Abstract
Plant microRNAs (miRNAs) and small interfering RNAs (siRNAs) bear a 2′-O-methyl group on the 3′-terminal nucleotide. This methyl group is post-synthetically added by the methyltransferase protein HEN1 and protects small RNAs from enzymatic activities that target the 3′-OH. A mutagenesis screen for suppressors of the partial loss-of-function hen1-2 allele in Arabidopsis identified second-site mutations that restore miRNA methylation. These mutations affect two subunits of the DNA-dependent RNA polymerase IV (Pol IV), which is essential for the biogenesis of 24 nt endogenous siRNAs. A mutation in RNA-dependent RNA polymerase 2, another essential gene for the biogenesis of endogenous 24-nt siRNAs, also rescued the defects in miRNA methylation of hen1-2, revealing a previously unsuspected, negative influence of siRNAs on HEN1-mediated miRNA methylation. In addition, our findings imply the existence of a negative modifier of HEN1 activity in the Columbia genetic background.
INTRODUCTION
In Arabidopsis, microRNAs (miRNAs) and small interfering RNAs (siRNAs) represent an average of 15% and 85% of cellular small RNAs, respectively (1–4). A subset of endogenous siRNAs is 21-nt trans-acting siRNAs (ta-siRNAs) derived from non-coding RNAs (5,6). The largest class of endogenous siRNAs representing 84% of the cellular small RNA population is that of 24-nt siRNAs, which tend to be derived from repeat sequences and transposons (1,2,4).
The biogenesis of endogenous 24-nt siRNAs requires RNA-dependent RNA polymerase 2 (RDR2) (7), and also two DNA-dependent RNA polymerases, Pol IV and Pol V (8–12). In Arabidopsis, NRPD1 and NRPE1 encode the largest subunits of Pol IV and Pol V, respectively, while NRPD2/NRPE2 (which we will hereafter refer to as NRPD2) encodes the shared, second largest subunit of the two polymerases. Pol IV is required for the biogenesis of almost all species of 24-nt siRNAs, while Pol V is only required for a subset of siRNAs, usually siRNAs from highly repeated sequences. It has been proposed that Pol IV transcribes all loci that give rise to siRNAs to generate precursors of siRNAs (8,11). Pol V generates non-coding transcripts at silenced loci (13) and is required for siRNA-mediated DNA methylation (9,12). It is thought that the role of Pol V in siRNA biogenesis is indirect such that Pol V-mediated DNA methylation at some loci leads to siRNA production in a feed-forward loop (12).
Plant miRNAs and siRNAs carry a 2′-O-methyl group on the 3′-terminal nucleotide, a modification introduced by the methyltransferase HEN1 (14,15). In plants carrying the severe hen1-1 allele, small RNAs lack methylation, accumulate at a lower level, and become heterogeneous in size due to the presence of one to six additional nucleotides, usually uridines, at the 3′-end of the small RNAs (14,16). siRNAs and piRNAs from animals also carry a 2′-O-methyl group, which is introduced by animal HEN1 homologs (17,18).
Here we report that loss-of-function nrpd1, nrpd2 and rdr2 alleles rescue miRNA methylation defects of hen1-2, a weak hen1 allele, suggesting that siRNAs compete with miRNAs for methylation in Arabidopsis when HEN1 function is compromised. Furthermore, our results with partial loss-of-function hen1 alleles from different ecotypes suggest the presence of a negative modulator of HEN1 activity in the Columbia (Col) genetic background.
MATERIALS AND METHODS
Plant strains
The mutants, rdr2-1 (7), nrpd1-4 (8), nrpe1-11 [formerly nrpd1b-1 (12) and later renamed nrpd1b-11] and hen1-8 are in the Col genetic background. hen1-1 and hen1-2 (19) are in the Ler genetic background.
To obtain the rdr2 hen1-2 double mutant in the Ler genetic background, we first constructed RDR2/rd2-1 HEN1/hen1-2 by crossing hen1-2 (Ler background) with rdr2-1 (Col background). Plants of this genotype were subjected to two rounds of crosses to Ler. In the F1 populations of each round of crosses, the double heterozygous mutants were determined through genotyping of rdr2-1 and hen1-2 (7,19). The three-time backcrossed RDR2/rdr2-1 HEN1/hen1-2 was crossed to hen1-2. In the F1 population, RDR2/rdr2-1 hen1-2/hen1-2 plants were identified through genotyping and allowed to self. In the F2 population, rdr2-1 hen1-2 double mutants were identified.
To construct hen1-1 nrpd1-8 or hen1-1 nrpd2-16, hen1-1 was crossed to hen1-2 nrpd1-8 or hen1-2 nrpd2-16. The desired double mutants were then identified through genotyping of hen1-1 (19) and nrpd1-8 or nrpd2-16 in the F2 populations. The nrpd1-8 mutation was genotyped through digestion of the F16M19-9F (5′-ggcgtttaatgccacaaact-3′)/F16M19-9R (5′-cagacatgttttgtttcgcttt -3′) PCR product with AccI, which could cut the PCR product from nrpd1-8 but not from wild type. For nrpd2-16 genotyping, the NRPD2-mobF (5′-caagagacgctcatgcagatt-3′)/NRPD2-mobR (5′-agccagttgcagacaggcag-3′) PCR product was digested with MoblII. nrpd2-16 resulted in the generation of a MoblII site.
To construct the nrpd1 hen1-8 or nrpe1 hen1-8 double mutant, hen1-8 was crossed to nrpd1-4 or nrpe1-11. The desired double mutants were identified in the F2 populations through genotyping nrpd1-4, nrpe1-11 and hen1-8 (8,12,19).
MAP-based cloning of NRPD1 and NRPD2
hen1-2 suppressors were crossed to hen1-8, which contains the same point mutation in HEN1 as hen1-2 but is in the Col genetic background. In the F2 population, plants with long siliques were collected as the mapping population. Initial mapping showed that the two suppressors were linked to the markers nga280 on chromosome 1 and nga162 on chromosome 3, respectively. New markers in these two regions were developed according to polymorphisms between Ler and Col (https://www.arabidopsis.org/cgi-bin/cereon/cereon_login.pl).
Complementation assay
A ∼10 kb genomic fragment containing the NRPD1 coding and promoter regions was amplified by PCR using primers 5′-gaggtaccttgctgaaatggtgattgaga-3′ and 5′-gaggatcctgggtcatcaagttgtcaaa-3′, and cloned into the pPZP211 binary vector to generate pPZP-NRPD1. Similarly, ∼7.7 kb genomic fragment containing the NRPD2 coding and promoter regions was amplified by PCR using primers 5′-cgggatccgtgtcccattgttgtgcaag-3′ and 5′-cgggatccggagcaaccccaactttgta-3′ and cloned into pPZP211 to generate pPZP-NRPD2. The pPZP-NRPD1 and pPZP-NRPD2 plasmids were transformed into nrpd1-8 hen1-2 and nrpd2-16 hen1-2, respectively. The T1 transgenic plants were selected on medium containing 50 µg ml−1 kanamycin.
RNA and protein analysis
RNA isolation and hybridization for miRNAs and endogenous siRNAs were carried out as described (16). Radioactive signals were detected with a phosphorimager. Sodium periodate treatment and β elimination were done as described (15). Western blotting to determine the levels of HEN1 in Ler and Col was performed with polyclonal anti-HEN1 antibodies generated in the Chen lab. The anti-Hsp73 mouse monoclonal antibody (Stressgen cat# SPA-818) was used to detect Hsp70 proteins from Arabidopsis as a loading control.
Bioinformatic analysis of miRNAs
The rdr2 and wild-type libraries were described previously (Nobuta et al., 2008). miRNAs annotated in miRBase were selected and their normalized abundance (TP2M, transcripts per 2 million) was determined in both libraries. The relative abundance of a miRNA in the total miRNA population was calculated as: individual miRNA count/total miRNA count.
RESULTS
nrpd1 and nrpd2 mutations suppress the hen1-2 fertility defects
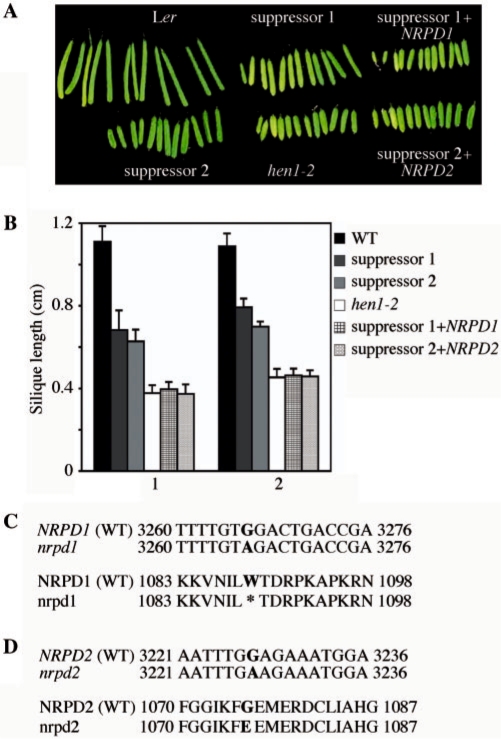
The hen1-2 mutation results in the substitution of an aspartic acid located close to the S-adenosyl methionine-binding site by an asparagine (19). Both in terms of morphological and molecular (i. e. miRNA accumulation) defects, hen1-2 is weaker than hen1-1 (14,19), suggesting that the hen1-2 protein is partially functional. hen1-2 plants have reduced fertility, as reflected by short fruits (siliques, Figure 1A). We carried out an EMS mutagenesis screen in hen1-2 and isolated two suppressors with longer siliques (Figure 1A). The average length of siliques in the two suppressors was increased by 50% compared with that of hen1-2 (Figure 1B). Backcrosses to hen1-2 showed that the two suppressors carry recessive, extragenic mutations. The two suppressors complemented each other, indicating that the two lines carry mutations in two genes. We mapped one suppressor mutation to an approximately 200 kb region of chromosome 1 that contains NRPD1. Sequencing NRPD1 revealed a G-to-A mutation that results in a premature stop codon at amino acid 1089 (Figure 1C). We mapped the second suppressor mutation to an approximately 200 kb region of chromosome 3 that contains NRPD2. Sequencing NRPD2 revealed a G-to-A mutation that results in the conversion of an invariant glycine among subunit II of RNA polymerase II and RNA polymerases IV and V to glutamic acid (Figure 1D). Introduction of NRPD1 and NRPD2 genomic sequences into the corresponding suppressor mutants reversed the fertility phenotype back to that of hen1-2 (Figure 1A and B). Therefore, mutations in NRPD1 and NRPD2 are responsible for the partial rescue of hen1-2 fertility. We named the new nrpd alleles nrpd1-8 and nrpd2-16, respectively. Three Pol IV-dependent siRNAs, siRNA1003, cluster 2 and AtSN1 (8,9,11,12), were absent in the two suppressor lines (Figure 2 and Supplementary Figure S1B), indicating that nrpd1-8 and nrpd2-16 are potentially null alleles.
Figure 1.
A mutation in NRPD1 or NRPD2 partially rescues the fertility defect of hen1-2. (A) The first 12 siliques from plants of the indicated genotypes. Ler, wild type; suppressor 1 and 2 harbor extragenic mutations in the hen1-2 background; suppressor 1+NRPD1, suppressor 1 harboring NRDP1 genomic DNA. suppressor 2+NRPD2, suppressor 2 harboring NRPD2 genomic DNA. (B) Quantification of silique length in various genotypes. 1, average length of the first five siliques; 2, average length of siliques 6–15. Eight plants from each genotype were included in the analysis. (C) Mutation in the NRPD1 gene and protein in suppressor 1. A G-to-A mutation at nucleotide position 3266 of NRPD1 results in a premature a stop codon at amino acid position 1089. (D) Mutation in the NRPD2 gene and protein in suppressor 2. A G-to-A mutation at nucleotide position 3227 of NRPD2 results in a glycine-to-glutamic acid conversion at amino acid position 1076. WT, wild type. The mutant gene and protein sequences are shown below the wild-type sequences.
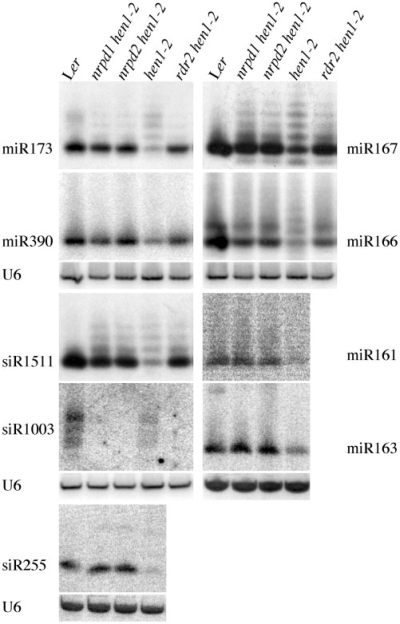
Figure 2.
Loss-of-function nrpd1, nrpd2 or rdr2 mutations result in an increase in miRNA and ta-siRNA levels in hen1-2. Six miRNAs and three endogenous siRNAs in various genotypes were monitored by filter hybridization. U6 was used as a loading control. Ler, wild type.
nrpd1 and nrpd2 mutations increase the levels of miRNAs and ta-siRNAs in hen1-2
As the fertility defect of hen1-2 is caused by the reduced accumulation of small RNAs, probably miRNAs, the nrpd1 and nrpd2 mutations may rescue the hen1-2 fertility defect by increasing the accumulation of miRNAs. We first examined the abundance of six miRNAs in nrpd1-8 hen1-2 and nrpd2-16 hen1-2 by RNA filter hybridization. In the hen1-2 mutant, the abundance of normal-sized miRNAs (i.e. miRNAs of the wild-type size) was greatly reduced as compared to wild type (Figure 2). In addition, uridylated (and therefore larger) species were detectable for miR173, miR167 and miR166 (Figure 2). In the two suppressor lines, the abundance of normal sized miRNAs was increased to a level similar to that of wild type. Furthermore, the proportion of uridylated forms of miR173, miR167 and miR166 was reduced in the two suppressor lines as compared to hen1-2 (Figure 2). The profiles of miR173 species were nearly identical between hen1-2 and the two suppressor lines rescued with NRPD1 or NRPD2 (Supplementary Figure S1A), demonstrating that the loss of NRPD1 and NRPD2 in hen1-2 caused the increased abundance of normal sized miRNAs and the reduced levels of uridylated miRNAs. In addition, we found that the levels of two ta-siRNAs (20), siRNA255, a ta-siRNA from the TAS1 locus and siRNA1511, a ta-siRNA from the TAS2 locus, were increased in the two suppressor lines (Figure 2). Uridylated siRNA1511 species were detected in hen1-2 and the two suppressor lines, but the proportion of uridylated siR1511 was reduced in the two suppressor lines.
nrpd1 and nrpd2 mutations enhance miRNA methylation in hen1-2
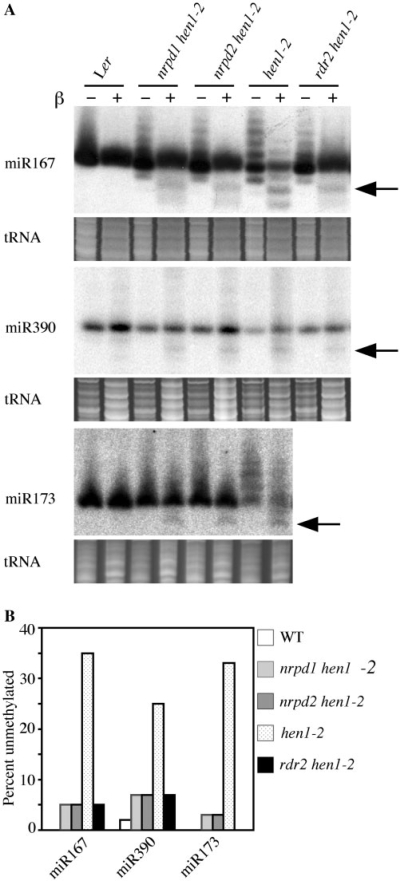
The increased accumulation and decreased uridylation of miRNAs in the two suppressor lines would be best explained by increased miRNA methylation because methylation protects miRNAs from degradation and uridylation (14,15). Therefore, we examined whether the nrpd1 or nrpd2 mutation in hen1-2 enhanced miRNA methylation using the periodate/β-elimination assay (15; Figure 3). Loss of methylation would result in faster migration of the RNA in this assay. After the chemical treatment, a band that migrated ∼2 nt faster than the normal sized miRNAs was detected (Figure 3A, arrow) in hen1-2, nrpd1 hen1-2, nrpd2 hen1-2 but not in wild type. The band represents the portion of the normal sized miRNAs that was unmethylated. We quantified the amount of the unmethylated miRNAs and calculated the proportion of the unmethylated miRNAs among total miRNAs of normal size. There was a clear reduction in the proportion of unmethylated miRNAs in hen1-2 nrpd1 and hen1-2 nrpd2 as compared to hen1-2 (Figure 3B), demonstrating that the nrpd mutations enhance miRNA methylation in hen1-2. Another formally possible explanation for the elevated miRNA accumulation in the two suppressor lines is increased transcription of the MIR genes. Real-time RT–PCR showed that the levels of pri-miR173 were similar in hen1-2, nrpd1 hen1-2 and nrpd2 hen1-2. Hence, it is unlikely that the elevated miRNA levels in the two suppressor lines resulted from increased transcription of MIR genes.
Figure 3.
Increased methylation of miRNAs in nrpd1-8 hen1-2, nrpd2-16 hen1-2 and rdr2-1 hen1-2. (A) Total RNAs treated (+) or not (−) with sodium periodate followed by β-elimination were separated on a 15% acrylamide gel and probed for various miRNAs by filter hybridization. Unmethylated miRNAs migrate ∼2 nt faster after the chemical treatment, while methylated miRNAs do not change mobility. The arrow in each panel marks the expected position where a normal sized and unmethylated miRNA would migrate after the chemical treatments. The ethidium-bromide stained gel in the region of tRNAs was shown below the corresponding miRNA blot to indicate the amount of RNAs used. (B) The proportion of unmethylated miRNAs within the population of normal sized miRNA species in each genotype. The numbers were derived from quantification of the intensity of the marked bands (unmethylated miRNAs) and the intensity of the major species in the ‘−’ lanes (total miRNAs of normal size) in Figure 3A and calculation of the ratio of the two intensities. Ler, wild type.
nrpd1 and nrpd2 mutations do not rescue miRNA defects of hen1-1
How do nrpd1 and nrpd2 mutations increase the methylation of miRNAs in hen1-2? One possibility is that Pol IV is required for the uridylation activity. If uridylation and methylation are competitive processes, reduced uridylation could lead to increased methylation. The second possibility is that Pol IV directly or indirectly inhibits HEN1-mediated miRNA methylation. Loss of Pol IV results in increased miRNA methylation, which protects miRNAs from uridylation and degradation.
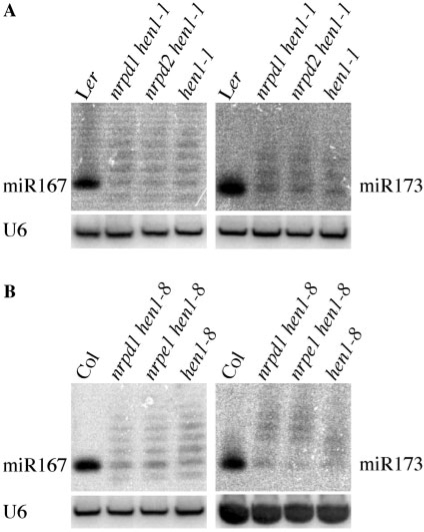
To distinguish these two possibilities, we evaluated the effect of the nrpd1 and nrpd2 mutations in hen1-1, a severe allele that leads to complete loss of miRNA methylation. The nrpd1-8 hen1-1 and nrpd2-16 hen1-1 double mutants appeared morphologically indistinguishable from hen1-1 plants. At the molecular level, nrpd1 and nrpd2 mutations were unable to rescue the miRNA defects of hen1-1 (Figure 4A). The levels of normal sized miRNAs as well as the uridylated miRNAs were similar in nrpd1-8 hen1-1, nrpd2-16 hen1-1 and hen1-1.
Figure 4.
The accumulation of miRNAs in hen1-1 or hen1-8 is not changed by loss-of-function of Pol IV subunits. (A) The levels of miRNAs in nrpd1-8 hen1-1 or nrpd2-16 hen1-1. Ler, wild type. (B) The levels of miRNAs in nrpd1-4 hen1-8 or nrpe1-11 hen1-8. U6 blots are loading controls.
The presence of uridylated miRNAs in nrpd1 (or 2) hen1-1 plants argues against a role of Pol IV in uridylating unmethylated miRNAs. The fact that Pol IV mutations lead to increased miRNA accumulation in hen1-2 but not hen1-1 suggests that Pol IV mutations suppress hen1-2 by allowing the hen1-2 protein to better methylate miRNAs.
The rdr2-1 mutation acts similarly to nrpd1 and nrpd2 mutations in hen1-2
How does Pol IV negatively impact miRNA methylation? One possibility is that Pol IV inhibits HEN1-mediated miRNA methylation by promoting the production of 24-nt siRNAs, which represent 84% of the cellular small RNA population and compete with the remaining small RNAs (miRNAs and tasiRNAs) for methylation by HEN1. If this were true, mutations in other genes essential for endogenous siRNA biogenesis would also result in increased abundance and methylation of miRNAs in hen1-2. We examined the abundance and methylation of miRNAs in rdr2-1 hen1-2, in which the Col rdr2-1 allele was introgressed into Ler by four backcrosses. Indeed, introducing rdr2-1 into hen1-2 increased the abundance of normal-sized miRNAs and reduced the proportion of uridylated miRNAs (Figure 2). In addition, like nrpd1 (or 2) hen1-2, the methylation of miRNAs was enhanced in rdr2-1 hen1-2 (Figure 3).
nrpd1 and nrpe1 mutations do not suppress hen1-8 defects
The fact that mutations in Pol IV suppress the hen1-2 defects prompted us to test whether mutations in Pol V might also suppress hen1-2. Because nrpe1 alleles are only available in the Col ecotype, we took advantage of the Col-derived hen1-8 allele (isolated from an independent genetic screen) that carries an identical molecular lesion as hen1-2 to address our question. Like hen1-2 but in contrast to the strong hen1-4 allele in Col (21), hen1-8 behaves as a weak hen1 allele in terms of both its ability to suppress sense transgene post-transcriptional gene silencing (PTGS) and its responses to viral infection (Supplementary Figure S2B and C).
We crossed nrpe1-11 [in the Col ecotype; formerly known as nrpd1b-1 (12)] to hen1-8. We also crossed nrpd1-4 (8) [formerly known as nrpd1a-4, also known as nrpd1a-1 (12)] to hen1-8 as a positive control. nrpe1-11 was unable to suppress the fertility defects of hen1-8. Surprisingly, nrpd1-4 was also unable to suppress the fertility defects of hen1-8. In addition, filter hybridization showed that the levels of normal-sized and uridylated miRNAs were the same in nrpd1-4 hen1-8, nrpe1-11 hen1-8 and hen1-8 (Figure 4B), indicating that loss-of-function of Pol IV or Pol V did not rescue the defects of hen1-8 in the Col background. This is in contrast to the fact that the loss of function of Pol IV suppresses hen1-2 in the Ler background. It is unknown whether loss of function of Pol V would suppress hen1-2 in the Ler background. The nrpe1-11 mutation in the Col background needs to be introgressed into Ler to test this.
DISCUSSION
In this study, we found that nrpd1 and nrpd2 mutations result in increased miRNA methylation in the hen1-2 background and partially rescue the hen1-2 fertility defects. The partial rescue of miRNA defects of hen1-2 by a mutation in RDR2, which is also required for endogenous 24-nt siRNA biogenesis, strongly supports the conclusion that endogenous siRNAs compete with miRNAs for methylation by the partially defective hen1-2 protein.
Such a competition between siRNAs and miRNAs may also occur when HEN1 activity is not compromised, albeit at a smaller scale. The levels of a number of miRNAs are unchanged in nrpd1 or nrpd2 mutants (8,9,11,12), suggesting that HEN1 activity is not limiting for these miRNAs. However, northern blots showed that the levels of nine miRNAs, including miR771, miR772 and others, are increased in the rdr2-1 mutant and a dcl2 dcl3 dcl4 triple mutant that lacks most endogenous siRNAs (2). Our analysis of high-throughput sequencing data on miRNAs from wild type and rdr2-1 (22) confirms that rare miRNAs including those detected by Lu et al. are increased in relative abundance among total miRNAs in rdr2-1 as compared to wild type (Supplementary Table S1). Variation among miRNA levels has also been demonstrated in sequence-based comparisons of the maize mop1-1 mutant to wild type (mop1 is the maize ortholog of the Arabidopsis RDR2; 22). One possible explanation is that without competition from siRNAs, these miRNAs can be more efficiently methylated and accumulate to higher levels. However, other causes for the increased accumulation of these miRNAs are also possible. Although not all miRNAs are affected (in terms of their abundance) by the competition from siRNAs under normal conditions, it is intriguing to speculate that such a competition for HEN1 activity between siRNAs and miRNAs could be augmented in certain cell types or under circumstances where a burst of small RNA synthesis occurs (such as under viral infection).
In the course of our studies, we unexpectedly discovered that loss-of-function mutations in NRPD1 suppress the Landsberg hen1-2 allele but not hen1-8, a Col allele that carries the same molecular lesion as hen1-2. This is not due to differences in the strength of the nrpd1 alleles since they result in the absence of siRNAs in both Ler (Figure 1) and Col (8,12). The most likely reason that nrpd1 rescues hen1-2 but not hen1-8 is that HEN1 has a stronger activity in Ler than in Col. In fact, hen1-8 exhibits more severe fertility defects than hen1-2. At the molecular level, hen1-8 exhibits similar levels of miRNA impairment as hen1-4 and hen1-5, two strong hen1 mutant alleles in the Col genetic background (21,23; Supplementary Figure S2A). While normal-sized miR167 accounts for the majority of miR167 species in hen1-2 (Figures 2 and 3), no apparent enrichment for normal-sized miR167 is found in hen1-8 (Figure 4B). The stronger methylation activity of hen1-2 relative to hen1-8 may be due to intrinsic differences between the Ler and Col HEN1 proteins, or due to the presence of a negative modulator of HEN1 expression or activity in Col. Western blotting showed that the levels of HEN1 were similar in Ler and Col inflorescences (Supplementary Figure S3). The HEN1 protein in Ler differs from that in Col by a single amino acid outside the methyltransferase domain (19). Although we cannot rule out that this single amino acid difference contributes to differences in HEN1 activity, genetic mapping pinpointed a locus on chromosome 1 that underlies the differences in phenotypic severity between hen1-2 and hen1-8 (Bin Yu, unpublished results). Since HEN1 resides on chromosome 4, it is likely that another gene modulates the activity of HEN1. Arabidopsis accessions exhibit natural variations in many processes, including flowering time, light response, lipid metabolism, and hormone responses (24). Our data indicate that natural genetic variation also modulates the biogenesis of small RNAs. Identification of this negative regulator of HEN1 will help elucidate the mechanisms controlling small RNA methylation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (GM61146 to X.C.); National Science Foundation (MCB-0718029 to X.C.); National Research Initiative of the USDA cooperative State Research, Education, and Extension Service (2007-35319-18325 to S-W.D.); National Science Foundation (DBI-0701745 to B.C.M.); Agence Nationale de la Recherche (ANR-06-BLAN-0203-02 to H.V.). Funding for open access charge: National Science Foundation (MCB-0718029).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Thierry Lagrange for nrpd1-4 nrpe1-11 seeds, David Bartel for helpful suggestions, and Theresa Dinh, Allison Mallory and Binglian Zheng for critical reading of the article.
REFERENCES
- 1.Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu C, Kulkarni K, Souret FF, MuthuValliappan R, Tej SS, Poethig RS, Henderson IR, Jacobsen SE, Wang W, Green PJ, et al. MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 2006;16:1276–1288. doi: 10.1101/gr.5530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE. Role of RNA polymerase IV in plant small RNA metabolism. Proc. Natl Acad. Sci. USA. 2007;104:4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 7.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 9.Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat. Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- 10.Mosher RA, Schwach F, Studholme D, Baulcombe DC. Pol IVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc. Natl Acad. Sci. USA. 2008;105:3145–3150. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 12.Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 18.Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Liu J, Cheng Y, Jia D. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development. 2002;129:1085–1094. doi: 10.1242/dev.129.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Boutet S, Vazquez F, Liu J, Beclin C, Fagard M, Gratias A, Morel JB, Crete P, Chen X, Vaucheret H. Arabidopsis HEN1: a genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 2003;13:843–848. doi: 10.1016/s0960-9822(03)00293-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nobuta K, Lu C, Shrivastava R, Pillay M, De Paoli E, Accerbi M, Arteaga-Vazquez M, Sidorenko L, Jeong DH, Yen Y, et al. Distinct size distribution of endogeneous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant. Proc. Natl Acad. Sci. USA. 2008;105:14958–14963. doi: 10.1073/pnas.0808066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vazquez F, Gasciolli V, Crete P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 2004;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 24.Holub EB. Natural variation in innate immunity of a pioneer species. Curr. Opin. Plant Biol. 2007;10:415–424. doi: 10.1016/j.pbi.2007.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.