Abstract
We have previously shown that the Leishmania genome possess two widespread families of extinct retroposons termed Short Interspersed DEgenerated Retroposons (SIDER1/2) that play a role in post-transcriptional regulation. Moreover, we have demonstrated that SIDER2 retroposons promote mRNA degradation. Here we provide new insights into the mechanism by which unstable Leishmania mRNAs harboring a SIDER2 retroposon in their 3′-untranslated region are degraded. We show that, unlike most eukaryotic transcripts, SIDER2-bearing mRNAs do not undergo poly(A) tail shortening prior to rapid turnover, but instead, they are targeted for degradation by a site-specific endonucleolytic cleavage. The main cleavage site was mapped in two randomly selected SIDER2-containing mRNAs in vivo between an AU dinucleotide at the 5′-end of the second 79-nt signature (signature II), which represents the most conserved sequence amongst SIDER2 retroposons. Deletion of signature II abolished endonucleolytic cleavage and deadenylation-independent decay and increased mRNA stability. Interestingly, we show that overexpression of SIDER2 anti-sense RNA can increase sense transcript abundance and stability, and that complementarity to the cleavage region is required for protecting SIDER2-containing transcripts from degradation. These results establish a new paradigm for how unstable mRNAs are degraded in Leishmania and could serve as the basis for a better understanding of mRNA decay pathways in general.
INTRODUCTION
Leishmania are unicellular protozoan parasites that cause a broad range of human diseases known as leishmaniases. Apart from being an important human pathogen, Leishmania is an evolutionary early branching unicellular eukaryote that displays unique features regarding genome organization and control of gene expression. In contrast to other eukaryotes, its ∼8300 genes are organized as large directional polycistronic clusters on the same DNA strand, comprising up to 100 functionally unrelated genes (1). Polycistronic transcription is initiated upstream of each cluster at the strand-switch region by a RNA polymerase II (pol II) in the absence of typical pol II promoter sequences and basal transcription factors (2). Individual mRNAs are produced from polycistronic pre-mRNAs by two co-transcriptional RNA-processing reactions, namely trans-splicing, which adds the spliced leader RNA (SL RNA) to the 5′-terminus of protein-encoding RNAs and 3′-cleavage/polyadenylation [reviewed in (3)]. Thus, regulation of gene expression in Leishmania and related kinetoplastid protozoa occurs almost exclusively at the post-transcriptional level, and in most cases, sequences within 3′-untranslated regions (3′-UTRs) have been shown to play a key role in controlling either the stability of mRNAs or the efficiency of their translation [reviewed in (4,5)].
Regulated mRNA turnover is a highly important process in the control of gene expression. For most eukaryotic mRNAs decay begins with a shortening of the poly(A) tail by a variety of mRNA deadenylases. Deadenylation can activate removal of the cap of transcripts by a protein complex composed of the decapping enzymes DCP1/2 and the RNA helicase DHH1 facilitating a processive degradation of the mRNA body by 5′–3′ exonucleases, including XRN1) (6). Alternatively, deadenylated mRNAs can then be degraded from the 3′-end by a large protein complex containing mainly 3′–5′ exonuclease activities, known as the exosome (7). In contrast to these exonuclease-based pathways, a small number of mRNAs are targeted for decay via endonucleolytic digestion or other specialized quality-control pathways (8). Although most homologs of eukaryotic mRNA decay enzymes have been identified in Leishmania and the related Trypanosoma species [reviewed in (4,5)], little is known however, about the mechanisms controlling mRNA degradation in these parasites. Decapping activity was detected in extracts of the trypanosome L. seymouri (9), but no homologs of the eukaryotic mRNA-decapping proteins DCP1 and DCP2 have been identified yet in any of the trypanosomatids (4,10). In T. brucei, it was shown that knockdown of the major mammalian deadenylase homolog CAF1 significantly delayed deadenylation and degradation of constitutively expressed mRNAs, while the levels of unstable transcripts remained unchanged (11). In contrast, XRNA, a putative XRN1-related 5′–3′ exonuclease in trypanosomes, was shown to play an important role in the degradation of highly unstable, developmentally regulated mRNAs (12). The exosome complex has been characterized in T. brucei and L. tarentolae and it resembles both the yeast and human exosomes in overall structure, although it appears to be of lesser complexity (13–15). In T. brucei, the exosome seems to play a minor role in mRNA degradation because RRP45 depletion, which probably disrupts exosome integrity, did not affect degradation of many stable RNA species (14,16). Destabilizing AU-rich (AREs) or U-rich (UREs) elements have been associated with rapid turnover of developmentally regulated transcripts in trypanosomes (4) and Leishmania (17).
We have recently reported the presence of small degenerated retroposons termed Short Interspersed DEgenerated Retroposons (SIDERs) in the 3′-UTRs of a large number of Leishmania mRNAs (18). The ∼2000 SIDER elements identified in Leishmania are truncated versions (∼0.55 kb) of formerly active retroposons of the ingi/L1Tc-related family that became extinct a long time ago (19). SIDERs constitute the largest family of transposable elements described so far in trypanosomatid genomes, they are ∼70 times more abundant in Leishmania compared to Trypanosoma species and can be divided into two phylogenetically distinct subfamilies, namely SIDER1 and SIDER2. The vast majority (95.4%) of SIDER elements are located in intergenic regions (IRs) between protein-coding genes and >75% are part of in silico-predicted 3′-UTRs (18,20,21). Given their privileged location in 3′-UTRs, SIDERs could act as cis-acting regulatory elements. We hypothesize that the considerable expansion of SIDERs within 3′-UTRs and their high-sequence diversity supply Leishmania with novel genetic material that contributes to diversifying regulatory functions and consequently help the parasite to gain an evolutionary edge. Interestingly, members of the SIDER1 subfamily, previously referred to as the 450-nt conserved element, were shown to play a role in translation regulation (22,23). More recent studies demonstrated that members of the SIDER2 subfamily promote mRNA destabilization (18). Several SIDER2-bearing mRNAs in L. major are expressed at lower levels compared to transcripts lacking SIDER2 and are short lived due to a rapid turnover conferred by sequences within SIDER2 (18).
This article provides a step forward towards our understanding of the mechanism underlying the rapid turnover of unstable SIDER2-containing mRNAs in Leishmania. We provide several lines of evidence that degradation of SIDER2-containing mRNAs is initiated by site-specific endonucleolytic cleavage within the second 79-nt signature sequence of SIDER2 without prior shortening of the poly(A) tail. Our data uncover a novel mechanism of mRNA decay in parasitic protozoa that targets short-lived mRNAs sharing a conserved retroposon signature sequence in their 3′-UTR.
MATERIALS AND METHODS
Leishmania culture
The L. major LV39 strain used in this experiment was described previously (24). Promastigotes were cultured at pH 7.0 and 25°C in SDM-79 medium supplemented with 10% heat-inactivated FCS (Wisent) and 5 µg/ml hemin.
Plasmid constructions and transfections
The parent plasmid pSPYNEOαLUC was described previously (22). The plasmids LUC-3′-UTR1270 and LUC-3′-UTR3810 expressing the luciferase (LUC) reporter gene under the control of the full-length 3′-UTR of LmjF08.1270 and LmjF36.3810, respectively, the plasmids LUC-ΔSIDER1270 and LUC-ΔSIDER3810 lacking the whole SIDER2 element, and the plasmids LUC-SIDER1270 and LUC-SIDER3810 with the SIDER2 element alone cloned downstream of the LUC gene were described elsewhere (18). All chimeric LUC deletion constructs lacking either the T-stretch, the first (I) or the second (II) 79-nt signature sequence or both signatures (I+II) from SIDER1270 and SIDER3810 or an 80-nt region from the end of SIDER3810 (Supplementary Table S1) were generated as follows. The 3′-UTR sequences were amplified by PCR as two separate halves leaving out the sequences to be deleted (size of the partial 3′-UTR sequences is indicated in Supplementary Table S1) using primers with inserted BamHI and PstI restriction sites. All obtained PCR products were first cloned into the pCR2.1 vector (Invitrogen) and then digested with BamHI and PstI (New England Biolabs). The two corresponding BamHI-PstI fragments were then subcloned into the BamHI site downstream of the LUC gene in vector pSPYNEOαLUC, thereby fusing the two partial sequences at the PstI site. The 435-nt-long IR of LmjF36.3810 and the SIDER2 lacking the T-rich stretch (SIDER-3810ΔT) were amplified by PCR using Taq DNA polymerase (Qiagen) and primers with inserted BamHI restriction sites (Supplementary Table S1). To engineer plasmids expressing either the full-length sense (SIDER1270s and SIDER3810s) or anti-sense SIDER2 elements [(SIDER1270as, SIDER3810as, SIDER1270(HYG)as, SIDER3810(HYG)as or SIDER3810(HYG)Δ79IIas lacking the second signature sequence (Supplementary Table S2)], SIDER2 sequences from LmjF36.3810 and LmjF08.1270 were PCR-amplified with primers containing BamHI restriction sites (Supplementary Table S1) and cloned in either orientation into the expression vector pSPαNEOα (25) or only in the anti-sense orientation into vector pSPYHYGα (C. Dumas unpublished data). The ORFmyc-3′-UTR3810 and ORFmyc-3′-UTR1270 constructs are described elsewhere (M. Müller et al., in revision). All constructs have been verified by sequencing. Purified plasmid DNA (10–20 µg, Qiagen Plasmid Mini Prep Kit) was transfected into Leishmania by electroporation as described previously (25). Stable transfectants were selected and subsequently cultivated with 0.04 mg/ml G-418 (Sigma) or 0.32 mg/ml of hygromycin B (Sigma). Stable transfectants were plated on SDM-79 (2X) medium with 1.5% agar and the appropriate drug selection and individual clones were obtained after 2 weeks. The plasmid copy number in each transfectant (Supplementary Table S2) was evaluated by Southern blot hybridization using the single copy BT1 (biopterin transporter 1) gene as a probe for normalization and quantification with the ImageQuant 5.2 software. The chimeric LUC mRNAs and the anti-sense SIDER2 transcripts in all the above vectors are processed at the 5′-end by sequences in the Leishmania α-tubulin IR (αIR). Correct processing at the 3′-end of the chimeric LUC reporter transcripts used in this study (Supplementary Table S2) was confirmed by 3′-RACE (rapid amplification of cDNA ends) using an oligo(dT) primer and specific primers complementary to the 3′-UTR sequences of transcripts 1270 and 3810. Amplified RT-PCR fragments were cloned into vector pCR2.1 and sequenced to map the poly(A)-addition sites.
Luciferase assays
The LUC activity of the recombinant parasites was determined as described previously (23). Briefly, 4 × 107- and 2 × 107-mid-log phase L. major promastigotes were harvested by centrifugation and the pellet was resuspended in 5× cell lysis buffer (Promega, http://www.promega.com) and frozen at −80°C to complete cell lysis. Twenty microlitres of each thawed lysate were then mixed with 100 µl luciferase assay buffer (Promega) containing D-luciferin potassium salt, and LUC activity was measured in 96-well plates using a Dynex MLX luminometer (Dynex MLX, http://www.dynextechnologies.com).
DNA, RNA and protein manipulations
Genomic DNA and total RNA of L. major promastigotes were extracted using the DNAzol and TRIzol reagents (Invitrogen), respectively, following the manufacturer’s instructions. Southern and northern blot hybridizations were performed following standard procedures. Double-stranded DNA probes were radioactively labeled with [α-32P]dCTP using random oligonucleotides and the Klenow enzyme (New England Biolabs). Strand-specific RNA probes (riboprobes) were prepared using the MAXIscript Kit (Ambion) including T7 polymerase, [α-32P]UTP and template DNA cloned into vector pCR2.1 (Invitrogen) downstream of a T7 promoter site. To prepare soluble protein lysates for western blots, L. major promastigotes were harvested by centrifugation, washed with ice-cold HEPES–NaCl buffer and the pellet was re-suspended in 1X Laemmli buffer. Proteins were quantified using Amido Black 10B (Bio-Rad), and 40 µg of total protein extracts were loaded onto 10% SDS–PAGE gels. After electrophoresis, gels were transferred onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore), and membranes were reacted with a goat-anti-LUC primary antibody (Promega) followed by a donkey-anti-goat secondary antibody (Santa Cruz Biotechnology). Protein loading was monitored by reincubating the membranes with a mouse anti-α-tubulin antibody (Sigma). All incubations were performed with a 1 : 10 000 dilution of the antibody in 5% milk. The immunoblot was visualized by chemiluminescence with an ECL+ Detection Kit (GE Healthcare). RNA and protein levels were estimated by densitometric analyses using the ImageQuant 5.2 software.
RNA stability assays
To determine the half-lives of SIDER2-containing transcripts, mid-log phase L. major promastigotes were incubated with 10 µg/ml of actinomycin D (ActD; Sigma) at different time points to arrest de novo transcription. All time points include 5 min of centrifugation times. Total RNA was extracted from each sample using Trizol reagent (Invitrogen) and subjected to northern blot hybridization. Quantitation of the different transcript intensities was done by densitometric analysis using the ImageQuant 5.2 software.
Deadenylation assay
For visualizing changes in the length of the poly(A) tail, 40 µg of total RNA from ActD-treated parasites were subjected to digestion with RNase H (Invitrogen), as described previously (17). Briefly, RNase H digestions were carried out in the presence of a specific DNA oligonucleotide complementary to a sequence within the 3′-UTR (for 3810, 5′-TGCAAAACACAGAGAGTTCCCAAAG-3′ and for 1270, 5′-GTGCGCCGTGTGAGGGCCCGGTCGG) located exactly at 300 nt from the 3′-end of the transcript with or without oligo(dT) (Invitrogen) and in the presence of an RNase inhibitor (RNaseOUT, Invitrogen). RNA products were then separated on 5% denaturing polyacrylamide gels (Sequagel, National Diagnostics) and transferred onto nylon membranes (Hybond XL, Amersham). Blots were probed with purified PCR fragments corresponding to the last 300 nt of transcripts 3810 and 1270.
RNase protection assay
Specific in vivo endonucleolytic cleavage products derived from the chimeric LUC reporter mRNAs in selected LUC transfectants were visualized by RNase protection assays (RPAs). Total RNA was isolated from L. major transfectants using Trizol reagent and was subsequently treated with RNase-free Turbo DNase (1 U/µg RNA; Ambion) for 1 h at 37°C to remove DNA contaminations. To generate riboprobes for detection of in vivo cleavage products, DNA fragments corresponding to the entire or partial sense SIDER1270 from gene LmjF08.1270 or SIDER3810 from gene LmjF36.3810 were either cloned into the pCR2.1 vector (Invitrogen) in reverse orientation upstream of a T7 promoter or amplified by PCR with primers containing a T7 promoter sequence. Anti-sense riboprobes were in vitro-transcribed and radiolabeled using the MAXIscript Kit (Ambion) with [α-32P]UTP and 500 ng purified DNA. The 480-nt SIDER1270 riboprobe contains the first 460 nt of SIDER1270 plus another 20 nt upstream of SIDER1270. The 600-nt SIDER1270 riboprobe contains the first 480 nt of SIDER1270 including the 20-nt upstream and an additional 127 nt of non-complementary sequence originating from the pCR2.1 vector (68 nt at the 5′-end and 59 nt at the 3′-end of the probe). The 400-nt SIDER3810 riboprobe contains the first 400 nt of SIDER3810. All these riboprobes were generated by PCR. Plasmids were linearized by digestion with HindIII and purified by gel extraction (Qiagen). Radiolabeled RNAs were gel-purified and then examined for RNase protection using the RPAIII kit (Ambion) following the manufacturer’s instructions. Briefly, radioactive riboprobes were mixed with total Leishmania RNA and incubated overnight at 45°C to allow annealing. Samples were subsequently digested with a mix of the RNases A/T1 to remove all single-stranded (ss) RNA sequences. Protected double-stranded RNA products were resolved on an 8% polyacrylamide urea gel and visualized by autoradiography. The RPA assays have been optimized according the manufacturer’s instructions to minimize the possibility of RNase A/T1 overdigestion. For the detection of anti-sense SIDER2 expression, the corresponding sense sequences were subjected to in vitro transcription, labeling and RNase protection. A radiolabeled control reaction was performed in parallel to confirm the size and integrity of the RNA on a 6% acrylamide gel.
Primer extension
Primer extension was used to confirm endonucleolytic cleavage and to map more precisely the putative cleavage sites within the SIDER2 signature II sequence. The size of the generated cDNA fragments reflects the number of nucleotides between the labeled primer and the 5′-end of the cleaved RNA, when the reverse transcriptase enzyme falls of its template. Different reverse primers of 20–40-nt length were designed complementary to a region 100–200-nt downstream of the predicted cleavage sites (Supplementary Table S1). Primers were labeled with [γ-32P]ATP following the polynucleotide kinase protocol (PNK; New England Biolabs). Total RNA was isolated from L. major LUC-transfectants and quantified with the bioanalyzer chip (Agilent Technologies). The 10 pmol of labeled primer and 30–70 µg of total RNA were used in all experiments. Primer extension reactions were carried out using the SuperScript™ III RT kit (Invitrogen) following the manufacturer’s recommendations. A X174 DNA/HinfI dephosphorylated DNA marker (Promega) was labeled with [γ-32P]ATP and PNK (New England Biolabs) according to the manufacturer’s recommendations and was used as a size marker. In addition, we used a dideoxy-sequencing to precisely estimate the cleavage sites at the nucleotide level. The ladder was generated using the sequenase 2.0 kit (USB) with 1 pmol of primer P1-3810 and 5 µg of ss plasmid DNA containing the SIDER3810 sequence. Primer extension fragments and markers were separated on 8% denaturating acrylamide gels (Sequagel, National Diagnostics) and visualized by autoradiography.
Reverse ligation-mediated PCR
Sixty micrograms of total RNA, isolated with the Trizol reagent and purified using the RNeasy clean-up kit (Qiagen) were subjected to DNase treatment with 40 U Turbo DNase I (Ambion) and 240 U of RNaseOUT (Invitrogen) for 60 min at 37°C. The reaction was terminated by phenol–chloroform extraction followed by ethanol precipitation. Circularization of ∼40 and ∼20 µg DNA-free RNA was performed overnight at 16°C in a reaction volume of 400 µl with 40 units of T4 ssRNA ligase (New England Biolabs) and 80 units of RNaseOUT as described (26). The reactions were terminated by phenol–chloroform extraction and ethanol precipitation. Reverse transcription was carried out with 50 pmol of primer P5-1270-RL and ∼10 µg RNA using the Superscript™ III RT kit following the manufacturer’s recommendations. PCR amplifications were performed during 30 cycles with 3 µl cDNA, 50 pmol of primers P5-1270-RL and P2-1270-RL using a Taq Polymerase PCR kit (Qiagen) in a 50-µl reaction volume. For nested PCRs, 2 µl PCR products were re-amplified in a 50-µl reaction during 30 cycles with 50 pmol for each of primers P6-1270-RL and P2-1270-RL. PCR products in the expected size range were gel-excised, purified and cloned into the TOPO® TA-cloning vector (Invitrogen). Twenty clones were subsequently chosen for sequencing (10 clones for each a and b; Supplementary Figure S4).
RESULTS
The highly conserved 79-nt signature II sequence of SIDER2 retroposons is essential for degradation of SIDER2-bearing mRNAs
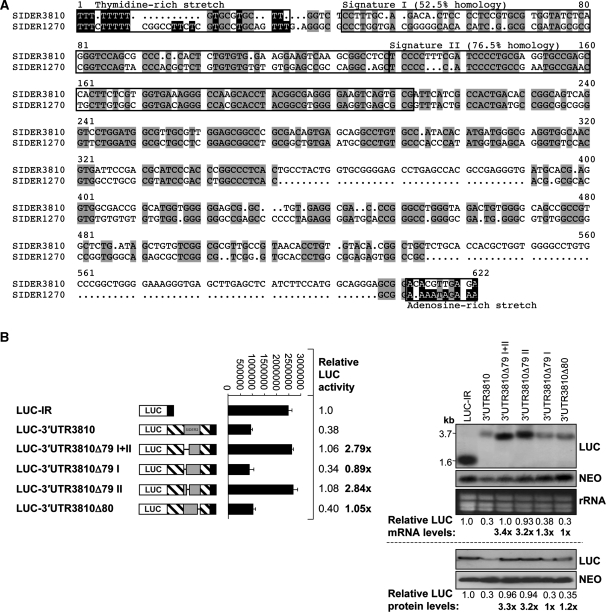
We have previously demonstrated that members of SIDER2 retroposons in L. major promote mRNA degradation and that transcripts containing a SIDER2 element in their 3′-UTR are often low abundant and short-lived (18). More than 1200 SIDER2 retroposons have been identified in L. major (18), but similar numbers were also found in L. infantum and L. braziliensis (21). Generally, SIDER2 retroposons share three conserved motifs. These include an ∼18-nt thymidine-rich stretch at the 5′-end corresponding to the former recognition site for an endonuclease encoded by the autonomous DIRE elements followed by two tandemly arranged boxes of 79 nt each (signatures I and II) representing the hallmark of trypanosomatid retroposons and by an adenosine-rich stretch at the 3′-end (18,19). Sequence alignment of SIDER3810 and SIDER1270 confirmed the presence of a T-rich stretch and the two 79-nt signatures (Figure 1A).
Figure 1.
The second highly conserved 79-nt signature sequence amongst SIDER2 retroposons is essential for mRNA degradation. (A) Sequence alignment of two selected SIDER2 sequences present in the 3′-UTRs of L. major LmjF36.3810 (3810) and LmjF08.1270 (1270) transcripts using the multiple sequence alignment program multialin (http://bioinfo.genotoul.fr/multalin/multalin.html). Conserved motifs within SIDER2 retroposons, including the thymidine-rich stretch, the two tandemly repeated signatures I and II at the 5′-end of SIDER2, and the adenosine-rich stretch at the 3′-end are highlighted. (B) Schematic representation of the chimeric luciferase (LUC) constructs tested and their corresponding names. The full-length 3′-UTR of 3810 (3′-UTR3810), a 3′-UTR lacking both conserved signatures (3′-UTR3810Δ79I+II), a 3′-UTR lacking either the first (3′-UTR3810Δ79I) or the second (3′-UTR3810Δ79II) signature and a 3′-UTR lacking the last 80 nt of SIDER3810 (3′-UTR3810Δ80) were placed downstream of the LUC reporter gene. The entire SIDER2 element or its truncated forms are illustrated as grey boxes. The plasmid LUC-IR, containing the IR of 3810 served as a control. LUC activity was measured in the different transfectants (middle panel). Fold differences in LUC activity were normalized with plasmid copy numbers (Supplementary Table S2) and the values indicated are relative to the LUC-IR control. Numbers in bold correspond to the relative fold changes compared to the full-length 3′-UTR3810. Each bar and value represents the mean and standard deviations of four independent experiments. Northern and western blot analyses of total RNA and protein extracts from recombinant L. major promastigotes were carried out using LUC-specific probes and antibodies (right panels). Expression levels (mRNA and protein) of the NEO marker present on all plasmids served as a control for loading and for determining differences in plasmid copy numbers. Signal intensities were quantified and normalized with respect to the loading controls and plasmid copy numbers.
To assess whether any of these conserved motifs plays a role in SIDER2-mediated mRNA decay, we generated a series of individual deletions within two distinct SIDER2 elements that are located in the 3′-UTR of the previously studied genes LmjF36.3810 (3810) encoding an aminomethyltransferase and LmjF08.1270 (1270) encoding a hypothetical protein (18). The effect of these deletions for both SIDER3810 and SIDER1270 on mRNA degradation was evaluated using a luciferase (LUC) reporter gene system (Figure 1; Supplementary Figures S1 and S2). The conserved stretch of uridines located in a GC-rich environment shares features with U-rich instability elements and might contribute to SIDER2-mediated mRNA degradation. Deletion of 31 nt corresponding to the T-stretch region either from the full-length 3′-UTR of transcript 3810 (LUC-3′-UTR3810ΔT) or from the SIDER3810 alone (LUC-SIDER3810ΔT) had neither effect on LUC mRNA abundance and its half-life nor on the amount and activity of LUC protein (Supplementary Figure S1A–D). In contrast, deletion of the whole SIDER2 sequence with (LUCΔSIDER3810) or without (LUCΔSIDER3810ΔT) the T-stretch greatly increased LUC mRNA accumulation and half-lives to a similar extent (242 ± 23 min versus 225 ± 7 min) (Supplementary Figure S1B and D).
We next assessed the contribution of the two 79-nt signature sequences to SIDER2-mediated mRNA degradation. Deletion of both signature sequences from either SIDER3810 (LUC-3′-UTR3810Δ79I+II) or SIDER1270 (LUC-3′-UTR1270Δ79I+II) retroposons increased LUC mRNA levels to those of stable transcripts lacking the whole SIDER2 (Supplementry Figure S2A–C). Higher accumulation of LUC-3′-UTR1270Δ79I+II and LUC-3′-UTR3810Δ79I+II transcripts was due to an increase in their half-lives from 50–65 min (full-length transcripts) to 150 ± 7 min and 245 ± 18 min, respectively (Supplementary Figure S2B and C). Deletion of signature I from the full-length 3810 3′-UTR (LUC-3′-UTR3810Δ79I) had no effect on LUC mRNA degradation or LUC protein levels. In contrast, deletion of signature II (LUC-3′-UTR3810Δ79II) increased mRNA and protein levels to a similar extent than deletion of the whole SIDER2 (Supplementary Figure S2A) or of both signatures (3.2- versus 3.3- and 3.4-fold, respectively) (Figure 1B). Deletion of signature II (79 nt) also increased the half-life of LUC-3′-UTR3810Δ79II mRNA by 3.7-fold (65 versus 242 min; Figure 2B and E). As an additional control, an independent deletion of the same length (80 nt) but from the 3′-end of SIDER3810 was made (LUC-3′-UTR3810Δ80), however this neither affected LUC mRNA nor LUC protein levels (Figure 1B). Altogether, these data indicate that signature II which is highly conserved amongst SIDER2 retroposons (21) is essential for mRNA degradation.
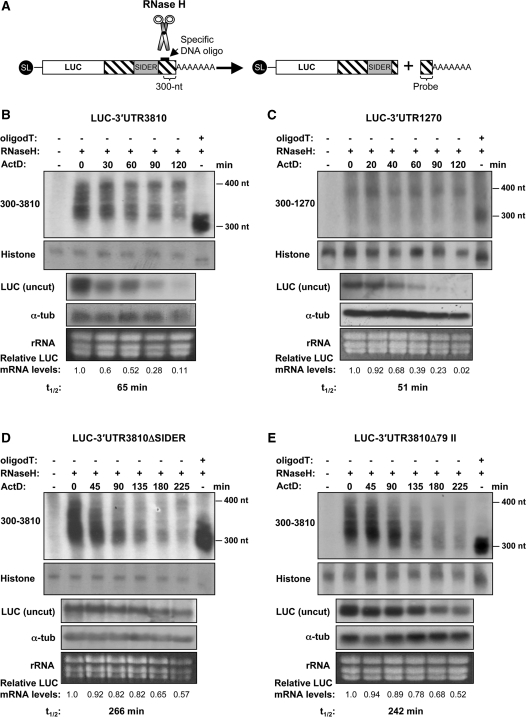
Figure 2.
Rapid degradation of unstable SIDER2-bearing transcripts is not initiated by a shortening of the poly(A) tail. (A) Schematic representation of the deadenylation assay. LUC transcripts are specifically cleaved at 300 nt from the poly(A) tail using oligonucleotide-directed RNase H cleavage. The resulting 3′-products containing the poly(A) tail are visualized by northern blot using a probe complementary to the last 300 nt of transcripts 3810 and 1270, respectively. Poly(A) tail lengths of chimeric LUC transcripts were analyzed at different time points after transcriptional arrest using ActD. In each experiment, one sample was treated with oligo(dT) and served as a control for a completely deadenylated mRNA species. Another RNA sample that was not treated with RNase H was used as negative control. Deadenylation profiles of the unstable SIDER2-containing transcripts (B) LUC-3′-UTR3810 and (C) LUC-3′-UTR1270 and of stable LUC chimeric mRNAs lacking either the complete (D) SIDER3810 or (E) signature II. Histone 4A mRNA was used as a loading control together with an ethidium bromide staining to visualize rRNA. Decay kinetics of the corresponding uncut LUC mRNAs (from identical samples) and their half-lives (t1/2) are shown below the blots to demonstrate the fate of the full-length LUC transcripts (uncut) in comparison to the cleaved 3′-ends including the poly(A) tails. LUC mRNA levels were normalized to those of the α-tubulin mRNA. The numbers indicated below the blots represent the relative LUC transcript abundance with respect to time point 0 (before addition of the transcription inhibitor Act D). Deadenylation assays shown here are representative of three independent experiments that yielded similar results.
Rapid turnover of unstable SIDER2-containing mRNAs is deadenylation independent
To investigate further the mechanism of mRNA degradation mediated by SIDER2 retroposons, we set out to determine which step of mRNA decay was targeted by these elements. There are two general pathways by which most eukaryotic mRNAs can be degraded [reviewed in (6)]. In both cases, mRNA degradation begins with the shortening of the poly(A) tail at the 3′-end of the mRNA. Deadenylation typically leads to a removal of the 5′-cap structure, which exposes the mRNA to 5′–3′ exonucleolytic degradation. Alternatively, following deadenylation, mRNAs can be degraded in a 3′–5′ direction by the exosome. To test whether turnover of SIDER2-containing transcripts is preceded by a rapid deadenylation, we monitored changes in the poly(A) tail lengths of LUC reporter transcripts harboring SIDER2 as part of the full-length 3′-UTR of transcripts 3810 and 1270 (LUC-3′-UTR3810, LUC-3′-UTR1270) at different time points after transcription arrest using actinomycin D (ActD). To do this, LUC transcripts were specifically cleaved at 300-nt upstream of the poly(A) tail via oligonucleotide-directed RNase H cleavage (Figure 2A). The resulting 3′-products containing the poly(A) tail were visualized by northern blot analysis using a probe complementary to the last 300 nt of transcripts 3810 and 1270 (Figure 2B and C). In addition, one sample in each experiment was treated with oligo(dT), which cleaves off the poly(A) tail and serves as a control for fully deadenylated mRNA species. Although the overall signal intensity of both uncut LUC-3′-UTR3810 and LUC-3′-UTR1270 transcripts decreased during the course of ActD treatment, there was no change in the apparent length of the poly(A) tails (Figure 2B and C). Similar deadenylation patterns were also observed with the endogenous L. major transcripts 3810 and 1270 (data not shown). In contrast, the poly(A) tails of the stable LUC transcripts lacking either the whole SIDER2 (LUC-ΔSIDER3810) or signature II only (LUC-3′-UTR3810Δ79II) were significantly shortened (from ∼100 to ∼10 or less adenosines), long before reaching their half-lives (Figure 2D and E). It is evident that in the case of the unstable SIDER2-containing transcripts, the cleaved 3′-end disappears much later than the full-length uncut LUC transcripts, confirming that mRNA decay does neither initiate nor proceed from this end. In contrast, both stable LUC transcripts lacking a functional SIDER2 showed a faster disappearance of their 3′-end compared to the full-length uncut LUC transcript, in line with our conclusion that these mRNAs are degraded progressively from the 3′-end after being deadenylated. Altogether, these data demonstrate that stable Leishmania transcripts (without a functional SIDER2) are slowly degraded after a progressive shortening of their poly(A) tails. In contrast, unstable SIDER2-bearing transcripts are degraded via a mechanism that seems to be deadenylation independent.
SIDER2-mediated mRNA degradation is initiated by a site-specific endonucleolytic cleavage within the cis-acting signature II sequence
Deadenylation-independent mechanisms of mRNA degradation in eukaryotes are rare and have been described so far only in yeast and mammalian cells. Such mechanisms initiate either by deadenylation-independent decapping or through cleavage by an endoribonuclease within a specific 3′-UTR sequence, which often leads to a rapid destruction of the remaining mRNA body by exoribonucleases (27–29). Although decapping activities have been described in trypanosome extracts (9), no genes encoding decapping enzymes could be identified in the genome of L. major (10). Therefore, we examined the possibility that rapid decay of SIDER2-containing mRNAs is initiated through endonucleolytic cleavage. We failed detecting cleavage products by northern blotting for the endogenous transcripts 1270 and 3810, but these are of low abundance. Hence, to increase the possibility of detecting in vivo cleavage products, we performed RPAs using chimeric LUC reporter transcripts expressed at higher levels from episomal vectors. Total RNA isolated from either L. major wild-type cells or from six L. major LUC transgenic cell lines (LUC-3′-UTR1270, LUC-3′-UTR1270Δ79I+II, LUC-3′-UTR3810, LUC-3′-UTR3810Δ79I+II, LUC-3′-UTR3810Δ79I and LUC-3′-UTR3810Δ79II) was independently incubated with in vitro-transcribed radiolabeled anti-sense RNA probes of different sizes complementary to the SIDER2 element in transcripts 3810 and 1270, and then subjected to RNase A/T1 treatment. Protected double-stranded RNA products were resolved on an 8% polyacrylamide urea gel and visualized by autoradiography.
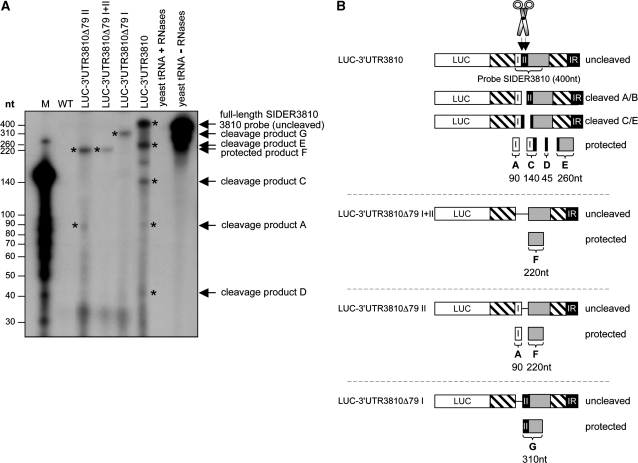
First, using RNA extracted from the LUC-3′-UTR3810 transfectant and a radiolabeled anti-sense RNA probe complementary to the first 400 nt of SIDER3810, we observed, in addition to the uncleaved protected 400-nt fragment, four smaller RNA products (Figure 3A; marked with asterisks). Fragment A (∼90 nt) is generated after cleavage at the beginning of signature II (cleavage A/B). The large product (310 nt; fragment B) derived from this cleavage was not detected, but instead, three fragments of 45, 140 and 260 nt were seen, indicating a second cleavage within signature II at ∼50-nt downstream of the first cleavage site (cleavage C/E; Figure 3A and B) (181–192 nt; Figure 1A). The 140- and 260-nt products add up to the size of the full-length SIDER3810 probe (400 nt) and are generated from a single cleavage reaction at the second site. The absence of the 310-nt cleavage fragment (first cleavage) together with the higher intensity of the 140 and 260-nt products (second cleavage) suggest that the second cleavage is more prominant. Deletion of signature I, shown to not alter mRNA decay (Figure 1B), seems to favor cleavage at the first position, as only the fragment of ∼310 nt (fragment G) was protected in the LUC-3′-UTR3810Δ79I mRNA (Figure 3A and B). Structural changes between these reporter mRNAs might explain differences in cleavage. In agreement with our deletion data demonstrating that signature II is essential for mRNA degradation (Figure 1B), RPA analysis of LUC-3′-UTR3810Δ79I+II and LUC-3′-UTR3810Δ79II mRNAs lacking either both signatures I and II or only signature II indicated no cleavage. Only a protected fragment of ∼220 nt corresponding to the size of the truncated SIDER3810 (fragment F) was detected. In the case of LUC-3′-UTR3810Δ79II mRNA, an additional product of ∼90 nt corresponding to signature I was detected (Figure 3A and B). This is the result of a ss cleavage due to a looping of the SIDER3810 probe at the position of the target RNA where signature II was deleted.
Figure 3.
Detection of in vivo-generated cleavage products derived from SIDER2-containing mRNAs by RPAs. (A) Total RNA was isolated from L. major wild-type cells (WT), the recombinant L. major expressing a chimeric LUC transcript carrying the full-length 3′-UTR of 3810 (LUC-3′-UTR3810) and the truncated mutants LUC-3′-UTR3810Δ79I+II, LUC-3′-UTR3810Δ79I and LUC-3′-UTR3810Δ79II lacking either both signatures (I+II) or only signature I or signature II, respectively. These RNAs were independently mixed with an in vitro-transcribed radiolabeled anti-sense SIDER3810 probe of 400 nt and thereafter subjected to RNase A/T1 treatment. Wild-type RNA and yeast RNA with (negative control) or without RNase treatment (positive control) were used as controls. A 3′-labeled RNA ladder was used to estimate the size of the fragments (M). (B) Schematic representation of the in vivo cleaved (unstable LUC-3′-UTR3810 and LUC-3′-UTR3810Δ79I) and uncleaved (stabilized LUC-3′-UTR3810Δ79I+II and LUC-3′-UTR3810Δ79II) SIDER2-containing mRNAs. The sizes of the observed cleavage products suggest that a first cleavage occurs at the beginning of SIDER3810 signature II (cleavage A/B) and that a second, more dominant, cleavage occurs at ∼50-nt downstream (cleavage C/E). In addition to the full-length protected band (400 nt), four cleavage products of ∼260, ∼140, ∼90 and ∼45 nt (indicated with asterisks) were detected from the LUC-3′-UTR3810 mRNA. The cleavage products of 260 and 140 nt correspond to the second cleavage and the 90-nt fragment to the first cleavage (the expected 310-nt fragment is not visible in this experiment, as it was further cleaved at the second position to generate the 45- and 260-nt fragments). The band observed between the cleavage fragments C and E in the LUC-3′-UTR3810 lane was not reproducible in the other experiments. In all cases, cleavage products sum up to the size of the full-length SIDER3810 probe (400 nt). In the truncated LUC-3′-UTR3810Δ79I mRNA-lacking signature I, only the first cleavage site was detected, as a single fragment of ∼310 nt was protected. In the case of LUC-3′-UTR3810Δ79II mRNA-lacking signature II, two cleavage fragments of 90 and 220 nt were detected, as expected, given that RNases could digest the ss region of the probe corresponding to signature II. Deletion of both signatures in LUC-3′-UTR3810Δ79I+II mRNA produced a 220-nt RNase-resistant fragment, as expected. The data shown here are representative of three independent experiments that generated the same RNase digestion patterns.
Furthermore, cleavage within signature II was confirmed by RPA using a distinct SIDER2 retroposon element, SIDER1270, which is part of the 3′-UTR of transcript 1270 sharing a ∼60% overall sequence identity with SIDER3810 (Figure 1A). Using total RNA extracted from L. major LUC-3′-UTR1270, we detected in addition to the protected 480-nt uncleaved fragment, two major RNA-protected products of ∼120 and ∼360 nt (marked with asterisks) whose sizes add up to the full-length RNA probe (480 nt) (Supplementary Figure S3A). These results are in agreement with cleavage at the beginning of signature II, as it was also shown for LUC-3′-UTR3810 mRNA (Figure 3). However, in the case of LUC-3′-UTR1270 mRNA, we did not detect any secondary clevage. Interestingly, no cleavage products were observed when using LUC-3′-UTR1270Δ79I+II mRNA lacking both signature sequences and a single fragment of ∼260 nt corresponding to the truncated SIDER1270 was protected from RNase treatment (Supplementary Figure S3A and B). This is a strong indication that these RNA intermediates are the products of an endoribonuclease activity and not of exoribonuclease pause sites at stable secondary structures (30). In order to distinguish the protected full-length fragments from undigested probe remaining in the sample, we used a 600-nt probe that is longer than the target RNA. This probe is complementary to the first 480 nt of the SIDER1270 sense RNA but contains an extension of 68 nt at the 5′-end and of 59 nt at the 3′-end originating from the plasmid vector (see Supplementary Figure S3C and ‘Materials and methods’ section). The longer probe detected the same protected RNA bands (Supplementary Figure S3B and C) than the fully complementary 480-nt probe (Supplementary Figure S3B and C), hence confirming the specificity of the protected uncleaved RNAs. RPAs have been optimized in order to minimize possible overdigestion by RNases, and all our control experiments support a specific cleavage. Indeed, no cleavage products were detected with the stabilized truncated RNAs (LUC-3′-UTR1270Δ79I+II, LUC-3′-UTR3810Δ79I+II and LUC-3′-UTR3810Δ79II) (Figure 3; Supplementary Figure S3). Furthermore, cleavage intermediates were only seen with the full-length LUC-3′-UTR1270 mRNA and were not detected in the control lanes containing wild type or yeast RNA treated or untreated with RNases.
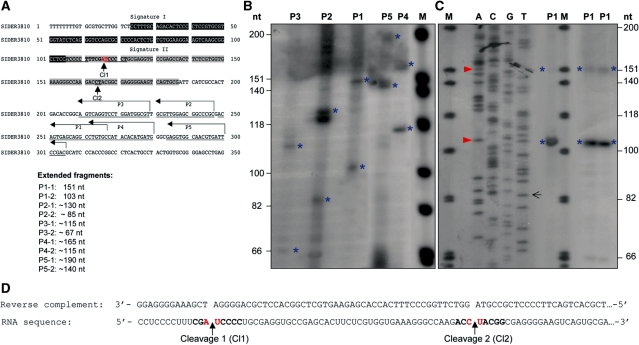
To map more precisely the putative cleavage site(s) within signature II and to confirm endonucleolytic cleavage by another method, LUC-3′-UTR3810 RNA samples were subjected to primer extension analysis. For this, five different reverse primers (P1–P5) complementary to a region between 100 and 200 nt downstream of the putative cleavage sites in SIDER3810 were designed (Figure 4A; Supplementary Table S1). To visualize specific cleavage products, the primers were 5′-end-labeled, annealed to total RNA isolated from the L. major LUC-3′-UTR3810 transfectant and extended via reverse transcription. The size of the generated cDNA fragments should correspond to the number of nucleotides between the labeled primer and the 5′-end of the cleaved RNA where reverse transcriptase falls of its template. Primer extension with all five primers produced in each case two specific extended fragments (1 and 2; marked with asterisks). The sizes of these fragments (Figure 4A and B) were in agreement with the RNase protection results (Figure 3) and confirmed the occurrence of two distinct cleavage events within SIDER3810 signature II. Using a dideoxy-sequencing strategy with primer P1 (Figure 4C), we mapped the first cleavage site between an AU dinucleotide surrounded by a C-rich motif (position 116-117 in Figure 4A) and the second cleavage site possibly between a CU (or a UA) dinucleotide at 50-nt downstream (position 164-166 in Figure 4A, C and D). Preliminary data using two different primers confirmed the occurrence of a major cleavage within the AU dinucleotide also for LUC-3′-UTR1270 mRNA (data not shown).
Figure 4.
Mapping of the cleavage site(s) in a SIDER2-containing mRNA by primer extension analysis. (A) Nucleotide sequence of SIDER3810 retroposon with signature I (black box) and II (grey box) sequences highlighted, and the five primers (P1–P5) indicated by arrows. The sizes of the expected primer extension products with full-length LUC-3′-UTR3810 mRNA, depending on whether cleavage occurs at the first (Cl1) or the second (Cl2) site within signature II, are listed below. (B) Primer extension assay with total RNA isolated from L. major LUC-3′-UTR3810 transfectant using five different reverse primers P1–P5 (Supplementary Table S1). Two specific extension fragments were obtained with each of the primers (indicated with blue asterisks) corresponding to both cleavages (1 and 2). Their sizes estimated with a radiolabeled DNA marker (M) are in agreement with the predicted cleavage regions. (C) A dideoxy-sequencing and primer extension with primer P1 allowed a precise mapping of the cleavage sites between an AU dinucleotide (cleavage 1) and a CU (or a UA; it is not easy to distinguish from the sequencing data) dinucleotide (cleavage 2) marked with a red arrow. (D) Reverse complement sequence (the black arrow indicates where the highlighted sequence starts) and the corresponding sequence of SIDER3810 RNA. Cleavage sites 1 and 2 are indicated with an arrow. The data shown here are representative of four independent experiments with similar results.
To further characterize endogenous cleavage products derived from SIDER2-bearing mRNAs, we used an RNA circularization and sequecing strategy referred to as reverse ligation-mediated PCR (RL-PCR) (26,31). Most endoribonucleases generate 5′-phosphate and 3′-hydroxyl termini after RNA cleavage, which can be ligated using T4 RNase ligase (31). In order to characterize the ends of the in vivo endonucleolytic cleavage products, total RNA from the L. major LUC-3′-UTR1270 transfectant was incubated with T4 ss RNA ligase at low concentrations that favor the ligation of ends from the same molecule, thereby generating circular RNAs. In our case, the cleavage sites (5′-end) should be joined directly to their poly(A)-tails (3′-end) (Supplementary Figure S4A). The full-length LUC-3′-UTR1270 transcript cannot be ligated, because it is capped at its 5′-end (Supplementary Figure S4A). Identical RNA samples without addition of T4 ssRNA ligase served as a negative control. After ligation, the 5′–3′ junction was reversed transcribed using primer P5 and then PCR-amplified with the primer pair P5/P2 (Supplementary Figure S4A and B; Supplementary Table S1). Due to the low abundance of endonucleolytic cleavage products, a nested PCR using the internal primers P6/P2 was performed (Supplementary Figure S4A and C; Supplementary Table S1). The results of the nested PCR using two different concentrations of total RNA (a = 20 µg; b = 40 µg) are shown in Supplementary Figure S4C. Amplification products without poly (A)-tail were predicted to be ∼90-bp long. All fragments within the expected size range (between 75 and 200 bp; marked with an asterisk) that were absent in the control lanes without ligase or reverse transcriptase were excised from the gel, purified, cloned and sequenced. A sequence alignment of nine unique clones (Supplementary Figure S4D) revealed that most PCR-products exhibit relatively short poly(A)-tails (∼10 nt) at the expected polyadenylation site. The detection of unusually short poly(A)-tails using the RL-PCR method was reported before (26,32) and is attributed to an intrinsic bias of this method against longer poly(A)-tails, which is further increased when nested PCR and cloning procedures are included. Interestingly, in the longest PCR fragment, poly(A)-tail was ligated with a T at the second TCCCCT repeat (Figure 1A), which corresponds to the first cleavage site between an AU dinucleotide (Supplementary Figure S4D), a region predicted also by primer extension (Figure 4; data not shown) and RPA (Figure 3; Supplementary Figure S3). The presence of other shorter fragments is expected, and indicates that the cleavage products may be subjected to rapid degradation by 5′–3′ exonucleases. No RNA-ligation products containing sequences beyond the predicted C-rich cleavage site were identified.
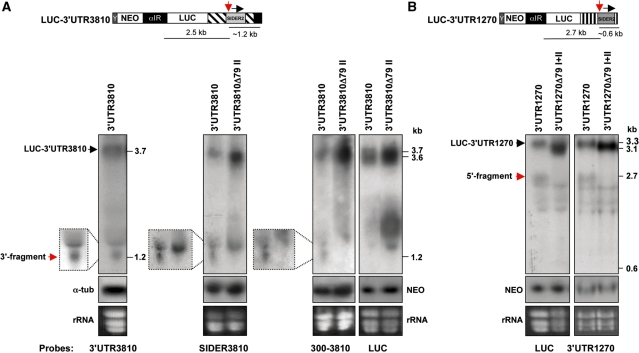
Interestingly, in vivo-generated 5′- and 3′-cleavage products were also detected by northern blotting. Northern blot hybridization is a less sensitive assay than RPA and primer extension, but represents the most reliable way to detect the cleavage fragments in vivo. DNA probes recognizing either the full-length 3′-UTR of transcript 3810 or the whole SIDER3810 sequence or the last 300 nt of the 3810 3′-UTR detected a fragment of ∼1.2 kb corresponding to the predicted 3′-cleavage product at the level of signature II from LUC-3′-UTR3810 mRNA (Figure 5A). The 5′-cleavage fragment (∼2.5 kb) was not detected with the 3′-UTR3810 or the LUC specific probes (Figure 5A), however, possibly because it was rapidly degraded by exonucleases. In the case of the LUC-3′-UTR1270 mRNA, a fragment of ∼2.7 kb corresponding to the expected 5′-cleavage product was detected by probes recognizing either the full-length 1270 3′-UTR or the LUC gene (Figure 5B). The 1270 3′-UTR probe failed to detect the corresponding 3′-cleavage fragment of ∼0.6 kb (Figure 5B), most likely due to the low abundance and faster exonucleolytic digestion of this smaller fragment. In agreement with our RPA and primer extension data, 5′- or 3′-cleavage products were absent in the stabilized RNAs lacking signature II (LUC-3′-UTR3810Δ79II and LUC-3′-UTR1270Δ79I+II) (Figure 5A and B). In addition, we have been able to detect 5′- and 3′-cleavage products by northern blotting in RNA preparations from another set of L. major transfectants expressing myc-tagged full-length 1270 and 3810 mRNAs (Supplementary Figure S5). In this experiment, both the 5′- and 3′-cleavage fragments derived from 3810ORFmyc-3′-UTR (∼2.0 and ∼1.2 kb) and 1270ORFmyc-3′-UTR (∼2.0 and ∼0.6 kb) mRNAs were detected by the full-length 3′-UTR probes. In summary, specific endonucleolytic cleavage products were detected from several distinct SIDER2-bearing mRNAs using four independent methods, only when the regulatory cis-acting region located within signature II was present. Altogether, these data support that decay of SIDER2-containing mRNAs is initiated by a site-specific endonucleolytic cleavage at the beginning of the conserved signature II sequence (137–147 nt; Figure 1A).
Figure 5.
Detection of in vivo-generated endonucleolytic cleavage products from reporter mRNAs harboring distinct SIDER2 retroposon elements by northern blotting. (A) Northern blots of total RNA from recombinant L. major LUC-3′-UTR3810 (LUC reporter gene under the control of the full-length 3′-UTR of transcripts 3810) and LUC-3′-UTR3810Δ79II (3′-UTR of 3810 lacking the SIDER2 signature II sequence) hybridized with four different DNA probes corresponding to the complete 3′-UTR3810 (left panel), the SIDER3810 sequence alone, the last 300 nt of the 3810 3′-UTR (middle panels) or the LUC gene (right panel). The full-length 3′-UTR3810, SIDER3810 and 300–3810 probes recognized a band of ∼1.2 kb, which corresponds to the expected 3′-cleavage product (indicated by a red arrow; see also schematic representation above the blots). This band is absent in the control RNA-lacking signature II (cleavage region). The 5′-cleavage fragment was not detected under these conditions using either the full-length 3′-UTR3810 or the LUC-specific probes. (B) Northern blots of total RNA from L. major transfectants LUC-3′-UTR1270 (LUC reporter gene under the control of the full-length 3′-UTR of transcripts 1270) and LUC-3′-UTR1270Δ79I+II (3′-UTR of 1270 lacking both signatures I and II) hybridized with two different DNA probes specific for LUC (left panel) and the 3′-UTR1270 (right panel). Both probes detected a band of ∼2.7 kb corresponding to the expected size of the 5′-cleavage product (indicated by a red arrow; see schematic representation above the blots), which is absent in the control RNA-lacking signatures I+II (cleavage region). The 3′-cleavage fragment was not detected under these conditions. Northern blots shown here are representative of three independent experiments yielding similar results. Expression levels of the NEO mRNA present on all plasmids served as a control for loading and for evaluating differences in plasmid copy numbers between the transfectants.
SIDER2 anti-sense RNA complementary to the endonucleolytic cleavage region protects SIDER2-containing transcripts from rapid degradation
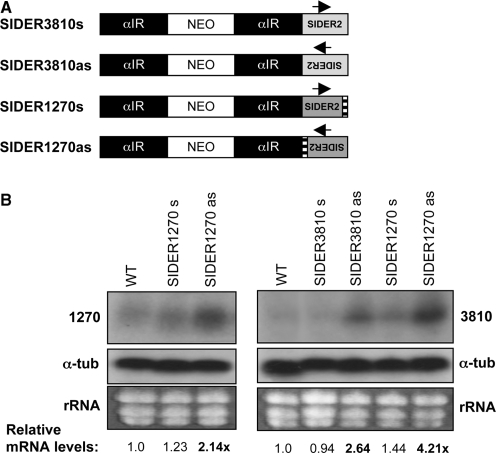
It is intringuing that while the vast majority of SIDER2 retroposons located in 3′-UTRs follows the direction of transcription, most of SIDER2 elements located in IRs are inserted in the opposite orientation relative to transcription (21). Moreover, SIDER2 elements were found in the anti-sense orientation of Leishmania directional gene clusters (21). Given that anti-sense transcription has previously been reported in Leishmania (2,33), we explored the possibility that SIDER2 anti-sense RNAs are naturally produced in this parasite. Strand-specific RT-PCR experiments confirmed indeed the presence of such RNA products (M. Müller et al., in revision; P.K. Padmanabhan, unpublished data). Hence, it was important to test whether SIDER2 anti-sense RNA could alter SIDER2-mediated mRNA degradation. To directly address this question, we first generated a series of transgenic L. major cell lines ectopically expressing either sense or anti-sense SIDER1270 and SIDER3810 RNAs (Figure 6A). As estimated by Southern blot analysis, the copy number of sense and anti-sense SIDER2-expressing vectors was comparable (Supplementary Table S2). Northern blots using SIDER2-specific probes confirmed that the ectopically provided SIDER2 RNAs were correctly processed and that were in average 6–10-fold more abundant than the endogenous transcripts 1270 and 3810, respectively (data not shown). Overexpression of sense SIDER1270 or SIDER3810 RNAs did not alter steady-state levels of the endogenous transcripts 1270 and 3810 (Figure 6B). In contrast, overexpression of an anti-sense RNA fully complementary to the SIDER2 sequence of transcripts 1270 or 3810 significantly increased accumulation of the latter by 2.14- and 2.64-fold, respectively (Figure 6B). Interestingly, even a heterologous anti-sense RNA (SIDER1270as) sharing high sequence similarity with the mapped SIDER3810 cleavage region (76% sequence identity; Figures 1A and 4) significantly increased accumulation of the endogenous transcript 3810 (Figure 6B).
Figure 6.
Ectopic expression of SIDER2 anti-sense RNA results in an increased accumulation of endogenous L. major SIDER2-containing transcripts. (A) Schematic representation of plasmids overexpressing SIDER2 in both orientations. SIDER2 sequences from transcripts 1270 or 3810 were cloned downstream of a NEO selection marker in the sense (s) or anti-sense (as) orientation and stably transfected into L. major wild-type cells (WT). αIR refers to the IR of the α-tubulin gene necessary for NEO mRNA processing. (B) Northern blot analysis with total RNA from L. major WT and transfectants. The endogenous transcripts 1270 or 3810 were detected with probes specific to the coding regions of 1270 (left panel) and 3810 (right panel). The copy number of the SIDER2 expression plasmids was estimated by Southern blot as indicated in Supplementary Table S2. The signal intensities were quantified and the mRNA abundance was normalized to α-tubulin mRNA and calculated with respect to WT mRNA levels.
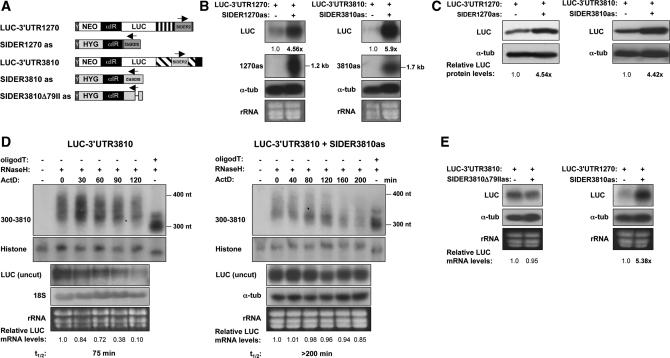
To investigate how a SIDER2 anti-sense RNA can interfere with the rapid decay of SIDER2-bearing transcripts, we generated L. major transgenic lines co-expressing LUC reporter constructs with either the full-length 3′-UTR1270 and SIDER3810 anti-sense RNA or the 3′-UTR3810 and SIDER1270 anti-sense RNA, as illustrated in Figure 7A. Expression and correct processing of SIDER2 anti-sense transcripts in the double transfectants was verified using strand-specific riboprobes (Figure 7B, middle panel) and RPAs (data not shown). Similarly to the single transfectants overexpressing only SIDER2 anti-sense RNA (Figure 6B), the steady-state levels of LUC-3′-UTR1270 and LUC-3′-UTR3810 transcripts were increased by 4.6- and 5.9-fold, respectively, in the double transfectants (Figure 7B). LUC protein levels were in accordance with LUC mRNA levels (Figure 7C). Higher accumulation of LUC-3′-UTR3810 mRNA in the presence of SIDER3810 anti-sense RNA was due to a 3-fold increase in mRNA stability (75 min versus >200 min; Figure 7D, three lower panels). Moreover, decay of the stabilized LUC-3′-UTR3810 transcript in the presence of SIDER3810 anti-sense RNA was initiated by poly(A)-tail shortening (Figure 7D, two upper panels), similarly to the non-regulated stable mRNAs (Figure 2), suggesting that an RNA complementary to SIDER2 sequences can interfere with the mechanism by which SIDER2-containing transcripts are normally degraded. As vectors harboring SIDER2 anti-sense RNA were generally ∼2-fold less expressed (∼2-fold less copies; Supplementary Table S2) than the reporter SIDER2 sense vectors in the double transfectants, we can exclude the possibility that mRNA stabilization is simply due to a sequestration of putative trans-acting factor(s) that might also bind to SIDER2 anti-sense RNA.
Figure 7.
SIDER2 anti-sense RNA complementary to the cleavage region blocks degradation of unstable SIDER2-containing reporter transcripts. (A) Schematic representation of the chimeric LUC reporter constructs bearing either a sense (s) or anti-sense (as) SIDER2 retroposon from transcripts 1270 or 3810. Pairs of sense/anti-sense vectors LUC-3′-UTR1270/SIDER1270as LUC-3′-UTR3810/SIDER3810as, LUC-3′-UTR3810/SIDER3810Δ79IIas and 3′-UTR1270/SIDER3810as were co-transfected into L. major. The plasmids expressing the full-length 3′-UTRs of 3810 and 1270 cloned downstream of the LUC gene harbor a neomycin phosphotransferase (NEO) gene as a selection marker. Anti-sense SIDER2 sequences were inserted downstream of a hygromycin B (HYG) selection marker. Arrows indicate the orientation of SIDER2 elements. Y corresponds to a 92-bp polypyrimidine stretch (34) and α-IR refers to the IR of α-tubulin, both necessary for NEO, HYG and SIDER2as RNA processing. (B) Northern blot analysis to compare LUC mRNA expression levels in single (−) and double (+) transfectants using a LUC-specific probe. Expression and correct processing of both fully complementary anti-sense SIDER2 transcripts (1270as and 3810as) was verified with riboprobes specific to each SIDER2 sequence. Their sizes of ∼1.2 kb indicate that the transcripts are spliced within the α-IR and do not contain HYG sequences. The LUC signal intensities were normalized with α-tubulin mRNA levels and calculated with respect to the LUC-plasmid copy numbers (Supplementary Table S2). (C) Western blot analysis using L. major protein lysates and a LUC-specific antibody. Protein loading was controlled with an anti-α-tubulin antibody. (D) Deadenylation profile and decay kinetics in single (LUC-3′-UTR3810) and double L. major transfectants (LUC-3′-UTR3810/SIDER3810as) as determined by northern blotting (identical samples). The numbers below the blots represent the relative LUC mRNA levels with respect to time point 0 (before addition of ActD). Histone 4A was used as a loading control. The mRNA half-lives of the uncut LUC transcripts are shown below the blots. (E) Northern blot hybridization to evaluate LUC-3′-UTR3810 mRNA accumulation in the absence or presence of a truncated SIDER3810 anti-sense RNA-lacking signature II (SIDER3810Δ79IIas) (left panel). Northern blot analysis to compare LUC-3′-UTR1270 mRNA abundance in the presence or absence of a heterologous SIDER3810 anti-sense RNA (right panel). SIDER3810 and SIDER1270 retroposons share a 76.5% sequence identity at the level of signature II and >80% identity within the cleavage site (Figure 1A). LUC signal intensities were normalized as indicated in (B).
To address whether an interaction between sense and anti-sense RNA at the endonucleolytic cleavage site contributes to the blockade of SIDER2-mediated mRNA degradation, we produced a truncated SIDER3810 anti-sense RNA lacking the second signature sequence, shown here to be essential for mRNA degradation and cleavage (Figures 1B, 2–5 and Supplementary Figure S3). As illustrated in Figure 7E (left panel), overexpression of SIDER3810 anti-sense RNA lacking signature II (LUC-SIDER3810Δ79IIas) failed to block degradation of LUC-3′-UTR3810 mRNA. Interestingly, overexpression of a heterologous anti-sense RNA (SIDER3810as) resulted in >5-fold accumulation of the LUC-3′-UTR1270 mRNA (Figure 7E, right panel), similarly to the effect seen with the homologous SIDER1270 anti-sense RNA (Figure 7B). In summary, these results suggest that SIDER2-bearing transcripts can be protected from degradation by anti-sense RNA, probably through base-pairing to a ss region within the cis-acting signature II sequence.
DISCUSSION
This article provides new insights into the underlying decay mechanism of unstable Leishmania transcripts involving extinct SIDER2 retroposons predominantly located within 3′-UTRs. We have provided evidence that (i) the second 79-nt signature, which is the most conserved sequence amongst SIDER2 retropososons (18,21) is the cis-acting element conferring degradation of SIDER2-bearing mRNAs; (ii) rapid turnover of SIDER2-containing transcripts is deadenylation-independent, as opposed to stable transcripts which are degraded through a progressive shortening of poly(A) tails; (iii) SIDER2-mediated decay is initiated by a site-specific endonucleolytic cleavage within the second 79-nt signature sequence, which is essential for degradation and (iv) endonucleolytic cleavage and subsequent rapid mRNA decay can be blocked by SIDER2 anti-sense RNA complementary to the cleavage site. These are original findings that establish a new paradigm for how short-lived mRNAs in Leishmania sharing a conserved retroposon signature sequence in their 3′-UTRs are degraded, and could serve as the basis for a better understanding of mRNA decay pathways not only in protozoan parasites but in general.
Our finding that the conserved signature II sequence of SIDER2 retroposons is essential for endonucleolytic cleavage and rapid mRNA decay provides also further explanations regarding the evolutionary divergence of SIDER1 and SIDER2 subfamilies to fulfill distinct biological functions. It is remarkable that SIDER1 retroposons, which have been associated with translational control rather than mRNA turnover (22,23) lack signature II (18,21). The fact that deletion of the first signature from SIDER2 did not change the fate of mRNA degradation is consistent with this hypothesis. Interestingly, analysis of the predicted secondary structure of selected SIDER2 retroposons using the RNAstructure program (35) indicated that the structure of signature II remained unchanged upon deletion of signature I (data not shown). A multiple sequence alignment of most Leishmania SIDER2 retroposons demonstrated that signature II is significantly more conserved than signature I (21). The high conservation of signature II sequences together with the remarkable expansion of SIDER2 elements throughout the Leishmania genome (>1300 copies) (21) and their proven involvement in mRNA decay suggest that a large number of Leishmania SIDER2-bearing transcripts may be under the same regulatory control. Several SIDER2-containing transcripts in Leishmania are short-lived (18) and/or differentially expressed in either life stage of the parasite (36) or preponderantly enriched among transcripts encoding metabolic functions (21). Therefore, it is possible that targeting of selected subsets of SIDER2-bearing transcripts for rapid degradation could permit their co-ordinated regulation in response to environmental changes throughout the digenetic life cycle of the parasite. Interestingly, the existence of post-transcriptional regulons has recently been reported in the related species T. brucei (37–40).
A major outcome of this experiment is the demonstration that rapid turnover of unstable SIDER2-containing mRNAs is initiated by a novel mechanism, which is deadenylation independent and involves a site-specific endonucleolytic cleavage. This is in sharp contrast to stable mRNAs in Leishmania lacking SIDER2 or carrying a truncated SIDER2 without signature II, which are degraded after progressive deadenylation, as it is the case for most yeast and mammalian mRNAs (6). Endonucleolytic digestion products from SIDER2-bearing mRNAs were detected in vivo by four different methods (e.g. RNase protection, primer extension, reverse ligation-mediated PCR and northern blotting) using distinct SIDER2 RNA substrates and a large number of test and control RNAs, which provided further support for the significance of endonucleolytic cleavage. Primer extension and reverse ligation-mediated PCR analyses indicated a preferential cleavage of N–pyrimidine (AU) or pyrimidine–pyrimidine (CU) bonds. The most prevalent endonucleolytic cleavage site was mapped between an AU dinucleotide flanked by a 5′-UCCCC-3′ duplicated motif at the 5′-extremity of the conserved 79-nt signature II sequence. However, in the case of LUC-3′-UTR3810 mRNA, we have detected a secondary predominant cleavage at ∼50-nt downstream the AU dinucleotide. This cleavage was also seen with the endogenous transcript 3810 (data not shown), suggesting that although the primary sequence of the putative cleavage site(s) is highly conserved, changes in the overall RNA secondary structure between the different SIDER2 elements might favor additional cleavages. The importance of these sequences for mRNA decay via endonucleolytic cleavage was further supported by deletion analysis, as removal of signature II from SIDER2 completely abolished cleavage, and turnover of the truncated stabilized RNAs was now initiated by deadenylation. Hence, combined data from different experimental approaches support that a site-specific endonucleolytic cleavage of SIDER2-bearing transcripts initiates their rapid degradation. Moreover, it is highly unlikely that the observed cleavages occur through spontaneous hydrolysis on hypersensitive sites after RNA was extracted from the cell. In this case and in the absence of RNases in the RNA sample, one would expect to detect equal amounts of the 5′- and 3′-cleavage fragments by northern blotting whose accumulation increased with time. However, in our experiments, either only one of the expected cleavage fragments was detected (Figure 5) or when both were visible in the blots, they were not equally abundant (Supplementary Figure S5) and did not exponentially accumulate with time (data not shown). This further argues for an in vivo cleavage of the RNAs (followed possibly by exonuclease activities) rather than an ex vivo RNA hydrolysis of phosphodiester bonds in RNA.
We favor a model of an endonucleolytic cleavage mediated through the action of an endoribonuclease. This model is supported by initial studies using general inhibitors of translation, e.g. puromycin and cycloheximide, that demonstrated a gradual increase in the half-lives of SIDER2-containing transcripts, but not of those lacking SIDER2 (data not shown) upon translational arrest, suggesting the action of a short-lived endoribonuclease. In addition, the circularization of SIDER2 cleavage products using T4 ssRNA ligase suggests that they exhibited 5′-monophosphate and 3′-hydroxyl ends, which are usually generated through the action of ribonucleases. However, catalytic RNAs can also generate 5′-phophate and 3′-hydroxyl as cleavage products, and at this point we cannot completely rule out the possibility that enzymatic cleavage requires an RNA co-factor as is the case of RNase P (41) and the eukaryotic RNase P MRP ribonucleases (42), but homologs of these enzymes are not encoded by the Leishmania genome. To date, only a few endoribonucleases, shown to cleave mRNAs in higher eukaryotes, have been indentified. These include PMR1, an estrogen-regulated polysomal endoribonuclease that destabilizes serum protein mRNAs (43), the Ras GTPase-activating protein-binding protein (G3BP) that cleaves c-myc and other mRNAs (44), the erythroid-enriched endoribonuclease (ErEN) involved in α-globin mRNA turnover (28), the inositol-requiring enzyme-1 (IRE1) that mediates rapid degradation of endoplasmic reticulum-localized mRNAs during the unfolded protein response (45), the mRNA processing endoribonuclease, which degrades mRNAs involved in the cell cycle regulation in Saccharomyces cerevisiae (46) and the RNAi components Dicer (47) and Argonaute 2 (48). Rrp44, a component of eukaryotic exosome (49,50), and SMG6, a protein involved in non-sense-mediated decay pathway (51) also possess an endonucleolytic activity. The potentially retrotransposition competent ingi and L1Tc elements from which SIDERs have apparently derived (21,52) encode a large protein containing the N-terminal apurinic/apyrimidinic-like endonuclease (APE), the reverse transcriptase, the RNase H and the C-terminal DNA-binding domains (19). A homolog of APE-type endonucleases encoded by all non-LTR retrotransposons (53) is present in the genome of Leishmania. Interestingly, APE1 was reported recently to cleave the coding region determinant of c-myc mRNA (54). However, to fully understand the mechanism and significance of endonucleolytic cleavage in the control of SIDER2-mediated mRNA decay, the responsible endoribonuclease must be identified and work in that direction is currently being pursued in our laboratory.
In this article, we also provide experimental evidence supporting a potential role of anti-sense RNA in preventing or altering rapid turnover of endogenous and reporter SIDER2-bearing mRNAs. We show that ectopically expressed SIDER2 anti-sense RNA targeting either the SIDER2-containing endogenous transcripts 3810 and 1270 or reporter transcripts harboring SIDER3810 and SIDER1270 elements in their 3′-UTR can protect these RNAs from endonucleolytic cleavage and deadenylation-independent decay, and consequently increase their half-lives. One plausible explanation of how anti-sense RNA could block mRNA degradation is through base-pairing, which could prevent the binding of an endonuclease. Although knowledge of the individual RNA structures would be required for validating this model, our data showed that a SIDER2 anti-sense RNA lacking signature II was unable to block degradation of its target RNA. In contrast, a heterologous SIDER2 (3810) anti-sense RNA sharing high-sequence similarity with the sense (1270) RNA, especially within the cleavage region, was able to prevent degradation as efficiently as the homologous SIDER2 anti-sense RNA. An interesting question is whether Leishmania, in the absence of a functional RNAi mechanism (55,56) known to silence widespread retrotransposon elements (57), uses an anti-sense RNA-based mechanism to mitigate regulation by SIDER2. A large number of SIDER2 retroposons are present in the anti-sense orientation within IRs of the three queried Leishmania genomes (15–20% of the SIDER2s) (21) and anti-sense RNA can in principle be produced following polycistronic transcription and pre-mRNA processing (3). Furthermore, several SIDER2 retroposons have been found in the anti-sense orientation of directional gene clusters (21), and as there is evidence for bidirectional transcription leading to anti-sense transcripts (2,33,58–60) in Leishmania, SIDER2 anti-sense RNA could be produced from that route as well. We have indeed detected anti-sense RNA products complementary to SIDER1270 by strand-specific RT-PCR and northern blotting using specific riboprobes, albeit at significantly lower levels than the sense RNA (data not shown). However, is not to be excluded that under certain conditions during its lifecycle, Leishmania could produce higher levels of SIDER2 anti-sense RNAs. In the majority of cases, anti-sense RNA action entails post-transcriptional inhibition of target RNA function (61), but, in a few cases, mRNA stabilizing effect by the anti-sense RNA has also been reported (62,63). Efficient targeting of an important mRNA decay pathway in Leishmania by anti-sense RNA represents a promising strategy towards the development of novel therapeutics against Leishmania infections. The use of anti-sense RNA to knockdown expression of individual genes in Leishmania (64,65) has been limited, with a moderate to good success.
The results reported here support a model in which rapid turnover of unstable or short-lived SIDER2-containing transcripts is mediated by a site-specific endonucleolytic cleavage without prior deadenylation. We had previously shown that rapid decay of developmentally regulated transcripts in Leishmania conferred by a U-rich 3′-UTR element (URE) is deadenylation-independent and probably not associated with elevated decapping activities (17). Consistent with this view, unstable transcripts in T. brucei regulated by UREs were not affected by depletion of the CAF1 deadenylase homolog (11). Instead, a putative 5′–3′ exonuclease, XRNA (12) or the exosome (16) was required for the initiation of rapid degradation of unstable mRNAs in T. brucei. Based on this knowledge, the current model for mRNA degradation in trypanosomatids involves at least two pathways: a regulated pathway that is rapid and seems to be deadenylation-independent [this study; (4,17)] and a constitutive pathway that is initiated with a progressive shortening of poly(A) tails (probably by the CAF1-NOT-complex) and operates at a slower kinetics during the degradation of stable mRNAs [this study; (5,17)].
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Operating grant from the Canadian Institutes of Health Research (CIHR) (MOP-12182) (to B.P.); PhD scholarship from Laval University (to M.M.); Centre for Host-Parasite Interactions (CHPI) (to M.M.); Postdoctoral fellowship from the CIHR STP-53924 Strategic Training Program (to P.K.P.). Funding for open access charge: Canadian Institutes of Health Research grant (MOP-12182 to B.P.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all members of the BP lab for useful discussions and Drs Marc Ouellette and François McNicoll for critical reading of the manuscript. B.P. is a member of a CIHR Group on Host-Pathogen Interactions and of a « Fonds Québecois de la Recherche sur la Nature et les Technologies » (FQRNT) Centre for Host-Parasite interactions.
REFERENCES
- 1.Myler PJ, Audleman L, deVos T, Hixson G, Kiser P, Lemley C, Magness C, Rickel E, Sisk E, Sunkin S, et al. Leishmania major Friedlin chromosome 1 has an unusual distribution of protein-coding genes. Proc. Natl Acad. Sci. USA. 1999;96:2902–2906. doi: 10.1073/pnas.96.6.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez-Calvillo S, Yan S, Nguyen D, Fox M, Stuart K, Myler PJ. Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol. Cell. 2003;11:1291–1299. doi: 10.1016/s1097-2765(03)00143-6. [DOI] [PubMed] [Google Scholar]
- 3.Liang XH, Haritan A, Uliel S, Michaeli S. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell. 2003;2:830–840. doi: 10.1128/EC.2.5.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clayton C, Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol. Biochem. Parasitol. 2007;156:93–101. doi: 10.1016/j.molbiopara.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Haile S, Papadopoulou B. Developmental regulation of gene expression in trypanosomatid parasitic protozoa. Curr. Opin. Microbiol. 2007;10:569–577. doi: 10.1016/j.mib.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 7.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell. Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 8.Coller J, Parker R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 9.Milone J, Wilusz J, Bellofatto V. Identification of mRNA decapping activities and an ARE-regulated 3′ to 5′ exonuclease activity in trypanosome extracts. Nucleic Acids Res. 2002;30:4040–4050. doi: 10.1093/nar/gkf521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwede A, Ellis L, Luther J, Carrington M, Stoecklin G, Clayton C. A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res. 2008;36:3374–3388. doi: 10.1093/nar/gkn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li CH, Irmer H, Gudjonsdottir-Planck D, Freese S, Salm H, Haile S, Estevez AM, Clayton C. Roles of a Trypanosoma brucei 5′->3′ exoribonuclease homolog in mRNA degradation. RNA. 2006;12:2171–2186. doi: 10.1261/rna.291506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cristodero M, Bottcher B, Diepholz M, Scheffzek K, Clayton C. The Leishmania tarentolae exosome: purification and structural analysis by electron microscopy. Mol. Biochem. Parasitol. 2008;159:24–29. doi: 10.1016/j.molbiopara.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Estevez AM, Kempf T, Clayton C. The exosome of Trypanosoma brucei. EMBO J. 2001;20:3831–3839. doi: 10.1093/emboj/20.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estevez AM, Lehner B, Sanderson CM, Ruppert T, Clayton C. The roles of intersubunit interactions in exosome stability. J. Biol. Chem. 2003;278:34943–34951. doi: 10.1074/jbc.M305333200. [DOI] [PubMed] [Google Scholar]
- 16.Haile S, Estevez AM, Clayton C. A role for the exosome in the in vivo degradation of unstable mRNAs. RNA. 2003;9:1491–1501. doi: 10.1261/rna.5940703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haile S, Dupe A, Papadopoulou B. Deadenylation-independent stage-specific mRNA degradation in Leishmania. Nucleic Acids Res. 2008;36:1634–1644. doi: 10.1093/nar/gkn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bringaud F, Muller M, Cerqueira GC, Smith M, Rochette A, El-Sayed NM, Papadopoulou B, Ghedin E. Members of a large retroposon family are determinants of post-transcriptional gene expression in Leishmania. PLoS Pathog. 2007;3:1291–1307. doi: 10.1371/journal.ppat.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bringaud F, Ghedin E, El-Sayed NM, Papadopoulou B. Role of transposable elements in trypanosomatids. Microbes. Infect. 2008;10:575–581. doi: 10.1016/j.micinf.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Smith M, Blanchette M, Papadopoulou B. Improving the prediction of mRNA extremities in the parasitic protozoan Leishmania. BMC Bioinformatics. 2008;9:158. doi: 10.1186/1471-2105-9-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith M, Bringaud F, Papadopoulou B. Organization and evolution of two SIDER retroposon subfamilies and their impact on the Leishmania genome. BMC Genomics. 2009;10:240. doi: 10.1186/1471-2164-10-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boucher N, Wu Y, Dumas C, Dube M, Sereno D, Breton M, Papadopoulou B. A common mechanism of stage-regulated gene expression in Leishmania mediated by a conserved 3′-untranslated region element. J. Biol. Chem. 2002;277:19511–19520. doi: 10.1074/jbc.M200500200. [DOI] [PubMed] [Google Scholar]
- 23.McNicoll F, Muller M, Cloutier S, Boilard N, Rochette A, Dube M, Papadopoulou B. Distinct 3′-untranslated region elements regulate stage-specific mRNA accumulation and translation in Leishmania. J. Biol. Chem. 2005;280:35238–35246. doi: 10.1074/jbc.M507511200. [DOI] [PubMed] [Google Scholar]
- 24.Roy G, Dumas C, Sereno D, Wu Y, Singh AK, Tremblay MJ, Ouellette M.MO, Papadopoulou B. Episomal and stable expression of reporter genes for quantifying Leishmania spp. infections in macrophages and in animal models. Mol. Biochem. Parasitol. 2000;110:195–206. doi: 10.1016/s0166-6851(00)00270-x. [DOI] [PubMed] [Google Scholar]
- 25.Papadopoulou B, Roy G, Ouellette M. A novel antifolate resistance gene on the amplified H circle of Leishmania. EMBO J. 1992;11:3601–3608. doi: 10.1002/j.1460-2075.1992.tb05444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwede A, Manful T, Jha BA, Helbig C, Bercovich N, Stewart M, Clayton C. The role of deadenylation in the degradation of unstable mRNAs in trypanosomes. Nucleic Acids Res. 2009;37:5511–5528. doi: 10.1093/nar/gkp571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Badis G, Saveanu C, Fromont-Racine M, Jacquier A. Targeted mRNA degradation by deadenylation-independent decapping. Mol. Cell. 2004;15:5–15. doi: 10.1016/j.molcel.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 28.Liu H, Kiledjian M. An erythroid-enriched endoribonuclease (ErEN) involved in alpha-globin mRNA turnover. Protein Pept. Lett. 2007;14:131–136. doi: 10.2174/092986607779816168. [DOI] [PubMed] [Google Scholar]
- 29.Muhlrad D, Parker R. The yeast EDC1 mRNA undergoes deadenylation-independent decapping stimulated by Not2p, Not4p, and Not5p. EMBO J. 2005;24:1033–1045. doi: 10.1038/sj.emboj.7600560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoenberg DR, Cunningham KS. Characterization of mRNA endonucleases. Methods. 1999;17:60–73. doi: 10.1006/meth.1998.0708. [DOI] [PubMed] [Google Scholar]
- 31.Grange T. Sensitive detection of mRNA decay products by use of reverse-ligation-mediated PCR (RL-PCR) Methods Enzymol. 2008;448:445–466. doi: 10.1016/S0076-6879(08)02622-0. [DOI] [PubMed] [Google Scholar]
- 32.Couttet P, Fromont-Racine M, Steel D, Pictet R, Grange T. Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc. Natl Acad. Sci. USA. 1997;27:5628–5633. doi: 10.1073/pnas.94.11.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monnerat S, Martinez-Calvillo S, Worthey E, Myler PJ, Stuart KD, Fasel N. Genomic organization and gene expression in a chromosomal region of Leishmania major. Mol. Biochem. Parasitol. 2004;134:233–243. doi: 10.1016/j.molbiopara.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 34.Papadopoulou B, Roy G, Ouellette M. Autonomous replication of bacterial DNA plasmid oligomers in Leishmania. Mol. Biochem. Parasitol. 1994;65:39–49. doi: 10.1016/0166-6851(94)90113-9. [DOI] [PubMed] [Google Scholar]
- 35.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl Acad. Sci. USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rochette A, Raymond F, Ubeda JM, Smith M, Messier N, Boisvert S, Rigault P, Corbeil J, Ouellette M, Papadopoulou B. Genome-wide gene expression profiling analysis of Leishmania major and Leishmania infantum developmental stages reveals substantial differences between the two species. BMC Genomics. 2008;9:255. doi: 10.1186/1471-2164-9-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen BC, Sivam D, Kifer CT, Myler PJ, Parsons M. Widespread variation in transcript abundance within and across developmental stages of Trypanosoma brucei. BMC Genomics. 2009;10:482. doi: 10.1186/1471-2164-10-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kabani S, Fenn K, Ross A, Ivens A, Smith TK, Ghazal P, Matthews K. Genome-wide expression profiling of in vivo-derived bloodstream parasite stages and dynamic analysis of mRNA alterations during synchronous differentiation in Trypanosoma brucei. BMC Genomics. 2009;10:427. doi: 10.1186/1471-2164-10-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ouellette M, Papadopoulou B. Coordinated gene expression by post-transcriptional regulons in African trypanosomes. J. Biol. 2009;8:100. doi: 10.1186/jbiol203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Queiroz R, Benz C, Fellenberg K, Hoheisel JD, Clayton C. Transcriptome analysis of differentiating trypanosomes reveals the existence of multiple post-transcriptional regulons. BMC Genomics. 2009;10:495. doi: 10.1186/1471-2164-10-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gopalan V, Vioque A, Altman S. RNase P: variations and uses. J. Biol. Chem. 2002;277:6759–6762. doi: 10.1074/jbc.R100067200. [DOI] [PubMed] [Google Scholar]
- 42.Xiao S, Scott F, Fierke CA, Engelke DR. Eukaryotic ribonuclease P: a plurality of ribonucleoprotein enzymes. Annu. Rev. Biochem. 2002;71:165–189. doi: 10.1146/annurev.biochem.71.110601.135352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang F, Schoenberg DR. Endonuclease-mediated mRNA decay involves the selective targeting of PMR1 to polyribosome-bound substrate mRNA. Mol. Cell. 2004;14:435–445. doi: 10.1016/j.molcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 44.Tourriere H, Gallouzi IE, Chebli K, Capony JP, Mouaikel J, van der Geer P, Tazi J. RasGAP-associated endoribonuclease G3Bp: selective RNA degradation and phosphorylation-dependent localization. Mol. Cell Biol. 2001;21:7747–7760. doi: 10.1128/MCB.21.22.7747-7760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 46.Gill T, Aulds J, Schmitt M. A specialized processing body that is temporally and asymmetrically regulated during the cell cycle in Saccharomyces cerevisiae. J. Cell Biol. 2006;173:35–45. doi: 10.1083/jcb.200512025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaskiewicz L, Filipowicz W. Role of Dicer in posttranscriptional RNA silencing. Curr. Top. Microbiol. Immunol. 2008;320:77–97. doi: 10.1007/978-3-540-75157-1_4. [DOI] [PubMed] [Google Scholar]
- 48.Lingel A, Izaurralde E. RNAi: finding the elusive endonuclease. RNA. 2004;10:1675–1679. doi: 10.1261/rna.7175704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lebreton A, Tomecki R, Dziembowski A, Seraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–996. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- 50.Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat. Struct. Mol. Biol. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat. Struct. Mol. Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 52.Bringaud F, Ghedin E, Blandin G, Bartholomeu DC, Caler E, Levin MJ, Baltz T, El-Sayed NM. Evolution of non-LTR retrotransposons in the trypanosomatid genomes: Leishmania major has lost the active elements. Mol. Biochem. Parasitol. 2006;145:158–170. doi: 10.1016/j.molbiopara.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 53.Zingler N, Weichenrieder O, Schumann GG. APE-type non-LTR retrotransposons: determinants involved in target site recognition. Cytogenet Genome Res. 2005;110:250–268. doi: 10.1159/000084959. [DOI] [PubMed] [Google Scholar]
- 54.Barnes T, Kim WC, Mantha AK, Kim SE, Izumi T, Mitra S, Lee CH. Identification of Apurinic/apyrimidinic endonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA. Nucleic Acids Res. 2009;37:3946–3958. doi: 10.1093/nar/gkp275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, Quail MA, Peters N, Adlem E, Tivey A, Aslett M, et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat. Genet. 2007;39:839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robinson KA, Beverley SM. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol. Biochem. Parasitol. 2003;128:217–228. doi: 10.1016/s0166-6851(03)00079-3. [DOI] [PubMed] [Google Scholar]
- 57.Siomi MC, Saito K, Siomi H. How selfish retrotransposons are silenced in Drosophila germline and somatic cells. FEBS Lett. 2008;582:2473–2478. doi: 10.1016/j.febslet.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 58.Belli SI, Monnerat S, Schaff C, Masina S, Noll T, Myler PJ, Stuart K, Fasel N. Sense and antisense transcripts in the histone H1 (HIS-1) locus of Leishmania major. Int. J. Parasitol. 2003;33:965–975. doi: 10.1016/s0020-7519(03)00126-7. [DOI] [PubMed] [Google Scholar]
- 59.Dumas C, Chow C, Muller M, Papadopoulou B. A novel class of developmentally regulated noncoding RNAs in Leishmania. Eukaryot. Cell. 2006;5:2033–2046. doi: 10.1128/EC.00147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kapler GM, Beverley SM. Transcriptional mapping of the amplified region encoding the dihydrofolate reductase-thymidylate synthase of Leishmania major reveals a high density of transcripts, including overlapping and antisense RNAs. Mol. Cell Biol. 1989;9:3959–3972. doi: 10.1128/mcb.9.9.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 2007;10:102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 62.Matsui K, Nishizawa M, Ozaki T, Kimura T, Hashimoto I, Yamada M, Kaibori M, Kamiyama Y, Ito S, Okumura T. Natural antisense transcript stabilizes inducible nitric oxide synthase messenger RNA in rat hepatocytes. Hepatology. 2008;47:686–697. doi: 10.1002/hep.22036. [DOI] [PubMed] [Google Scholar]
- 63.Opdyke J, Kang J, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen DQ, Kolli BK, Yadava N, Lu HG, Gilman-Sachs A, Peterson DA, Chang KP. Episomal expression of specific sense and antisense mRNAs in Leishmania amazonensis: modulation of gp63 level in promastigotes and their infection of macrophages in vitro. Infect. Immun. 2000;68:80–86. doi: 10.1128/iai.68.1.80-86.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang WW, Matlashewski G. Loss of virulence in Leishmania donovani deficient in an amastigote- specific protein, A2. Proc. Natl Acad. Sci. USA. 1997;94:8807–8811. doi: 10.1073/pnas.94.16.8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.