Abstract
Several periodic motifs have been implicated in facilitating the bending of DNA around the histone core of the nucleosome. For example, di-nucleotides AA/TT/TA and GC at ∼10-bp periods, but offset by 5 bp, are found with higher-than-expected occurrences in aligned nucleosomal DNAs in vitro and in vivo. Additionally, regularly oscillating period-10 trinucleotide motifs non-T, A/T, G and their complements have been implicated in the formation of regular nucleosome arrays. The effects of these periodic motifs on nucleosome formation have not been systematically tested directly by competitive reconstitution assays. We show that, in general, none of these period-10 motifs, except TA, in certain sequence contexts, facilitates nucleosome formation. The influence of periodic TAs on nucleosome formation is appreciable; with some of the 200-bp DNAs out-competing bulk nucleosomal DNA by more than 400-fold. Only the nucleotides immediately flanking TA influence its nucleosome-forming ability. Period-10 TA, when flanked by a pair of permissive nucleotides, facilitates DNA bending through compression of the minor groove. The free energy change for nucleosome formation decreases linearly with the number of consecutive TAs, up to eight. We suggest how these data can be reconciled with previous findings.
INTRODUCTION
Little is known about the possible function of the bulk of the human genome. It is likely that some functionally influential non-coding DNA exerts its influence through chromatin structure. Nucleosomes appear to be positioned with respect to the DNA sequence to some extent in most regions of genomic DNA in vivo (1).
It has been known for some time that nucleosomes prefer to form on certain DNA sequences (2), and tend to avoid others in vitro (3). The nucleosome arrangement over large DNA regions is expected to influence the chromatin higher-order structure, according to some models (4,5). If DNA sequence information can be used to predict which regions of DNA prefer to form nucleosomes in vivo, it would provide a valuable resource for understanding DNA packaging and chromatin structure in nuclei. The accessibility of particular regions of chromatin to histone modifications and the effects of chromatin remodeling might also be influenced by the DNA sequence.
Several periodic motifs have been implicated in the preferential formation of nucleosomes. Periodic di-nucleotides have been found in nucleosomal DNA (6–8). It has also been shown that some synthetic DNAs having AT-rich regions alternating with GC-rich regions approximately every 5 bp have a high preference for forming nucleosomes in vitro (9). In addition, it has been shown that synthetic DNAs possessing period-10 A-tracts have an increased ability to form nucleosomes (10). The preferred compression of the DNA minor groove at the AT-rich motifs and the preferred compression of the major groove at the GC-rich motifs are thought to facilitate bending around the histone core, when the AT-rich and 5-bp offset GC-rich motifs occur at multiples of 10 bp, the periodicity of the DNA double helix (7,11,12).
The periodic di-nucleotide motifs AA/TT/TA, with GC 5 bp away, has recently been implicated in a ‘DNA code’ for nucleosome positioning (11,13). In addition, the periodic tri-nucleotide motifs non-T, A/T, G (VWG) with complement C, T/A, non-A (CWB) (14) have been shown to correlate with nucleosome arrangements in arrays (15) and with chromosome function (16) in vivo. It is not clear why both of these two apparently unrelated periodic motifs (AA/TT/TA and VWG/CWB) appear to influence nucleosome formation. Moreover, no direct unambiguous measurements of the relative abilities of most of these DNA sequence motifs to form nucleosomes exist. The evidence that these motifs preferentially form nucleosomes is mostly correlative. For the di-nucleotide motifs, relatively weak 10-bp periodicities, over a background, were found in DNA isolated from nucleosomes, when the sequences were aligned and the total number of occurrences of the motif at each position was counted (7,11–13). In the few cases where competitive reconstitutions were performed, the studies were not systematic, and it was not clear exactly which motif, or which combination of motifs, were responsible for preferential nucleosome formation. For the tri-nucleotide motifs, no direct tests of nucleosome-forming ability exist at all.
We have begun a systematic experimental investigation of the nucleosome-forming abilities (in vitro) of various periodic di- and tri-nucleotide motifs using radio-labeled 200-bp synthetic DNA fragments in competitive reconstitution assays with bulk nucleosomal DNA. It is important to point out that this simple in vitro assay may be measuring a different aspect of nucleosome formation potential for a particular DNA sequence than what exists in vivo. However, we used this method because it is well established, readily quantifiable, the magnitudes of the effects that we observed were large and the thermodynamic property of histone–DNA interactions measured likely contributes to nucleosome preferences for particular DNA sequences in vivo. The results turned out to be surprising. Neither perfect period-10 AA/TT, even with GC 5 bp away, nor perfect period-10 VWG/CWB have increased nucleosome-forming abilities. In contrast, 200-bp sequences having periodic TA, in some sequence contexts, have very high nucleosome-forming abilities. We suggest how this result can be reconciled with previous findings. Periodic TA is abundant in human genomic DNA, and these data should permit more reliable computational predictions about chromatin structure to be made.
MATERIALS AND METHODS
Preparation and labeling of synthetic DNA
The preparation and the radiolabeling of the periodic synthetic sequences used in this study (listed in Table 1) are described in Supplementary Figure S1. Sequences, except for p-10 CTACN6 (mix) and p-10 TAN8 (mix), were cloned into pJAZZ-OC linear vector (BIGEASY, Lucigen), and DNA sequenced by SL1 and NZrevC primers (Table S1). The 195-bp p-10 CTACN6 (mix) and the p-10 TAN8 (mix) mixtures containing random nucleotides were prepared by a single-stranded (ss) DNA-based polymerase chain reaction (PCR) method (Figure S1E). Briefly, a 195-nt synthetic single-stranded DNA containing the SL1 primer sequence at the 5′ end, 15 multiples of the 10-nt N-containing motif (one DNA with CTACN6 and another with TAN8, where N is a random nucleotide) and the complement to the TevC primer sequence at the 3′ end was purchased. PCR was performed in the presence of the SL1 and the TevC primers. Only the TevC primer anneals to the ssDNA template at the first PCR cycle, converting the template to a double-stranded DNA. For subsequent cycles, both primers anneal and PCR amplification occurs. The 195-bp double-stranded DNA product, which is a mixture of many thousands of different sequences, was gel purified before use. See Supplementary Figure S1E for an illustration and for experimental details.
Table 1.
Competitive reconstitution results and the sequences of repeating 20-bp units that were used in this study
| *Sequence name | †Sequence (20 bp) | ‡Fold out-competition |
|---|---|---|
| 601 sequence | 950 ± 380 | |
| 1. p-10 AAn3GC | 5′ AAGCTGCCGGAAGCTGCCGG 3′ | 2.65 ± 0.17 |
| 3′ TTCGACGGCCTTCGACGGCC 5′ | ||
| 2. p-10 GTAC (GTAC) | 5′ GTACGGGCGGGTACGGGCGG 3′ | 1.15 ± 0.08 |
| 3′ CATGCCCGCCCATGCCCGCC 5′ | ||
| 3. p-10 ATAG (CTAT) | 5′ ATAGGGCAGCATAGGGCAGC 3′ | 0.65 ± 0.16 |
| 3′ TATCCCGTCGTATCCCGTCG 5′ | ||
| 4. p-10 ATAT (ATAT) | 5′ ATATGGCAGCATATGGCAGC 3′ | 0.77 ± 0.32 |
| 3′ TATACCGTCGTATACCGTCG 5′ | ||
| 5. p-10 TTAT (ATAA) | 5′ TTATGGCAGCTTATGGCAGC 3′ | 0.83 ± 0.21 |
| 3′ AATACCGTCGAATACCGTCG 5′ | ||
| 6. p-10 AT | 5′ ATCGGCAGCCATCGGCAGCC 3′ | 0.37 ± 0.04 |
| 3′ TAGCCGTCGGTAGCCGTCGG 5′ | ||
| 7. p-10 TAn3AA | 5′ TACGGAATCCTACGGAATCC 3′ | 326 ± 22 |
| 3′ ATGCCTTAGGATGCCTTAGG 5′ | ||
| 8. p-10 CTAC (GTAG) | 5′ CTACGGCAGCCTACGGCAGC 3′ | 447 ± 63 |
| 3′ GATGCCGTCGGATGCCGTCG 5′ | ||
| 9. p-10 TTAC (GTAA) | 5′ TTACGGCAGCTTACGGCAGC 3′ | 415 ± 116 |
| 3′ AATGCCGTCGAATGCCGTCG 5′ | ||
| 10. p-10 TTAA (TTAA) | 5′ TTAAGGCAGCTTAAGGCAGC 3′ | 64 ± 11 |
| 3′ AATTCCGTCGAATTCCGTCG 5′ | ||
| 11. p-10 CTAA (TTAG) | 5′ CTAAGGCAGCCTAAGGCAGC 3′ | 132 ± 79 |
| 3′ GATTCCGTCGGATTCCGTCG 5′ | ||
| 12. p-10 ATAC (GTAT) | 5′ ATACCGGCAGATACCGGCAG 3′ | 104 ± 79 |
| 3′ TATGGCCGTCTATGGCCGTC 5′ | ||
| 13. p-10 CTAG (CTAG) | 5′ CTAGGCAGCCCTAGGCAGCC 3′ | 494 ± 148 |
| 3′ GATCCGTCGGGATCCGTCGG 5′ | ||
| 14. p-10 CTACN6 (mix) | 5′ CTACNNNNNNCTACNNNNNN 3′ | 355 ± 45 |
| 3′ GATGXXXXXXGATGXXXXXX 5′ | ||
| 15. p-10 TAN8 (mix) | 5′ TANNNNNNNNTANNNNNNNN 3′ | 42 ± 14 |
| 3′ ATXXXXXXXXATXXXXXXXX 5′ | ||
| 16. p-10 TA/AT | 5′ TACGGCAGCCATCGGCAGCC 3′ | 0.66 ± 0.08 |
| 3′ ATGCCGTCGGTAGCCGTCGG 5′ | ||
| 17. p-10 TA/GG | 5′ TACGGCAGCCGGCGGCAGCC 3′ | 1.08 ± 0.36 |
| 3′ ATGCCGTCGGCCGCCGTCGG 5′ | ||
| 18. p-10 TA/AA | 5′ TACGGCAGCCAACGGCAGCC 3′ | 118 ± 16 |
| 3′ ATGCCGTCGGTTGCCGTCGG 5′ |
*n3 indicates the presence of three specified nucleotides.
(mix) indicates a mixture of randomly chosen nucleotides.
Nucleotides in parenthesis represent the complementary nucleotides on the other strand.
†N and X stand for randomly chosen nucleotides in one strand, and its complement, respectively.
Bold di- and tri-nucleotides represent the periodic sequence motif of interest.
‡Fold that the 200-bp synthetic DNAs out-compete nucleosomal DNA. Standard deviations shown are calculated from three independent measurements (n = 3), except for the 601 sequence (n = 6).
In vitro competitive nucleosome reconstitution method
Core histones and mononucleosomal DNA (147–210 bp) were prepared as previously described (17). Competitive nucleosome reconstitution was carried out by a modified stepwise dilution method, summarized in Figure S2 (9). Briefly, for high nucleosome-forming ability sequences, the reaction mixture [1 M NaCl, 10 mM Tris–HCl pH 8.0, 0.2 mM ethylenediaminetetraacetic acid (EDTA)] containing 1 ug of core histones, a tracer amount of the labeled synthetic sequence and 3.4 µg of mononucleosomal DNA competitor in a total volume of 10 ul were incubated for 30 min at 4°C. For average nucleosome-forming-ability sequences, the core histones to DNA ratio was increased; 3 ug of histone octamers and 2.6 µg of mononucleosomal DNA were mixed. Stepwise salt dilution was performed by adding 5 mM Tris–HCl pH 8.0, 0.5 mM EDTA (TE) buffer for the first dilution to 0.8 M NaCl for 1 h at 4°C, and the second dilution to 0.6 M NaCl for 15 min at 4°C. Finally, samples were dialyzed against TE buffer for 3 h at 4°C using Millipore (0.025 µm) dialysis disks.
Polyacrylamide gel electrophoresis
Reconstituted samples were analyzed on a 4% native polyacrylamide gel. Gels were pre-run for 1 h at 150 V, and samples were run at 150 V for 3.5 h at 4°C. Gels were stained with ethidium bromide (EtBr), and digitized for the bulk mononucleosomal DNA. Subsequently, the gels were dried, and exposed to MR film (Kodak) to detect the labeled DNA sequence of interest. An optimum film exposure for quantitation was obtained by examining several different exposure times; results were consistent with those obtained by phosphoimager analysis. Nucleosome-forming ability was measured as described in Supplementary Figure S2. The 601 sequence was used as a reference for the sequences that possessed high nucleosome-forming ability of more than 300-fold relative to bulk DNA (ΔΔG° < −3.3 kcal/mol). Otherwise, nucleosome-forming ability of sequences were measured relative to bulk DNA from EtBr-stained gels. Using (low specific activity) radiolabeled bulk nucleosomal DNA in control experiments, we determined that the fraction of the DNA that reconstitutes into nucleosomes in a given reconstitution was essentially identical when measured by EtBr fluorescence or by radiolabeling (data not shown). The stated standard deviations were calculated from at least three independent experiments.
Hydroxyl radical footprinting assay
Nucleosomes were reconstituted using 5′-32P-labeled p-10 CTAC (GTAG) sequence as described in Supplementary Figure S1F. By appropriately increasing the ratio of core histones to competitor DNA in the reaction mixture, described in Supplementary Figure S2, the percentage of labeled DNA fragment incorporated into nucleosomes was adjusted to >90% to minimize the signal from contaminating naked DNA. Hydroxyl radical footprinting was performed as described by Tullius et al. (18). The reaction was initiated by adding 10 µM (NH4)2Fe(SO4)2•6H2O, 20 µM EDTA, 1 mM ascorbic acid and 0.03% H2O2 for naked DNA and nucleosome-formed DNA samples. The reaction was terminated by adding 10 mM thiourea, and 50 mM EDTA; then samples were deproteinized by phenol–chloroform extraction, and ethanol precipitated in the presence of 60 µg/ml tRNA (18). A G/A chemical reaction was also performed as described (19). Samples were subjected to 8% polyacrylamide sequencing gel (acrylamide: bisacrylamide ratio of 19:1) containing 7 M urea and 10% formamide on 33 × 42 × 0.04 cm with 1X Tris–borate–EDTA buffer (90 mM Tris/borate, 2 mM EDTA). Gels were pre-run for 1 h at 45 mA constant current, and samples preheated at 90°C for 10 min were run for 3 h. Gels were dried, and exposed to 35 × 43 cm classic blue autoradiography film (MidSci). Autoradiograms were digitalized by means of a standard scanner (Epson Perfection 4990). Thereafter, lane scans were performed for each lane using IPLab Gel software (Signal Analytics Corp.).
Other methods
Counting of total TA or VWG/CWB motifs in a sliding 100-bp window along the mouse adenosine deaminase (MADA) gene locus was performed computationally as described (20). Analysis of DNA fragments for macroscopic curvature was performed on 10% polyacrylamide gels at 5°C (10).
RESULTS AND DISCUSSION
Direct determination of the nucleosome-forming abilities of 200-bp synthetic DNAs with periodic motifs
The competitive reconstitution method is depicted schematically in Supplementary Figure S2. This method allows the direct measurement of the relative nucleosome-forming abilities of any sequence relative to bulk nucleosomal DNA, or the relative nucleosome-forming abilities of any two sequences (9). We chose bulk nucleosomal DNA (147–210 bp) as our reference state. After reconstituting a mixture containing a relatively large amount of bulk nucleosomal DNA and a small (tracer) amount of radiolabeled DNA at an appropriate core histone to DNA ratio, nucleosomes are separated from the unreacted DNA on a polyacrylamide gel, and the relative amounts of DNA in each band are quantitated on the stained gel and on the autoradiogram. From the ratios of the intensities of the nucleosome and DNA bands, the nucleosome-forming ability of the sequence of interest relative to bulk nucleosomal DNA (and the ΔΔG° value) is calculated (as described in Supplementary Figure S2). The bulk nucleosomal DNA serves as an internal control. In addition, because of the relatively large quantity of bulk nucleosomal DNA present, reactions using different radiolabeled DNAs are highly reproducible, allowing for the comparison of the nucleosome-forming ability of one synthetic DNA with another, when the same amounts of core histones and bulk nucleosomal DNA are used. This method has been extensively tested by Widom and co-workers (21). The conditions used here have been shown to reliably reflect the relative nucleosome-forming abilities of different DNA sequences in vitro.
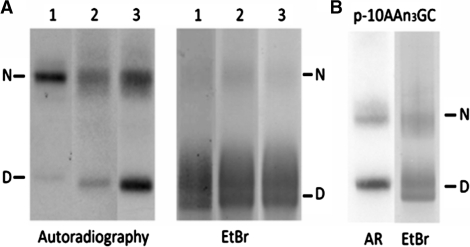
Figure 1 shows a sampling of our competitive reconstitution data. In Figure 1, we included DNA sequences that exhibited a wide range of nucleosome-forming abilities. It is evident by comparing the autoradiogram with the stained gel (EtBr) that all three of the synthetic DNAs in Figure 1A form nucleosomes more efficiently than bulk nucleosomal DNA. Lane 1 shows the ‘601’ sequence (22), which has the highest nucleosome-forming ability known (∼1000-fold that of bulk nucleosomal DNA). This sequence was useful for more precisely quantitating those sequences (such as the one shown in lane 2) that had nucleosome forming abilities >300 times that of bulk nucleosomal DNA, as described in ‘Materials and Methods’. The sequence used in lane 3 had a nucleosome-forming ability of ∼40 times that of bulk nucleosomal DNA. In Figure 1B, we show the data for a periodic synthetic DNA with an average nucleosome-forming ability, one that is not significantly different from that of bulk nucleosomal DNA. The N-to-D band intensities were similar for the bulk DNA (EtBr) and the labeled DNA (AR). The nucleosome-forming abilities of 18 different periodic synthetic DNAs are reported in Table 1 and Supplementary Table S2. Twenty base pairs of sequence, encompassing the repeating unit, of each 200-bp DNA is shown, with the motifs of interest indicated. The result found for each synthetic DNA is discussed subsequently. A value greater than one for the fold that the sequence outcompetes bulk nucleosomal DNA indicates that the sequence forms nucleosomes more readily than bulk nucleosomal DNA. This information, including ΔΔG° values, is reported in Supplementary Table S2.
Figure 1.
Examples of the direct measurements of the nucleosome-forming abilities of DNA sequences by competitive reconstitution. (A) Three DNAs, each with high nucleosome-forming ability, were reconstituted (separately) using a low core histone to bulk DNA ratio. Lanes 1–3, respectively, show the 601 sequence, the p-10 CTACN6(mix) and the p-10 TAN8 (mix). PAGE autoradiogram (left) and corresponding EtBr-stained gel (right) are shown. N and D denote nucleosomes and free DNA, respectively. (B) PAGE autoradiogram (AR) and EtBr-stained gel are shown for the p-10 AAn3GC sequence, which had an average nucleosome-forming ability. This sequence was reconstituted using a higher core histone to bulk DNA ratio than used in (A).
‘Established’ nucleosome-forming motifs do not form nucleosomes preferentially
It has been thought for some time that DNA sequence regions containing period-10 AA/TT and period-10 GC, offset by 5 bp, should readily bend around the histone cores of nucleosomes (7). This claim is even illustrated in a popular textbook (23). More recently, period-10 AA/TT/TA with period-10 GC, offset by 5 bp, has been referred to as a sequence motif that is known to facilitate the bending of DNA around the histone core of the nucleosome (11). Sequence 1 (Table 1) contains 20 perfectly periodic (period-10) AA/TTs and GCs offset by 5 bp. The increase in nucleosome-forming ability over bulk nucleosomal DNA is only about 2-fold, a very small increase when compared to the ∼1000-fold increase of the 601 sequence (Table 1, top of the list). In addition, sequence 2, which contains 20 perfectly periodic (period-10) TAs and GCs, offset by 5 bp, exhibits no appreciable increase in nucleosome-forming ability over bulk nucleosomal DNA. These direct experimental measurements give results that are inconsistent with the ‘established’ preferred DNA motifs for nucleosome formation.
It has also been proposed that the period-10 triplet motifs non-T, A/T, G (VWG) and their complements (CWB) preferentially form nucleosomes by having high bending potentials of the in-phase triplets toward the major groove (14). The triplet pair CAG/CTG was proposed to have the highest bendability. Sequences 3–6 (Table 1) all contain 20 perfectly periodic (period-10) CAG/CTG motifs, yet they do not form nucleosomes more readily than bulk nucleosomal DNA. Sequences 3–5 contain period-10 TA in addition to period-10 CAG/CTG. These two motifs, offset by 5 bp, should have synergized, according to the expected minor groove compression for TA and major groove compression for CAG/CTG. Sequence 6 did not have TA or AA/TT di-nucleotides, but it had AT every 10 bp. All of these results (sequences 1–6) are inconsistent with the idea that ‘established’ period-10 di-nucleotide motifs or period-10 VWG/CWB motifs facilitate nucleosome formation.
Some period-10 TA sequences form nucleosomes very efficiently
Some period-10 TA sequences 7-13 (Table 1) form nucleosomes very efficiently. For example, sequences 8, 9 and 13 have affinities for core histones that approach that of the 601 sequence. It is interesting that only one of these seven sequences (sequence 12) has period-10 GC offset by 5 bp from TA. Thus, the period-10 GC motif offset by 5 bp from TA does not generally increase nucleosome-forming ability, going against the ‘established’ rule. Moreover, sequence 7 has period-10 AA offset by 5 bp from TA, which would be expected to interfere with DNA bending by minor groove compression at AA/TT.
The period-10 TA sequence context for nucleosome formation depends primarily on nearest-neighbor flanking nucleotides
In our competitive reconstitution studies, we observed that many period-10 CTAC sequences having various combinations of nucleotides at the other six positions all had very high nucleosome-forming abilities (see Supplementary Table S3). These results suggested that the other 6 nt in the repeating unit might not be important. To test this idea, we prepared a mixture of different 200-bp sequences that were all period-10 CTAC, but had randomly chosen nucleotides (Ns) inserted at the other positions (sequence 14). After competitive reconstitution, we obtained a very high nucleosome-forming preference of 355 ± 45 relative to bulk nucleosomal DNA, which is not statistically different from the value of 447 ± 63, obtained for sequence 8 (P-value of 0.11 for two-tailed t-test). Thus, the 2 nt immediately flanking TA (one on each side) are major contributors to the nucleosome-forming ability of the periodic DNA.
Determination of which nucleotide combinations flanking TA are necessary for high nucleosome-forming ability
There are 16 possible pairs of nucleotides to flank TA with one on the left and one on the right. In double-stranded DNA, for each nucleotide pair on the top strand, the complementary pair is present (flanking TA) on the bottom strand. For example, ATAA provides TTAT. Therefore, generally, two complementary pairs of TA-flanking nucleotides are provided from each sequence. However, 4 of the 16 di-nucleotides (TA, AT, GC, CG) are self-complementary, and these flanking di-nucleotides therefore only provide one pair. Thus, there are 10 [(16 – 4)/2 + 4] sequences needed to provide all possible TA-flanking nucleotide pairs. In Table 1, these sequences are numbers 2–5 and 8–13; the tetra-nucleotides in parentheses (Table 1, first column) show the left and right nucleotide flanks of TA on the other strand. For example, TTAC (GTAA), sequence number 9 in Table 1, has (left, right) flanking nucleotides TC/GA, denoting that T and C flank TA on one strand, while, G and A flank TA on the other strand. It can be seen from Table 1 that the six (left, right) TA-flanking nucleotide (/complement) pairs, CC/GG, TC/GA, TA, CA/TG, AC/GT and CG, confer high nucleosome-forming ability to period-10 TA, whereas the four TA-flanking nucleotide (/complement) pairs, GC, AG/CT, AT and TT/AA, confer average or low nucleosome-forming ability to period-10 TA. The structural reasons why these particular flanking nucleotide pairs confer high or average/low nucleosome-forming abilities are unknown. Sequence 15 (Table 1) was a mixture of different 195-bp sequences that were all period-10 TA, but had randomly chosen nucleotides (Ns) inserted at the other positions. Our determined nucleosome-forming ability of 42 ± 14 times that of bulk nucleosomal DNA is consistent with the value of 34 calculated for a hypothetical mixture of the 10 sequences (2–5, 8–13) listed in Table 1 with the individual values listed.
An AA/TT located 10 nt from a TA can in part substitute for a TA
We also investigated whether DNAs with periodic TAs every 20 bp had high nucleosome-forming abilities. To achieve this arrangement, we substituted every other period-10 TA in sequence 8 with another di-nucleotide. Substituting every other TA for either AT or GG (Table 1, sequences 16 and 17, respectively) abolished the high nucleosome-forming ability. This result suggests that, generally, at least two TAs that are 10 bp apart may be required. However, substitution of every other TA with AA/TT (Table 1, sequence 18) maintained a high nucleosome-forming ability, although it was approximately four times lower than that of the parent construct, sequence 8. This result suggests that the AA/TT motif can, under some circumstances, contribute to nucleosome formation.
The number of consecutive period-10 TAs needed for preferential nucleosome formation
Our synthetic DNA molecules that had high nucleosome-forming abilities contained 20 perfectly periodic TAs. Very few, if any, genomic DNA regions contain 20 period-10 TAs. However, there is evidence that <100 bp determines the histone octamer preference (24), and it is expected that periodic anisotropically flexible nucleotide motifs such as TA incrementally contribute to the free energy of nucleosome formation, with a roughly constant increase per TA (9). We therefore examined how the free energy of nucleosome formation depended upon the number of consecutive TAs present in the center of a 200-bp DNA fragment.
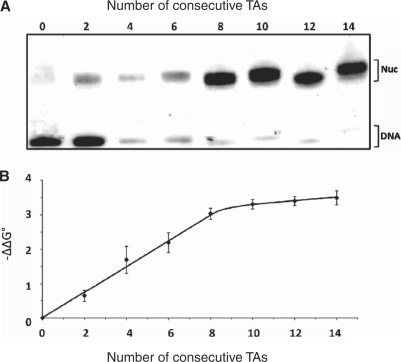
Figure 2 shows the effect of increasing the number of consecutive TAs from 0 (no TA) to 14 in competitive reconstitutions versus bulk mononucleosome DNA, as in our other experiments. The sequence context was CTAC, as in sequence 8 of Table 1. The consecutive period-10 TAs were located in the centers of 200-bp fragments; the regions flanking the consecutive period-10 TAs did not contain any period-10 TAs. Figure 2A shows the autoradiogram, while Figure 2B shows the ΔΔG° value as a function of the number of consecutive TAs, using the zero TA construct as the reference (ΔΔG° = 0). The ΔΔG° value becomes increasingly more negative in a linear fashion until about eight consecutive TAs (80 bp); then it approaches saturation. Table S4 reports the fold that each of the sequences outcompetes the zero TA sequence and the corresponding ΔΔG° values. This experiment confirms that the free energy increment is roughly constant with the number of periodic TAs, and shows that only four (consecutive) TAs is necessary to achieve nearly 50% of the maximum free energy change possible. This ΔΔG° value (–1.7 kcal/mol), for four consecutive TAs, corresponds to about a 12-fold preference for nucleosome formation over the zero TA sequence. This value is still appreciable.
Figure 2.
Variation of the free energy of nucleosome formation with the number of consecutive period-10 TAs. (A) PAGE autoradiogram of different labeled 200-bp DNA fragments, having from 0 to14 period-10 TAs located in the center, reconstituted competitively using a low core histone to bulk DNA ratio. (B) The x-axis is the number of consecutive TAs in the center of each DNA fragment, and the y-axis is the calculated value of –ΔΔG°, obtained from analysis of the data shown in (A), as described in the text and Supplementary Data. Error bars represent the SD from three independent experiments.
We do not currently know to what extents the ΔΔG° values change when there are four or more TAs in a 100-bp window in different non-consecutive periodic arrangements; however, 100-bp regions of genomic DNA having four periodic TAs are fairly common (unpublished data). Moreover, we have found (data not included) that a mix of TA periods of 9 bp and 11 bp is almost as effective as period 10, suggesting that there should be considerable modulation of TA within this period range in genomic DNA. It is very plausible that this degree of modulation could influence nucleosome positioning on DNA.
The rotational orientation of period-10 TA in nucleosomes and the preferred bending of TA in DNA
DNA sequences having period-10 motifs that exhibit a high preference for nucleosome formation are expected to bend anisotropically. The periodicity of the DNA double helix is ∼10 bp, and sequence-dependent DNA distortions will be in phase, leading to DNA bending. The current view is that A/T-rich regions prefer to compress their minor grooves, which face in toward the histone octamer, while G/C-rich regions prefer to have their minor grooves widened and facing out (7,11,12). This type of ‘rotational positioning’ is well documented in nucleosomes, but usually it is not possible to separate the effects of TA from those of the other A/T-rich di-nucleotides (9,12). Moreover, there is evidence that TA could preferentially compress its major groove (12,25–36). We therefore, examined the rotational positioning of our sequence 8 (Table 1), which had a very high nucleosome-forming ability, and for which the only A/T-rich di-nucleotide was TA, existing every 10 bp. Determining whether TA compressed its minor groove or its major groove could be readily accomplished by hydroxyl radical footprinting. Since hydroxyl radicals attack the DNA via the minor groove, compressed minor grooves should be cut less frequently, whereas expanded minor grooves should be cut more frequently than those of the (unbent) DNA in solution (37).
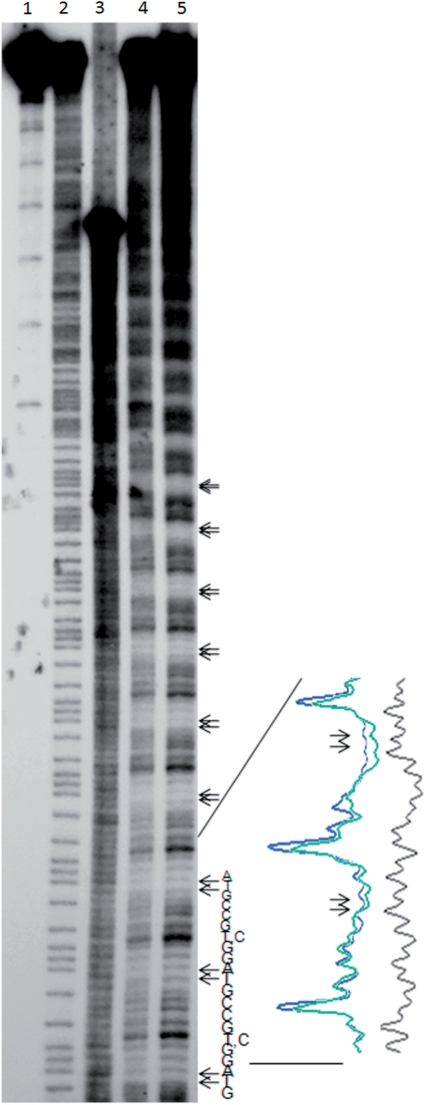
Figure 3 shows that the TAs are cut less frequently than the other di-nucleotides both in the nucleosome and in the naked DNA in solution. There is clearly a period-10 modulation in the cutting intensities for the synthetic DNA that is not present in a non-periodic fragment from pUC 19 DNA (lane 3). Lane scans are shown to the right of the autoradiogram for two periods of the synthetic DNA sequence. The hydroxyl radical cleavage patterns of the periodic synthetic DNA free in solution and in the nucleosome are very similar. On the one hand, this result seems remarkable considering that the minor groove accessibilities free in solution presumably arise from transient local bending. There is no macroscopic DNA curvature that we can detect by anomalously mobility on a polyacrylamide gel (data not shown). In the nucleosome, the TA minor grooves should be stably bent or kinked inward toward the histone octamer. It has been suggested that the TA step is highly flexible, and is well suited for minor groove kinking through large positive slide and negative roll (38). On the other hand, the similarity between the hydroxyl radical cutting patterns of the naked and the nucleosomal synthetic DNA perhaps should not be that surprising, because it has recently been shown that hydroxyl radical cleavage patterns of naked human genomic DNA correlate with chromatin function (39).
Figure 3.
Hydroxyl radical cutting patterns of p-10 CTAC (GTAG) nucleosomes and DNA in solution. The autoradiogram from a sequencing gel is shown. Lane 1 shows the 0-time point for the p-10 CTAC (GTAG) DNA. Lane 2 shows a G/A chemical cleavage reaction on p-10 CTAC (GTAG) DNA in solution. Lane 3 shows a 5-min hydroxyl radical cutting reaction on a 240-bp pUC 19 DNA fragment in solution. Lanes 4 and 5 show 5-min hydroxyl radical cutting reactions of p-10 CTAC (GTAG) naked DNA in solution and nucleosomal DNA, respectively. The TA di-nucleotides, occurring every 10 bp, are indicated by pairs of arrows. The sequence of 23 nt of p-10 CTAC (GTAG) from the bottom of the autoradiogram is shown. Lane scans are shown to the right of the autoradiogram for two periods of the synthetic p-10 CTAC (GTAG) DNA sequence: naked DNA (lane 4, green), the nucleosomal DNA (lane 5, blue) and the pUC 19 naked DNA (lane 3, black).
An alternative interpretation is that the TA steps, for the sequence contexts determined here that enhance nucleosome formation, simply have narrow minor grooves without base pair roll bending into the minor groove (40). It has recently been shown that the binding of arginine residues to narrow minor grooves is a widely used mechanism for protein–DNA recognition (36). Therefore, rather than ‘allowed’ TA steps preferentially flexing or kinking to narrow their minor grooves, sequence-specific contacts between histone arginine residues may occur periodically at ‘allowed’ TAs in the nucleosome. Narrow minor grooves have been suggested to enhance the stability of arginine contacts through electrostatic effects (36).
How these results can be reconciled with other findings
We have shown directly that neither of the period-10 motifs AA/TT or VWG/CWB contributes to nucleosome formation, whereas period-10 TA, in certain sequence contexts, strongly promotes nucleosome formation in vitro. Yet, the seemingly unrelated motifs period-10 motifs AA/TT/TA and VWG/CWB appear to correlate with nucleosome positioning (11,13) and chromatin structure/function (15,16), respectively. How might these findings be reconciled?
We suggest that period-10 AA/TT/TA di-nucleotides are able to predict nucleosome positions to some extent predominantly because of the TA (our result), and the fact that DNA sequences containing period-10 A-tracts (which contain AA/TT/TA/AT di-nucleotides) generally have high nucleosome-forming abilities (10). Recently, it has been shown that yeast nucleosomes are enriched in A-tracts of at least length of three (36). Taken together with our finding that AA/TT is influential when it is 10 bp away from a TA, these results suggest how period-10 AA/TT might have been mistaken for a high nucleosome-forming motif based only upon the correlative evidence obtained from the alignment of nucleosomal DNAs (7) and of selected DNAs possessing high nucleosome-forming abilities (22).
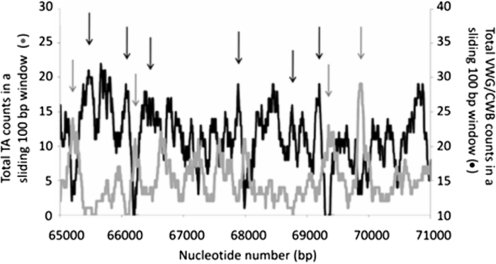
An explanation for the predictive power of period-10 VWG/CWB is provided by Figure 4, which shows that VWG/CWB strongly anti-correlates with TA. Generally, wherever the count of VWG/CWB in a 100-bp window is high, the count of TA is low, and vice versa. Thus, in large regions of DNA where period-10 VWG/CWB oscillated regularly, shown to correlate with the formation of regular nucleosome arrays (15), period-10 TA also oscillated regularly.
Figure 4.
Anti-correlation of the VWG/CWB and the TA signals in the MADA gene DNA sequence. The oscillation of the total TA counts in a sliding 100-bp window (gray curve) and the VWG/CWB counts (black curve) are shown along the DNA sequence. The black arrows indicate where high VWG/CWB counts anti-correlate with low TA counts, and the gray arrows indicate where high TA counts anti-correlate with low VWG/CWB counts.
Although it is likely that many different DNA sequence motifs will be found that serve to facilitate nucleosome formation as our understanding of DNA increases, we suggest that periodic TA motifs, which are abundant in genomic DNA, are major contributors to preferential nucleosome formation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Department of Biological Sciences, Purdue University.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 2.Simpson RT, Stafford DW. Structural features of a phased nucleosome core particle. Proc. Natl Acad. Sci. USA. 1983;80:51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prunell A. Nucleosome reconstitution on plasmid-inserted poly(dA) poly(dT) EMBO J. 1982;1:173–179. doi: 10.1002/j.1460-2075.1982.tb01143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engelhardt M. Choreography for nucleosomes: the conformational freedom of the nucleosomal filament and its limitations. Nucleic Acids Res. 2007;35:e106. doi: 10.1093/nar/gkm560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woodcock CL, Grigoryev SA, Horowitz RA, Whitaker N. A chromatin folding model that incorporates linker variability generates fibers resembling the native structures. Proc. Natl Acad. Sci. USA. 1993;90:9021–9025. doi: 10.1073/pnas.90.19.9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drew HR, Travers AA. DNA bending and its relation to nucleosome positioning. J. Mol. Biol. 1985;186:773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- 7.Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 8.Trifonov EN, Sussman JL. The pitch of chromatin DNA is reflected in its nucleotide sequence. Proc. Natl Acad. Sci. USA. 1980;77:3816–3820. doi: 10.1073/pnas.77.7.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shrader TE, Crothers DM. Artificial nucleosome positioning sequences. Proc. Natl Acad. Sci. USA. 1989;86:7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzgerald DJ, Anderson JN. Unique translational positioning of nucleosomes on synthetic DNAs. Nucleic Acids Res. 1998;26:2526–2535. doi: 10.1093/nar/26.11.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Widom J. Role of DNA sequence in nucleosome stability and dynamics. Q Rev. Biophys. 2001;34:269–324. doi: 10.1017/s0033583501003699. [DOI] [PubMed] [Google Scholar]
- 13.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baldi P, Brunak S, Chauvin Y, Krogh A. Naturally occurring nucleosome positioning signals in human exons and introns. J. Mol. Biol. 1996;263:503–510. doi: 10.1006/jmbi.1996.0592. [DOI] [PubMed] [Google Scholar]
- 15.Cioffi A, Fleury TJ, Stein A. Aspects of large-scale chromatin structures in mouse liver nuclei can be predicted from the DNA sequence. Nucleic Acids Res. 2006;34:1974–1981. doi: 10.1093/nar/gkl078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takasuka TE, Cioffi A, Stein A. Sequence information encoded in DNA that may influence long-range chromatin structure correlates with human chromosome functions. PLoS ONE. 2008;3:e2643. doi: 10.1371/journal.pone.0002643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong SW, Lauderdale JD, Stein A. Chromatin assembly on plasmid DNA in vitro. Apparent spreading of nucleosome alignment from one region of pBR327 by histone H5. J. Mol. Biol. 1991;222:1131–1147. doi: 10.1016/0022-2836(91)90597-y. [DOI] [PubMed] [Google Scholar]
- 18.Tullius TD, Dombroski BA, Churchill ME, Kam L. Hydroxyl radical footprinting: a high-resolution method for mapping protein-DNA contacts. Methods Enzymol. 1987;155:537–558. doi: 10.1016/0076-6879(87)55035-2. [DOI] [PubMed] [Google Scholar]
- 19.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. N.Y: Cold Spring Harbor Laboratory, Cold Spring Harbor; 1982. [Google Scholar]
- 20.Dalal Y, Fleury TJ, Cioffi A, Stein A. Long-range oscillation in a periodic DNA sequence motif may influence nucleosome array formation. Nucleic Acids Res. 2005;33:934–945. doi: 10.1093/nar/gki224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thastrom A, Lowary PT, Widlund HR, Cao H, Kubista M, Widom J. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J. Mol. Biol. 1999;288:213–229. doi: 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- 22.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 23.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 5th edn. New York: Garland Science; 2008. [Google Scholar]
- 24.Thastrom A, Bingham LM, Widom J. Nucleosomal locations of dominant DNA sequence motifs for histone–DNA interactions and nucleosome positioning. J. Mol. Biol. 2004;338:695–709. doi: 10.1016/j.jmb.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 25.Zhurkin VB. Specific alignment of nucleosomes on DNA correlates with periodic distribution of purine-pyrimidine and pyrimidine-purine dimers. FEBS Lett. 1983;158:293–297. doi: 10.1016/0014-5793(83)80598-5. [DOI] [PubMed] [Google Scholar]
- 26.Ulanovsky L, Bodner M, Trifonov EN, Choder M. Curved DNA: design, synthesis, and circularization. Proc. Natl Acad. Sci. USA. 1986;83:862–866. doi: 10.1073/pnas.83.4.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagerman PJ. Sequence-directed curvature of DNA. Nature. 1986;321:449–450. doi: 10.1038/321449a0. [DOI] [PubMed] [Google Scholar]
- 28.Travers AA, Klug A. The bending of DNA in nucleosomes and its wider implications. Philos. Trans. Roy. Soc. Lond. B Biol. Sci. 1987;317:537–561. doi: 10.1098/rstb.1987.0080. [DOI] [PubMed] [Google Scholar]
- 29.Burkhoff AM, Tullius TD. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987;48:935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- 30.Goodsell DS, Kaczor-Grzeskowiak M, Dickerson RE. The crystal structure of C-C-A-T-T-A-A-T-G-G Implications for bending of B-DNA at T-A steps. J. Mol. Biol. 1994;239:79–96. doi: 10.1006/jmbi.1994.1352. [DOI] [PubMed] [Google Scholar]
- 31.El Hassan MA, Calladine CR. Conformational characteristics of DNA: empirical classifications and a hypothesis for the conformational behaviour of dinucleotide steps. Philos. Trans. Roy. Soc. Lond. A. 1997;355:43–100. [Google Scholar]
- 32.Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl Acad. Sci. USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anselmi C, Bocchinfuso G, De Santis P, Savino M, Scipioni A. A theoretical model for the prediction of sequence-dependent nucleosome thermodynamic stability. Biophys. J. 2000;79:601–613. doi: 10.1016/S0006-3495(00)76319-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stefl R, Wu H, Ravindranathan S, Sklenar V, Feigon J. DNA A-tract bending in three dimensions: solving the dA4T4 vs dT4A4 conundrum. Proc. Natl Acad. Sci. USA. 2004;101:1177–1182. doi: 10.1073/pnas.0308143100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caserta M, Agricola E, Churcher M, Hiriart E, Verdone L, Di Mauro E, Travers A. A translational signature for nucleosome positioning in vivo. Nucleic Acids Res. 2009;37:5309–5321. doi: 10.1093/nar/gkp574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein–DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tullius TD, Dombroski BA. Iron(II) EDTA used to measure the helical twist along any DNA molecule. Science. 1985;230:679–681. doi: 10.1126/science.2996145. [DOI] [PubMed] [Google Scholar]
- 38.Tolstorukov MY, Colasanti AV, McCandlish DM, Olson WK, Zhurkin VB. A novel roll-and-slide mechanism of DNA folding in chromatin: implications for nucleosome positioning. J. Mol. Biol. 2007;371:725–738. doi: 10.1016/j.jmb.2007.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parker SC, Hansen L, Abaan HO, Tullius TD, Margulies EH. Local DNA topography correlates with functional noncoding regions of the human genome. Science. 2009;324:389–392. doi: 10.1126/science.1169050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mack DR, Chiu TK, Dickerson RE. Intrinsic bending and deformability at the T-A step of CCTTTAAAGG: a comparative analysis of T-A and A-T steps within A-tracts. J. Mol. Biol. 2001;312:1037–1049. doi: 10.1006/jmbi.2001.4994. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.