Abstract
β-amyloid (Aβ) is the main protein component of the amyloid plaques associated with Alzheimer’s disease. Transthyretin (TTR) is a homotetramer that circulates in both blood and cerebrospinal fluid. Wild-type transthyretin (TTR) amyloid deposits are linked to senile systemic amyloidosis, a common disease of aging, while several TTR mutants are linked to familial amyloid polyneuropathy. Several recent studies provide support for the hypothesis that these two amyloidogenic proteins interact, and that this interaction is biologically relevant. For example, upregulation of TTR expression in Tg2576 mice was linked to protection from toxic effects of Aβ deposition [Stein, T.D. and Johnson, J.A. (2002) J. Neurosci. 22: 7380–7388]. We examined the interaction of Aβ with wt TTR as well as two mutants: F87M/L110M, engineered to be a stable monomer, and T119M, a naturally occurring mutant with higher tetrameric stability than wildtype. Based on enzyme-linked immunoassays as well as crosslinking experiments, we conclude that Aβ monomers bind more strongly to TTR monomers than to TTR tetramers. The data further suggest that TTR tetramers interact preferably with Aβ aggregates rather than Aβ monomers. Through tandem mass spectrometry analysis of crosslinked TTR-Aβ fragments, we identified the A strand, in the inner β-sheet of TTR, as well as the EF helix, as regions of TTR that are involved with Aβ association. Light scattering and electron microscopy studies demonstrate that the outcome of the TTR-Aβ interaction strongly depends on TTR quaternary structure. While TTR tetramers may modestly enhance aggregation, TTR monomers decidedly arrest Aβ aggregate growth. These data provide important new insights into the nature of TTR-Aβ interactions. Such interactions may regulate TTR-mediated protection against Aβ toxicity.
Alzheimer’s disease (AD), the most common age-associated neurodegenerative disease, affects approximately 5 million Americans. Amyloid plaques are one of the primary characteristic features of AD, with β-amyloid (Aβ) ase the main protein component of these amyloid plaques. The two common isoforms, Aβ(1–40) and Aβ(1–42), are generated from cleavage of the transmembrane amyloid precursor protein (APP) by β- and γ–secretases (1, 2). Both transgenic animal studies and numerous in vitro studies indicate that Aβ aggregation is causally linked to cellular toxicity, although the exact relationship between Aβ and neuronal toxicity in the progression of AD remains an area of controversy (3–5). Multiple studies indicate that toxicity arises early, and is attributable to soluble Aβ oligomers and/or protofibrils (6, 7).
Aβ is just one of many amyloidogenic proteins which will, under certain conditions, associate into aggregates with a cross-β structure and fibrillar morphology. Another amyloidogenic protein is transthyretin (TTR): TTR amyloid fibrils are found in patients with familial amyloid polyneuropathy (FAP) or senile systemic amyloidosis (SSA). SSA, characterized by deposition of wild-type TTR, may affect as much as 25% of population over age 80 (8). FAP is an inherited disorder, associated with one of numerous TTR mutants (9).
TTR is a homotetramer that circulates in both blood and cerebrospinal fluid, with each 14kDa subunit composed of 127 residues (10). Each monomer is a β-pleat sandwich of two four-stranded β-sheets - an inner sheet of strands D, A, G, and H, and an outer sheet of strands C, B, E and F - plus one short helix between the E and F strands. Monomers assemble into dimers through extensive hydrogen bonding involving the F and H strands. The tetramer is formed by hydrophobically driven association of two dimers in a face-to-face manner, held together by loops which project from the edge of the dimer (10, 11). This creates a central channel lined by the inner sheets of the four monomers, where thyroxine (T4) is known to bind (12). TTR is also a transporter for retinol binding protein (13). In vitro evidence supports a mechanism of amyloid formation whereby TTR tetramers first dissociate into monomers, which then undergo a slight conformational re-arrangement and re-assemble into fibrils (14, 15). Wt TTR is normally quite stable at neutral pH; dissociation to monomer and subsequent fibril assembly is facilitated at moderately acidic pH. Disease-associated TTR mutants generally have less stable quaternary structures and are more prone to dissociation (14, 16). Hydrogen-deuterium exchange studies along with NMR data indicate that the ‘edge’ strands C and D are labile and likely unfold during TTR fibrillogenesis, while strands A, B, E and G form a stable core that is resistant to unfolding (17, 18). FAP mutants have a less stable core (19). TTR fibrils may arise by forming a non-native interface between A and B strands on different monomers, centered on residues 13 and 31, respectively (18).
Increasing evidence supports the hypothesis that TTR and Aβ interact, and that this interaction is biologically relevant. An early study demonstrated that cerebrospinal fluid (CSF), where TTR is the major protein component, inhibited Aβ amyloid fibril formation (20). Several studies have now demonstrated binding between TTR and Aβ, and in some cases it was also shown that TTR influences Aβ aggregation, and reduces Aβ toxicity in vitro (21–25). Interestingly, some groups have reported that TTR levels in the CSF of AD patients are lower than in healthy controls (26–30), although other researchers have detected no differences (31). Relevant studies have been conducted with Tg2576 transgenic mice, which express the Swedish mutation of APP (APPSw) and produce extensive Aβ deposits but lack the paired helical filaments and extensive neuronal cell loss one would see in AD (32). Stein and Johnson discovered that expression of TTR is greatly upregulated in Tg2576 mice and, importantly, were able to link increased TTR expression to neuroprotection (33, 34). Increased TTR expression has been recently confirmed by two other groups (35, 36); interestingly, in one of these studies, decreased TTR levels in aged Tg2576 mice were reported (35). Furthermore, Choi et al. observed accelerated Aβ deposition in APPSw mice with heterozygous TTR deletions (37), and Buxbaum and coworkers demonstrated that TTR expression was protective in an APP23 transgenic mouse model (38). In contrast, Wati et al. reported that TTR accelerated vascular Aβ deposits (39), while Doggui et al. observed essentially no effect of TTR deletion on plaque deposition in an aggressive mouse AD model (40). In sum, much, but not all, of the data point to a biologically relevant interaction between TTR and Aβ, thus motivating further investigation into the molecular-level nature of this interaction.
In this study we aim to further characterize the association between Aβ and TTR. Experiments were conducted with both Aβ(1–40) and Aβ(1–42), and with wild-type (wt) TTR as well as two mutants: T119M and F87M/L110M (M-TTR). T119M is a naturally occurring mutant with a higher tetrameric stability than wt (41); much of the increased stability appears to be due to subunit contacts rather than tertiary structural changes (42, 43). T119M acts to suppress aggregation when it is incorporated with aggregation-prone mutants into TTR heterotetramers (43, 44). M-TTR is a double mutant engineered to be stable as a monomer at neutral pH while fully retaining the native secondary and tertiary structure of wt TTR (45).
EXPERIMENTAL PROCEDURES
Expression and purification of recombinant TTR
A recombinant plasmid of human transthyretin (pTWIN1-TTR) was constructed as described previously (46). The IMPACT-TWIN system was chosen because it allows for expression of protein with fully human sequence with native N- and C-termini and purification by single-step affinity adsorption without the need for proteases. TTR mutants T119M and F87M/L110M (M-TTR) were prepared with the QuikChange Site-Directed Mutagenesis Kit (Stratagene, LaJolla, CA) using pTWIN1-TTR (Met−1) as the template. Plasmids were transformed into ER2566 cells (Strategene, LaJolla, CA) and cells were grown on LB media supplemented with 0.1 mg/mL ampicillin. Protein expression was induced with isopropyl-β-D-thiogalatopyranoside. Cells were harvested by centrifugation and then lysed (20 mM Tris-HCl, 500 mM NaCl, 1 mM EDTA, 20 µM PMSF, 1 mM DTT, pH 9.0, containing 8 M urea) as described previously (46). Clarified cell lysate was applied to chitin beads (NEB, Beverly, MA) equilibrated with column buffer (20 mM Tris-HCl, 500 mM NaCl, 1 mM EDTA, 1 mM DTT, pH 9.0), and protein was cleaved from the column and eluted as described (46), dialyzed against PBSA (0.01 M Na2HPO4/NaH2PO4, 0.15 M NaCl, 0.02% w/v NaN3, pH 7.4), and stored at 4°C. Protein concentration was determined by absorbance at 280 nm, assuming an extinction coefficient of 77,600 M−1cm−1.
Characterization of TTR
Wt and mutant proteins were analyzed by a linear trap/FT-ICR MS (LTQ FT Ultra) hybrid mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). A single peak was observed for each sample, with molecular masses of 13761.11, 13791.02 and 13763.01 Da for wt TTR, T119M and M-TTR, respectively, compared to theoretical values of 13761.04, 13791.12 and 13763.08, respectively. SDS gel electrophoresis on boiled samples revealed single bands eluting at 14 kDa; if samples were not boiled, wt TTR and T119M ran as single bands of 55 kDa, indicating formation of tetramers, while M-TTR ran as a 14 kDa (monomer) band. By size exclusion chromatography on a calibrated TSK-GEL G3000SWXL column (TOSOH Bioscience, LLC, PA), M-TTR eluted as a monomer peak while wt and T119M eluted as tetramers. Molecular weight determination by static light scattering further established that M-TTR was monomeric in solution while wt and T119M were tetramers. An ANS binding assay demonstrated that recombinant wt TTR bound thyroxine, indicating assembly into functional tetramers (46). Circular dichroism spectra were obtained to confirm that the secondary structure of the recombinant proteins was virtually identical to commercially available TTR purified from plasma. Tryptophan fluorescence emission spectra, with excitation at 290 nm, was measured; maximum fluorescence was observed at 338 nm, which is consistent with attainment of correct tertiary structure.
Sample preparation
All chemicals were purchased from Fisher (Fair Lawn, NJ) unless otherwise stated. PBSA was double-filtered through 0.22-µm filters (Millex); 8 M urea was prepared in 10 mM glycine-NaOH buffer, pH 10, then filtered through 0.22-µm filters. Aβ(1–40) (AnaSpec, San Jose, CA) and Aβ(1–42) (American Peptide, Sunnyvale, CA) were used for this study. Aβ(1–40) stock solution was prepared by directly dissolving lyophilized Aβ(1–40) in 8 M urea to a final concentration of 16 mg/mL (47). Although this treatment was sufficient to dissociate preformed Aβ(1–40) aggregates, we found it was not sufficient for Aβ(1–42). Therefore, Aβ(1–42) was pretreated with TFA/HFIP (48). Briefly, 0.1 mg Aβ(1–42) was dissolved in 100 µL TFA, and sonicated at room temperature for 15 min, then an additional 1 mL TFA was added to achieve complete solubilization. The solvent was evaporated under nitrogen, 1 mL HFIP was added to redissolve Aβ and then removed under nitrogen. HFIP treatment was repeated 3 times to completely remove all residual TFA. The vial was dried overnight under vacuum, leaving a thin film of Aβ on the vial wall. Aβ(1–42) stock was prepared by dissolving the film in 8 M urea to a final concentration of 6 mg/mL. Native gel electrophoresis confirmed that Aβ(1–40) and Aβ(1–42) stock solutions were in monomeric/dimeric state.
ELISA
Mouse monoclonal anti-Aβ antibodies 6E10 and 4G8 (Covance Inc, Emeryville, CA) were selected for this study. These antibodies recognize both monomeric and aggregated Aβ. 6E10 recognizes the N-terminus of Aβ; its epitope is believed to be residues 4–10 (49) while 4G8 is reactive to a central sequence, residues 17–24 (50, 51). In preliminary experiments, 6E10 and 4G8 were immobilized on 96-well plates. Detection with anti-mouse antibody produced uniformly strong color development, as expected; if TTR and anti-TTR antibody were added to these plates, no non-specific binding was detected.
ELISA plates (Corning Inc, Corning, NY) were coated with 5 µg/mL wt or mutant TTR (100 µL/well) in coating buffer (10mM sodium carbonate, 30mM sodium bicarbonate, 0.05% NaN3, pH 9.6) overnight at room temperature. For positive controls, 6E10 and 4G8 were coated onto the plate (2.5 µg/mL and 5 µg/mL respectively); while for negative controls, only coating buffer was added (blank wells). The plate was washed three times with wash buffer (PBSA with 0.05% Tween 20) and incubated with blocking buffer (5% non-fat dry milk in wash buffer) for 1 hr at room temperature. Freshly prepared Aβ(1–40) or Aβ(1–42) was diluted to 5 µg/mL in PBSA and then immediately added to TTR-coated and blank wells (50 µL/well). For background, PBSA was added into TTR-coated wells. The plate was incubated at room temperature or at 37°C for 1 hr with gently shaking. After washing the plate, 6E10 or 4G8 was diluted per manufacturer’s instruction in wash buffer, added to each well (100 µL/well), and the plate was incubated at room temperature for 1 hr. After washing, anti-mouse HRP antibody (Pierce, Rockford, IL) was added to each well and the plate was incubated for 1 hr. The plate was washed three times with wash buffer, and then 100 µL 3,3’,5,5’ – tetramethylbenzidine (TMB) substrate solution (Pierce, Rockford, IL) was added to each well. After further incubation for 15–30 min, color development was stopped by adding 100 µL 2 M sulfuric acid to each well. Absorbance was measured at 450 nm with EL800 Universal Microplate Reader (Bio-tek Instruments Inc, Winooski, VT). Aβ binding intensity was reported as relative absorbance (Ar) by subtracting the background from sample absorbance.
In some experiments, freshly prepared Aβ(1–40) or Aβ(1–42) was added into TTR-precoated plates and incubated at room temperature for various time intervals (0, 4, 8, and 24 hr for Aβ(1–40) or 0 and 24 hr for Aβ(1–42)). Alternatively, aggregated Aβ samples were prepared by 20-fold dilution of Aβ(1–40) stock into PBSA (to 0.8 mg/mL) followed by overnight incubation at room temperature. Pre-aggregated Aβ(1–40) was diluted to 5 µg/mL in PBSA right before adding to TTR pre-coated wells; plates were processed as described.
Crosslinking and analysis by gel electrophoresis and Western blotting
2.5 µL Aβ(1–40) stock (16 mg/mL in 8 M urea) or Aβ(1–42) stock (6 mg/mL in 8 M urea) was added into 47.5 µL PBSA (control) or TTR solution (0.1 mg/mL in PBSA) and incubated at room temperature for 2 hr. 50 µL of Aβ+TTR or TTR alone was mixed with 2 µL of 25% glutaraldehyde solution and incubated at room temperature for 2 min. The cross-linking reaction was terminated by the addition of 2 µL of 7% (w/v) sodium borohydride in 0.5 M sodium hydroxide. Crosslinked proteins were precipitated with 2 µL of 78% trichloroacetic acid and centrifuged at 14000 rpm for 10 min, then the supernatant was removed by suction. The precipitate was resuspended in 5% SDS and boiled at 95 °C for 5 min. The samples were analyzed on a Precise™ 4–20% polyacrylamide gradient gel with 4% stacking gel (Pierce, Rockford, IL) under constant voltage conditions. In some experiments, Aβ was incubated with TTR overnight before cross-linking and processing as described.
To test whether acid-monomerized TTR would correctly re-assemble to tetramers in the presence of Aβ, 34 µL wt or T119M (0.15 mg/mL, in PBSA) was mixed with 13.5 µL 100 mM acetic acid (final concentration 0.1 mg/mL, pH 4.2) and then incubated at 4 °C for 1 day. For samples with TTR alone, 15 µL 100 mM NaOH was added to adjust the final pH back to neutral; while for TTR+Aβ samples, 2.5 µL Aβ(1–40) stock (16 mg/mL in 8 M urea) was added into acidified TTR solution right before readjusting pH with NaOH. TTR or TTR+Aβ solutions were then incubated at room temperature for 2 hr before glutaraldehyde cross-linking as described above.
Cross-linked Aβ+TTR samples were separated by SDS-PAGE and then transferred onto 0.45 µm nitrocellulose membrane (Pierce, Rockford, IL) at 60 V for 1hr. Membranes were blocked with 5% nonfat dry milk in TBST (20 mmol/L Tris, 150 mmol/L NaCl, 0.05% (v/v) Tween-20, pH 7.6) for 1h at room temperature or at 4 °C over night and then reacted with polyclonal rabbit anti-human TTR antibody (DAKO, Glostrup, Denmark) at 1:1500 dilution in the same TBST buffer for 1 hr at room temperature. The membrane was subsequently treated with anti-rabbit immunoglobulins/HRP (DAKO, Glostrup, Denmark) at a 1:1500 dilution in TBST at room temperature for 1 hr. TTR was visualized by means of ECL™ Western Blotting Analysis System (GE Healthcare, Buckinghamshire, UK).
Crosslinking and tandem mass spectrometry analysis
BS3 is a homobifunctional N-hydroxysuccinimide (NHS) ester with a spacer arm length of 11.4 Å, which reacts with primary amines at pH 7–9. TTR and Aβ were crosslinked with BS3-d0/d4 (Pierce, Rockford, IL). Briefly, for M-TTR+Aβ, 10 µL Aβ(1–40) stock (16 mg/mL in 8 M urea, final concentration 93 µM) was added to 390 µL M-TTR solution (0.14 mg/mL or 10 µM, in PBSA). After incubating at room temperature for 2 hr, 19 µL aliquots were taken and cross-linking was initiated by adding 2 µL BS3-d0/d4 solution (in a 1:1 mixture of d0 and d4 reagent at concentration of 10 mM or 20mM in DMSO), thus yielding 100-, and 200-fold molar excess of cross-linker over M-TTR. The reaction mixture was incubated at room temperature for 30 min or 60 min, and then terminated by adding 1 M NH4CO3 to final concentration of 20 mM. Samples were boiled at 95°C for 5 min followed by separation with SDS-PAGE. Western blotting with anti-TTR antibody was also performed as described previously. For wt TTR+Aβ, 2 µL Aβ(1–40) stock (16 mg/mL in 8 M urea, final concentration 93 µM) was added to 78 µL wtTTR (0.18 mg/mL or 3.3 µM, in PBSA), and the mixture was incubated at room temperature for 2 hr. Then, 2 µL BS3-d0/d4 (in a 1:1 mixture of d0 and d4 reagent at 350 mM in DMSO) was added to initiate crosslinking (~2650-fold molar excess of cross-linker over wt TTR). Control samples of cross-linked wt TTR alone were also prepared. The reaction mixtures were incubated at room temperature for 60 min, and then the reaction was terminated with 1 M NH4CO3. Samples were boiled at 95 °C for 5 min before loading onto gel.
Cross-linked gel bands of interest were excised and enzymatic digestion was performed with In-gel tryptic digestion kit (Pierce, Rockford, IL) following manufacturer’s instruction. Reduction and alkylation were conducted to improve the recovery of cystine-containing peptides and minimize the appearance of unknown masses from disulfide bond formation and side chain modification. Trypsin (~100 ng per digestion) in ammonium bicarbonate was used for digestion at 30°C overnight. Peptide fragments were separated by 2-dimensional LC on an Eksigent system (nano2DLC) with 2 independent binary gradient pumps. The sample was injected into the loading pump, with water/acetonitrile/ethanol (98:1:1, with 0.1% formic acid) as solvent, and the flow rate was set as 7 µL/min, with 2 minute steps. The peptides were separated with a total run of 55 min using a gradient with the following conditions: 10–40% solvent B from 1 to 40 minutes at a constant flow of 400 nL/min, hold at 40% solvent B for 5 minutes, ramp for 5 minutes to 90% solvent B and hold for 5 minutes (solvent A: water with 0.1% formic acid; solvent B: acetonitrile/ethanol (50:50) with 0.1% formic acid). Agilent Zorbax 300SB C8 trap (300Å pore, 5µm particle) was used as first dimension column, and the second dimension column was Magic C18 column (0.075 mm × 120 mm, 300Å pore, 5µm particle).
Nano LC was on-line coupled with LTQ mass spectrometer (Thermo Scientific, Inc., Bremen, Germany). The LTQ mass spectrometer was operated in a data-dependent triple play mode in which each full MS scan (400 to 2000 m/z, centroid) was followed by zoom scan (5000NL minimum, 10 m/z window, +1 charge rejection) where the centroid ions were automatically selected and fragmented by collision-induced dissociation (CID) using a normalized collision energy of 35% (5000NL minimum, 2.4 m/z isolation, wideband activation). All LTQ spectra were collected by Xcalibur (version 2.0.5) (Thermo Scientific Inc., Bremen, Germany). The isotopic pairs were searched and validated manually. Crosslinked peptides were identified using the XQuest software (available at: http://prottools.ethz.ch/orinner/public/htdocs/xquest/). Mass tolerance (m/z) for precursor ions was 1.0 and mass tolerance (m/z) for MS/MS was 0.8.
Alternatively, an in-solution digested sample was prepared. BS3-d0/d4- crosslinked M-TTR+Aβ was prepared as described previously except that the Aβ concentration was 47 µM. After terminating the cross-linking reaction, 4–5 vol of cold acetone (−20 °C) was added, and the sample was kept at − 20°C for at least 2 hr followed by centrifugation at 14,000 rpm for 10–15 min. After removing supernatant, the sample was dried at room temperature before reconstituting in 100 µL 8 M urea buffer (8 M urea in 25 mM ammonium bicarbonate). Reducing reagent (5 µL of 200 mM DTT in 25 mM ammonium bicarbonate) was added, then the sample was incubated at 37 °C for 1 hr. Alkylation was followed by addition of 20 µL 200 mM iodoacetamide (IAA) and incubation for 1 hr at room temperature in the dark. 20 µL reducing reagent was added to consume any leftover IAA. The sample was then diluted to 0.6 M urea with ammonium bicarbonate and digested with trypsin (~1:20 trypsin:protein) at 37 °C overnight. After centrifugation, the supernatant was transferred to a new tube, and ~ 250 µL ammonium bicarbonate was added to lower the urea concentration to 0.4 M before adding GluC (~1:20 GluC:protein). Digestion was performed by incubating the sample at 37 °C overnight.
A model 4800 MALDI TOF/TOF analyzer (Applied Biosystems, Framingham, MA) equipped with a 200 Hz, 355 nm Nd:YAG laser was used for direct peptide profiling of in-solution digested M-TTR+Aβ sample. Acquisitions were performed in positive ion reflectron mode. Instrument parameters were set using the 4000 Series Explorer software (Applied Biosystems). Mass spectra were obtained by averaging 1000 laser shots covering mass range m/z 700–4000. A 6 mg/mL solution of α-cyano-4-hydroxycinnamic acid (CHCA) in 50% (v/v) acetonitrile was used as matrix. For sample spotting, 0.4 µl of sample was spotted on MALDI plate and allowed to dry, followed by application of 0.4 µl matrix solution.
Peptides of M-TTR+Aβ from in-solution double digest were acidified to pH 2 with TFA and analyzed by nano-LC-MS/MS using the Agilent 1100 nanoflow system (Agilent, Palo Alto, CA) connected to a hybrid linear ion trap–orbitrap mass spectrometer (LTQ-Orbitrap, Thermo Fisher Scientific, San Jose, CA) equipped with a nano-electrospray ion source. HPLC was performed using an in-house fabricated column with integrated electrospray emitter made from 360 µm outer diameter × 75 µm inner diameter fused silica tubing. The column was packed with 3 µm C18 particles (Column Engineering, Ontario, CA) to approximately 15 cm. Sample loading and desalting were achieved using a trapping column in line with the autosampler (Zorbax 300SB-C18, 3 µm, 5 × 0.3 mm, Agilent). The sample was desalted over 20 min at a flow rate of 15 µL/min using an isocratic HPLC pump to deliver 0.1 M acetic acid, 1% acetonitrile. Peptides were gradient eluted using a binary solvent system with solvent A (0.1 M acetic acid in water) and solvent B (0.1 M acetic acid, 95% acetonitrile (v/v) in water) at 200 nL/min. The gradient elution was achieved by increasing solvent B from 1% to 40% over 95 min, 40% to 60% over 20 min, and 60% to 100% over 5 min, The LTQ-Orbitrap was set to acquire MS/MS spectra in data-dependent mode as follows: MS survey scans from m/z 300 to 2000 were collected in centroid mode at a resolving power of 100,000. MS/MS spectra were collected on the five most-abundant signals in each survey scan requiring that precursors be present in the 2+ or higher charge state, and that they pass the dynamic exclusion criteria. Crosslinked peptides were identified using XQuest (available at: http://prottools.ethz.ch/orinner/public/htdocs/xquest/). Mass tolerance (m/z) for precursor ions was 1.0 and mass tolerance (m/z) for MS/MS was 0.8.
Light Scattering of Aβ-TTR
Aβ(1–40) stock was diluted 20-fold (final concentration 0.8 mg/mL) into filtered PBSA, or PBSA containing 0.1 mg/mL TTR. The samples were rapidly filtered through 0.45 µm filter directly into a light scattering cuvette placed in a temperature-controlled (25°C) bath of the index-matching solvent decahydronaphthalene. A Coherent (Santa Clara, CA) argon ion laser at 488 nm was focused on the cuvette and light scattering data were collected via Malvern 4700c system (Southborough, MA), as described in more detail elsewhere (52). The average scattered intensity of the sample IS(90°) was measured at 90° scattering angle repeatedly over a 24 hr interval. Scattering from toluene, Itol(90°), was measured to account for changes in laser power or aperture, as was background scattering from the solvent, Ib(90°). Data are normalized to the total mass concentration of protein, ctot:
| (1) |
Transmission Electron Microscopy (TEM)
Aβ with M-TTR, wt TTR and T119M was prepared as for light scattering analysis and incubated for 2 weeks at room temperature, then stained with NanoW negative stain (Nanoprobes.com, Yaphank, NY) and placed on a pioloform coating grid support film (Ted Pella Inc., Redding, CA). Images of fibrils were taken with a Philips CM120 Transmission Electron Microscope (FEI Corp., Eindhoven, The Netherlands). Fibril number and length were counted and measured manually with ImageJ Software (Open source, Public domain). Fibril length ranged from 100 nm to 2250 nm. An array of 10 evenly spaced bins (215 nm each) was chosen initially to count the fibril distribution. Since only a few long fibrils were observed, we grouped the larger bins (530–2250 nm) together.
RESULTS
Binding interactions between Aβ and TTR
We looked for direct evidence of binding between Aβ and TTR using ELISA’s. We used recombinant human wt TTR as well as two mutants: M-TTR (F87M/L110M), an engineered mutant that retains native structure but is a stable monomer (45), and T119M, a naturally occurring mutant with greater tetramer stability than wt (43, 44). We were interested in determining whether there were differences between binding of Aβ to monomer versus tetrameric TTR, or differences in binding of freshly-prepared versus pre-aggregated Aβ to TTR. We used two different antibodies to Aβ, with distinct epitopes, to probe for Aβ binding to TTR, reasoning that differences in antibody binding to Aβ-TTR complexes may shed light on the nature and/or site of any binding interaction. 6E10 and 4G8 react with monomers, oligomers and fibrils of Aβ (53, 54). 6E10 recognizes the hydrophilic N-terminal sequence 1–16 of Aβ; its epitope is reportedly residues 4–10 (49), while 4G8 is reactive to Aβ residues 17–24 (50).
Freshly prepared Aβ was added to wells pre-coated with TTR (wt or mutant). After 1 h incubation, binding of Aβ to immobilized TTR was probed with 6E10 or 4G8 (Figure 1A). With 6E10 as the probe, binding was observed for all three TTRs, but the signal was significantly higher when M-TTR was immobilized than when either wt or T119M was coated on the wells. With 4G8 as the probe, weakly positive binding was observed for all three TTRs. We repeated this experiment with Aβ(1–42) and obtained results that were very similar to those with Aβ(1–40) (Figure 1B): specifically, the strongest signal was obtained for Aβ binding to M-TTR when probing with 6E10. We repeated the experiment at 37°C rather than room temperature, and obtained virtually identical results (data not shown).
Figure 1.
ELISA analysis of TTR-Aβ association. Wt TTR, T119M or M-TTR was immobilized on ELISA plates. (A) Freshly prepared Aβ(1–40) was added to each well; after 1 h incubation and washing to remove unbound material, anti-Aβ antibody 6E10 or 4G8 was used to detect bound Aβ. * p < 0.05, n = 6. (B) Freshly prepared Aβ(1–42) was added to each well; after 1 h incubation and washing to remove unbound material, anti-Aβ antibody 6E10 or 4G8 was used to detect bound Aβ. * p < 0.05, n = 4. (C) The binding of Aβ(1–40) to immobilized TTR as a function of time was measured with anti-Aβ antibody 6E10. * p < 0.05, n = 6. (D) Pre-aggregated Aβ(1–40) was added to TTR coated ELISA plates. Anti-Aβ antibody 6E10 or 4G8 was used to detect bound Aβ. * p < 0.05, n = 6. When Aβ was added to blank wells (negative controls), no absorbance was detected above empty wells (data not shown); these wells were used as “zero” absorbance.
We repeated the experiments, except after Aβ addition the plates were incubated for longer periods of time before the probe antibody was added. With both Aβ(1–40) (Figure 1C) and Aβ(1–42) (not shown) we observed a substantial (3-fold to 12-fold) increase in the amount of 6E10 bound over 24 h. Again, the greatest binding was detected when M-TTR was immobilized.
In another experiment, Aβ(1–40) was pre-aggregated (1 day at room temperature) prior to adding it to TTR-coated wells (Figure 1D). In all cases, the signal was substantially stronger with pre-aggregated Aβ compared to freshly prepared Aβ. (Quantitative comparison between fresh and aggregated Aβ is not possible because the difference was greater than the dynamic range of the experiment. When prepared and processed at otherwise identical conditions, color development was very strong and immediate with pre-aggregated Aβ while it took several minutes with fresh Aβ.) Aβ binding was detected for wt and mutant-coated wells, with the strongest signal again observed with M-TTR-coated wells. However, in contrast to the case with freshly prepared Aβ, detection of pre-aggregated Aβ binding with 4G8 and 6E10 were not statistically different.
Cross-linking of TTR-Aβ complexes
To further investigate the specific interaction of TTR with Aβ, we crosslinked solutions with glutaraldehyde and then analyzed the resulting complexes by SDS-PAGE and Western blots.
In the first set of experiments, TTR was mixed with freshly prepared Aβ(1–40) (1:8 mass ratio) and incubated at room temperature for 2 hr before cross-linking. Samples were boiled and then analyzed by gel electrophoresis (not shown) and by Western blot with anti-TTR antibody detection (Figure 2). Under these conditions, uncrosslinked TTR and Aβ were both monomeric (not shown). On Coomassie-stained gels, both wt and T119M ran as tetramers, while the M-TTR ran primarily as monomer (not shown), demonstrating that the crosslinking conditions retained the native quaternary structure without inducing excessive aggregation. Crosslinked Aβ exhibited a mix of monomers (~4kDa) and multimers, with a strong dimer band and what appeared to be weaker trimer, tetramer and/or pentamer bands. Samples containing Aβ crosslinked with wt TTR or T119M appeared to be a simple additive of TTR and Aβ alone, while with M-TTR+Aβ samples, a series of protein bands with molecular weights in the ~20 kDa to ~30 kDa range were observed. We analyzed these samples further by Western blot, probing with antibodies to TTR (Figure 2A). No signal was detected with the Aβ sample, as expected. We observed a small dimer band along with the strong tetramer band for wt and T119M alone. As with the Coomassie-stained gel, for wt and T119M there was no clear difference between TTR alone and with Aβ, while distinct differences were evident when comparing M-TTR alone versus M-TTR with Aβ. With M-TTR alone, a dimer band was detected, suggesting that M-TTR formed transient dimers that were locked into place by the crosslinker. Interestingly, when M-TTR was mixed with Aβ, we obtained a ladder of bands. The spacing of the bands suggests that these crosslinked species are a series of M-TTR+(Aβ)n complexes. Of additional note is the disappearance of the dimer band from the crosslinked M-TTR+Aβ sample compared to M-TTR alone. These data suggest that Aβ competes with TTR monomer for self-association. We repeated these experiments with TTR and Aβ(1–42), with similar results (Figure 2B). As with Aβ(1–40), there were no obvious differences between wt or T119M alone versus with Aβ, and the dimer band detected was much weaker in crosslinked M-TTR+Aβ than in in crosslinked M-TTR.
Figure 2.
Crosslinking of TTR and Aβ. Aβ was incubated with TTR for 2 hr and then crosslinked with glutaraldehyde. Samples were boiled and analyzed by SDS-PAGE. Samples were then transferred to membrane and detected with anti-TTR antibody. The gels were heavily stained in order to detect minor species. Notations: wt TTR alone (W) or with Aβ (W+A), T119M alone (T) or with Aβ (T+A), M-TTR alone (M) or with Aβ (M+A), and Aβ alone (A). (A) Aβ(1–40) cross-linked with TTR. (B) Aβ(1–42) cross-linked with TTR.
Next, TTR+Aβ samples were preincubated for 1 day, crosslinked, and then analyzed by SDS-PAGE and Coomassie staining (Figure 3). With M-TTR, we again observed a ladder of complexes upon addition of Aβ. When wt and T119M were incubated for 1 day with Aβ, there was a subtle but measurable and reproducible shift towards higher molecular weight species, relative to the TTR tetramer band. Furthermore, the TTR+Aβ band was reproducibly “streaky”. Such streaks on the gel are frequently considered diagnostic of protein aggregates (55). The shift upward in molecular weight and the streaking in the band were observed only with the longer incubation period, suggesting either that Aβ associates only very slowly with TTR tetramers, or that Aβ must be allowed to aggregate before it will associate with TTR tetramers.
Figure 3.
Crosslinking of TTR and Aβ. Aβ(1–40) was incubated with TTR for 1 day and then crosslinked with glutaraldehyde. Samples were boiled and then analyzed by SDS-PAGE and detected with Coomassie stain. Notations: wt TTR alone (W) or with Aβ (W+A), T119M alone (T) or with Aβ (T+A), M-TTR alone (M) or with Aβ (M+A), and Aβ alone (A).
Reassembly of TTR tetramers after acid-induced dissociation in the presence of Aβ
Given the distinctly different interaction of monomeric vs. tetrameric TTR with Aβ, we wondered if this was simply a function of the introduced mutations in M-TTR, or was related to the loss of quaternary structure. At neutral pH, wt TTR tetramer assembly is strongly favored: the half-life for tetramer dissociation is ~24 h while re-assembly occurs within seconds (56). Wt TTR dissociates slowly to monomer at moderately acidic pH, and will remain monomeric if the sample is held at 4°C (14). Much of the native tertiary structure is retained at acidic pH, although there is evidence for some conformational changes involving the EF helix and adjacent loop (15). We obtained monomers from wt TTR by acid dissociation, and then tested their re-assembly to tetramer in the absence and presence of Aβ. Specifically, we first dissociated TTR tetramers by incubating a solution at pH 4.2 and 4°C overnight. Native gels confirmed complete dissociation into monomers under our conditions (data not shown). We then added Aβ and immediately adjusted the pH back to 7.4. Samples were crosslinked after 2 hr incubation. In the absence of Aβ, a return to neutral pH allowed TTR tetramers to reassemble, as assessed by both SDS-PAGE and Western blotting (Figure 4). Over-exposure of the Western blot revealed a strong tetramer smear as well as dimer and monomer bands (the latter bands were not visible on the Coomassie-stained gel likely due to their low concentration). There were no apparent differences in the intensity of the tetramer, dimer and monomer bands between the dissociated and re-assembled TTR compared to TTR that had not been acidified at all. This demonstrates that both wt and T119M will fully re-assemble into tetramers after acid dissociation followed by return to neutral pH. With Aβ present during re-assembly, most of the TTR again reassembled into tetramers; however, there were faint but distinct bands detectable at ~18 kDa and ~22 kDa. These bands were not present if Aβ was absent; since the bands were detected by anti-TTR antibody they contain TTR. The molecular weight of these bands corresponded closely to those obtained when M-TTR was crosslinked with Aβ. These data indicate that complexes between wt TTR monomer and one or two Aβ (TTRmon-Aβ, 18 kDa; TTRmon-Aβ2, 22 kDa) will form if the TTR tetramer is first dissociated. In other words, Aβ binding to TTR monomers competes with re-assembly to tetramer.
Figure 4.
Acid-dissociation and re-assembly of TTR with Aβ. Wt TTR was acid-dissociated into monomers and then re-assembled at neutral pH in the absence or presence of Aβ(1–40). Samples were cross-linked by glutaraldehyde and then analyzed by SDS-PAGE. Notations: wt TTR (W), acid-dissociated and re-assembled wt TTR alone (diW) or with Aβ (diW+A), T119M (T), acid-dissociated and re-assembled T119M alone (diT) or with Aβ (diT+A), M-TTR with Aβ (M+A), and Aβ alone (A).
Mass spectrometry analysis of crosslinked TTR-Aβ fragments
To identify region(s) of TTR that interact with Aβ, we prepared crosslinked samples (wt or M-TTR with Aβ), isolated the crosslinked material by gel electrophoresis, in-gel digested the sample, and analyzed fragments by mass spectrometry. The homobifunctional, amine-reactive cross-linker BS3-d0/d4 was used. BS3 forms amide-bond coupled peptides that cause a characteristic mass shift of 138.067 Da (BS3-d0), whereas the partially hydrolyzed cross-linker causes mass shift of 156.078 Da (BS3-d0). BS3-d0/d4 was used to facilitate the identification of crosslinked peptides based on the appearance of isotopic pairs with 4.025 Da mass difference in the mass spectra.
Figure 5A shows a Western blot of BS3-d0/d4 cross-linked M-TTR and Aβ. At 100-fold molar excess crosslinker and 30 min reaction time, 1:1 and 1:2 M-TTR/Aβ complexes readily formed. No obvious differences were obtained when further increasing the quantity of crosslinker or the reaction time. The gel bands corresponding to 1:1 complexes were excised and subjected to in-gel tryptic digestion.
Figure 5.
Crosslinked samples used in LTQ MS/MS analysis. Aβ(1–40) was incubated with TTR for 2 hr and then crosslinked with BS3-d0/d4. Boxes indicate the gel bands that were excised for enzymatic digestion. (A) M-TTR and Aβ were cross-linked with 100-, or 200-fold molar excess of BS3-d0/d4, then the reaction was terminated after 30 or 60 min. Samples were analyzed by SDS-PAGE, transferred to membrane, and detected with anti-TTR antibody. (B) wt TTR and Aβ were cross-linked with ~2650-fold molar excess of BS3-d0/d4, then the reaction was terminated after 60 min. Samples were analyzed by SDS-PAGE. Dash lines indicate the center of protein bands.
For BS3-d0/d4 crosslinked wt TTR+Aβ, a higher molar ratio of crosslinker to TTR (~2650:1) was required to maintain TTR’s tetrameric structure when denaturing samples. Even with excess crosslinker, some TTR dimers and monomers were observed (Figure 5B). Further adjusting the quantity of crosslinker or reaction time did not improve the crosslinking efficiency (data not shown). Compared to wt TTR alone, the center of wt TTR+Aβ bands exhibited a slight shift towards higher mass. Given the fact that the mass of wt TTR+Aβ complex (1:1) is just 4 kDa higher than wt TTR itself, this small mass shift suggests the formation of crosslinked wt TTR+Aβ complex (1:1). This band was excised and subjected to in-gel tryptic digestion.
The digested peptides were analyzed by LC/MS/MS. On each TTR monomer unit there are 8 lysines that can participate in crosslinking (residues 9, 15, 35, 48, 70, 76, 80, and 126). On Aβ there are two lysines (residues 16 and 28). After enzymatic fragmentation of crosslinked materials, intra- and intermolecular cross-linking products as well as ‘dead-end’ peptides (modified by a partially hydrolyzed cross-linker) are obtained. In addition, the mixture contains many non-crosslinked fragments. Since we are interested in finding the interacting sites on TTR and Aβ, the intermolecular crosslinked species are the targeted peptides. To find these, we manually screened for precursor ions with isotopic patterns, indicating the presence of the crosslinker. The corresponding MS/MS data of each isotopic pair was analyzed with xQuest software in order to identify the crosslinking type as well as to unambiguously identify the cross-linking sites. Since reduction and alkylation were performed before enzymatic digestion, the mass increase (57.02 Da) caused by alkylation with iodoacetamide was taken into consideration for each MS/MS calculation.
For M-TTR+Aβ, one (and only one) intermolecular cross-linked product was detected. Intramolecular and dead-end cross-linked peptides were also observed, but not further analyzed. Figure 6A gives the CID spectrum at m/z 1023.29 of the intermolecular cross-linked product ([M+4H]4+). A series of y type and b type ions were observed, with the fragmentation patterns identifying the cross-linking site on Lys-9 of M-TTR and Lys-28 of Aβ (Scheme 1A). For wt TTR+Aβ, only one intermolecular cross-linked peptide was identified (Figure 6B). MS/MS data analysis indicated that Lys-15 of wt TTR was linked to Lys-16 of Aβ as shown in Scheme 1B. Lys-9 is located near the start of the A strand of TTR while Lys-15 is near the center of the A strand. Thus, analysis of both M-TTR and wt TTR clearly implicates residues in the A strand of TTR in interactions with Aβ.
Figure 6.
MS/MS spectra of cross-linked peptides from trypsin in-gel digestion. (A) CID spectrum of the signal at m/z 1023.29 ([M+4H]4+) corresponding to a cross-linked product between M-TTR (residues 1–15) and Aβ (residues 17–40) cross-linked with 100-fold excess BS3-d0/d4, reaction time 30 min. (B) CID spectrum of the signal at m/z 1382.72 ([M+4H]4+) corresponding to a cross-linked product between wt TTR (residues 10–34) and Aβ (residues 6–28) cross-linked with 2650-fold excess BS3-d0/d4, reaction time 60 min.
Scheme 1.
Fragmentation pattern of cross-linked peptides (LTQ). (A) cross-linked M-TTR+Aβ peptides, (B) cross-linked wt TTR+Aβ peptides,
The advantage of in-gel digestion is that the crosslinked sample is isolated from non-crosslinked materials, thus reducing the number of fragments present. A possible disadvantage, however, is that some fragments, particularly crosslinked materials, can be difficult to elute from the excised gel band. We decided therefore to conduct in-solution digestion of crosslinked M-TTR+Aβ as an alternative approach, to improve sample recovery. A second enzyme, GluC, was introduced after trypsin in order to reduce the size of digested fragments. Because of the anticipated additional complexities of the mixture when tetrameric rather than monomeric TTR is used, particularly the large number of intramolecular crosslinks, we chose not to attempt this procedure with wt TTR. MALDI and LC/Orbitrap with higher resolution than LTQ were combined as complementary techniques to increase the sequence coverage.
As shown in Figure 7, two intermolecular cross-linked peptides were identified for M-TTR+Aβ. In both cases, Lys-28 was confirmed as the cross-linking site on Aβ (Scheme 2). Figure 7A gives the product ion spectrum at m/z of 983.16 ([M+H]4+). In this fragment, Lys-15 on TTR is identified as carrying the crosslinker. Thus, consistent with the LTQ studies, the A strand of TTR is identified as a region involved with binding to Aβ. Figure 7B shows the CID spectrum of precursor ion at m/z 949.99 ([M+H]4+). This fragmentation pattern identifies a crosslinked peptide involving Lys-76 of TTR and Lys-28 of Aβ (Scheme 2). Lys-76 lies near the beginning of the EF helix, the sole helix in TTR.
Figure 7.
MS/MS spectra of cross-linked peptides from trypsin/GluC in-solution digestion. (A) CID spectrum of the signal at m/z 983.16 ([M+4H]4+) corresponding to a cross-linked product between M-TTR (residues 10–21) and Aβ (residues 17–40). (B) CID spectrum of the signal at m/z 949.99 ([M+4H]4+) corresponding to a cross-linked product between M-TTR (residues 71–80) and Aβ (residues 17–40).
Scheme 2.
Fragmentation pattern of cross-linked M-TTR+Aβ peptides (Orbitrap). Aβ residues 17–40 were cross-linked to (A) M-TTR residues 10–21; and (B) M-TTR residues 71–80.
Effects of TTR on Aβ aggregation kinetics and Aβ aggregate morphology
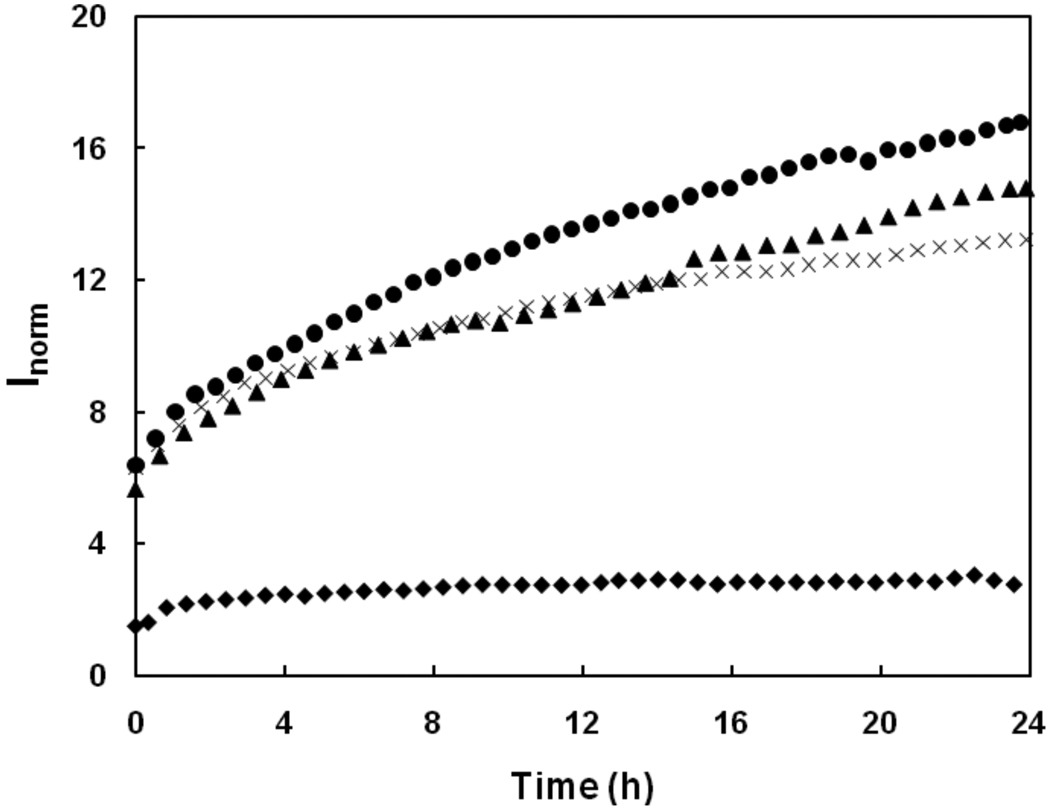
We next examined whether the interaction of TTR with Aβ affected Aβ aggregation. The aggregation rate of Aβ in the presence of each TTR mutant was studied via laser light scattering. Scattered intensity at a 90° scattering angle was measured over time, and normalized for the total protein concentration. The scattered intensity is approximately proportional to the average molecular weight of the particles in solution. Experiments were carried out at a low TTR:Aβ ratio (1:8 mass ratio or 1:110 molar ratio), to minimize the contribution of TTR to the total signal. Aβ alone aggregated at a moderate rate over time, as indicated by the increase in intensity. Wt TTR had very little effect on the Aβ aggregation rate, while T119M moderately increased Aβ aggregation rate over the time course of measurement (Figure 8). The strongest effect was detected with M-TTR, which significantly suppressed Aβ aggregation, lowering both the total intensity and the rate of increase over ~20 hr.
Figure 8.
Normalized scattering intensity at 90° angle Inorm for Aβ alone (✕), or with 0.1 mg/mL wt TTR (▲), T119M (□) or M-TTR (♦). Scattering due to the solvent was subtracted, and results were normalized to the scattering intensity of toluene, to account for changes in laser strength and aperture, and to the mass concentration of peptide/protein.
Aβ+TTR samples were incubated at room temperature for 2 weeks and then examined by TEM (Figure 9). With Aβ alone, a few long, rigid fibrils were observed while the majority of the aggregates were shorter and semi-flexible, with lengths typically around ~100–350nm. EM images of Aβ alone or with wt TTR were very similar to each other (Figure 9A and 9B). With T119M+Aβ (Figure 9C), there appeared to be relatively more long fibrils. The most striking difference was with M-TTR+Aβ (Figure 9D), where very few long fibrils were observed. We measured the length of 100 to 200 aggregates for each sample and plotted a histogram of the results to obtain quantitative estimates of the fibril length distribution (Figure 10). This quantitative data confirmed the qualitative observations made from the images. Approximately 15–20% of aggregates of Aβ, Aβ+wt TTR and Aβ+T119M were long (530–2250 nm), with a slight increase in the fraction of these longest aggregates in the order Aβ < Aβ+wt < Aβ+T119M. However, virtually none of the Aβ+M-TTR aggregates fell into this category. At the other end of the size spectrum, only a few percent of the aggregates of Aβ, Aβ+wt TTR and Aβ+T119M were shorter than 100 nm, while roughly 50% of the Aβ+M-TTR aggregates were this short. More than 95% of the Aβ+M-TTR aggregates were shorter than 315 nm. Thus, these TEM results are consistent with our light scattering observations, specifically, that T119M modestly enhanced Aβ aggregation while M-TTR strongly suppressed aggregation.
Figure 9.
TEM images of (A) Aβ alone or with (B) wt TTR, (C) T119M, or (D) M-TTR. Samples were prepared with 0.8 mg/mL Aβ and 0.1 mg/mL TTR in PBSA. Images were recorded 2 weeks after sample preparation.
Figure 10.
Histogram of Aβ fibrils distribution of Aβ alone (A), wt TTR+Aβ (W+A), T119M +Aβ (T+A) and M-TTR+Aβ (M+A).
DISCUSSION
Our ELISA data demonstrate that Aβ binds to a greater extent to monomeric (M-TTR) than to tetrameric TTR. Interestingly, detection of binding of freshly prepared Aβ to M-TTR was much stronger with 6E10 than with 4G8. 6E10 binds to the N-terminus of Aβ while the 4G8 epitope lies within residues 17–24. Assuming that 6E10 and 4G8 bind equally to monomeric unbound Aβ, this result suggests that the central domain of Aβ (4G8 epitope) but not the N-terminus (6E10 epitope) is partially blocked by binding to M-TTR. Thus, it is likely that residues in or near the central hydrophobic domain (17–24) of Aβ are involved in initial binding of Aβ to M-TTR.
Further confirmation that freshly prepared Aβ binds more readily to monomeric than to tetrameric TTR is obtained through crosslinking studies, where TTR-Aβ complexes were more readily detected with M-TTR than with wt or T119M. Crosslinking of M-TTR alone yielded some dimers, likely due to trapping of transient dimers. Interestingly, these dimers disappeared when Aβ was co-incubated with M-TTR. In its place appeared a ladder of bands with molecular weights corresponding to complexes of M-TTR with one or more Aβ. We checked to see if similar behavior could be detected with wt TTR monomers. Acid-dissociated wt monomers rapidly re-assembled into tetramers when the pH was shifted back to neutral. However, if Aβ was present during re-assembly, we found that a small fraction of the TTR monomers associated with Aβ, forming a TTR-Aβn ladder that was virtually identical to that made with M-TTR and Aβ. Thus, we conclude that it is not the mutations in M-TTR per se that are required for binding of Aβ to TTR monomers; rather, it is the exposure of residues in dissociated TTR that would be sterically blocked or otherwise less accessible in the assembled tetramer. Furthermore, our data demonstrate that Aβ competes, albeit inefficiently, with TTR monomers for assembly into multimers.
Pre-aggregated Aβ bound to a greater extent to M-TTR than freshly prepared Aβ, but there was no difference between 4G8 and 6E10 detection. Since other researchers have demonstrated that both epitopes are exposed in Aβ aggregates (54), we suspect that the 4G8 but not the 6E10 epitope is involved with initial binding of Aβ to M-TTR, that initially-bound Aβ provides a template upon which more Aβ is slowly deposited (57), that there are multiple Aβ bound per TTR, and that the deposited Aβ aggregates have exposed 4G8 and 6E10 epitopes. In other words, the 4G8:6E10 ratio could be a marker for the relative amount of aggregated versus monomeric Aβ: as this ratio approaches 1:1, the fraction of Aβ bound that is aggregated increases. Our data do not allow us to distinguish between formation of aggregates in solution and subsequent deposition, versus deposition of monomers onto a growing aggregate; most likely both mechanisms are in play.
From the ELISA results we found that fresh Aβ bound poorly to tetrameric TTR (either wt or T119M), while binding of pre-aggregated Aβ was greater. The difference in binding of pre-aggregated Aβ to TTR tetramers versus M-TTR, although still significant, was less than the difference with fresh Aβ. In almost all cases (Aβ(1–40), Aβ(1–42), freshly prepared, pre-aggregated), there was no statistically significant difference in 6E10 vis-à-vis 4G8 detection of bound Aβ to TTR tetramers, in contrast to the different 6E10/4G8 pattern with M-TTR. There are several possible explanations. First, there may be no special orientation of Aβ binding to tetrameric TTR, unlike the case with M-TTR: both 6E10 and 4G8 epitopes could be partially blocked. Second, perhaps a different part of Aβ, distant from both epitopes, is involved with binding of Aβ to TTR tetramers, and there is simply less total Aβ bound to tetrameric than to monomeric TTR. Third, there could be binding of Aβ aggregates but not Aβ monomers to tetrameric TTR; this idea is consistent with the notion that the 4G8:6E10 ratio correlates with the degree of aggregation of the bound Aβ.
Consistent with the ELISA results, in crosslinking studies in which Aβ was incubated with wt or T119M, no Aβ-TTR complex was observed when the incubation time was short. However, when samples were incubated for a full day, we noticed a small shift towards higher molecular weights in the tetramer band as well as streaking (Figure 3), usually taken as an indicator of protein aggregates. We did not observe the low-molecular weight ladder that was seen with M-TTR (Figure 2) or with acid-dissociated TTR (Figure 4); in other words, there was no evidence that Aβ strips monomers from stable TTR tetramers. Taken together with the ELISA data, these data suggest that Aβ binds only slowly and relatively weakly to TTR tetramer, and that the binding is mediated primarily through Aβ aggregates rather than through Aβ monomers.
Costas et al. (25) reported no differences in binding of TTR to Aβ monomers, oligomers, or fibrils, in apparent contrast to our findings that Aβ aggregates bind more than monomers. In their experiments, Aβ was prepared at different stages of aggregation and then coated onto wells at pH 9.6, and binding of TTR (at neutral pH) was measured using a competitive inhibition assay. There are several differences between their study and ours that may explain this apparent discrepancy. First, we immobilized a constant quantity of TTR and measured the difference in quantity of pre-aggregated versus fresh Aβ bound to TTR, whereas Costas et al. immobilized a constant quantity of Aβ and measured the binding affinity of TTR by varying its concentration. There is also the possibility that the high pH required to coat Aβ in the Costas et al. study leads to dissociation of preformed aggregates (58). Another possibility is that there are differences in TTR stability arising from different preparation methods: this group previously reported that a significant fraction of recombinant wt TTR dissociated into monomers at 0.3 µM (59), whereas we observed no dissociation of tetramers under similar conditions (not shown). Or, the discrepancies between their work and ours could be due to other differences in methodology or materials.
We used crosslinking combined with tandem mass spectrometry in an attempt to identify the region(s) on TTR involved with Aβ association. Since our focus was on finding heterogeneous intermolecular TTR-Aβ crosslinks, we tried to select conditions that would yield TTR-Aβ complexes at a 1:1 ratio and maximize the number of these heterogeneous crosslinks compared to homogeneous crosslinks (e.g., between TTR monomers or between Aβ monomers). The mixed isotope crosslinker was chosen to facilitate identification of crosslinked peptides in the presence of a large number of other fragments. Gel electrophoresis and excision of the appropriate band was used to isolate the 1:1 TTR-Aβ complex prior to in-gel digestion. Both wt and M-TTR were used in this experiment. Because larger fragments sometimes fail to elute after in-gel digestion, we also used in-solution digestion on a crosslinked mixture of M-TTR and Aβ as a complementary approach.
Using these methods we identified four crosslinked peptides containing fragments from Aβ and TTR (Scheme 1 and Scheme 2). MS/MS analysis localized the crosslinker on TTR to Lys-9 (M-TTR, in-gel digestion), Lys-15 (wt, in-gel digestion), Lys-15 (M-TTR, in-solution digestion) and Lys-76 (M-TTR, in-solution digestion). The position of these lysines are highlighted (Figure 11). No other sites were detected.
Figure 11.
(A) Crystal structure of TTR monomer (pdb entry 1DVQ) with the side chains of Lys9, Lys15 and Lys76 indicated that were found to be cross-linked with the Aβ peptide. The AB loop is highlighted in yellow. Since the N-terminal 10 residues were not in the crystal structure because they are disordered, we added this strand by hand to indicate its relative position. (B) Crystal structure of TTR tetramer where the side chain of Lys15 of each monomer were shown.
These data clearly identify the A strand as a major site of binding between Aβ and TTR, since both Lys-15 and Lys-9 are on or next to this strand. A Kyte-Doolittle analysis identifies this region as a sequence with high hydrophobicity. The A strand is on the ‘inner’ β-sheet of TTR, lining the channel in the center of the assembled tetramer (Figure 11B). This ‘inner’ sheet is solvent-exposed in M-TTR, but access would be sterically restricted in the TTR tetramer. This finding could explain why we observed greater binding of Aβ to M-TTR than to wt. It also explains why Aβ binding to acid-dissociated TTR monomers could compete with re-assembly into tetramers.
Strands A, B, E and G form a highly stable protein core that is resistant to unfolding, while strands C and D are ‘edge’ strands that are fairly labile (17, 18). NMR structural analysis of TTR fibrils support the hypothesis that the C and D strands become more exposed during fibrillogenesis and that a novel interface forms between A strands of different monomers, centered on residue 13, as monomers assemble into fibrils (18). Given this, it is tempting to speculate that the A strand of TTR is an ‘amyloidogenic’ domain, that is recognized by a similar amyloidogenic domain on Aβ.
Involvement of the EF helix in TTR-Aβ association is suggested by the MS/MS data in Figure 8B. In the tetramer, this region protrudes out from the protein surface (Figure 11B). In contrast to the A strand, the EF helix is highly solvent exposed and relatively labile. At acidic conditions (conditions that support tetramer dissociation and subsequent fibrillogenesis), the EF helix and adjacent loop begin to unfold, becoming completed disordered as the pH drops (15). Based on strictly steric considerations, one can easily imagine that Aβ binding to or near the EF helices of the TTR tetramer would be much less restricted than binding to the A strand. Furthermore, one can speculate that there may be opportunities for multivalent attachment of Aβ aggregates to two helices along one ‘side’ of the tetramer.
We next examined the effect of TTR on Aβ aggregation, with a focus on the relative size of aggregates. Our light scattering and electron microscopy studies show conclusively that M-TTR arrests Aβ aggregate growth, even at sub-stoichiometric ratios. The long fibrils that appear in samples containing only Aβ disappeared in the presence of M-TTR. In contrast, wt TTR and T119M did not prevent the appearance of fibrillar aggregates and did not arrest Aβ growth; rather, they may have enhanced aggregation slightly.
Previously we have shown that TTR may accelerate or inhibit Aβ aggregation, depending on the source (purified from plasma versus recombinant), and the degree and nature of modification at Cys-10 (24, 46). Post-translational modification of Cys-10 in plasma and cerebrospinal fluid is common (60, 61). In this study we were careful to prevent Cys-10 modification, confirmed by mass spectrometry, to reduce this source of variability. We are currently conducting a systematic investigation of the effect of specific Cys-10 modification on TTR- Aβ association.
Based on these combined findings, we propose the following scenario. Briefly, we hypothesize that TTR monomers bind to Aβ, with the interaction involving the residues on or near the A strand on TTR, and the central hydrophobic domain on Aβ (near residues 17–24, the 4G8 epitope). This central domain is well known to be a critical structural element in fibrillar Aβ aggregates (62–64), and both Lys-16 and Lys-28 of Aβ flank this domain. If a small Aβ aggregate, with an exposed hydrophobic domain, encounters a TTR monomer rather than another Aβ monomer, then binding between the Aβ hydrophobic domain and the A strand of TTR monomer would effectively arrest further growth of Aβ aggregates. This outcome is indeed what we observed by light scattering and EM (Figure 8 and Figure 9).
MS/MS data indicate that TTR tetramers can also bind to Aβ via residues on or near the A strand. However, ELISA and crosslinking studies support the conclusion that Aβ does not bind as much to TTR tetramers as it does to TTR monomers. Because the A strand lies in the inner sheet of the TTR tetramer, steric constraints would likely limit the extent to which Aβ could be accommodated inside the channel. Data from the experiment in which TTR tetramers were acid-dissociated and then reassembled (Figure 4) suggest that Aβ can compete with TTR monomers for binding during tetramer reassembly, although not particularly effectively. This is reasonable as the rate of TTR tetramer reassembly is extremely fast (56).
We tentatively identified the EF helix as another site on TTR that may be involved with TTR-Aβ association. This helix is solvent-exposed in both monomer and tetramer, and one can imagine that binding of both Aβ monomers and aggregates could be easily accommodated. The driving force for association is unknown. The helix is relatively labile, and Lys-76 forms a weak salt bridge with Glu-89 that is lost at low pH (15). A highly speculative possibility is that this weak intramolecular salt bridge is replaced with an intermolecular salt bridge between Lys-76 (and possibly Lys-80) on TTR and acidic residues on Aβ, of which there are several in the unstructured N-terminus of Aβ (e.g., Asp-1, Glu-3). Structural models of Aβ fibrils indicate that the N-terminus remains disordered and is not incorporated into the fibril (65). Thus, it is plausible that association between TTR tetramers and Aβ aggregates mediated through the EF helix would not interfere with continued growth of Aβ aggregates, consistent with light scattering and EM data. Also, the limited effect of TTR tetramers compared to monomers on Aβ aggregation could be simply due to less binding. One caveat to this analysis is that we have not yet obtained quantitative data as to the relative importance of each site on TTR (the A strand and the EF helix) in mediating TTR-Aβ association.
The biological significance of our results remains to be established. There are several intriguing studies suggesting that TTR plays a neuroprotective role by binding to Aβ, and that the loss of such protection is linked to the onset of Alzheimer’s disease (33, 34, 38). If true, then characterization of the molecular basis for TTR-Aβ association is needed to elucidate the mechanism of neuroprotection, and to understand why such protection might be lost.
ACKNOWLEDGMENT
The authors thank Matt Tobelmann and Jie Hou for their work in designing, producing, and characterizing the transthyretin mutants, Randall Massey for assistance with TEM, Matt Lawrence, Amy Harms and Greg Wilt for assistance with mass spectrometry, and Jeff Johnson and Lingjun Lin for helpful discussions. MS data were collected at the Human Proteomics Program Mass Spectrometry Facility and at the Biotechnology Program Mass Spectrometry Facility, both at the University of Wisconsin-Madison.
Abbreviations
- Aβ
beta-amyloid
- AD
Alzheimer’s disease
- APP
amyloid precursor protein
- BS3
bis(sulfosuccinimidyl) suberate
- CID
collision-induced dissociation
- CSF
cerebrospinal fluid
- ELISA
enzyme-linked immunoassay
- FAP
familial amyloid polyneuropathy
- HRP
horseradish peroxidase
- LC
liquid chromatography
- PBSA
phosphate-buffered saline with azide
- SSA
senile systemic amyloidosis
- T4
thyroxine
- TBST
Tris-buffered saline with Tween
- TMB
3,3’,5,5’ – tetramethylbenzidine
- TTR
transthyretin
Footnotes
This work was supported by funding from the National Institutes of Health (R01 AG033493).
REFERENCES
- 1.Tabaton M, Tamagno E. The molecular link between beta- and gamma-secretase activity on the amyloid beta precursor protein. Cell Mol Life Sci. 2007;64:2211–2218. doi: 10.1007/s00018-007-7219-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gralle M, Ferreira ST. Structure and functions of the human amyloid precursor protein: The whole is more than the sum of its parts. Prog Neurobiol. 2007;82:11–32. doi: 10.1016/j.pneurobio.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Hardy J. Testing times for the "amyloid cascade hypothesis". Neurobiol Aging. 2002;23:1073–1074. doi: 10.1016/s0197-4580(02)00042-8. [DOI] [PubMed] [Google Scholar]
- 4.Wirths O, Multhaup G, Bayer TA. A modified beta-amyloid hypothesis: intraneuronal accumulation of the beta-amyloid peptide - the first step of a fatal cascade. J Neurochem. 2004;91:513–520. doi: 10.1111/j.1471-4159.2004.02737.x. [DOI] [PubMed] [Google Scholar]
- 5.Lee HG, Casadesus G, Zhu XW, Takeda A, Perry G, Smith MA. Challenging the amyloid cascade hypothesis - Senile plaques and amyloid-beta as protective adaptations to Alzheimer disease. Strategies for Engineered Negligible Senescence: Why Genuine Control of Aging May Be Foreseeable. 2004;1019:1–4. doi: 10.1196/annals.1297.001. [DOI] [PubMed] [Google Scholar]
- 6.Ferreira ST, Vieira MNN, De Felice FG. Soluble protein oligomers as emerging toxins in Alzheimer's and other amyloid diseases. IUBMB Life. 2007;59:332–345. doi: 10.1080/15216540701283882. [DOI] [PubMed] [Google Scholar]
- 7.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nature Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 8.Westermark P, Sletten K, Johansson B, Cornwell GG. Fibril in Senile Systemic Amyloidosis Is Derived from Normal Transthyretin. Proc Natl Acad Sci USA. 1990;87:2843–2845. doi: 10.1073/pnas.87.7.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ando Y, Nakamura M, Araki S. Transthyretin-related familial amyloidotic polyneuropathy. Arch Neurol. 2005;62:1057–1062. doi: 10.1001/archneur.62.7.1057. [DOI] [PubMed] [Google Scholar]
- 10.Hamilton JA, Benson MD. Review. Transthyretin: a review from a structural perspective. Cell Mol Life Sci. 2001;58:1491–1521. doi: 10.1007/PL00000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wojtczak A. Crystal structure of rat transthyretin at 2.5 A resolution: first report on a unique tetrameric structure. Acta Biochim Pol. 1997;44:505–517. [PubMed] [Google Scholar]
- 12.Chanoine JP, Alex S, Fang SL, Stone S, Leonard JL, Korhle J, Braverman LE. Role of transthyretin in the transport of thyroxine from the blood to the choroid plexus, the cerebrospinal fluid, and the brain. Endocrinology. 1992;130:933–938. doi: 10.1210/endo.130.2.1733735. [DOI] [PubMed] [Google Scholar]
- 13.Power DM, Elias NP, Richardson SJ, Mendes J, Soares CM, Santos CRA. Evolution of the thyroid hormone-binding protein, transthyretin. Gen Comp Endocrinol. 2000;119:241–255. doi: 10.1006/gcen.2000.7520. [DOI] [PubMed] [Google Scholar]
- 14.Lashuel HA, Lai ZH, Kelly JW. Characterization of the transthyretin acid denaturation pathways by analytical ultracentrifugation: Implications for wild-type, V30M, and L55P amyloid fibril formation. Biochemistry. 1998;37:17851–17864. doi: 10.1021/bi981876+. [DOI] [PubMed] [Google Scholar]
- 15.Palaninathan SK, Mohamedmohaideen NN, Snee WC, Kelly JW, Sacchettini JC. Structural Insight into pH-induced conformational changes within the native human transthyretin tetramer. J Mol Biol. 2008;382:1157–1167. doi: 10.1016/j.jmb.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 16.Babbes ARH, Powers ET, Kelly JW. Quantification of the thermodynamically linked quaternary and tertiary structural stabilities of transthyretin and its disease-associated variants: The relationship between stability and amyloidosis. Biochemistry. 2008;47:6969–6984. doi: 10.1021/bi800636q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu K, Cho HS, Hoyt DW, Nguyen TN, Olds P, Kelly JW, Wemmer DE. Deuterium-proton exchange on the native wild-type transthyretin tetramer identifies the stable core of the individual subunits and indicates mobility at the subunit interface. J Mol Biol. 2000;303:555–565. doi: 10.1006/jmbi.2000.4164. [DOI] [PubMed] [Google Scholar]
- 18.Olofsson A, Ippel JH, Wijmenga SS, Lundgren E, Ohman A. Probing solvent accessibility of transthyretin amyloid by solution NMR spectroscopy. J Biol Chem. 2004;279:5699–5707. doi: 10.1074/jbc.M310605200. [DOI] [PubMed] [Google Scholar]
- 19.Liu K, Kelly JW, Wemmer DE. Native state hydrogen exchange study of suppressor and pathogenic variants of transthyretin. J Mol Biol. 2002;320:821–832. doi: 10.1016/s0022-2836(02)00471-0. [DOI] [PubMed] [Google Scholar]
- 20.Schwarzman AL, Goldgaber D, Palha, Buxbaum JD, Saraiva, Frangione, Kelly, Blake, Kirschner, Arvinte Interaction of transthyretin with amyloid beta-protein: Binding and inhibition of amyloid formation. Nature and Origin of Amyloid Fibrils. 1996;199:146–164. doi: 10.1002/9780470514924.ch10. [DOI] [PubMed] [Google Scholar]
- 21.Schwarzman AL, Gregori L, Vitek MP, Lyubski S, Strittmatter WJ, Enghilde JJ, Bhasin R, Silverman J, Weisgraber KH, Coyle PK. Transthyretin sequesters amyloid beta protein and prevents amyloid formation. Proc Natl Acad Sci USA. 1994;91:8368–8372. doi: 10.1073/pnas.91.18.8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsuzuki K, Fukatsu R, Hayashi Y, Yoshida T, Sasaki N, Takamaru Y, Yamaguchi H, Tateno M, Fujii N, Takahata N. Amyloid beta protein and transthyretin, sequestrating protein colocalize in normal human kidney. Neurosci Lett. 1997;222:163–166. doi: 10.1016/s0304-3940(97)13369-9. [DOI] [PubMed] [Google Scholar]
- 23.Giunta S, Valli MB, Galeazzi R, Fattoretti P, Corder EH, Galeazzi L. Transthyretin inhibition of amyloid beta aggregation and toxicity. Clinical Biochemistry. 2005;38:1112–1119. doi: 10.1016/j.clinbiochem.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Liu L, Murphy RM. Kinetics of inhibition of beta-amyloid aggregation by transthyretin. Biochemistry. 2006;45:15702–15709. doi: 10.1021/bi0618520. [DOI] [PubMed] [Google Scholar]
- 25.Costa R, Goncalves A, Saralva MJ, Cardoso I. Transthyretin binding to A-Beta peptide - Impact on A-Beta fibrillogenesis and toxicity. FEBS Letters. 2008;582:936–942. doi: 10.1016/j.febslet.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 26.Riisoen H. Reduced prealbumin (transthyretin) in CSF of severely demented patients with Alzheimer’s disease. Acta Neurol Scand. 1988;78:455–459. doi: 10.1111/j.1600-0404.1988.tb03687.x. [DOI] [PubMed] [Google Scholar]
- 27.Serot J-M, Christmann D, Dubost T, Couturier M. Cerebrospinal fluid transthyretin: aging and late onset Alzheimer’s disease. J Neurol Neurosurg Psychiatry. 1997;63:506–508. doi: 10.1136/jnnp.63.4.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castaño EM, Roher AE, Esh CL, Kokjohn TA, Beach T. Comparative proteomics of cerebrospinal fluid in neuropathologically-confirmed Alzheimer's disease and non-demented elderly subjects. Neurol Res. 2006;28:155–163. doi: 10.1179/016164106X98035. [DOI] [PubMed] [Google Scholar]
- 29.Gloeckner SF, Meyne F, Wagner F, Heinemann U, Krasnianski A, Meissner B, Zerr I. Quantitative analysis of transthyretin, tau and amyloid-beta in patients with dementia. J Alzheimer's Disease. 2008;14:17–25. doi: 10.3233/jad-2008-14102. [DOI] [PubMed] [Google Scholar]
- 30.Hansson SF, Andreasson U, Wall M, Skoog I, Andreasen N, Wallin A, Zetterberg H, Blennow K. Reduced Levels of Amyloid-beta-Binding Proteins in Cerebrospinal Fluid from Alzheimer's Disease Patients. J Alzheimers Dis. 2009;16:389–397. doi: 10.3233/JAD-2009-0966. [DOI] [PubMed] [Google Scholar]
- 31.Schultz K, Nilsson K, Nielsen JE, Lindquist SG, Hjermind LE, Andersen BB, Wallin A, Nilsson C, Petersen A. Transthyretin as a potential CSF biomarker for Alzheimer's disease and dementia with Lewy bodies: effects of treatment with cholinesterase inhibitors. Eur J Neurol. 17:456–460. doi: 10.1111/j.1468-1331.2009.02841.x. [DOI] [PubMed] [Google Scholar]
- 32.Rockenstein E, Crews L, Masliah E. Transgenic animal models of neurodegenerative diseases and their application to treatment development. Adv Drug Delivery Rev. 2007;59:1093–1102. doi: 10.1016/j.addr.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 33.Stein TD, Anders NJ, DeCarli C, Chan SL, Mattson MP, Johnson JA. Neutralization of transthyretin reverses the neuroprotective effects of secreted amyloid precursor protein (APP) in APP(Sw) mice resulting in tau phosphorylation and loss of hippocampal neurons: Support for the amyloid hypothesis. J Neurosci. 2004;24:7707–7717. doi: 10.1523/JNEUROSCI.2211-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stein TD, Johnson JA. Lack of neurodegeneration in transgenic mice overexpressing mutant amyloid precursor protein is associated with increased levels of transthyretin and the activation of cell survival pathways. J Neurosci. 2002;22:7380–7388. doi: 10.1523/JNEUROSCI.22-17-07380.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai KJ, Yang CH, Lee PC, Wang WT, Chiu MJ, Shen CKJ. Asymmetric expression patterns of brain transthyretin in normal mice and a transgenic mouse model of Alzheimer's disease. Neuroscience. 2009;159:638–646. doi: 10.1016/j.neuroscience.2008.12.045. [DOI] [PubMed] [Google Scholar]
- 36.Wu ZL, Ciallella JR, Flood DG, O'Kane TM, Bozyczko-Coyne D, Savage MJ. Comparative analysis of cortical gene expression in mouse models of Alzheimer's disease. Neurobiol Aging. 2006;27:377–386. doi: 10.1016/j.neurobiolaging.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 37.Choi SH, Leight S, Lee VM-Y, Li T, Wong PC, Johnson JA, Saraiva M, Sisodia SS. Accelerated Abeta deposition in APPswe/PS1DE9 mice with hemizygous deletions of TTR (Transthyretin) J. Neurosci. 2007;27:7006–7010. doi: 10.1523/JNEUROSCI.1919-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buxbaum JN, Ye Z, Reixach N, Friske L, Levy C, Das P, Golde T, Masliah E, Roberts AR, Bartfai T. Transthyretin protects Alzheimer's mice from behavioral and biochemical effects of Aβ toxicity. Proc Natl Acad Sci USA. 2008;105:2681–2686. doi: 10.1073/pnas.0712197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wati H, Kawarabayashi T, Matsubara E, Kasai A, Hirasawa T, Kubota T, Harigaya Y, Shoji M, Maeda S. Transthyretin Accelerates Vascular A beta Deposition in a Mouse Model of Alzheimer's Disease. Brain Pathol. 2009;19:48–57. doi: 10.1111/j.1750-3639.2008.00166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doggui S, Brouillette J, Chabot JG, Farso M, Quirion R. Possible involvement of transthyretin in hippocampal beta-amyloid burden and learning behaviors in a mouse model of Alzheimer's Disease (TgCRND8) Neurodegenerative Diseases. 2010;7:88–95. doi: 10.1159/000285513. [DOI] [PubMed] [Google Scholar]
- 41.Quintas A, Saraiva MJM, Brito RMM. The amyloidogenic potential of transthyretin variants correlates with their tendency to aggregate in solution. FEBS Letters. 1997;418:297–300. doi: 10.1016/s0014-5793(97)01398-7. [DOI] [PubMed] [Google Scholar]
- 42.Hammarström P, Jiang X, Hurshman AR, Powers ET, Kelly JW. Sequence-dependent denaturation energetics: A major determinant in amyloid disease diversity. Proc Natl Acad Sci USA. 2002;99:16427–16432. doi: 10.1073/pnas.202495199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palhano FL, Leme LP, Busnardo RG, Foguel D. Trapping the monomer of a non-amyloidogenic variant of transthyretin: Exploring its possible use as a therapeutic strategy against transthyretine amyloidogenic diseases. J Biol Chem. 2009;284:1443–1453. doi: 10.1074/jbc.M807100200. [DOI] [PubMed] [Google Scholar]
- 44.Hammarstrom P, Schneider F, Kelly JW. Trans-suppression of misfolding in an amyloid disease. Science. 2001;293:2459–2462. doi: 10.1126/science.1062245. [DOI] [PubMed] [Google Scholar]
- 45.Jiang X, Smith CS, Petrassi HM, Hammarstrom P, White JT, Sacchettini JC, Kelly JW. An engineered transthyretin monomer that is nonamyloidogenic, unless it is partially denatured. Biochemistry. 2001;40:11442–11452. doi: 10.1021/bi011194d. [DOI] [PubMed] [Google Scholar]
- 46.Liu L, Hou J, Du J, Chumanov RS, Xu Q, Ge Y, Johnson JA, Murphy RM. Differential modification of Cys10 alters transthyretin's effect on beta-amyloid aggregation and toxicity. Protein Eng Des Sel. 2009;22:479–488. doi: 10.1093/protein/gzp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pallitto MM, Murphy RM. A mathematical model of the kinetics of beta-amyloid fibril growth from the denatured state. Biophys J. 2001;81:1805–1822. doi: 10.1016/S0006-3495(01)75831-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jao SC, Ma K, Talafous J, Orlando R, Zagorski MG. Trifluoroacetic acid pretreatment reproducibly disaggregates the amyloid beta-peptide. Amyloid-Intl J Exp Clin Invest. 1997;4:240–252. [Google Scholar]
- 49.Perdivara I, Deterding LJ, Cozma C, Tomer KB, Przybylski M. Glycosylation profiles of epitope-specific anti-beta-amyloid antibodies revealed by liquid chromatography-mass spectrometry. Glycobiology. 2009;19:958–970. doi: 10.1093/glycob/cwp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim KS, Wen GY, Bancher C, Chen CMJ, Sapienza VJ, Hong H, Wisniewski HM. Detection and quantitation of amyloid β-peptide with 2 monoclonal-antibodies. Neurosci Res Commun. 1990;7:113–122. [Google Scholar]
- 51.Thakker DR, Weatherspoon MR, Harrison J, Keene TE, Lane DS, Kaemmerer WF, Stewart GR, Shafer LL. Intracerebroventricular amyloid-beta antibodies reduce cerebral amyloid angiopathy and associated micro-hemorrhages in aged Tg2576 mice. Proc Natl Acad Sci USA. 2009;106:4501–4506. doi: 10.1073/pnas.0813404106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lowe TL, Strzelec A, Kiessling LL, Murphy RM. Structure-function relationships for inhibitors of beta-amyloid toxicity containing the recognition sequence KLVFF. Biochemistry. 2001;40:7882–7889. doi: 10.1021/bi002734u. [DOI] [PubMed] [Google Scholar]
- 53.Ramakrishnan M, Kandimalla KK, Wengenack TM, Howell KG, Poduslo JF. Surface plasmon resonance binding kinetics of Alzheimer's Disease amyloid beta peptide-capturing and plaque-binding monoclonal antibodies. Biochemistry. 2009;48:10405–10415. doi: 10.1021/bi900523q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Necula M, Kayed R, Milton S, Glabe CG. Small molecule inhibitors of aggregation indicate that amyloid β oligomerization and fibrillization pathways are independent and distinct. J Biol Chem. 2007;282:10311–10324. doi: 10.1074/jbc.M608207200. [DOI] [PubMed] [Google Scholar]
- 55.Gallagher SR. One-dimensional SDS gel electrophoresis of proteins. Curr Protoc Protein Sci. Chapter. 2001;10 doi: 10.1002/0471140864.ps1001s00. Unit 10.11 -Unit 10.11. [DOI] [PubMed] [Google Scholar]
- 56.Wiseman RL, Green NS, Kelly JW. Kinetic stabilization of an oligomeric protein under physiological conditions demonstrated by a lack of subunit exchange: Implications for transthyretin amyloidosis. Biochemistry. 2005;44:9265–9274. doi: 10.1021/bi050352o. [DOI] [PubMed] [Google Scholar]
- 57.Esler WP, Stimson ER, Jennings JM, Vinters HV, Ghilardi JR, Lee JP, Mantyh PW, Maggio JE. Alzheimer's disease amyloid propagation by a template-dependent dock-lock mechanism. Biochemistry. 2000;39:6288–6295. doi: 10.1021/bi992933h. [DOI] [PubMed] [Google Scholar]
- 58.Edwin NJ, Hammer RP, McCarley RL, Russo PS. Reversibility of beta-amyloid self-assembly: effects of pH and added salts assessed by fluorescence photobleaching recovery. Biomacromolecules. 2010;11:341–347. doi: 10.1021/bm900833b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quintas A, Saraiva MJM, Brito RMM. The tetrameric protein transthyretin dissociates to a non-native monomer in solution - A novel model for amyloidogenesis. J Biol Chem. 1999;274:32943–32949. doi: 10.1074/jbc.274.46.32943. [DOI] [PubMed] [Google Scholar]
- 60.Biroccio A, del Boccio P, Panella M, Bernardini S, Di Ilio C, Gambi D, Stanzione P, Sacchetta P, Bernardi G, Martorana A, Federici G, Stefani A, Urbani A. Differential post-translational modifications of transthyretin in Alzheimer's disease: A study of the cerebral spinal fluid. Proteomics. 2006;6:2305–2313. doi: 10.1002/pmic.200500285. [DOI] [PubMed] [Google Scholar]
- 61.Lim A, Prokaeva T, McComb ME, Connors LH, Skinner M, Costello CE. Identification of S-sulfonation and S-thiolation of a novel transthyretin Phe33Cys variant from a patient diagnosed with familial transthyretin amyloidosis. Protein Sci. 2003;12:1775–1785. doi: 10.1110/ps.0349703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams AD, Portelius E, Kheterpal I, Guo JT, Cook KD, Xu Y, Wetzel R. Mapping A beta amyloid fibril secondary structure using scanning proline mutagenesis. J Mol Biol. 2004;335:833–842. doi: 10.1016/j.jmb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 63.Kheterpal I, Chen M, Cook KD, Wetzel R. Structural differences in A beta amyloid protofibrils and fibrils mapped by hydrogen exchange - Mass spectrometry with on-line proteolytic fragmentation. J Mol Biol. 2006;361:785–795. doi: 10.1016/j.jmb.2006.06.066. [DOI] [PubMed] [Google Scholar]
- 64.Tjernberg LO, Naslund J, Lindqvist F, Johansson J, Karlstrom AR, Thyberg J, Terenius L, Nordstedt C. Arrest of beta-amyloid fibril formation by a pentapeptide ligand. J Biol Chem. 1996;271:8545–8548. doi: 10.1074/jbc.271.15.8545. [DOI] [PubMed] [Google Scholar]
- 65.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer's beta-amyloid fibrils. Proc Natl Acad Sci USA. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]