Summary
There is a clear need for an instrument-free molecular diagnostic system for detecting HIV-1 RNA or DNA that can be used in developing countries. Such a test could find utility in the early diagnosis of HIV-1 infection in infancy, as well as serve as a surrogate end-point for vaccine trials. We have developed an IsoAmp® HIV-1 assay (BioHelix Corp, Beverly, MA) targeting the HIV-1 gag gene using our isothermal reverse transcription helicase dependent amplification chemistry (RT-HDA). This IsoAmp® HIV assay uses a disposable amplicon containment device with an embedded vertical-flow DNA detection strip to detect the presence of HIV-1 amplicons. The vertical-flow DNA detection strip has a control line to validate the performance of the device as well as a test line to detect the analyte. The analyte is detected by a sandwich immunoassay for reporter moieties on a capture probe and a detection probe. The control line consists of the detection probe reporter moiety conjugated to the vertical-flow DNA detection strip. The preliminary limit of detection of the IsoAmp HIV assay was evaluated by testing serial dilutions of the HIV-1 Armored® RNA (Assuragen, Austin TX). We found that twenty-one out of twenty-eight (75%) assays were positive when 50 copies of the HIV-1 Armored RNA were input into the IsoAmp HIV reaction.
Keywords: HIV, isothermal amplification, vertical-flow DNA detection strip, helicase dependent amplification
Introduction
Early Human immunodeficiency virus (HIV) infection is accompanied by a rapid transient decrease in peripheral blood CD4+ T cell count and relatively high RNA viral load. For example, Schacker et al. [1] report that median plasma HIV-1 RNA level within the first 30 days of HIV infection is 235,000 copies/mL. Viral load decreases to an average of 46,000 copies/mL after 60 days, and after 120 days viral load ranges between 717 000 and 200 copies/mL. This acute phase is followed by an asymptomatic period that may last several years, and which begins with a rapid partial recovery of the loss in CD4+ cells, as well as a reduction in viral RNA load. If infected asymptomatic patients are left untreated, they will experience a progressive reduction in immune function, as well as a resurgence of RNA viral load [2]. The high risk of death before the age of 2 years among HIV-infected infants, and the fact that maternal antibodies complicate diagnosis for the first 18 months of the infants life, argues in favor of the early detection of HIV infection using molecular tests [3]. Current rapid assays, (e.g., OraQuick ™), based on antibody detection, are relatively insensitive in the early stages of infection; i.e., a 14 to 16 day window must pass before antigen tests can give a positive result, and a 21 day window must pass before antibody tests can yield positive results [4]. On the other hand, false negative results of HIV rapid antibody tests have been reported in infants with AIDS [5]. In contrast, molecular tests are sensitive, and shorten the window of detection period to 8–10 days [4].
Implementing molecular testing for HIV in developing countries can be very challenging because of the high degree of complexity of the current tests used in developed countries as well as the high cost of the instruments used to perform these tests. For example, the branched DNA HIV test (Quantiplex HIV-1 version 3.0; Abbott Laboratories, Abbott Park, IL) is sold in Brazil for $9 (Dr. C. Plicher, personal communication); i.e., a price in line with estimates for the material costs of real-time reverse transcriptase polymerase chain reaction (RT-PCR) assays [6]. The Quantiplex HIV-1, and the other commercial HIV-1 molecular assays (Roche Amplicor HIV-1 Monitor Test, version 1.5, bioMérieux NucliSens HIV-1 QT Assay, Abbott RealTime, Primagen Retina Rainbow/NucliSens EasyQ) all suffer one major limitation: a need for expensive instrumentation and skilled personnel [7]. There are few affordable systems for performing HIV molecular tests in poorer developing countries [7]. Existing low cost CD4+ monitoring tests [8] cannot replace viral RNA as a means of detection for early HIV infection because only the central memory pool of CD4+ T cells (representing less than 1% of the circulating CD4+ T cells) is affected very early in HIV infection. Therefore, there is a clear need for an instrument-free molecular diagnostic system for HIV-1 RNA to support vaccination trials. In addition, the early diagnosis of HIV-1 infection in infancy is not feasible without molecular testing due to the persistence of passively transferred maternal antibodies for up to 18 months after birth [9, 10].
Some nucleic acid detection platforms have potential applications as “instrument-free” means of detecting HIV nucleic acid amplification reaction products. Among these are assays that detect amplification by increases in liquid turbidity due to the formation of magnesium pyrophosphate [11], colorimetric dot blot hybridization assays [12], and immuno-chromatographic detection of amplification products [13, 14]. In addition, isothermal amplification technologies are clearly preferable for “equipment-free” HIV molecular diagnostics in developing countries. Unfortunately most isothermal reactions rely on complicated sets of primers and biochemical manipulations [15–20]. Unlike these other technologies, helicase dependent amplification (HDA) is relatively simple [21–23]. Indeed, HDA is the system most like PCR because it uses helicases to separate DNA strands rather than heat, such that it simply relies on DNA polymerase to amplify DNA rather than on the combinations of polymerases with other enzymes used in other isothermal nucleic acid amplification methods. This greatly simplifies the enzymology involved in the amplification process, while keeping the advantage of all isothermal amplification technologies. In addition, HDA use of probes for detecting amplification products and can readily accommodate the use of internal controls. In the case of HIV detection where accuracy is essential, both of the aforementioned characteristics of HDA are preferable to non-specific means of detection, like turbidity due to the formation of magnesium pyrophosphate.
We have developed an HIV-1 assay targeting the HIV-1 gag gene using our reverse transcription HDA [22]. This assay uses an innovative amplicon containment device with an embedded vertical-flow DNA detection strip [13, 14] to detect the presence of HIV-1 amplicons. The vertical-flow DNA detection strip has a control line to validate the performance of the device as well as a test line to detect the analyte.
Materials and Methods
Oligonucleotides
Multiple-sequence alignment of a 456-base fragment of the HIV-1 gag gene was carried out by analyzing a set of different HIV-1 subtype sequences that are available from public database with the Megalign program of Lasergene (DNASTAR Inc, Madison, WI). A conserved region was chosen to design the RT-HDA primer pair GagF11 (ACCATGCTAAACACAGTGGGGGGACA), GagR5 (ATCCCATTCTGCAGCTTCCTCATTGAT), the capture probe GagP4FI (CAAGCAGCCATGCAAATGTTA-FITC), and the detection probe GagP4Bio (ACCATGCTAAACACAGTGGG-biotin) with the help of Primer3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The melting temperature of the primers and the probes was selected at 57–62 °C and 50–55 °C respectively from the nearest neighbor calculation using the Oligonucleotide Properties Calculator program (http://www.basic.northwestern.edu/biotools/oligocalc.htm) to suit RT-HDA and cassette detection. The specificity of the primers and probes were analyzed with BLASTN (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). Probes GagP4FI and GagP4Bio were labeled with fluorescein isothiocyanate (FITC) and biotin at their 3′ ends respectively. All oligonucleotides were purchased from Operon Biotechnologies, Inc. (Huntsville, AL).
Sample preparation
Sample preparation was performed in triplicate using the QIAamp® Viral RNA Mini Kit according to the manufacturer’s protocol (Qiagen, Valencia, CA). We created simulated clinical samples by spiking 140 μL of human plasma spiked with either 1,400 or 700 or 140 copies of HIV Armored RNA (Assuragen, Austin, TX). A negative control consisted of 140 μL of HIV-negative, pooled unspiked plasma. Extractions were performed by adding 560 μL of Viral Lysis Buffer (Qiagen, Valencia CA) with carrier RNA in a 1.5-mL microcentrifuge tube, adding 140 μL of human plasma, and incubated at room temperature for 10 min. After the incubation 560 μL of 100% ethanol was added to the mixture, and the samples were applied to QIAamp spin columns. Columns were centrifuged at 6000 × g for 1 min., washed using Buffers AW1 by centrifuging at 6000 × g for 1 min, and washed with AW2 by centrifuging at 20,000 × g for 3 min. RNA was eluted from the column with 60 μL of Buffer AVE by incubating the solution in the column at room temperature for 1 min and then centrifuging at 6000 × g for 1 min. After sample preparation, RT-HDA was carried out using 5 μL of the eluates in a final volume of 50 μL. Note: according to the QIAamp® Viral RNA Mini Kit Handbook, this Kit can also be performed using the QIAvac® 24 Plus (Qiagen, Valencia, CA) if a centrifuge is not available.
RT-HDA
RT-HDA was performed using HIV-1 Armored RNA (Asuragen, Inc., Austin, TX) as template. RT-HDA conditions and reagent concentrations were optimized to obtain the final parameters described hereafter. RT-HDA was set up by using an IsoAmp® Rapid HIV Detection Kit (BioHelix Corporation, Beverly, MA). To generate single-stranded amplicon for probe hybridization, asymmetric RT-HDA was performed. In brief, 25 μl of Mix A containing RNA template, primers, and probes was mixed with 21.5 μl IsoAmp® HIV Reaction Mix and 3.5 μl IsoAmp® Enzyme Mix in a 200-μl PCR reaction tube to provide a final concentration of 20 mM Tris-HCl (pH 8.8 at 25°C), 10 mM KCl, 40 mM NaCl, 3.5 mM MgSO4, 400 μM of each of the four deoxynucleoside triphosphates, 3 mM dATP, 20 units/ml ThermoScript reverse transcriptase (Invitrogen, Carlsbad, CA), 50 nM gag gene primer GagF11, 100 nM gag gene primer GagR5, 30 nM capture probe GagP4FI, and 30 nM detection probe GagP4Bio in the RT-HDA reaction. After a 75-minute incubation at 65°C in a water bath or heat block, the reaction tube was directly placed into the BioHelix® Express Strip (BESt™) Cassette (BioHelix Corporation, Beverly, MA) for amplicon detection.
BESt™ Cassette detection
The dual-labeled probe-amplicon products generated from the asymmetric RT-HDA were detected by using a Type I BESt™ Cassette with the test (T) line for capturing FITC/biotin-labeled gag gene probe-amplicon and the control (C) line for capturing the excess streptavidin-conjugated color particles. The biotin labels in the detection probe attract streptavidin-conjugated color particles for visualization and the test results are shown as a colored line visible by the naked eye. In brief, the RT-HDA reaction tube was placed in an amplicon cartridge of a Type I BESt™ Cassette immediately after RT-HDA. The cartridge was then closed and inserted into the detection chamber. The handle of the detection chamber was closed to start amplicon detection. The assay result was read by eye to score from the detection window of the chamber after 10 to 15 minutes. A positive read was scored when both the T line and the C line were visible through the detection window of the cassette. A negative read was scored when only the C line was displayed. The assay was regarded as invalid when neither the T line nor the C line was displayed.
Results
Development of the IsoAmp® HIV Assay
A one-step asymmetric RT-HDA assay was developed to amplify a conserved region in the gag gene among HIV-1 subtypes. As illustrated in Figure 1, an excess of the reverse primer is used to generate single stranded DNA that can be hybridized to the capture and detection probes, and probes are labeled with different 3′ modifications such as to allow for a sandwich immuno-assay detection of amplicons. The detection probe of gag gene was labeled with biotin for binding the streptavidin-conjugated color particles, and the capture probe was labeled with FITC for the capture at the T line on the vertical-flow DNA detection strip. The C line in the vertical-flow DNA detection strip is a biotin conjugate that can capture latex beads conjugated to streptavidin. The RT-HDA reaction was carried out in one-step in one tube. Single-stranded DNA (ssDNA) was generated from the excess primer in the asymmetric RT-HDA, and no denaturation was required to make the ssDNA available for binding by the FITC and the biotin labeled probes (Figure 1). The concentrations of the primers and the probes were optimized to achieve the best performance of asymmetric RT-HDA and BESt™ Cassette detection.
Figure 1.
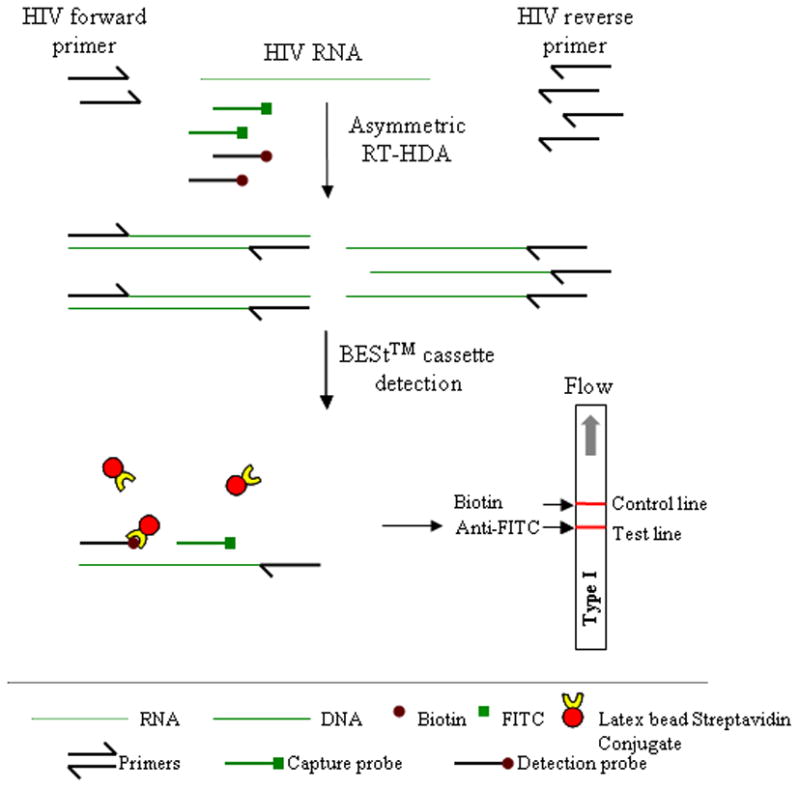
Schematic diagram of the mechanisms for IsoAmp® HIV Assay. By using an asymmetric RT-HDA method, single-stranded DNA (ssDNA) is generated from the amplification of HIV-1 gag gene. The biotin labeled detection probe and the FITC labeled capture probe can bind to the ssDNA after the RT-HDA reaction. These probe-target hybrids then bind to streptavidin-conjugated color particles and are captured by an anti-FITC antibody striped on the DNA detection strip in the BESt™ Cassette detection.
A Type I BESt™ Cassette was used for end-point analysis of the RT-HDA products. Figure 2, shows the individual steps involved in the post-amplification detection of amplicons with BESt cassettes as well as the constituent elements of the BESt cassette in a blow up diagram. The self-contained cassette is comprised of two individual components: an amplicon cartridge that holds the running buffer and a single 0.2-ml thin wall PCR tube containing the amplified product; and the detection chamber which houses the amplicon cartridge and a vertical-flow DNA detection strip embedded into the cassette. The DNA detection strip is striped with an anti-fluorescein isothiocyanate (FITC) antibody and biotin and serves as the T line and the C line respectively in the assay. A razor blade and a plastic pin lodged at the bottom of the detection chamber cut open the PCR tube and the running buffer bulb when the handle of the detection chamber is closed. The mixture flows through a fiberglass paper connected with the DNA detection strip that is attached with a fiberglass pad pre-loaded with streptavidin-conjugated color particles for color visualization.
Figure 2.
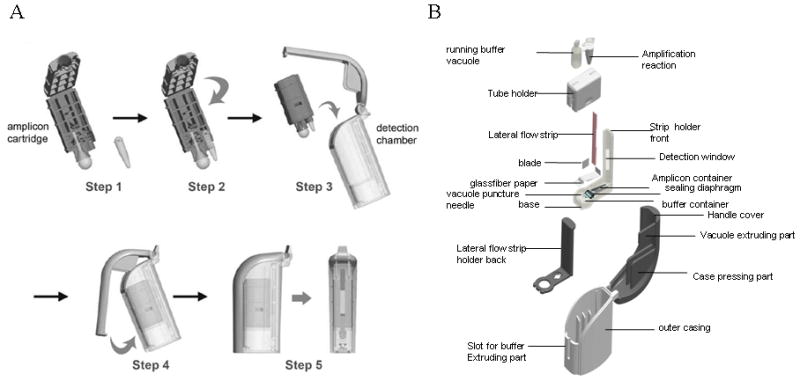
A. Operation of the BESt™ cassette. The amplification reaction vessel is placed in an amplicon cartridge (step 1), the cartridge is closed to immobilize the reaction vessel (step 2), the amplicon cartridge is inserted into the detection chamber (step 3), the handle of the detection chamber is closed to seal the vessel into the chamber, and to cut open the running buffer reservoir, and the reaction vessel (step 4), after 15 minutes the detection window of the chamber is read by eye to score the assay result (step 5). B. Blow-up view of the BESt cassette.
Detection sensitivity test
The detection sensitivity of the IsoAmp® HIV Assay was evaluated by testing a serial dilutions of 500, 50, and 5 copies of the HIV-1 Armored RNA. Eight out of eight (100%) IsoAmp® HIV Assay detected 500 copies of HIV-1 Armored RNA. When the copy number was reduced to 50, twenty-one out of twenty-eight (75%) assays were tested positive. With the copy number at 5, none of the three assays was tested positive. These test results suggest our assay has a detection sensitivity of approximately 50 copies of HIV Armored RNA per assay. Note: Assuragen’s HIV Armored RNAis derived from subtype B of the M clade of HIV-1 and the sensitivity of our assay with other subtypes has not been determined. An example from the detection sensitivity study of the IsoAmp® HIV Assay is shown in Figure 3.
Figure 3.

Determining the detection sensitivity of IsoAmp® HIV Assay. Different amounts of HIV-1 Armored RNA were first amplified by RT-HDA and then applied to BESt™ Cassette detection. The amounts of HIV-1 Armored RNA per reaction used in the assay are: 1, 500 copies; 2, 50 copies; 3, 5 copies; 4, 0 copy.
Integrated assays
HIV-1 Armored RNA was spiked in human plasma at concentrations ranging from 1,000 to 10,000 copies/mL. Total RNA was extracted from the spiked plasma samples using the QIAamp® Viral RNA Mini Kit and 1/12 of the eluate (17 to 117 copies) was input into IsoAmp® HIV Assays. Figure 4 shows a sampling of the data obtained. Three out 3 from plasma samples containing 10,000 copies, 5,000 copies, and 1,000 copies of HIV-1 Armored RNA/mL all tested positive in the IsoAmp® HIV Assays (Figure 4).
Figure 4.

Integrated assays using plasma spiked with Armored RNA HIV. Armored RNA HIV was spiked into human plasma at concentrations ranging from 10,000 copies/mL to 1,000 copies/mL amd extracted with a QIAamp® Viral RNA Mini Kit according to the manufacturer’s protocol. Cassettes #1 to #3, show results obtained with reactions performed with samples containing 10,000 copies HIV Armored RNA/mL plasma; cassettes #4 to #6, are results of amplification with samples containing 5,000 copies HIV Armored RNA/mL of plasma; cassettes #7 to #9, are results obtained with amplification of samples containing 1,000 copies HIV Armored RNA/mL plasma; cassette #10, is the result of a reaction performed with unspiked plasma; cassette #11, is a reaction performed with 500 copies HIV Armored RNA; cassette #12, is a no template control reaction; and cassette #13, is a no reverse transcriptase control performed with 500 copies HIV Armored RNA.
Discussion
This is the first report on the use of RT-HDA in combination with a vertical-flow DNA detection strip embedded in an amplicon containment cassette. The vertical-flow DNA detection strip uses streptavidin conjugated to latex beads and antibodies that bind to the reporter groups incorporated into probes (biotin, FITC, or digoxigenin). As several different reporters can be used in combination with biotin, the device can detect multiple amplicons providing capture zones with the appropriate antibodies have been conjugated to the strip. Takada et al. [24] reported using a similar sandwich immunoassay amplicon detection format for PCR assays. Embedding the vertical-flow DNA detection strip in a closed device protects the laboratory from being contaminated with the amplicons that would be released from the amplification reaction vessel if it were opened after the reaction.
In addition, we are currently developing dry reagent formulations for our assays that may facilitate the deployment of IsoAmp DNA amplification chemistry in the field. The primers and probes described in this article can also be used to amplify HIV DNA from dry blood spots as described by Patton et al. [25], and Zhang et al. [10]. By combining this device with simple sample extraction method, and dry reagents, we feel this technology has the potential to greatly simplify the detection of early HIV infection of seroconverted patients; i.e., detect HIV in newborns of HIV positive mothers as well as in candidates of HIV immunization trials.
Another key requirement for the assay is to couple it with a moderate complexity sample preparation. We have tested the use of FTA filters as a means of extracting “Armored RNA” from human plasma that had been spiked with known quantities of virions prior to extraction. Extraction of RNA with FTA filters is a relatively simple process requiring no precise volumetric measurement prior to the elution of nucleic acids by boiling. We have shown that our assay can detect ~103 copies of HIV RNA in 1 mL of plasma, therefore we anticipate that a relatively low volume of plasma could be used to screen patients for HIV infection if the screening if performed frequently enough to detect early infection (i.e., within the first 30 days when viral titers are in the range of 105/mL of plasma). A more complete investigation of the sensitivity of the integrated assay will be needed to determine if this assay is sensitive enough to detect the resurgence of HIV RNA after the asymptomatic period.
The cost of goods for the IsoAmp HIV test is relatively modest, and thus may be in right price range for developing countries. The BESt cassette is manufactured in China and has a transfer cost of $2 to $3.75. RT-HDA reaction kits have a cost-of-goods of $5, and the FTA filter based sample preparation method a sale price of $1/disc. Therefore, we believe we can offer a “moderate complexity”, and “instrument free” molecular test for under $10. The cost purchasing controls, performing quality assurance as per CLIA regulation, and technician time are not factored in this list price estimate.
Acknowledgments
This study was supported by National Institutes of Health grants 1R43AI066487-01, 1R43AI77418-01, and by the Department of Homeland Security. All the authors, except for Dr. Yi-Wei Tang, were employees of BioHelix Corporation when the research described here was performed. Dr. Yi-Wei Tang is a member of the scientific advisory board at BioHelix Corporation.
Funding statement:
Supported, in part, by National Institutes of Health grants 1R43AI066487-01, 1R43AI77418-01, a contract from the Department of Homeland Security, and by BioHelix Corporation.
Footnotes
Conflict of interest statement:
The IsoAmp® HIV assay is sold as a research use only (RUO) reagent by BioHelix Corporation
References
- 1.Schacker TW, Hughes JP, Shea T, Coombs RW, Corey L. Biological and virologic characteristics of primary HIV infection. Ann Intern Med. 1998;128:613–20. doi: 10.7326/0003-4819-128-8-199804150-00001. [DOI] [PubMed] [Google Scholar]
- 2.Mellors JW, Rinaldo CR, Jr, Gupta P, White RM, Todd JA, Kingsley LA. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–70. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 3.Palumbo PE, Raskino C, Fiscus S, Pahwa S, Fowler MG, Spector SA, Englund JA, Baker CJ. Predictive value of quantitative plasma HIV RNA and CD4+ lymphocyte count in HIV-infected infants and children. JAMA. 1998;279:756–61. doi: 10.1001/jama.279.10.756. [DOI] [PubMed] [Google Scholar]
- 4.Grant PR, Busch MP. Nucleic acid amplification technology methods used in blood donor screening. Transfus Med. 2002;12:229–42. doi: 10.1046/j.1365-3148.2002.00382.x. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Wang J, Wilson GJ, Tang YW, Lu HZ. Negative results of a rapid antibody test for HIV in a 16-month-old infant with AIDS. Ann Clin Lab Sci. 2008;38:293–5. [PubMed] [Google Scholar]
- 6.Drosten C, Panning M, Drexler JF, Hansel F, Pedroso C, Yeats J, de Souza Luna LK, Samuel M, Liedigk B, Lippert U, Sturmer M, Doerr HW, Brites C, Preiser W. Ultrasensitive monitoring of HIV-1 viral load by a low-cost real-time reverse transcription-PCR assay with internal control for the 5′ long terminal repeat domain. Clin Chem. 2006;52:1258–66. doi: 10.1373/clinchem.2006.066498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiscus SA, Cheng B, Crowe SM, Demeter L, Jennings C, Miller V, Respess R, Stevens W Forum for Collaborative HIV Research Alternative Viral Load Assay Working Group. HIV-1 viral load assays for resource-limited settings. PLoS Med. 2006;3:e417. doi: 10.1371/journal.pmed.0030417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez WR, Christodoulides N, Floriano PN, Graham S, Mohanty S, Dixon M, Hsiang M, Peter T, Zavahir S, Thior I, Romanovicz D, Bernard B, Goodey AP, Walker BD, McDevitt JT. A microchip CD4 counting method for HIV monitoring in resource-poor settings. PLoS Med. 2005;2:e182. doi: 10.1371/journal.pmed.0020182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sison AV, Campos JM. Laboratory methods for early detection of human immunodeficiency virus type 1 in newborns and infants. Clin Microbiol Rev. 1992;5:238–47. doi: 10.1128/cmr.5.3.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Q, Wang L, Jiang Y, et al. Early infant human immunodeficiency virus type 1 detection suitable for resource-limited settings with multiple circulating subtypes by use of nested, three-monoplex DNA PCR and dried blood spots. J Clin Microbiol. 2008;46:721–6. doi: 10.1128/JCM.01539-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasaki Y, Nagumo S. Rapid identification of Curcuma longa and C. aromatica by LAMP. Biol Pharm Bull. 2007;30:2229–30. doi: 10.1248/bpb.30.2229. [DOI] [PubMed] [Google Scholar]
- 12.Teng PH, Chen CL, Sung PF, Lee FC, Ou BR, Lee PY. Specific detection of reverse transcription-loop-mediated isothermal amplification amplicons for Taura syndrome virus by colorimetric dot-blot hybridization. J Virol Methods. 2007;146:317–26. doi: 10.1016/j.jviromet.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 13.Kong H, Ranalli T, Lemieux B. New isothermal molecular diagnostic platforms IVD Technology. 2007 November/December; (on line publication at http://www.devicelink.com/ivdt/archive/07/11/009.html)
- 14.Goldmeyer J, Li H, McCormac M, Cook S, Stratton C, Lemieux B, Kong H, Tang W, Tang Y-W. Identification of Staphylococcus aureus and Determination of Methicillin Resistance Directly from Positive Blood Cultures by Isothermal Amplification and Disposable Detection Device. J Clin Microbiol. 2008 doi: 10.1128/JCM.02234–07. published ahead of print on 30 January 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanco L, Lazaro JM, de Vega M, Bonnin A, Salas M. Terminal protein-primed DNA amplification. Proc Natl Acad Sci U S A. 1994;91:12198–202. doi: 10.1073/pnas.91.25.12198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guatelli JC, Whitfield KM, Kwoh DY, Barringer KJ, Richman DD, Gingeras TR. Isothermal, in vitro amplification of nucleic acids by a multienzyme reaction modeled after retroviral replication. Proc Natl Acad Sci U S A. 1990;87:1874–8. doi: 10.1073/pnas.87.5.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lizardi PM, Huang X, Zhu Z, Bray-Ward P, Thomas DC, Ward DC. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat Genet. 1998;19:225–32. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- 18.Tyagi S, Landegren U, Tazi M, Lizardi PM, Kramer FR. Extremely sensitive, background-free gene detection using binary probes and beta replicase. Proc Natl Acad Sci U S A. 1996;93:5395–400. doi: 10.1073/pnas.93.11.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker GT, Fraiser MS, Schram JL, Little MC, Nadeau JG, Malinowski DP. Strand displacement amplification--an isothermal, in vitro DNA amplification technique. Nucleic Acids Res. 1992;20:1691–6. doi: 10.1093/nar/20.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang DY, Brandwein M, Hsuih TC, Li H. Amplification of target-specific, ligation-dependent circular probe. Gene. 1998;211:277–85. doi: 10.1016/s0378-1119(98)00113-9. [DOI] [PubMed] [Google Scholar]
- 21.Vincent M, Xu Y, Kong H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004;5:795–800. doi: 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.An L, Tang W, Ranalli TA, Kim HJ, Wytiaz J, Kong H. Characterization of a thermostable UvrD helicase and its participation in helicase-dependent amplification. J Biol Chem. 2005;280:28952–8. doi: 10.1074/jbc.M503096200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldmeyer J, Kong H, Tang W. Development of a novel one-tube isothermal reverse transcription thermophilic helicase-dependent amplification platform for rapid RNA detection. J Mol Diagn. 2007;9:639–44. doi: 10.2353/jmoldx.2007.070012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takada K, Sakaguchi Y, Oka C, Hirasawa M. New rapid polymerase chain reaction-immunochromatographic assay for Porphyromonas gingivalis. J Periodontol. 2005;76:508–12. doi: 10.1902/jop.2005.76.4.508. [DOI] [PubMed] [Google Scholar]
- 25.Patton JC, Sherman GG, Coovadia AH, Stevens WS, Meyers TM. Ultrasensitive HIV-1 p24 antigen assay modifi ed for use on dried whole blood spots as a reliable, affordable test for infant diagnosis. Clin Vaccine Immunol. 2006;13:152–155. doi: 10.1128/CVI.13.1.152-155.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]


