Abstract
The combination of RNA interference (RNAi) with the tetracycline-controlled transcription activation (tet) system promises to become a powerful method for conditional gene inactivation in cultured cells and in whole organisms. Here, we tested critical sequence elements that originated from miRNA mR-30 for optimal efficiency of RNAi-based gene knockdown in mammalian cells. Rationally designed miRNAs, expressed conditionally via the tet system, led to an efficient knockdown of the expression of both reporter genes and the endogenous mitotic spindle protein TPX2 in HeLa cells. Quantitative studies of the tet-controlled gene inactivation revealed that the residual expression of the target gene is an intrinsic attribute of all cells that cannot be eliminated either by increasing the miRNA to target mRNA ratio or by simultaneous expression of miRNAs targeting different sequences within the transcript. The kinetic analysis of the reversibility of the miRNA mediated knockdown suggests that the recovery of target gene expression is primarily driven by cell division. Our miRNA design provides a useful tool for conditional gene inactivation in combination with the RNA-polymerase II based tet system. The identified characteristics of the conditional RNAi-mediated knockdown need to be considered for its application in cell culture or in vivo.
INTRODUCTION
Despite deciphering the complete sequence information of more than 2300 eukaryotic genomes (Entrez Genome Project), determining gene functions within living organisms remains challenging. The discovery of RNA interference (RNAi) (1) in the nematode Caenorhabditis elegans provided a simple but specific tool for down-regulation (knockdown) of gene expression. As such, it was applied in the functional analysis of genes from a complete chromosome (2) or the entire genome (3,4) of C. elegans. More recently, the use of the RNAi technology was expanded to mammalian cells, initially through transient transfection of siRNAs (5) and later on through stable expression of shRNAs from RNA polymerase III (pol III) promoters (6).
To link a gene sequence to its function in a living organism, one method of choice is the combination of the RNAi technology with a conditional gene expression system that as a result provides spatial and temporal control over the gene function. The tetracycline-controlled transcription activation system (tet system) (7) is the most widely used tool for conditional, yet fully reversible regulation of gene expression. Tet-regulated shRNA expression was used successfully for temporal gene knockdown in cell culture (8,9). However, these systems are not suitable for cell-type specific gene inactivation since they rely on a repression principle. The most promising approaches applicable for the use in living organisms consist of either the expression of a shRNA under an activator dependent, tet-regulated pol III system (10) or the expression of a redesigned microRNA (miRNA) under control of the RNA polymerase II (pol II)-based minimal promoter of the original tet system (Ptet–1) (11,12).
Endogenous miRNAs are small, non-coding RNAs expressed in most tissues and involved in many developmental processes (13). They are typically transcribed by pol II (14), released as a pre-form from their primary transcripts by the action of the Drosha–DGCR8 complex (15,16) and exported from the nucleus to the cytoplasm by Exportin 5 (17). The pre-miRNAs are subsequently processed by Dicer to generate mature miRNAs (18), these are then transferred to and processed by the protein Argonaute 2, the catalytical engine of the ‘RNA-interfering silencing complex’ (RISC) [reviewed in (19)]. A miRNA equipped with a double-stranded sequence fully complementary to the target mRNA will trigger its RISC-dependent degradation (20). Such a miRNA driven by a pol II promoter is a suitable tool for inducing RNAi-mediated gene inactivation in vivo.
The knockdown of gene expression by miRNA-mediated RNAi expressed under the control of the tet system promises to become a versatile method for tissue specific, conditional inhibition of selected gene activity in cell culture as well as in living organisms. Additionally, this approach allows the exploitation of the large repertoire of already existing mouse lines expressing the tetracycline activator (tTA or rtTA) in a cell-type specific manner (21).
In this study, we describe a design of minimized miRNA applicable for the knockdown of any gene of interest via the tet system. Within the rationally designed miRNAs, the functions of the individual sequence elements are systematically characterized. The most potent miRNAs in combination with pol II promoters match the efficacy of knockdown achieved by a pol III driven shRNA. We also provide a detailed description of the dynamic range of gene regulation that is possible with tet-regulated miRNAs. The rational basis for the application of this technology is prepared by the quantitative kinetic analysis of the miRNA mediated gene knockdown and the reinduction of the expression of targeted mRNA after termination of miRNA expression.
MATERIAL AND METHODS
Generation of plasmid vectors
All miRNAs described herein were generated from two end-to-end matching oligonucleotide primers (Supplementary Table S1; biomers.net, Germany) by PCR using Pwo-polymerase (Roche, Germany). Resulting DNAs, coding for the respective miRNA targeting either firefly (fluc) or renilla luciferase (rluc), were subsequently cloned into pSKII+ mod2 (Supplementary Data) as EcoRI/PstI fragments. This yielded respective pSKII+ mod2 vectors containing a single miRNA. For the construction of plasmids, in which miRNAs are under the control of either the CMV promoter (PCMV) or the tet-regulatable promoter (Ptet–1), respective miRNA-coding DNA was released from the pSKII+ mod2 vector by EcoRI and XbaI and the resulting fragment was cloned into either pUHD 10-1 [PCMV; (22)] or pUHD10-3 [Ptet–1; (23)]. For the generation of vectors, in which the miRNA is expressed under the control of the promoter of the RNA component of RNase P, H1 (PH1), the respective miRNA-coding DNA fragment was excised from the pSKII+ mod2 vector using BglII and XhoI and cloned into pSUPER (6).
All shRNAs described herein (Supplementary Table S1) were generated by the annealing of two complementary oligonucleotides (biomers.net, Germany), designed as described in the guidelines of Brummelkamp et al. (6). For vectors, in which a respective shRNA is expressed under the control of PH1, annealed oligonucleotides forming shRNA(FL-B) or shRNA(RL-A) were integrated into pSUPER by the use of the restriction sites BglII and HindIII. For the generation of vectors, in which the respective shRNA is expressed under control of PCMV, the corresponding oligonucleotides (Supplementary Table S1) were annealed and cloned into pUHD10-1 via EcoRI and XbaI.
The vector pBI-11 miRNA3(FL-B), in which the bidirectional tet-regulated promoter (Ptet bi–1) (24) controls the expression of miRNA3(FL-B) and rluc, was generated in a two-step process. First, miRNA3(FL-B) was released as a fragment having a blunt EcoRI and a NheI site from its pSKII+ mod2 vector and then cloned into pBI-4 (24) cut with PvuII and NheI yielding pBI-4 miRNA3(FL-B). Next, the reading frame of rluc, released from pRL-SV40 (Promega, Germany) as a fragment with a blunt SacI and a SalI site, was ligated with a vector fragment from pBI4 miRNA3(FL-B) with a blunt NotI site and a SalI site, yielding pBI-11 miRNA3(FL-B).
Plasmid pBI-9 miRNA3(TPX2-C), in which Ptet bi–1 controls the expression of eGFP and miRNA3(TPX2-C) was constructed by integrating the PCR-synthesized DNA of this miRNA3 (Supplementary Table S1), digested with PvuII and NheI, into pBI-eGFP (Clontech, USA). In a subsequent step, the tet-regulated bidirectional expression cassette was released from pBI-9 miRNA3(TPX2-C) by HpaI and BglII and cloned into pBI-F3.M.F (25) cut with the same restriction sites. The resulting plasmid [pBI-9F.miRNA3(TPX2-C)] enables Flp-mediated recombinase-mediated cassette exchange (RMCE) of the tet-regulated expression cassette in a well defined locus of cell line HeLa EM2-11 (25) with optimal regulatory properties.
The construction of the vectors, in which a fluc fusion protein is expressed under the control of either PCMV [pUHD(NR1_opt3)-131-1] or the promoter of elongation factor 1α (PEF1α) [pUHD(NR1_opt3)-131-EF1α], is described in the Supplementary Data.
Cell culture and transient transfections
Wild-type HeLa cells (CCL-2, American Type Culture Collection, USA) and all derived HeLa cell lines were cultured in DMEM-medium (Invitrogen, Germany) supplemented with 10% tet-approved FCS (Clontech, USA) at 37°C and 5% CO2. For transient expression experiments, 15–20 µg of a respective DNA mixture was added to 2–3 million cells in 300 µl serum free DMEM medium in a 4-mm GenePulser cuvette (BioRad). The cells were electroporated with an ElectroCellManipulator ECM630 (BTX, USA) at 200 V, 25 Ω and 975 µF at room temperature and spread on adhesion-enhanced six-well tissue cultures plates (Cell+, Sarstedt, Germany). The DNA mixture consisted of equal amounts of a vector for constitutive fluc production (pCMV-luc; D. Bartsch, unpublished data), a vector for constitutive expression of the rluc gene (pRL-SV40; Promega, Germany) and the vector that expresses the respective interfering RNA from either PCMV or PH1. For the transient expression of tet-regulated miRNAs equal amounts of plasmids pCMV-luc, pRL-SV40, pUHT61-1 and the respective miRNA vector were used. pUHT61-1 is a plasmid, in which the gene of the optimized tTA2 is expressed by PCMV (26). For the experiments, in which the expression of both fluc and rluc was inactivated by simultaneous production of respective miRNAs, a proportionate amount of vector pUHD34-1, having the gene for eGFP under the control of PCMV, was added in addition to the above described DNA mixture.
After 36 h of growth in regular or doxycycline (dox) (Sigma, Germany) containing medium, the cells were washed twice with PBS and lysed on tissue culture plate surface with passive lysis buffer (Promega, Germany). From the lysates, following centrifugation at 14 000 rpm for 5 min at 4°C, the fluc and rluc activities were measured in the supernatant using the Dual-Luciferase Reporter Assay system (Promega, Germany) according to the manufacturer's recommendations in combination with Wallac Victor 2 multilabel counter (PerkinElmer, USA). Relative activity of fluc was obtained by normalization of fluc activity to rluc; relative rluc activity was normalized to fluc activity. In the case of the experiments that demonstrated simultaneous knockdown of fluc and rluc by co-expression of both miRNAs, eGFP fluorescence was determined in the lysate using the Wallac Victor 2 multilabel counter and used for normalization of transfection efficiency.
Generation of stably transfected cell lines
HtTA 15-1, HtTA 16-1 and HtTA 16-2 triple stable cells were obtained by transfecting the cell lines HtTA 15 and HtTA 16 (generation described in Supplementary Data) with 20 µg SapI-linearized pBI-11 miRNA3(FL-B) and 2 µg EcoRI-linearized pPUR (Clontech, USA) via electroporation as described above. One day after transfection, the cells were treated with G418 (200 µg/ml), hygromycin (200 µg/ml) and puromycin (2 µg/ml) for selection of respective transgenic expression cassettes and 100 ng/ml dox to keep tet-regulated expression turned off. For clonal analysis, cells were washed twice with PBS and then cultivated in dox-free medium for 3 days. Cell lysis and determination of fluc and rluc activities was carried out as described above. The protein concentration of the sample, used for normalization of luciferase activities, was determined using the BioRad Protein assay according to the manufacturer's protocol.
For the stable cell line with rtTA-inducible miRNA mediated TPX2 knockdown, the plasmid pBI-9F miRNA3(TPX2-C) was used for stable integration into cell line HeLa EM2-11ht via recombinase-mediated cassette exchange (RMCE) as described in Weidenfeld et al. (25). The HeLa EM2-11-TPX2 cells were grown to 25% confluence and the addition of 200 ng/ml dox was used to induce expression of miRNA3(TPX2-C) and eGFP at time point 0. Samples were taken at desired time points and either fixed with 3% paraformaldehyde for immunofluorescence analysis, 70% Ethanol for flow cytometry or directly resuspended in SDS sample buffer for quantitative western blot analysis.
Quantification of mRNA concentrations by real-time RT–PCR
Total RNA of the respective cells was isolated using TRIzol Reagent (Invitrogen, Germany) according to the manufacturer's recommendations. To remove any traces of contaminating genomic or plasmid DNA, the RNA was reconstituted in RNase-free water and incubated with 50 U RNase-free DNase I (Roche, Germany) under recommended conditions for 15 min at 37°C. To terminate DNase I digestion, 500 µl TRIzol was added to the reaction mixture. Subsequent RNA extraction followed according to the manufacturer's protocol.
Next, 1 µg of total RNA was used for reverse transcription with SuperScript III and oligo(dT)20 primer (Invitrogen, Germany) according to manufacturer's protocol. Resulting cDNAs were diluted 1/2 and 1/4 and 1 µl of either undiluted or respectively diluted cDNA subjected to real-time PCR analysis as duplicates.
Real-time PCR reactions were run on an Applied Biosystems 7900 HT fast real-time PCR system. For simultaneous detection of fluc and rluc mRNA, the following primers and probes were used: fluc forward primer 5′-TGTGGACGAAGTACCGAAAGGT-3′; fluc reverse primer 5′-CTTCTTGGCCTTTATGAGGATCTC-3′; fluc probe 5′-FAM-CCGGAAAACTC-GACGCAAGAAAAATCAG-TAMRA-3′; rluc forward primer 5′-AAAGGTGAAGTTCGT-CGTCCA-3′; rluc reverse primer 5′-CAACGTCAGGTTTACCACCTTTT-3′; rluc probe 5′-HEX-CATTATCATGGCCTCGTGAAATCCCGT-TAMRA-3′ (FAM: 6-carboxy-fluoresceine; HEX: hexachloro-fluoresceine; TAMRA: 6-carboxy-tetramethyl-rhodamine; biomers.net). Real-time PCR was performed in a total volume of 20 µl using the Taqman Universal PCR master mix (Applied Biosystems, Germany) according to the manufacturer's protocol. Final concentrations of primers were 300 nM, those of probes 250 nM. For detection of human β-actin, we used the following primers: Actin forward primer: 5′-AGCAC-AGAGCCTCGCCTTT-3′; Actin reverse primer: 5′-AGGGTGAGGATGCCTTCTCTT-3′. In this case, real-time PCR was conducted with the Power SYBR-Green PCR master mix (Applied Biosystems, Germany) according to the manufacturer's guidelines. Final concentrations of primers were 300 nM.
Quantification of mature miRNA
Detection of processed miRNA was performed using a Custom Taqman Small RNA Assay (AssayID: CCN0QSF, Applied Biosystems, Germany) according to the manufacturer's protocol. Briefly, 10 ng of total RNA was dissolved in 5 µl RNase free water and then mixed with 7 µl of recommended RT-master mix (containing Superscript III reverse transcriptase) and 3 µl of the 5 × RT primer provided with the Taqman Small RNA Assay. RT reaction was run in a thermocycler, programmed for 30 min at 16°C, 30 min at 42°C and 5 min at 80°C. Real-time PCR reactions were run with either undiluted, 1/2 or 1/4 diluted RT reactions using a Applied Biosystems 7900 HT fast real-time PCR system in accordance with the recommended protocol. Each PCR reaction was performed in triplicates and consisted of 1 µl RT reaction, 1 µl Custom Taqman Small RNA Assay and 10 µl Taqman Universal PCR Master Mix (Applied Biosystems, Germany) in a total volume of 20 µl.
Immunofluorescence, immunoblotting and flow cytometry
Immunofluorescent detection was performed using polyclonal antibodies against full length human TPX2 (27), or GFP (a kind gift from Dirk Görlich) or a monoclonal antibody against α-Tubulin (Sigma, DM1a). To quantify nuclear TPX2 signals, cells were imaged and TPX2 fluorescence was determined over background signal using the Image J software. Flow cytometry was performed as described (28). Western blot analysis was performed as described (29) using a LICOR infrared scanning system and the corresponding software.
Tumour growth analysis with the nude mouse xenograft model
Female nude mice (Crl:CD1-Foxn1nu) were obtained from Charles River (Sulzfeld, Germany) at an age of 6–8 weeks. They were housed at specific pathogen-free conditions in a mini barrier system of the central animal facility of the German Cancer Research Center and kept under controlled conditions (21 ± 2°C room temperature, 60% humidity, and 12-h Light–dark rhythm). Autoclaved food and water were given ad libitum to the animals.
Experiments were approved by the responsible governmental animal ethics committee (Regierungspräsidium Karlsruhe, Germany). Sub-confluent cells of the line HeLa EM2-11- mCherry (Kai Schönig and Dusan Bartsch, unpublished) or HeLa EM2-11-TPX2 were harvested using 2 mM EDTA in PBS (phosphate buffered saline without Ca2+ and Mg2+), counted in a Neubauer's chamber and suspended in RPMI 1640-medium in the respective dilution. An amount of 75 µl of the medium containing either 104 or 105 cells was augmented with 25 µl Geltrex (Invitrogen, Germany) and subsequently injected subcutaneously along the mammary line of the nude mice. The horizontal and vertical diameter of the tumour was determined using external calipers and the volume calculated according to a2 × b/2 (a ≤ b).
RESULTS
Gene inactivation efficiencies of miRNAs expressed under control of RNA polymerase II or III promoters
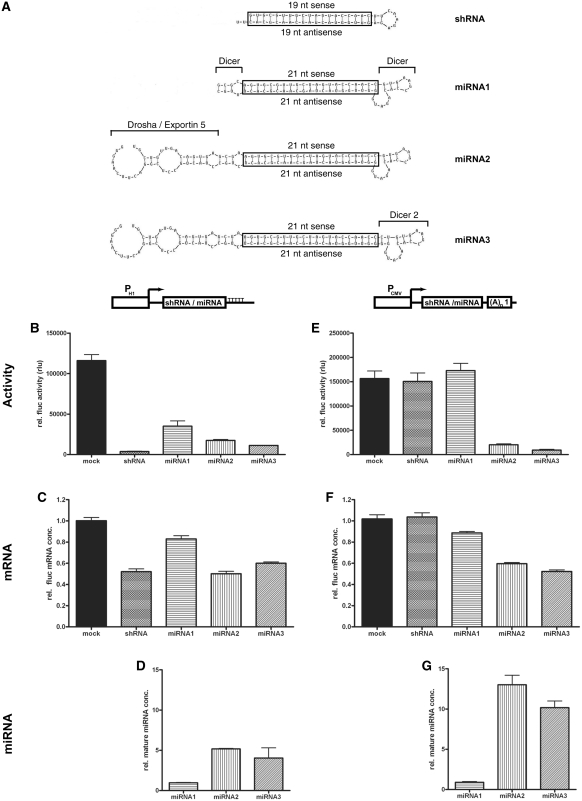
To identify the miRNA core sequences necessary for efficient gene knockdown, we tested three different miRNA designs (Figure 1A). The central block of all miRNAs was a double-stranded sequence of 21 nt completely homologous to the targeted mRNA (boxed sequences in Figure 1A). In miRNA1 this central double-stranded region was flanked by sequences necessary for Dicer cleavage (30), which were derived from endogenous miRNA mR-30. In miRNA2, additional sequences containing the Drosha cleavage site and the nuclear export signal of mR-30 (15) were added to the 5′-end and the 3′-end of the miRNA1, respectively. In miRNA3, the loop connecting the sense and the antisense targeting sequence was exchanged for an improved version originally described by Boden et al. (31).
Figure 1.
Knockdown efficiencies of different miRNA designs. (A) Sequences and secondary structures of different miRNA designs. Boxed sequences are the double-stranded RNA sequences matching the target site within the firefly luciferase (fluc) mRNA. Added or modified sequence elements from miRNA1 to miRNA3 are explicitly labelled. Dicer: Dicer cleavage sequences (30). Drosha/Exportin 5: Sequences for cleavage by Drosha and nuclear export signal by Exportin 5 (15). Dicer 2: Dicer cleavage loop as used in miB-tat in Boden et al. (31). (B–G) Quantification of knockdown efficiencies of differently designed miRNAs, expressed from either the pol III promoter PH1 (B–D) or the pol II promoter PCMV (E–G). Relative fluc activities (B and E), relative fluc mRNA concentrations (C and F) and the concentrations of mature miRNA (D and G) were determined in cell extracts from individual transient expression experiments. The results in B and E were normalized to activity and in C and F to mRNA concentration of rluc, produced from the SV40 promoter within the co-transfected expression vector. Figures B–G show the means and SEM from multiple samples of at least two independent transfection experiments. (A)n1: SV40 polyadenylation signal. TTTTT: pol III termination signal.
For the functional and quantitative evaluation of the different miRNA designs, a 21-bp firefly luciferase (fluc) target sequence (FL-B) was included, which has been previously shown to confer efficient fluc knockdown (32). The respective miRNAs were cloned into vectors, in which their expression was controlled either by the pol III dependent promoter of the RNA component of RNase P, H1 (PH1), or by the pol II dependent human cytomegalovirus immediate early promoter (PCMV). For reference, a shRNA sharing the same fluc targeting sequence was constructed according to Brummelkamp et al. (6) and cloned into both pol II and pol III expression vectors.
The knockdown efficacy of the different miRNA designs was evaluated in transient expression experiments, in which the particular expression vector for each miRNA was transfected into HeLa cells together with vectors constitutively expressing fluc (targeted mRNA) and renilla luciferase (rluc), the latter serving as an internal standard for transfection efficiency. The mRNA concentrations and the enzymatic activities of both fluc and rluc were quantified from total RNA and protein lysates yielded from transfected cells. In addition, the concentration of mature miRNA yielded from each individual miRNA design was determined.
The comparison of efficacy between PH1 expressed shRNA and miRNAs is shown in Figure 1B–D. The best reduction of fluc activity was achieved by the conventional shRNA (residual fluc activity of 3%), although significant knockdown efficiencies were also obtained with miRNA2(FL-B) (residual fluc activity 15%) and miRNA3 (residual fluc activity 10%) (Figure 1B). The least efficient downregulation of fluc activity in this experiment was observed upon transfection with miRNA1(FL-B) (residual fluc activity 30%), indicating that sequences for Drosha cleavage and nuclear export in miRNA2 and miRNA3 contribute to some degree to the knockdown potency of pol III produced miRNAs. The knockdown efficiency measured by luciferase activities were also reflected by the relative fluc mRNA concentrations, although relative residual mRNA concentrations were higher (Figure 1C). The higher cellular concentration of mature miRNA generated from miRNA2 and miRNA3 compared to miRNA1 (Figure 1D) supports again the beneficial effect of Drosha cleavage and nuclear export sequences on miRNA processing.
When the inhibitory RNAs were expressed from a pol II dependent promoter, no impediment of fluc activity was observed with either shRNA(FL-B) or miRNA1(FL-B) (Figure 1E). Drosha and nuclear export sequences, present in miRNA2 and miRNA3, increased knockdown efficiencies by 10- to 20-fold [miRNA2(FL-B), residual fluc activity 13%; miRNA3(FL-B), residual fluc activity 6%]. The relative fluc mRNA concentrations upon transfection also reflect the fluc activity measurements, although the relative residual mRNA concentrations were again much higher than the relative residual luciferase activity (Figure 1F). The concentration of mature miRNAs in transfected cells (Figure 1G) was highest with miRNA2(FL-B) and miRNA3(FL-B), demonstrating the importance of Drosha and nuclear export sequences for the efficacy of pol II driven miRNAs.
The concentration of mature miRNA2(FL-B) and miRNA3(FL-B) produced by pol II is superior to that yielded by miRNAs expressed from the very strong pol III promoter PH1. Correspondingly, the resulting knockdown of fluc activity is most efficient by pol II driven miRNA2(FL-B) and miRNA3(FL-B), with an efficacy comparable to the pol III expressed shRNA. Interestingly, all knockdown efficiencies determined by mRNA quantification are lower than the efficiencies determined by measuring reporter gene activity.
General applicability of the most efficient miRNA design
Next, we tested whether the high efficacy observed with miRNA2 and miRNA3 produced by pol II is applicable for other mRNAs than fluc. Therefore, we tested a 21-bp sequence (RL-A) targeting renilla luciferase (rluc), with no homology to fluc site FL-B, in the context of all three miRNA designs. As seen in Supplementary Figure S1, the relative knockdown efficiencies achieved with the respective shRNA and miRNAs targeting rluc at sequence RL-A are comparable with those targeting fluc at sequence FL-B (Figure 1B and E), providing evidence for the general applicability of the identified designs. Although we did not observe a significantly higher knockdown efficiency with miRNA3 compared to miRNA2, we still think that the improved loop may provide efficient Dicer processing for a wider range of target sequences. For this reason, we have selected miRNA3 as the design of choice for further experiments.
In the next experiment, we investigated whether the minimized miRNA3 sustains its high knockdown efficacy independently of its surrounding sequences within the pol II generated transcript. We therefore cloned miRNA1(FL-B) and miRNA3(FL-B) into the expression vector pIntron (Supplementary Figure S2A). In these plasmids, the respective miRNA is located within an artificial intron that precedes the ORF of eGFP. The expression of the entire mRNA, including the intron, is controlled by PCMV. The knockdown efficacies of the intronic miRNAs were compared in transient transfection experiments with efficacies of the same miRNAs cloned into the pCMV vector used in previous experiments. Our results show that knockdown efficacy of miRNA3(FL-B) is independent of the expression vector used (Supplementary Figure S2B). Therefore, we conclude that the miRNA3 can operate as a functional unit independent from the integration sequences in the expression vector.
In an alternative RNAi-capable miRNA, used within the BLOCK-iT pol II miR RNAi expression system (Invitrogen, USA), the antisense sequence of the mRNA target site is present on the 5′-side and the corresponding sense sequence on the 3′-side of the Dicer cleavage loop, which is the opposite orientation of the sequences compared to our miRNA designs. To clarify the influence of sense and antisense sequence orientation within the miRNA on gene knockdown efficiency, we generated miRNA3-rev(FL-B), in which the 21 nt antisense sequence of the target site is placed 5′- and the 21-nt sense sequence 3′ of the Dicer cleavage loop, similar to the BLOCK-iT design. In a transient transfection experiment, both the miRNA3-rev(FL-B) and miRNA3(FL-B), produced from PCMV, had identical efficacy. The residual fluc activity upon transfection of miRNA3-rev(FL-B) was 8.4 ± 1.1% and upon transfection of miRNA3(FL-B) was 6.1 ± 0.5%.
Efficacy of gene inactivation in stable cell lines with tet-regulated miRNA expression
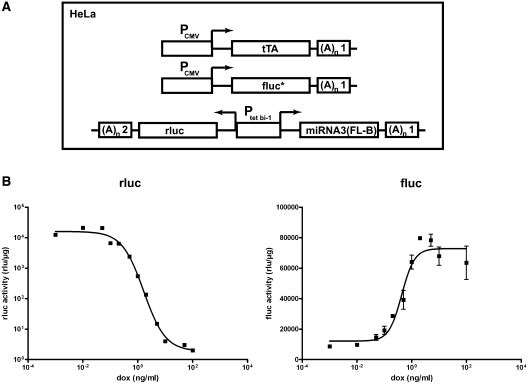
As the miRNA3 design was most effective in transient transfection experiments, we established stably transfected cell lines with the tet-regulated miRNA3(FL-B) construct for detailed kinetic analyses of miRNA activity. First, the genome of the original Tet-Off HeLa cell line HtTA, bearing a cassette for constitutive expression of tTA (7), was augmented with a modified fluc gene, constitutively expressed from PCMV (33). Two stable cell clones with both transgenes (HtTA 15 and 16) that showed constitutive and high fluc activity were selected for further use. In a second step, the conditional expression cassette, in which the bidirectional tet-regulated promoter Ptet bi–1 (23) controlled the expression of both miRNA3(FL-B) and rluc gene, was randomly integrated into the genomes of both HtTA 15 and 16. In the resulting triple transgenic Tet-Off cell lines (Figure 2A), rluc activity mirrored the kinetics of miRNA production from the tet-regulatable promoter, whereas changes in fluc activity reflected the degree of miRNA-mediated RNAi.
Figure 2.
Dose–response analysis of tet-regulated miRNA3-mediated inactivation of firefly luciferase in the Tet-Off cell line HtTA 16-1. (A) Schematic representation of sequence elements within the transgenic constructs used for the generation of triple stable HeLa cell lines. PCMV, CMV-Promoter; tTA, tet-dependent transactivator; Ptet bi–1, bidirectional tet-regulated promoter; fluc*, firefly luciferase fusion protein; rluc, renilla luciferase; (A)n1, SV40 polyadenylation signal; (A)n2, β-globin polyadenylation signal. (B) Dose–response analysis of miRNA target gene firefly luciferase (fluc) and renilla luciferase (rluc) activity, mirroring miRNA transcription at various doxycycline (dox) concentrations in cell line HtTA 16-1, measured 72 h after induction with dox and normalized to total protein concentration. Figure B shows means and SEM from multiple samples of at least two independent experiments.
From a total of 29 triple stable clones, three clones were selected (Table 1), in which rluc activity can be regulated in the range of more than three orders of magnitude by doxycycline (dox). Interestingly, this resulted only in an 11-fold reduction of fluc production at most. In the clone HtTA 16-1, the knockdown efficiency of miRNA3(FL-B) determined by either fluc activity (Table 1) or fluc mRNA concentration (Supplementary Table S2) is highly comparable. Figure 2B shows the dose–response curve of HtTA 16-1. As deduced from the curve, the expression of miRNA3(FL-B) had to reach ∼13% of the induction maximum, achieved with dox concentrations of ∼0.5 ng/ml, to elicit RNAi.
Table 1.
Knockdown efficiency of tet-regulated miRNA3(FL-B) in stable cell lines
| Cell line | cDNA expression |
miRNA-mediated knockdown |
||||
|---|---|---|---|---|---|---|
| Renilla luciferase activity (rlu/µg) |
Firefly luciferase activity (rlu/µg) |
|||||
| –dox | +dox | fold regulation | –dox | +dox | fold regulation | |
| HtTA 15-1 | 53 911 ± 4291 | 54 ± 19 | 1007 | 5104 ± 536 | 35 813 ± 4166 | 7.0 |
| HtTA 16-1 | 113 063 ± 6201 | 37 ± 19 | 3087 | 6075 ± 1129 | 68 871 ± 11 679 | 11.2 |
| HtTA 16-2 | 49 210 ± 14046 | 89 ± 10 | 550 | 6585 ± 502 | 55 381 ± 11 521 | 8.4 |
Fluc and rluc luciferase activities, normalized to total protein concentration, obtained from cells of indicated triple stable HeLa Tet-Off lines, that were grown in the absence (–dox) or presence (+dox) of 500 ng/ml dox in the tissue culture medium for 72 h.
These data favoured the conclusion that miRNA3(FL-B) expression was rate limiting for the knockdown of fluc activity. However, the efficiency of final steady state gene inactivation in three analysed clones (residual fluc activity: HtTA 15-1: 14%; HtTA 16-1: 9%; HtTA 16-2: 12%;) seemed to be largely independent of the maximal induction of miRNA3(FL-B), measured by rluc activity. In fact, in the HtTA 16-1 line, transcribing twice as much miRNA3(FL-B) in comparison to the other cell clones, only results in a knockdown efficiency that is minimally higher. To address this discrepancy in more detail, further experiments were performed.
Knockdown efficiency is largely independent from the degree of target gene expression
Next, we tested whether the knockdown efficiency of miRNA3(FL-B) was dependent on the initial concentration of the target mRNA. Therefore, we performed a co-transfection experiment, in which miRNA3(FL-B) interfered with the expression of a fluc gene that was expressed either by PCMV or by the promoter of elongation factor 1α (PEF1α). Fluc activities obtained in control transfections (Table 2) demonstrated that expression from PEF1α was ∼30 times lower than expression driven by PCMV. Surprisingly, the resulting relative knockdown efficiencies generated by Ptet–1 produced miRNA3(FL-B) were identical in regard to the experimental error (residual fluc activity: CMV: 12%; EF1α: 8%). This possibly indicates that a subpopulation of target gene mRNAs escapes miRNA-mediated degradation independent of the primary concentration of the targeted mRNA.
Table 2.
Knockdown efficiency of miRNA3(FL-B) at different target gene concentrations
| Promoter | Firefly luciferase activity (rlu/µg) |
||
|---|---|---|---|
| Mock | miRNA3(FL-B) | fold regulation | |
| CMV | 32 089 ± 7964 | 4068 ± 394 | 7.9 |
| EF1α | 1015 ± 26 | 84 ± 26 | 12.1 |
In transient expression experiments, either a vector expressing miRNA3(FL-B) under the control of Ptet–1 [miRNA(FL-B)] or an empty expression vector pUHD10-1 (mock) was co-transfected with a plasmid constitutively producing a firefly luciferase protein, a plasmid constitutively producing rluc and a vector constitutively expressing tTA. In one case (CMV) the fluc protein production was controlled by the strong promoter PCMV, in the other case by the weaker promoter of elongation factor 1 α PEF1α. From these transfection experiments, fluc activities were quantified and normalized to yielded rluc activities.
Efficiency of simultaneous inactivation of multiple genes by tandem-arrayed miRNAs
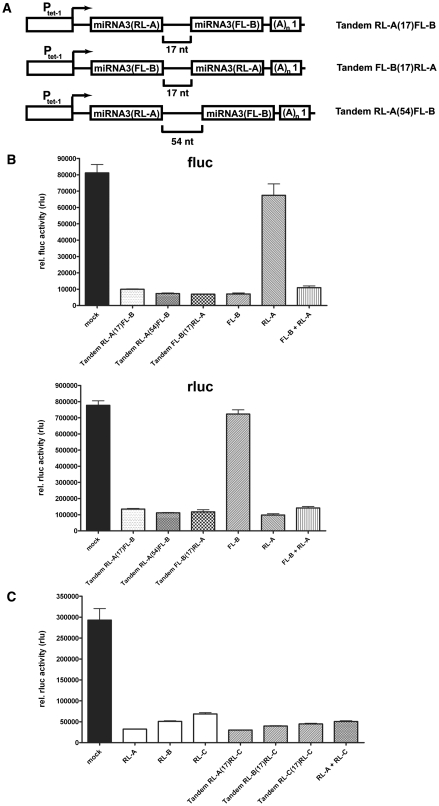
About 40% of miRNA genes discovered so far are organized in polycistronic clusters, from which they are co-expressed by pol II as a single transcript (14,34). From this primary transcript, pre-miRNAs are liberated by RNase III cleavage activity of Drosha (15). Therefore, arraying multiple rationally designed miRNAs offers the possibility of either the simultaneous inactivation of multiple genes or the targeting of a single gene by synergistic action of multiple miRNAs targeting the mRNA at different sequence sites.
To model the simultaneous inactivation of multiple genes, miRNA3(RL-A) was cloned upstream of miRNA3(FL-B) into the expression vector, in which the miRNA3(FL-B) was already under Ptet–1 control. The spacer sequence between the two miRNAs was 17 nt for tandem RL-A(17)FL-B and 54 nt for tandem RL-A(54)FL-B (Figure 3A). These two vectors were then tested for their capability to knockdown both fluc and rluc in transient transfections. As shown in Figure 3B, both tandem arrays of miRNAs reduced activities of fluc and rluc with the same efficiency as individually expressed miRNAs. There was no difference between the tandem RL-A(17)FL-B and the tandem RL-A(54)FL-B, thus demonstrating that a spacing of 17 nt is sufficient for independent processing of both miRNAs by Drosha. Cloning and testing of construct tandem FL-B(17)RL-A, where miRNA3(RL-A) was inserted downstream of miRNA3(FL-B) (Figure 3A), revealed no positional advantage for miRNA within the bicistron.
Figure 3.
Simultaneous gene inactivation of multiple genes by tandem-arrayed miRNAs. (A) Schematic representation of sequences present in expression vectors with tandem-arrayed miRNAs. Ptet–1, tet-regulatable promoter; (A)n1, SV40 polyadenylation signal. Brackets show the size of the non-coding linker sequence separating the individual miRNAs of the type miRNA3. (B) Firefly (fluc) and renilla (rluc) luciferase activities were quantified in a co-transfection experiment, in which the constitutive production of firefly and renilla luciferase from respective expression vectors was inhibited by the indicated expression vector bearing a tet-regulated miRNA3 cassette. Measured luciferase activities were normalized to the present fluorescence intensity of eGFP, which was expressed under control of PCMV from an additional co-transfected expression plasmid. Structure of tandem expression vectors is described in (A) FL-B: expression vector with miRNA3(FL-B) under control of Ptet–1. RL-A, expression vector with miRNA3(RL-A) under control of Ptet–1; FL-B + RL-A, co-transfection of two expression vectors containing either miRNA3(FL-B) or miRNA3(RL-A) under control of Ptet–1. (C) Rluc activities were determined in a co-transfection experiment, in which the constitutive production of renilla luciferase was inhibited by the indicated vectors and normalized to fluc activity. RL-A, RL-B, RL-C: expression vectors with a single miRNA3 targeting rluc at a particular sequence (A or B or C) under control of Ptet–1. Tandem bicistronic constructs follow the nomenclature as described in (A). RL-A+ RL-C: co-transfection of two expression vectors containing either a single miRNA3(RL-A) or a single miRNA3(RL-C) under the control of Ptet–1. Figures B and C show means and SEM from multiple samples taken from at least two independent transfection experiments.
Given the option to target one transcript with two or more different miRNAs, we next investigated whether the simultaneous expression of miRNAs targeting the same transcript at different target sites could further decrease the residual activity upon knockdown achieved with only a single miRNA. As seen in Figure 3C, individual miRNAs targeting rluc mRNA at different sequences (RL-A, RL-B, RL-C) reduced the production of rluc to residual activities ranging from 11% to 23%. Next, bicistronic DNA expression units expressed under control of Ptet–1 were generated. Here, the first cistron consisted of one of the miRNA3 targeting rluc at RL-A or RL-B or RL-C. The second cistron was always miRNA3(RL-C). If the effect of the additional miRNA3(RL-C) is linearly additive, the residual rluc activity observed with one rluc targeting miRNA should be lowered by additional 77% because of the expression of miRNA3(RL-C). However, for miRNA3(RL-A) and miRNA3(RL-B), no further reduction of residual rluc activity was accomplished by the additional miRNA (Figure 3C). In addition, the expression of a tandem array of two miRNA3(RL-C) had only a minor effect on further reducing residual activity compared to a single cassette with only one miRNA.
Comparable residual activity after miRNA-mediated knockdown is present in all affected cells
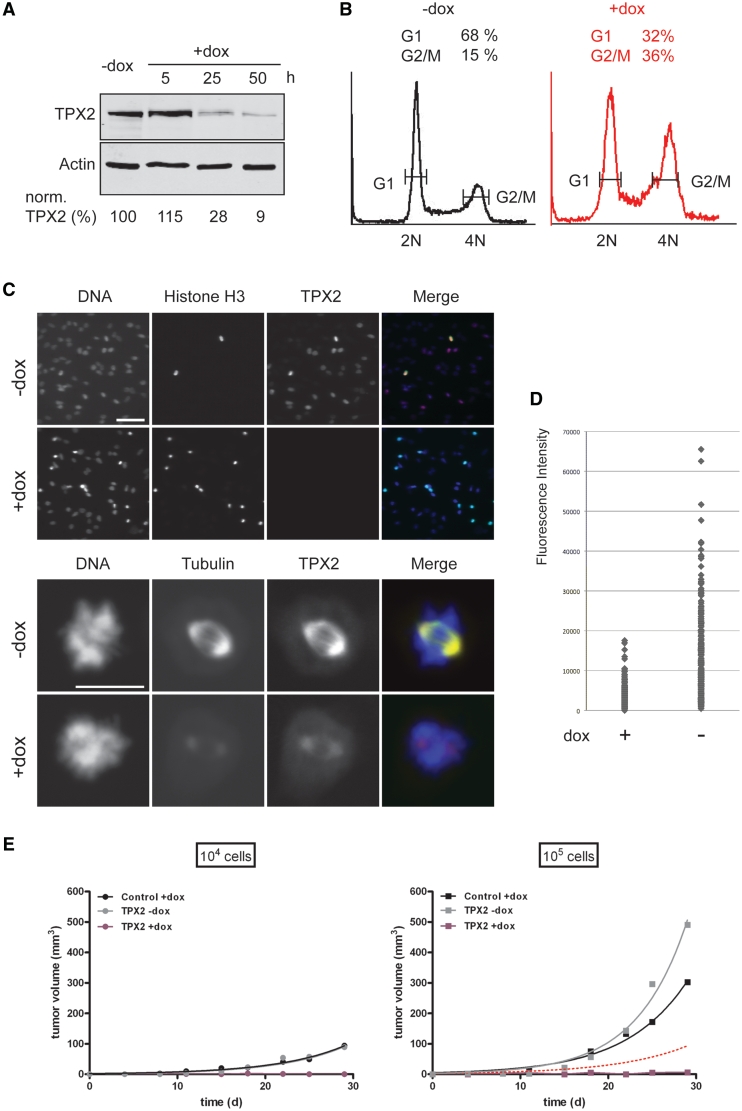
Furthermore, we addressed the nature of the residual gene activity present after miRNA-mediated knockdown. We asked whether it originated from a subpopulation of cells deficient for RNAi mediated knockdown or from a pool of endogenous mRNAs that escaped the miRNA-triggered degradation uniformly in most or all cells. As a target we selected the TPX2 (targeting protein for Xklp2) gene product, a protein recently identified to play a key role in mitotic spindle formation in HeLa cells (27,35). Suppression of TPX2 expression by transient transfection of siRNAs led to a block of mitotic spindle formation and subsequently to a persistent cell-cycle arrest (27). Therefore, a subpopulation of cells deficient in RNAi machinery should be identifiable as dividing cells with normal TPX2 expression.
To knockdown TPX2, we generated a stable cell line, in which the expression of a miRNA3 targeting TPX2 and the reporter gene eGFP was regulated by the Tet-On system. Therefore, an expression cassette, in which the bidirectional tet-dependent promoter controlled both the expression of eGFP and miRNA3(TPX2-C), was recombineered into the genome of a precursor cell line HeLa EM2-11ht (25) by recombinase-mediated cassette exchange, yielding cell line HeLa EM2-11-TPX2. To determine the miRNA-mediated knockdown efficiency of the endogenous TPX2, HeLa EM2-11-TPX2 cells were cultured either in the absence or in the presence of dox, harvested at different time points and analysed. Expression of TPX2 protein could be reduced by miRNA3(TPX2-C) to a residual level of below 10% within 50 h (Figure 4A). This reduction of TPX2 protein concentration led to a dramatic reduction of spindle microtubule density, as shown by the immunofluorescence staining of TPX2 and microtubules on a single cell level in Figure 4C. Consistent with that and with previously published results (27,35), knockdown of TPX2 caused an accumulation of cells in mitosis as visualized by indirect immunofluorescence using an antibody against the mitosis-specific phosphorylation of histone H3 on serine 10 (Figure 4C, upper panels). Moreover, flow cytometry revealed a higher percentage of cells bearing G2/M DNA content (4N) after TPX2 knockdown (Figure 4B, 15% in controls without dox, as compared to 36% after addition of dox and TPX2 knockdown). Low concentration of TPX2 could still be detected in miRNA3(TPX2-C) expressing cells. This was consistent with the analysis of TPX2 knockdown efficiencies in a population of cells using immunofluorescence staining (Figure 4C, upper panels): Variations in TPX2 protein concentrations were observed in control cells (Figure 4C, TPX2, –dox) consistent with changing protein concentration of TPX2 during the cell cycle in human cells (27,36). In contrast, no cells could be identified with a strong TPX2 signal upon expression of the TPX2-targeting miRNA (Figure 4C, TPX2, +dox). To analyse this more quantitatively, we imaged ∼200 cells from control and knockdown samples and quantified the nuclear TPX2 signals by indirect immunofluorescence. While the average fluorescence signal was significantly reduced upon expression of the TPX2-targeting miRNA, the cells showed a similar relative signal distribution. Most importantly, we did not observe any cells with high TPX2 concentrations upon miRNA expression (Figure 4D). This strongly argues against the existence of a subpopulation of cells still expressing original TPX2 protein concentrations in the presence of the miRNA but favours the idea of a homogenous gene knockdown, with homogenous residual activity in all cells.
Figure 4.
Tet-regulated gene knockdown of endogenous mitotic spindle assembly factor TPX-2 in Tet-On cell line HeLa EM2-11-TPX2. (A) Immunoblot analysis of TPX2 concentrations in controls or 5, 25 and 50 h after induction of tet-regulated expression of miRNA3 targeting TPX2 by dox. TPX2 concentrations were normalized to present β-actin. (B) Flow cytometry analysis of cells grown for 44 h in the absence (–dox) or presence (+dox) of tet-induced expression of miRNA targeting TPX2. The y axis shows the number of cells stained by propidium iodide. The x axis indicates the intensity of the cellular staining and thus the DNA content. 2N: diploid, 4N: tetraploid. The relative percentage of cells in either the G1 or G2/M cell-cycle stage is indicated above the peaks. (C) Immunofluorescence analysis of cells as in (B). Cells expressing miRNA3(TPX2) (+dox) or uninduced controls (–dox) were stained with either DAPI (DNA, blue in merge), an antibody against TPX2 (TPX2, red in merge), an antibody against phosphoserine 10 in histone H3 (Histone H3, upper panel, green in merge), or an antibody against α-Tubulin (Tubulin, lower panel, green in merge). Scale bars: 100 µm (upper panels) and 10 µm (lower panels). (D) A total of 200 cells from both control samples (–dox) and after TPX2 knockdown (+dox) were labelled using TPX2 antibodies. The intensity of each individual nuclear TPX2 signal was quantified and plotted as a dot. (E) Tumour growth analysis of cell line HeLa EM2-11-TPX2 in the nude mouse xenograft model. 104 (points) or 105 cells (squares) of cell line HeLa EM2-11-TPX2 (TPX2) or HeLa EM2-11 (Control) were injected subcutaneously along the right or left mammary line of CD1 nude mice. The volume of developing tumours was monitored in vivo using an external caliper over a period of 30 days. One group of mice (+dox) was treated with 2 mg/ml dox in drinking water starting from Day 7 after inoculation of cells. The other group (–dox) never received dox treatment. Black lines: Average tumour volume (n = 5) of control cell line in the presence of dox. Gray lines: Average tumour volume (n = 5) of line HeLa EM2-11-TPX2 in the absence of dox. Purple lines: in the presence of dox. The dotted red line displays the tumour growth curve of 104 cells of line HeLa EM2-11-TPX2 in the absence of dox.
Supporting the evidence for the absence of any cells showing normal, unmitigated expression of TPX2 in the presence of miRNA3(TPX2-C), we tested the effect of the miRNA-mediated TPX2 knockdown on tumour growth of HeLa EM2-11-TPX2 in the nude mouse xenograft model. Therefore, either 104 or 105 cells of line HeLa EM2-11-TPX2 were injected subcutaneously along the mammary line of nude mice. One group of mice (n = 5) never received dox treatment [and therefore did not express miRNA3(TPX2-C)] and served as a control for the tumourigenic properties of the cell line. The effect of the miRNA3(TPX2-C) mediated suppression of TPX2 expression on tumour growth was studied in a second group of mice (n = 5), in which expression of miRNA3(TPX2-C) in the inoculated cells was activated from day 7 p.i. onwards by treating the mice with dox in drinking water (2 mg/ml). To exclude any therapeutic effect of dox on tumour growth itself, a third group of mice had received corresponding quantities of cells of line HeLa EM2-11-mCherry, in which the production of fluc and the red fluorescent protein mCherry is controlled via the Tet-On system (Kai Schönig and Dusan Bartsch, unpublished). These mice were treated with dox in drinking water the same way as the animals of the second group. Figure 4E shows tumour growth in all three groups of mice within a period of 30 day. Upon inoculation of 104 cells, switched off HeLa EM2-11-TPX2 as well as switched on HeLa EM2-11-mCherry cells developed into measurable tumours. No tumours appeared in mice from cells of line HeLa EM2-11-TPX2, where tet-regulated expression of miRNA3(TPX2-C) had been activated through dox treatment. If a subpopulation of ∼10% RNAi defective cells were present in line HeLa EM2-11-TPX2, injection of 105 cells should yield tumours in similar size to those observed with 104 switched off cells of the respective line (indicated by the red dotted line). While injection of either 105 switched off HeLa EM2-11-TPX2 cells or 105 switched on HeLa EM2-11-mCherry cells gave rise to larger tumours compared to injected 104 cells, still no tumours developed from 105 cells with tet-activated miRNA3(TPX2-C) expression. This finding further supports the view that there are no RNAi resistant cells in the injected population.
Kinetic properties of tet-mediated gene inactivation by miRNA3
A very important parameter of any conditional gene inactivation strategy is the knowledge of the on- and off kinetics of the respective system. Therefore, the onset and the decay of miRNA-mediated RNAi was investigated using the Tet-Off cell line HtTA 16-1.
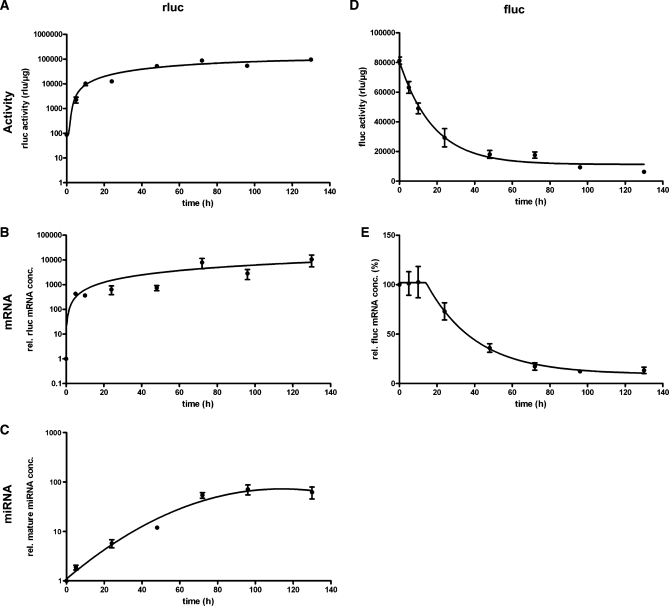
Figure 5 shows the time course of the appearance of miRNA-mediated gene inactivation in relation to the onset of tet-regulated gene expression and mature miRNA generation. As seen from rluc activity (Figure 5A) and mRNA concentration (Figure 5B), tet-regulated expression has a very fast onset, reaching almost its maximal activity within the first 24 h after the Tet-Off system has been activated by removal of dox. The generation of mature miRNAs initiates more slowly and stabilizes reaching a plateau 72 h after miRNA expression has been started (Figure 5C). The appearance of mature miRNAs occurs in parallel with the decay of the targeted gene, deduced from the exponential decay curves of fluc activity (Figure 5D) and mRNA concentration (Figure 5E), that show a half-life time of ∼15 h in both cases. Interestingly, exponential decay of target gene mRNA initiated after a delay period of 10 h.
Figure 5.
Activation kinetic of tet-regulated miRNA3-mediated gene knockdown in Tet-Off cell line HtTA 16-1. Tet-regulated expression of miRNA3(FL-B) and rluc was started in cell line HtTA16-1 (utilizing the Tet Off system) by removal of dox from the culture medium. Activation of tet-regulated transcription of miRNA3(FL-B) and miRNA-mediated knockdown of fluc was monitored over a period of 130 h by quantifying rluc (A) and fluc (D) activity, relative rluc (B) and fluc (E) mRNA concentration and the concentration of mature miRNA (C) in the samples of HtTA 16-1 cells collected at the indicated time points. Both luciferase activities were normalized to total protein concentration, mRNA and miRNA concentrations to the concentration of β-actin mRNA. Figures show means with SEM of multiple samples from two independent experiments.
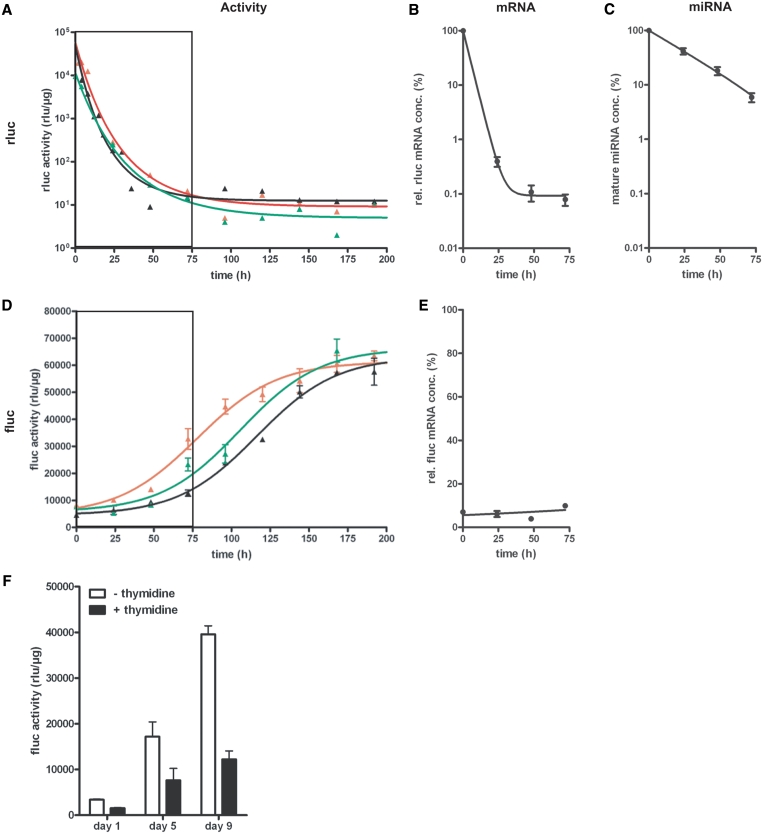
Multiple studies have shown the capability of siRNAs to induce epigenetic silencing through the assembly of heterochromatin in mammalian cells (37,38). To clarify whether an extended time of expression of an RNAi-capable miRNA has any long lasting, miRNA-independent effect on the expression of the target gene after expression of the interfering RNA has ceased, we analysed the restoration of target gene expression after various times of persistent miRNA production. Therefore, miRNA3(FL-B) expression was induced in HtTA 16-1 cells for 1, 2 or 4 weeks. Subsequently tet-regulated miRNA production was switched off by adding dox to the cell culture medium and the kinetics of rluc decay (mirroring miRNA transcription) and the fluc reinduction were determined over extended times (Figure 6A–E). After dox addition, the transcription from the bidirectional tet-regulatable promoter ceased very fast, independently of duration of activation, as depicted by exponential decay curves for rluc activity (Figure 6A) with an average half-life time of 5.2 h. The herein determined half-life of rluc activity is consistent with previously reported values (39). Exponential decay of rluc mRNA revealed a half-life time of 2.8 h (Figure 6B). In contrast to this fast process, complete reestablishment of basal fluc activity was not achieved until 5 days after 1 week (red line), and 7 days after 2 weeks (black line) or 4 weeks (green line) of persistent miRNA3 mediated knockdown (Figure 6D). However, since comparable basal fluc activities were reached after all investigated times, an epigenetic modification of reporter gene expression is unlikely to have occurred. Interestingly, computational analysis of sigmoidal reinduction curves uncovered a doubling time of fluc activity recovery to be ∼24 h. In addition, the concentration of mature miRNA decayed exponentially with a half-life time of ∼20 h in an experiment, where a sustained production of the respective miRNA over 2 weeks was shut off (Figure 6C). Since HeLa cells divide every 24 h, these two observations indicated that the recovery of targeted reporter gene expression to basal levels might be simply due to the progressive dilution of miRNA-activated RISCs by cell division.
Figure 6.
Recovery kinetics of target gene expression after the termination of the tet-regulated miRNA3-mediated gene knockdown in the Tet-Off cell line HtTA 16-1. (A+D) Tet-regulated co-expression of miRNA3(FL-B) and rluc in the cell line HtTA16-1 (utilizing the Tet Off system) was kept activated for 1 week (red), 2 weeks (black) and 4 weeks (green). Then, tet-regulated gene expression was switched off, by addition of dox (500 ng/ml), and rluc (A) and fluc (D) activity measured at indicated time points over a period of 192 h. Both luciferase activities were normalized to total protein concentration. The boxed area marks the time period analysed in B, C and E. (B+C+E) Quantification of relative rluc (B) and fluc (E) mRNA concentrations and the concentration of mature miRNA (C) within a period of 72 h after tet-regulated gene expression, that has been activated for 2 weeks, was switched off. The mRNA and miRNA concentrations were normalized to the concentration of β-actin mRNA In (E), 100% represents the mRNA concentration in cells expressing no miRNA. Therefore, the rel. fluc mRNA concentration mirrors the knockdown efficiency at respective time points. Figures show means and SEM of multiple samples from two independent experiments. (F) Recovery of fluc activity after miRNA3(FL-B) knockdown in cell populations with different division rates. In cell line HtTA 16-1, miRNA expression was activated by growing the cells in the absence of dox for 10 days. After subsequent addition of dox to the tissue culture medium (500 ng/ml) to terminate tet-regulated miRNA3(FL-B) expression, cells were divided into two populations. In one population, the cell cycle was artificially elongated to 48 h by the use of S-phase blocker thymidine (+thymidine, black). In the other population (–thymidine, white), cell division rate was left unaltered at 24 h, serving as a control population. Figure shows fluc activities (rlu/µg total protein) at Days 1, 5 and 9 normalized to a control experiment, in which fluc activities of the same time points were determined in both respective cell populations, that were never subjected to a miRNA-based knockdown of fluc expression.
To investigate this assumption in more detail, we conducted an experiment, in which the doubling time of HtTA 16-1 was artificially extended to 48 h by a thymidine-mediated S-phase block. miRNA3(FL-B) expression was turned on for 10 days and as soon as miRNA production was switched off, the respective cells were split into two population, one control population, and one with an elongated cell cycle of 48 h. As seen in Figure 6F, recovery of fluc took much longer in cells with an extended cell division time in comparison to cells with an unaltered division rate. This provided further evidence that a substantial component responsible for the herein and previously observed (11,12) reinstatement of basal target gene expression after miRNA-mediated knockdown is cell division.
DISCUSSION
In the present study, we systematically characterized a tet-regulated miRNA expression system, which is applicable for tissue specific, temporal down-regulation of any gene of interest in mammalian cells and tissues. Our study shows that the minimized miRNA design miRNA3 with defined miRNA core elements, when utilized in combination with a pol II promoter, promoted gene knockdowns with a comparable efficiency to those achieved by the most potent interfering RNA expressed from the pol III promoter PH1. Pol II promoter driven expression of RNAi-active transcripts has several benefits, in particular as a tool for a tissue specific, conditional gene inactivation in transgenic animals. Not only can all the previously developed molecular components of the original tet system be used, but also the large repertoire of already existing characterized transactivator mouse lines (21) can be exploited. Previously described tet-regulatable pol III systems (8,9) rely on a repression principle and are thus unsuitable for reversible tissue-specific gene inactivation in transgenic animals. The default state in these systems is ubiquitous shRNA production and thus global inactivation of the target gene in the entire animal, which can only be suspended in those tissues, which express a tet-sensitive transcriptional repressor or silencer. For the establishment of a tissue-specific, tet-regulatable knockdown an activator dependent, tet-regulated pol III system developed recently (10) is a choice. Nevertheless, several desired features like the simultaneous expression of reporter genes together with the miRNA cannot be easily realized with pol III based systems. Therefore, in our view, the expression of miRNAs from pol II promoters is preferable for studies of tissue-specific gene function in living animals using RNAi knockdowns.
Although the use of tet-regulated miRNAs has been described previously (11,12), our study now for the first time systematically analysed the critical sequence elements necessary for efficient gene knockdown. Through the comparison of different miRNA designs, we demonstrated that sequences encoding for the cleavage site of the Drosha nuclease and the nuclear export signal for Exportin 5 are essential for the efficiency of a designer miRNA, when expressed from a pol II promoter. The general applicability of the proposed miRNA design was further supported by demonstrating high knockdown efficiency independent of the target sequence used and the vector sequence in which the miRNA was integrated. The designation of minimal essential sequences reduced the total size of a highly effective miRNA to 106 nt, which requires a spacer of just 17 nt to be functionally recognized as an individual unit. The miRNA described by Stegmeier and colleagues (12) worked only when integrated into the 3′-UTR of an open reading frame possibly suggesting a deficiency in Drosha processing and/or nuclear export could be compensated by an open reading frame upstream of a miRNA.
Studying the regulatory properties of miRNA3 in a quantitative way revealed three interesting aspects: first, the knockdown of gene expression via the miRNA was not complete and the expression of the targeted gene persisted above a certain threshold, independently of the miRNA to target mRNA ratio. This enduring residual activity of the targeted mRNA was neither affected by increasing the miRNA concentration nor by lowering the concentration of target mRNA. In addition, the indefatigable activity could not be eliminated by the simultaneous expression of second miRNAs targeting different sequence sites within the transcript. In our TPX2 knockdown experiments, the residual activity was shown to be homogenously distributed to all cells and did not originate from a distinct, potentially RNAi-deficient subpopulation of cells. An RNAi-resistant gene activity within the same range was also observed for synthetic siRNAs (40). Consistent with this, in many miRNA or siRNA experiments even higher residual activities compared to our experiments were reported. But since this residual activity is distributed to all cells, many biological experiments utilizing this system can be conducted, as long as the endogenous inactivation of expression is not compensated and sufficient to elicit a phenotype. Here, we were able to demonstrate the suitability of stable transgenic cell lines utilizing this regulatory system to study the biology of tumour formation.
Second, the use of quantifiable reporter genes expressed from the bidirectional tet-regulated promoter in our study allowed us to directly compare for the first time the kinetic features of the RNAi-mediated gene inactivation with induction kinetics of the tet system itself. As expected, miRNA-dependent mRNA degradation accompanies the emergence of processed miRNAs and follows the rapid induction of tet-regulated reporter gene expression with a short delay. Interestingly, within the first 10 h after initiation of tet-regulated miRNA expression, we see a decrease in target gene activity without a reduction of its mRNA concentration. Both the activation of tet-regulated gene expression (41) or the inducer doxycycline in doses used (42) have no inhibitory effects on the cellular translation machinery. Therefore, we assume that within this initial phase, inhibition of mRNA translation by miRNA-loaded RISC complexes precedes mRNA degradation. In fact, it has been shown recently that a substantial proportion of the RNAi-mediated gene down-regulation, elicited by siRNAs or miRNAs fully complementary to the respective target site, is mediated by translational repression (43). Interestingly, these data support well our assessment of the transient miRNA efficacies. As seen in Figure 1, knockdown efficiencies are higher when calculated from reporter gene activities compared to those deduced from mRNA concentrations. However, in stable cell line HtTA16-1, 72 h after activation of miRNA expression, translational repression is not detectable and knockdown efficiencies calculated from target gene activities (Table 1) or mRNA concentrations (Supplementary Table S2) are comparable. We suggest that substantial translational repression by fully complementary miRNAs is present either within the initial phase (10 h) of the RNAi process, where all components necessary for RISC-dependent mRNA degradation might not be present or properly assembled, or for much longer periods (3 days) in transient transfection experiments, in which a massive overexpression of the target gene is commonly taking place. The results from transient experiments also suggest that RISC-mediated mRNA degradation may be the time limiting step of RNAi process.
Third, we show that the assumption regarding the generally fast reversibility of a miRNA-mediated knockdown (11,12) may be an overestimation. In contrast to the relatively fast onset, reversal of induced miRNA-mediated gene knockdown was observed here within only 6–8 days, consistent with other published data (11,12). Given a 1-day doubling time of HeLa cells, the simplest explanation for the observed half-life times of 24 h for fluc activity recovery and of 20 h for the decay of mature miRNA concentration is the dilution of RNAi-active complexes in several rounds of cell division, which drives most of the process. This point of view is further supported by the fact, that the artificial elongation of HeLa cell division to 48 h slows down the recovery of target gene expression mediated by previous miRNA expression. Supposing that all detected mature miRNAs are part of an active RISC complex, a mathematical correction of the half-life time of mature miRNA concentration determined herein for the cell division rate would yield a half-life time for the pure decay of miRNA-loaded RISC complexes of ∼140 h. If this was the case, then a return to 95% of original expression level of the targeted gene in non-dividing cell types would take around 30 days. Therefore, reversal of miRNA-mediated gene knockdown within one cell cycle, as seen in the case of tet-regulated gene expression, is unlikely. This interpretation is consistent with recently published data, showing prolonged duration of transient shRNA knockdown after cell-cycle arrest (44). Although recent publications show reversibility of miRNA-mediated gene inactivation in transgenic mice within a period of days, this was observed in highly proliferative, oncogenic B-lymphocytes (45). For the faithful assignment of half-life times of RNAi-active RISCs and to address the question of general reversibility of miRNA action in living animals, in our opinion quantitative data for gene knockdown in less proliferating cell types are needed.
Despite the numerous potential benefits of the RNAi technology, its limitations have to be taken seriously. We predict that residual activity of the target gene is an intrinsic feature of miRNA knockdown and will therefore remain a major limitation for the use of RNAi-mediated gene inactivation, in particular where more substantive down-regulation is necessary. In fact, the knockdown of the enzyme mgcRACGAP to a residual 10–20% concentration by a specific miRNA3 did not result in the expected cellular phenotype (data not shown). The other important issue is the expected slow reversibility of the system in slowly dividing or non-dividing cells and the risk that the competition between the exogenous miRNA and the endogenous miRNAs for RISC formation would eventually be deleterious to the cellular metabolism, arising after an estimated intrinsic lag phase of 4 weeks. In fact, strong overexpression of shRNAs in the liver led to severe toxicity and high mortality rate in mice only after a period of ∼4 weeks (46).
In conclusion, our data define the frame within which conditional gene inactivation via tet-regulated miRNAs is effective. The use of Ptet bi–1 allows the simultaneous expression of miRNAs with a reporter function for identifying and quantitatively monitoring cells expressing the miRNAs. Even though miRNA technology has its limitations, this approach allows auspicious experimental designs. For example, co-expression of two or more miRNAs allows simultaneous targeting of multiple genes at once. Such approaches will be fruitful for targeting highly redundant regulatory pathways. Furthermore, a cDNA encoding a mutant of a gene of interest can be co-expressed with a miRNA targeting the endogenous unaltered gene generating a conditional gene replacement. In the absence of totipotent, germline competent embryonic stem cells from organisms other than mice, the use of the inducible gene knockdown system will enable targeted gene inactivation in species, where pronuclear injection is technically feasible. This includes not only the rat model, the most widely used model in physiology and neurobiology, but almost all commercially important livestock.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Research Foundation (DFG) [collaborative Grant SFB 636 to D.B.]; German Federal Ministry of Education and Research [PNS-01GS08152-7 (NGFN-plus) to D.B. and S.B.]; European Union [MRTN-CT-2006-035367 (CAVNET) to D.B., HEALTH-F2-2007-201714 (DEVANX) to D.B.]; University of Heidelberg [Frontier Grant to D.B., S.B. and K.S.]; federal state Baden-Württemberg [collaborative Grant ‘Network Aging Research’ to D.B. and K.S.]. Funding for open access charge: German Research Foundation (SFB 636).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Reuven Agami for providing plasmid pSUPER, Rolf Sprengel and Peter Seeburg for providing plasmid pJN18-1 and Ronald de Pinho for providing plasmid pEF1prtTA. The authors are grateful to Dirk Görlich for the α-GFP antibody and his support and to Hermann Bujard for inspiring discussions. The authors would like to thank Shamila Ahmed for critical reading of the manuscript.
REFERENCES
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 3.Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- 4.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 5.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 6.Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- 7.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van de Wetering M, Oving I, Muncan V, Pon Fong MT, Brantjes H, van Leenen D, Holstege FC, Brummelkamp TR, Agami R, Clevers H. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 2003;4:609–615. doi: 10.1038/sj.embor.embor865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J. Virol. 2003;77:8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amar L, Desclaux M, Faucon-Biguet N, Mallet J, Vogel R. Control of small inhibitory RNA levels and RNA interference by doxycycline induced activation of a minimal RNA polymerase III promoter. Nucleic Acids Res. 2006;34:e37. doi: 10.1093/nar/gkl034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat. Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 12.Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc. Natl Acad. Sci. USA. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 14.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 16.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 18.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 19.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat. Rev. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 20.Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl Acad. Sci. USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berger S, Bujard H. Novel mouse models in biomedical research: the power of dissecting pathways by quantitative control of gene activities. In: Offermanns S, Hein L, editors. Handbook of Experimental Pharmacology. Vol. 159. Heidelberg: Springer; 2004. pp. 3–30. [Google Scholar]
- 22.Deuschle U, Pepperkok R, Wang FB, Giordano TJ, McAllister WT, Ansorge W, Bujard H. Regulated expression of foreign genes in mammalian cells under the control of coliphage T3 RNA polymerase and lac repressor. Proc. Natl Acad. Sci. USA. 1989;86:5400–5404. doi: 10.1073/pnas.86.14.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baron U, Bujard H. Tet repressor-based system for regulated gene expression in eukaryotic cells: principles and advances. Methods Enzymol. 2000;327:401–421. doi: 10.1016/s0076-6879(00)27292-3. [DOI] [PubMed] [Google Scholar]
- 24.Baron U, Freundlieb S, Gossen M, Bujard H. Co-regulation of two gene activities by tetracycline via a bidirectional promoter. Nucleic Acids Res. 1995;23:3605–3606. doi: 10.1093/nar/23.17.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weidenfeld I, Gossen M, Low R, Kentner D, Berger S, Görlich D, Bartsch D, Bujard H, Schönig K. Inducible expression of coding and inhibitory RNAs from retargetable genomic loci. Nucleic Acids Res. 2009;37:e50. doi: 10.1093/nar/gkp108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, Hillen W. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl Acad. Sci. USA. 2000;97:7963–7968. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gruss OJ, Wittmann M, Yokoyama H, Pepperkok R, Kufer T, Sillje H, Karsenti E, Mattaj IW, Vernos I. Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell. Biol. 2002;4:871–879. doi: 10.1038/ncb870. [DOI] [PubMed] [Google Scholar]
- 28.Godl K, Gruss OJ, Eickhoff J, Wissing J, Blencke S, Weber M, Degen H, Brehmer D, Orfi L, Horvath Z, et al. Proteomic characterization of the angiogenesis inhibitor SU6668 reveals multiple impacts on cellular kinase signaling. Cancer Res. 2005;65:6919–6926. doi: 10.1158/0008-5472.CAN-05-0574. [DOI] [PubMed] [Google Scholar]
- 29.Dumont J, Petri S, Pellegrin F, Terret ME, Bohnsack MT, Rassinier P, Georget V, Kalab P, Gruss OJ, Verlhac MH. A centriole- and RanGTP-independent spindle assembly pathway in meiosis I of vertebrate oocytes. J. Cell. Biol. 2007;176:295–305. doi: 10.1083/jcb.200605199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 31.Boden D, Pusch O, Silbermann R, Lee F, Tucker L, Ramratnam B. Enhanced gene silencing of HIV-1 specific siRNA using microRNA designed hairpins. Nucleic Acids Res. 2004;32:1154–1158. doi: 10.1093/nar/gkh278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyagishi M, Taira K. U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol. 2002;20:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- 33.Berger S. Untersuchungen zur nichtinvasiven Tetrazyklin-abhängigen Kontrolle von Genaktivitäten in vivo. PhD thesis. 2003 University of Heidelberg. http://archiv.ub.uni-heidelberg.de/volltextserver/frontdoor.php?source_opus=4142 (13 July 2010, date last accessed) [Google Scholar]
- 34.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garrett S, Auer K, Compton DA, Kapoor TM. hTPX2 is required for normal spindle morphology and centrosome integrity during vertebrate cell division. Curr. Biol. 2002;12:2055–2059. doi: 10.1016/s0960-9822(02)01277-0. [DOI] [PubMed] [Google Scholar]
- 36.Heidebrecht HJ, Buck F, Steinmann J, Sprenger R, Wacker HH, Parwaresch R. p100: a novel proliferation-associated nuclear protein specifically restricted to cell cycle phases S, G2, and M. Blood. 1997;90:226–233. [PubMed] [Google Scholar]
- 37.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 38.Ting AH, Schuebel KE, Herman JG, Baylin SB. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat. Genet. 2005;37:906–910. doi: 10.1038/ng1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl Acad. Sci. USA. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsui K, Horiuchi S, Sando S, Sera T, Aoyama Y. RNAi silencing of exogenous and endogenous reporter genes using a macrocyclic octaamine as a “compact” siRNA carrier. Studies on the nonsilenced residual activity. Bioconjug. Chem. 2006;17:132–138. doi: 10.1021/bc050112l. [DOI] [PubMed] [Google Scholar]
- 41.Gossen M, Bujard H. Studying gene function in eukaryotes by conditional gene inactivation. Annu. Rev. Genet. 2002;36:153–173. doi: 10.1146/annurev.genet.36.041002.120114. [DOI] [PubMed] [Google Scholar]
- 42.Wishart JA, Hayes A, Wardleworth L, Zhang N, Oliver SG. Doxycycline, the drug used to control the tet-regulatable promoter system, has no effect on global gene expression in Saccharomyces cerevisiae. Yeast. 2005;22:565–569. doi: 10.1002/yea.1225. [DOI] [PubMed] [Google Scholar]
- 43.Wu L, Fan J, Belasco JG. Importance of translation and nonnucleolytic ago proteins for on-target RNA interference. Curr. Biol. 2008;18:1327–1332. doi: 10.1016/j.cub.2008.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maliyekkel A, Davis BM, Roninson IB. Cell cycle arrest drastically extends the duration of gene silencing after transient expression of short hairpin RNA. Cell Cycle. 2006;5:2390–2395. doi: 10.4161/cc.5.20.3363. [DOI] [PubMed] [Google Scholar]
- 45.Dickins RA, McJunkin K, Hernando E, Premsrirut PK, Krizhanovsky V, Burgess DJ, Kim SY, Cordon-Cardo C, Zender L, Hannon GJ, et al. Tissue-specific and reversible RNA interference in transgenic mice. Nat. Genet. 2007;39:914–921. doi: 10.1038/ng2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.