006;25:263–276. doi: 10.1152/physiolgenomics.00214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]