Abstract
A recent genome-wide association study (PanScan) identified significant associations at the ABO gene locus with pancreatic cancer risk; however, the mechanisms underlying these associations and the influence of specific ABO genotypes remain unknown. We determined ABO genotypes (OO, AO, AA, AB, BO, BB) in 1534 cases and 1583 controls from 12 prospective cohort studies participating in PanScan. We also grouped participants by genotype-derived serologic blood type (O, A, AB, B). Adjusted odds ratios (ORs) for pancreatic cancer by ABO alleles were calculated using logistic regression. Compared to blood type O, the ORs for pancreatic cancer in subjects with types A, AB, and B were 1.38 (95% confidence interval [CI], 1.18-1.62), 1.47 (95% CI, 1.07-2.02), and 1.53 (95% CI, 1.21-1.92), respectively. The incidence rates (cases per 100,000 subjects per year) for blood types O, A, AB, and B were 28.9, 39.9, 41.8, and 44.5. An increase in risk was noted with the addition of each non-O allele. Compared to OO, subjects with AO and AA had ORs of 1.33 (95% CI, 1.13-1.58) and 1.61 (95% CI, 1.22-2.18), while subjects with BO and BB had ORs of 1.45 (95% CI, 1.14-1.85) and 2.42 (1.28-4.57). The population attributable fraction for non-O blood type was 19.5%. In a joint model with smoking, current smokers with non-O blood type had an adjusted OR of 2.68 (95% CI, 2.03-3.54), compared with non-smokers with blood type O. Among participants in a large prospective cohort consortium, ABO genotypes were significantly associated with pancreatic cancer risk.
Keywords: Pancreatic cancer, ABO blood group, prospective cohort study
Introduction
Pancreatic cancer is the fourth leading cause of cancer-related mortality in Western societies, and more than 95% of patients who develop pancreatic cancer will die from the disease (1). Several highly penetrant but rare genetic alterations have been identified that predispose individuals to pancreatic cancer, but predisposing genetic alterations remain unknown for the vast majority of patients who develop this disease (2).
Work by Dr. Karl Landsteiner in the early 20th century led to the identification of three blood groups, forming the basis of transfusion medicine (3). A single gene on chromosome 9q34 was ultimately found to define a person's blood group, and its nucleotide sequence was elucidated in 1990 (4, 5). The ABO gene encodes a glycosyltransferase with three main variant alleles (A, B, and O), with different substrate specificities (6). The A, B and O glycosyltransferases transfer N-acetylgalactosamine, D-galactose, and no sugar residue, respectively, to a protein backbone, known as the H antigen, which is expressed on the surface of red blood cells and numerous other tissues throughout the body (7). A role for ABO blood group antigens in human disease has been suspected for several decades (8, 9), although an association with pancreatic cancer risk has been inconsistent (9-14).
A recent genome-wide association study (GWAS) among pancreatic cancer cases and controls (PanScan) found that several single nucleotide polymorphisms (SNPs) at the ABO gene locus were among the most statistically significant associations with pancreatic cancer risk (15). However, the nature of this association and the influence of specific ABO genotypes on the risk of pancreatic cancer remain unknown. We hypothesized that defining a subject's ABO blood group alleles might provide additional risk information and provide supportive evidence for the role of ABO glycosyltransferase specificity in pancreatic tumorigenesis. Therefore, we utilized genotype data from over 3000 subjects in 12 prospective cohort studies participating in PanScan to impute individual ABO alleles and determine their association with pancreatic cancer risk. This allowed us to investigate the full range of genotypic variation at the ABO gene locus (OO, AO, AA, AB, BO, BB), as well as ABO serotype (O, A, B, AB) inferred from ABO genotypes. In addition, our nested prospective design allowed for rigorous investigation of interactions between known risk factors for pancreatic cancer and ABO blood group alleles.
Materials and Methods
Study Population
The Pancreatic Cancer Cohort Consortium includes nested case-control studies from 12 prospective cohorts: Alpha-Tocopherol Beta-Carotene Cancer Prevention Study (ATBC), CLUE II, American Cancer Society Cancer Prevention Study-II (CPS II); European Prospective Investigation into Cancer and Nutrition Study (EPIC); Health Professional's Follow-up Study (HPFS); New York University Women's Health Study (NYUWHS); Nurses' Health Study (NHS); Physicians' Health Study I (PHS I); Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO); Shanghai Men's and Women's Health Study (SMWHS); Women's Health Initiative (WHI); and Women's Health Study (WHS). See Supplementary Table 1 for list of participating cohorts with corresponding methodologic references. In each cohort, a defined population of subjects was followed prospectively with repeated assessments of lifestyle factors and ascertainment of cancer diagnoses. Cases included subjects with incident primary pancreatic adenocarcinoma (ICD-O-3 code C250-C259 or C25.0-C25.3, C25.7-C25.9). All subjects with non-exocrine pancreatic tumors (C25.4, histology type, 8150, 8151, 8153, 8155, 8240, 8246) were excluded. Each cohort study selected participants with blood or buccal cells collected prior to cancer diagnosis. Incident pancreatic cancer cases identified by self-report, report of next-of-kin or through national death indices were confirmed by subsequent medical record review, linkage with a cancer registry, or both, without prior knowledge of genetic data.
One control was selected per case within each cohort. Controls were matched on year of birth (+/-5 years), gender, self-reported race/ethnicity, and source of DNA (peripheral blood or buccal cells). Controls were alive without pancreatic cancer on the incidence date of the matched case. Four cohorts (HPFS, NHS, PHS, and WHS) were additionally matched on smoking status (never, former, current), and some cohorts were also matched on age at baseline (+/-5 years), age at blood draw (+/-5 years), date/time of day of blood draw, or fasting status at blood draw. Each cohort obtained informed consent from study participants and approval from its Institutional Review Board (IRB). The Special Studies Institutional Review Board (SSIRB) of the National Cancer Institute approved the pooled PanScan study.
Assessment of ABO Blood Group Alleles
Detailed methods for genotyping by PanScan can be found elsewhere (15). Haplotypes of rs687289 and rs8176746 are perfectly correlated (r2=1) with the O and B alleles, respectively, in the 60 HapMap Phase 2 CEPH European founders (16). Since rs687289 was not genotyped as part of PanScan, we used rs505922, which is a perfect surrogate for rs687289 in all HapMap Phase 2 samples. Using rs505922 and rs8176746, all subjects' phased haplotype pairs could be inferred using an EM algorithm with greater than 99% posterior probability (17). Since each subject has two ABO alleles, six genotypes were possible, OO, AO, AA, AB, BO, and BB.
In the HPFS and NHS, participants self-reported ABO status as defined serologically (O, A, AB or B) in 1996. In a validation study of these two cohorts, self-reported ABO blood type was concordant with laboratory assessed serologic blood type in 91% of participants (14). We further examined the concordance of genotype-derived and self-reported blood type in 187 NHS and HPFS participants included in PanScan. In 92% of subjects, the self-reported blood type and the genotype-derived blood type were identical; this proportion is within the limit of error expected from the prior validation of self-report, and strongly supports the accuracy of the genotype-defined blood group alleles.
Assessment of Covariates
Across all 12 participating cohorts, covariates were collected though written questionnaires or in-person interviews. Detailed descriptions of data collection methods have been published previously (see Supplemental Table 1 for references). We obtained data from each cohort on participants' age, gender, race/ethnicity (White, Asian, African, other), body-mass index (BMI) smoking status (current, past, never), and history of diabetes (yes, no).
Statistical Analyses
Participant characteristics were examined for cases and controls, and by blood type among controls. We compared the distribution of blood type alleles in our study with the distributions seen in other comparable populations. We used unconditional logistic regression to calculate odds ratios (ORs) and 95% confidence intervals (CI) for pancreatic cancer by ABO alleles, adjusted for age, gender, race/ethnicity, cohort, smoking status, BMI, and history of diabetes. We repeated our analyses limited to White subjects and after excluding the 74 cases and their matched controls that were included in our prior analysis of self-reported ABO serotype and pancreatic cancer risk (14). In addition, to adjust for detectable differences in population substructure, a principal component analysis of all DNA samples used in PanScan was performed using the entire set of the nearly 550,000 SNPs with the EIGENSTRAT program (15, 18). Five principal components were effective for distinguishing significant population groups and we performed an additional analysis that included these principal components in our models to correct for genetic admixture. We used the likelihood ratio test for nested models to assess whether a model based on ABO genotypes improved model fit relative to a model using rs505922 alone.
We assessed effect-measure modification by conducting analyses stratified by other risk factors for pancreatic cancer, including age (≤ 65, 66-75, > 75 years), gender (male, female), race/ethnicity (White, non-White), smoking (current/recent quitter, never/distant quitter), and BMI (<25, 25-29.9, >30 kg/m2). Tests for modification by other risk factors were assessed by entering the cross-product of blood type and the covariate into the model. We jointly assessed the effect on pancreatic cancer risk of ABO genotype and the three best characterized modifiable risk factors for pancreatic cancer: tobacco use, obesity, and diabetes mellitus. Participants from the HPFS, NHS, PHS, and WHS were matched on smoking status, and therefore were excluded from the joint analyses with smoking status.
The population attributable fraction (PAF) for pancreatic cancer due to non-O blood groups was calculated using the following equation: PAF = Pd ([OR − 1] / OR), where Pd = prevalence of exposure among pancreatic cancer cases and OR = multivariable-adjusted OR calculated by logistic regression models (19). Using the PAF and population average incidence rate λ, we calculated absolute incidence rates of pancreatic cancer in Whites by ABO blood group G as (1-PAF) λ ORG (20). Average age-adjusted population rates weighted by gender for U.S. Whites 50 years and older were obtained from Surveillance, Epidemiology and End Results (SEER) data.
We assessed heterogeneity in the association between blood type and pancreatic cancer risk across the cohorts using Cochran's Q-statistic (21). All statistical analyses were performed using the SAS 9.1 statistical package (SAS Institute, Cary, North Carolina), and all P-values were two-sided.
Results
From the 12 participating cohorts, 1534 pancreatic cancer cases and 1583 controls were available for analysis. As expected, a higher proportion of cases than controls were current smokers or reported a history of diabetes (Supplemental Table 2). Characteristics of control participants were similar among the ABO blood types, except that participants with blood types AB and B were less likely to be White, a pattern consistent with the increased frequency of B alleles among Asians (Table 1). The frequency distributions of ABO alleles were highly similar among our control participants and subjects in previous studies (14, 16, 22-25) (Table 2). The frequencies of blood types O, A, AB, and B were 41.5%, 40.6%, 5.6%, and 12.3%, respectively, among our control participants, which were also consistent with previously reported studies (14, 16, 22-25).
Table 1. Characteristics by genotype-derived ABO blood type among control participants.
| Characteristic | Genotype-derived ABO Blood Type | |||
|---|---|---|---|---|
| O | A | AB | B | |
| No. of control participants | 657 | 642 | 89 | 195 |
| Mean age (years, +/- SD) | 69.6 +/- 7.5 | 68.9 +/- 8.4 | 70.3 +/- 5.9 | 69.3 +/- 7.6 |
| Female gender (%) | 54.0 | 50.2 | 48.3 | 50.8 |
| White race/ethnicity (%) | 92.9 | 94.1 | 86.5 | 84.1 |
| Smoking status (%) | ||||
| Current | 19.8 | 25.2 | 21.4 | 26.7 |
| Past | 37.9 | 31.2 | 44.9 | 28.7 |
| Never | 41.3 | 42.1 | 31.5 | 42.6 |
| Unknown | 1.1 | 1.6 | 2.3 | 2.1 |
| Mean body-mass index (kg/m2, +/- SD) | 26.5 +/- 4.5 | 26.4 +/- 4.6 | 25.2 +/- 4.2 | 26.1 +/- 4.4 |
| History of diabetes mellitus (%) | 6.2 | 7.3 | 5.6 | 5.1 |
Abbreviations: SD, standard deviation
Table 2. Distributions of ABO alleles among control participants and comparable reference populations.
We estimated the risk of pancreatic cancer according to genotype-derived ABO blood type among all study participants (Table 3). Compared to subjects with blood type O, those with blood types A, AB and B were at greater risk of developing pancreatic cancer. Moreover, when we limited the analysis to Whites only (Table 3), removed the 74 cases and matched controls who were included in a previous analysis of self-reported ABO blood type and pancreatic cancer risk (14), or included the five principal components of genetic structure in our model, the adjusted ORs for blood types A, AB and B were essentially unchanged. The incidence rates for pancreatic cancer (cases per 100,000 persons at risk) among White participants with blood types O, A, AB, and B were 28.9, 39.9, 41.8, and 44.5, respectively.
Table 3. Age-adjusted and multivariable-adjusted odds ratios (95% confidence intervals) for incident pancreatic cancer by genotype-derived ABO blood type.
| O | A | AB | B | |
|---|---|---|---|---|
| No. of cases / controls | 511 / 657 | 700 / 642 | 97 / 89 | 226 / 195 |
| Age-adjusted OR | 1.0 | 1.39 (1.19-1.63) | 1.43 (1.05-1.95) | 1.51 (1.21-1.89) |
| Multivariable-adjusted OR* | 1.0 | 1.38 (1.18-1.62) | 1.47 (1.07-2.02) | 1.53 (1.21-1.92) |
| Multivariable-adjusted OR† | 1.0 | 1.38 (1.17-1.63) | 1.45 (1.03-2.04) | 1.54 (1.20-1.97) |
Abbreviations: OR, odds ratio (95% confidence interval)
Multivariable adjustment by age, gender, race/ethnicity, cohort, smoking status, body-mass index, and history of diabetes mellitus, in the entire study population.
Multivariable adjustment by age, gender, cohort, smoking status, body-mass index, and history of diabetes mellitus, in White participants only.
We further examined pancreatic cancer risk according to ABO genotype. An increased risk was observed with the addition of each non-O allele (OR, 1.29; 95% CI, 1.16-1.42). Compared to subjects with OO, those with AO and AA had ORs of 1.33 (95% CI, 1.13-1.58) and 1.61 (95% CI, 1.22-2.18), respectively, while subjects with BO and BB had ORs of 1.45 (95% CI, 1.14-1.85) and 2.42 (95% CI, 1.28-4.57), respectively (Table 4). The comparison of a model including full genotypic variability at the ABO locus and a model with only genotypic variation at rs505922, the most statistically significant SNP from the GWAS, resulted in a p-value of 0.48.
Table 4. Age-adjusted and multivariable-adjusted odds ratios (95% confidence intervals) for incident pancreatic cancer by ABO blood group alleles.
| Second Allele | First Allele | |||
|---|---|---|---|---|
| O | A | B | ||
| O | No. of subjects | 1168 | 1080 | 377 |
| Age-adjusted OR | 1.0 | 1.35 (1.14-1.59) | 1.43 (1.14-1.81) | |
| Multivariable-adjusted OR* | 1.0 | 1.33 (1.13-1.58) | 1.45 (1.14-1.85) | |
| Multivariable-adjusted OR† | 1.0 | 1.32 (1.11-1.57) | 1.47 (1.14-1.90) | |
| A | No. of subjects | - | 262 | 186 |
| Age-adjusted OR | 1.61 (1.23-2.11) | 1.43 (1.05-1.95) | ||
| Multivariable-adjusted OR* | 1.61 (1.22-2.18) | 1.47 (1.07-2.02) | ||
| Multivariable-adjusted OR† | 1.68 (1.27-2.24) | 1.45 (1.03-2.04) | ||
| B | No. of subjects | - | - | 44 |
| Age-adjusted OR | 2.35 (1.26-4.39) | |||
| Multivariable-adjusted OR* | 2.42 (1.28-4.57) | |||
| Multivariable-adjusted OR† | 2.54 (1.19-5.39) | |||
Abbreviations: OR, odds ratio (95% confidence interval)
Multivariable adjustment by age, gender, race/ethnicity, cohort, smoking status, body-mass index, and history of diabetes mellitus, in the entire study population.
Multivariable adjustment by age, gender, cohort, smoking status, body-mass index, and history of diabetes mellitus, in White participants only.
We calculated the population attributable fraction for participants with non-O blood groups (i.e. blood types A, AB, or B). White participants with non-O blood group had an adjusted OR for pancreatic cancer of 1.42 (95% CI, 1.21-1.66). Based on this OR and the prevalence of non-O blood types in these cases (66.2%), 19.5% of all pancreatic cancers in our European ancestry population were attributable to the inheritance of a non-O blood group.
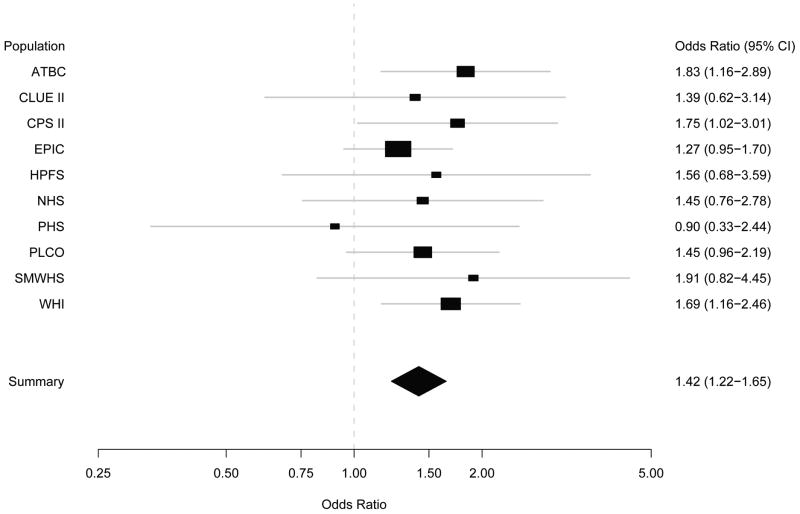
The odds ratios comparing subjects with non-O blood groups to those with blood group O were highly similar across cohorts (Cochran's Q-statistic P = 0.91 for the comparison of non-O blood type [i.e. A, AB or B] to O blood type across cohorts; Figure 1). In addition, the odds ratios comparing subjects with non-O blood groups to those with blood group O were not modified to a significant extent by age, gender, race/ethnicity, smoking status or BMI (Supplemental Table 3). As tobacco use, obesity and diabetes mellitus are the best-defined modifiable risk factors for pancreatic cancer, we also evaluated these covariates and ABO blood type in joint models (Table 5). In combination with smoking, overweight or diabetes, the non-O blood type was associated with ORs of 2.68, 1.66, and 2.29, respectively, compared to subjects who had O blood type and lacked the exposure. These odds ratios are compatible with a multiplicative odds ratio model for the joint effects of these factors and non-O blood type; there was no evidence for statistically significant interactions.
Figure 1. Risk of pancreatic cancer in non-O blood type versus O blood type by prospective cohort study*.
Abbreviations: ATBC, Alpha-Tocopherol Beta-Carotene, Cancer Prevention Study; CI, confidence interval; CPS II, American Cancer Society Cancer Prevention Study-II; EPIC, European Prospective Investigation into Cancer and Nutrition Study; HPFS, Health Professional's Follow-up Study; NHS, Nurses' Health Study; PHS, Physicians' Health Study I; PLCO, Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial; SMWHS, Shanghai Men's and Women's Health Study; WHI, Women's Health Initiative; CI, confidence interval.
* Two participating cohorts (the New York University Women's Health Study and Women's Health Study) were not included in this figure due to unstable estimates with small numbers of contributed subjects (≤ 25 cases).
Table 5. Odds ratios (95% confidence intervals) and incidence rates for pancreatic cancer by genotype-derived ABO blood type and selected covariates among White participants.
| Predisposing Factor | Genotype-Derived ABO Blood Type | P-interaction | |
|---|---|---|---|
| O | Non-O | ||
| Smoking Status* | 0.33 | ||
| Never / Quit date ≥ 5 years | |||
| No. of cases / controls | 241 / 357 | 433 / 449 | |
| Adjusted OR† (95% CI) | 1.0 | 1.44 (1.17-1.78) |
|
| Incidence rate (cases / 100,000 subjects) | 24.5 | 35.3 | |
| Current / Quit date < 5 years or unknown | |||
| No. of cases / controls | 136 / 150 | 355 / 263 | |
| Adjusted OR† (95% CI) | 1.66 (1.22-2.27) |
2.68 (2.03-3.54) |
|
| Incidence rate (cases / 100,000 subjects) | 40.7 | 65.6 | |
| Body-Mass Index | 0.95 | ||
| < 25 kg/m2 | |||
| No. of cases / controls | 171 / 252 | 347 / 350 | |
| Adjusted OR† (95% CI) | 1.0 | 1.43 (1.11-1.83) |
|
| Incidence rate (cases / 100,000 subjects) | 26.1 | 37.4 | |
| ≥ 25 kg/m2 | |||
| No. of cases / controls | 295 / 353 | 560 / 490 | |
| Adjusted OR† (95% CI) | 1.20 (0.93-1.54) |
1.66 (1.31-2.10) |
|
| Incidence rate (cases / 100,000 subjects) | 31.3 | 43.2 | |
| History of Diabetes Mellitus | 0.44 | ||
| No | |||
| No. of cases / controls | 382 / 550 | 772 / 753 | |
| Adjusted OR† (95% CI) | 1.0 | 1.46 (1.24-1.73) |
|
| Incidence rate (cases / 100,000 subjects) | 27.1 | 39.6 | |
| Yes | |||
| No. of cases / controls | 50 / 35 | 75 / 50 | |
| Adjusted OR† (95% CI) | 2.15 (1.35-3.41) |
2.29 (1.55-3.39) |
|
| Incidence rate (cases / 100,000 subjects) | 58.1 | 62.0 | |
Abbreviations: OR, odds ratio; CI, confidence interval
Participants from the Health Professional's Follow-up Study, Nurses' Health Study, Physicians' Health Study, and Women's Health Study were matched on smoking status, and therefore were excluded from the joint analyses with smoking status.
Multivariable adjustment by age, gender, cohort, smoking status, history of diabetes mellitus, and body-mass index, excluding the covariate included in the joint analysis.
Discussion
Among pancreatic cancer cases and controls from 12 large prospective cohort studies, we observed a significantly elevated risk for incident pancreatic cancer among those with blood group alleles A or B compared to those with blood group O. Importantly, an increased risk was noted with the addition of each non-O allele, with a large increase in risk noted for participants with blood type BB. These data suggest that additional useful risk information may be provided by determining the full genotypic variability at the ABO locus; however, further studies are needed of ABO alleles and pancreatic cancer risk to confirm these findings. Joint models demonstrated further increases in risk, when ABO status was evaluated jointly with known risk factors for pancreatic cancer; statistically significant interactions were not observed between known predisposing factors and ABO blood type, indicating that this risk factor may be relevant across populations with diverse clinical characteristics. We estimate that 19.5% of all cases of pancreatic cancer in European ancestry populations are attributable to inheriting a non-O blood group.
Studies examining serotype-defined blood type or tumoral expression of ABO antigens have suggested a role for ABO blood groups in the development and progression of cancer for several decades (26). However, studies examining serologic blood type and pancreatic cancer risk have been somewhat inconsistent (9-13), likely due to small case numbers, retrospective data collection, heterogeneous pathology review, and the use of poorly matched control populations.
We previously performed a prospective cohort study of participants in the NHS and HPFS to examine the influence of serologic-defined ABO blood type on the subsequent development of pancreatic cancer (14), Compared with participants with blood group O, those with blood groups A, AB, or B were more likely to develop pancreatic cancer, with adjusted hazard ratios for incident pancreatic cancer of 1.32 (95% CI, 1.02-1.72), 1.51 (95% CI, 1.02-2.23), and 1.72 (95% CI = 1.25 to 2.38), respectively. However, this analysis included only 316 pancreatic cancer cases and was unable to examine the full range of blood type allelic variation (i.e. OO, AO, AA, AB, BO, and BB), as it was based on serotype.
Cases and controls in the current analysis were drawn from the Pancreatic Cancer Cohort Consortium, which recently completed a GWAS, involving nearly 550,000 SNPs across the human genome (15). The GWAS identified four SNPs at the ABO gene locus (rs505922, rs495828, rs657152 and rs630014) as among the most statistically significant associations with pancreatic cancer risk (P < 10-5); however, the mechanism by which these SNPs influence risk is unknown. A possible explanation for the association of these SNPs with disease risk is their linkage to ABO glycosyltransferase specificity, i.e. with the polymorphisms that define glycosyltransferase O, A, and B. The current study would support this explanation, given the increase in cancer risk with increasing the number of non-O alleles, and the suggested differences in risk for subjects with blood types AA and BB. Alternatively, SNPs at the ABO locus could modulate gene expression, such that glycosyltransferase specificity is of lesser importance than levels of ABO gene expression. Finally, our study cannot rule out that these SNPs may act as markers of allelic variants in nearby genes, and the ABO antigens may not be directly involved in pancreatic carcinogenesis.
Glycoconjugates are important mediators of intercellular adhesion and membrane signaling, two processes integral to malignant progression and spread (7). In addition, these surface molecules are recognized by the host immune response and may have a role in facilitating immunosurveillance for malignant cells (27). Nevertheless, little data are available to directly link these processes to an association between blood type and pancreatic cancer risk.
Chronic inflammation is a predisposing factor for pancreatic carcinogenesis; pancreatic cancer induces a strong desmoplastic reaction that acts as an abundant source of inflammatory mediators, supporting tumor growth and metastases (28, 29). Interestingly, two recent GWAS suggest that ABO blood group antigens may affect the systemic inflammatory state (16, 30). SNPs at the ABO locus were associated with two serum markers of inflammation, tumor necrosis factor-alpha (TNF-α) (30) and soluble intercellular adhesion molecule 1 (sICAM-1) (16). TNF-α is a pro-inflammatory cytokine known to modulate rates of pancreatic ductal cell apoptosis (28), while plasma levels of sICAM-1 are associated with the risk of incident diabetes (31), a known predisposing factor for pancreatic cancer. These results raise the possibility that blood group antigens may alter the systemic inflammatory state, thereby influencing the risk of developing pancreatic cancer.
Our study has several possible limitations. Blood type in this study was derived from genotype data, not determined serologically, leading to the possibility of measurement error and exposure misclassification. However, the ABO gene was cloned nearly 20 years ago (4, 5), and methods for determining blood type from a subject's DNA are well established (22, 24, 25). In addition, the genotype-derived blood types imputed in the current study were highly concordant with validated blood type in NHS and HPFS participants. Moreover, any resultant misclassification due to measurement error is likely to be non-differential in nature, and therefore attenuate, rather than exaggerate our findings.
Our study population was composed primarily of White participants, which somewhat limits the generalizability of our results. However, other risk factors for pancreatic cancer do not appear to differ substantially by race/ethnicity, and the association between ABO blood type and risk did not differ materially between the White and non-White subjects in this study. Nonetheless, further investigations that include more diverse study populations are warranted. Finally, we cannot definitively rule out the presence of residual confounding or that our risk estimates are higher than those that will be confirmed in other populations due to the winner's curse (32).
Our study has several notable strengths. The Pancreatic Cancer Cohort Consortium provided a large number of pancreatic cancer cases from 12 cohort studies, and the prospective design of these cohorts minimized the potential for survival or selection biases. The risk of detecting a false association due to population stratification was relatively low, given the inclusion of prospective cohorts with homogeneous ethnic compositions, the primarily non-Hispanic European ancestry of the full study population, and the paucity of evidence for variation in pancreatic cancer risk in the ancestral population (33, 34). In addition, after adjusting for potential population stratification bias by including the top 5 principal components of genetic variation as covariates in the logistic regression models, our results were unchanged. Finally, prospectively collected covariate information was not subject to recall bias or the need for proxy interviews, and thus improved our ability to control for confounding and evaluate effect modification.
There remains only a limited understanding of the genetic determinants of pancreatic cancer risk. Our results suggest that ABO blood group alleles represent a common, partially penetrant genetic determinant for pancreatic cancer. Additional investigation is necessary to elaborate mechanisms by which ABO antigens may influence pancreatic cancer risk. In the future, it is possible that ABO blood type could be incorporated into predictive models for this disease, together with other genetic loci and currently identified risk factors, such as tobacco use, obesity and diabetes mellitus.
Supplementary Material
Acknowledgments
Role of Funding Source:
The funding sources for this study did not participate in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Funding/Support:
The NYU Women's Health Study is supported by research grants R01CA034588, R01CA098661, center grant P30CA016087 from the NCI and the center grant ES000260 from the National Institute of Environmental Health Sciences.
The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts N01WH22110, 24152, 32100-2, 32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26, 42129-32, and 44221. The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found at: http://www.whiscience.org/publications/WHI_investigators_shortlist.pdf.”
The PHS, NHS, HPFS and WHS at Harvard were supported by the NCI, NIH (Grants No. P01 CA87969, P01 CA55075, P50 CA127003, R01 CA124908).
The Shanghai Men's Health Study was supported by the National Cancer Institute extramural research grant [R01 CA82729]. The Shanghai Women's Health Study was supported by the National Cancer Institute extramural research grant [R01 CA70867] and, partially for biological sample collection, by the Intramural Research Program of National Cancer Institute (Division of Cancer Epidemiology and Genetics).
We are in debt to the contributions of Drs. Yu-Tang Gao and Yong-Bing Xiang in these two cohort studies. The studies would not be possible without the continuing support and devotion from the study participants and staff of the SMHS and SWHS.
PLCO was supported by individual contracts from the NCI to the University of Colorado Denver NO1-CN-25514, Georgetown University NO1-CN-25522, Pacific Health Research Institute NO1-CN-25515, Henry Ford Health System NO1-CN-25512, University of Minnesota, NO1-CN-25513, Washington University NO1-CN-25516, University of Pittsburgh NO1-CN-25511, University of Utah NO1-CN-25524, Marshfield Clinic Research Foundation NO1-CN-25518, University of Alabama at Birmingham NO1-CN-75022, Westat, Inc. NO1-CN-25476, University of California, Los Angeles NO1-CN-25404
The ATBC Study was supported by funding provided by the Intramural Research Program of the NCI, NIH, and through U.S. Public Health Service contracts (N01-CN-45165, N01-RC-45035, and N01-RC-37004) from the NCI.
For the EPIC cohorts, all coauthors coordinated the initial recruitment and management of the studies. All authors contributed to the final paper. The authors thank all of the participants who took part in this research and the funders and support and technical staff who made this study possible. The work described in this paper was carried out with the support of the European Commission: Public Health and Consumer Protection Directorate 1993-2004; Research Directorate-General 2005-.”; Ligue contre le Cancer, Societé 3M, Mutuelle Générale de l'Education Nationale, Institut National de la Santé et de la Recherche Médicale (INSERM) (France); German Cancer Aid, German Cancer Research Center, Federal Ministry of Education and Research (Germany); Danish Cancer Society (Denmark); Health Research Fund (FIS) of the Spanish Ministry of Health, The participating regional governments and institutions (Spain); Cancer Research UK, Medical Research Council, Stroke Association, British Heart Foundation, Department of Health, Food Standards Agency, the Wellcome Trust (United Kingdom); Greek Ministry of Health and Social Solidarity, Hellenic Health Foundation and Stavros Niarchos Foundation (Greece); Italian Association for Research on Cancer (AIRC) (Italy); Dutch Ministry of Public Health, Welfare and Sports, Dutch Prevention Funds, LK Research Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF) (the Netherlands); Swedish Cancer Society, Swedish Scientific Council, Regional Government of Skane and Västerbotten (Sweden).
CLUE II was supported by National Institute of Aging grant (5U01AG018033) and National Cancer Institute grants (CA105069, CA73790). The authors express their appreciation to the participants of the CLUE II cohort and thank the staff at the George W. Comstock Center for Public Health Research and Prevention for their dedication and contributions to the study: Judy Hoffman-Bolton, Clara Krumpe, Kitty Spoonire and Betty Miner.
The Cancer Prevention Study II Nutrition Cohort is supported by the American Cancer Society. The authors thank all of the men and women in the Cancer Prevention Study II Nutrition Cohort for their many years of dedicated participation in the study.
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Average age-adjusted population rates weighted by gender for U.S. Whites 50 years and older were obtained from Surveillance, Epidemiology and End Results (SEER) data. http://seer.cancer.gov/statistics/. Accessed: January 22, 2009.
Financial Support: Please see Acknowledgments section, as each prospective cohort has individual support mechanisms
Conflicts of Interest: None
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–49. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 3.Owen R. Karl Landsteiner and the first human marker locus. Genetics. 2000;155:995–8. doi: 10.1093/genetics/155.3.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamoto F, Clausen H, White T, Marken J, Hakomori S. Molecular genetic basis of the histo-blood group ABO system. Nature. 1990;345:229–33. doi: 10.1038/345229a0. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto F, Marken J, Tsuji T, White T, Clausen H, Hakomori S. Cloning and characterization of DNA complementary to human UDP-GalNAc: Fuc alpha 1---2Gal alpha 1----3GalNAc transferase (histo-blood group A transferase) mRNA. J Biol Chem. 1990;265:1146–51. [PubMed] [Google Scholar]
- 6.Reid ME, Mohandas N. Red blood cell blood group antigens: structure and function. Semin Hematol. 2004;41:93–117. doi: 10.1053/j.seminhematol.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Hakomori S. Antigen structure and genetic basis of histo-blood groups A, B and O: their changes associated with human cancer. Biochim Biophys Acta. 1999;1473:247–66. doi: 10.1016/s0304-4165(99)00183-x. [DOI] [PubMed] [Google Scholar]
- 8.Hoskins LC, Loux HA, Britten A, Zamcheck N. Distribution of ABO blood groups in patients with pernicious anemia, gastric carcinoma and gastric carcinoma associated with pernicious anemia. N Engl J Med. 1965;273:633–7. doi: 10.1056/NEJM196509162731204. [DOI] [PubMed] [Google Scholar]
- 9.Vogel F. Controversy in human genetics. ABO blood groups and disease. Am J Hum Genet. 1970;22:464–75. [PMC free article] [PubMed] [Google Scholar]
- 10.Aird I, Lee DR, Roberts JA. ABO blood groups and cancer of oesophagus, cancer of pancreas, and pituitary adenoma. Br Med J. 1960;1:1163–6. doi: 10.1136/bmj.1.5180.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newell GR, Gordon JE, Monlezun AP, Horwitz JS. ABO blood groups and cancer. J Natl Cancer Inst. 1974;52:1425–30. doi: 10.1093/jnci/52.5.1425. [DOI] [PubMed] [Google Scholar]
- 12.Annese V, Minervini M, Gabbrielli A, Gambassi G, Manna R. ABO blood groups and cancer of the pancreas. Int J Pancreatol. 1990;6:81–8. doi: 10.1007/BF02933042. [DOI] [PubMed] [Google Scholar]
- 13.Vioque J, Walker AM. Pancreatic cancer and ABO blood types: a study of cases and controls. Med Clin (Barc) 1991;96:761–4. [PubMed] [Google Scholar]
- 14.Wolpin BM, Chan AT, Hartge P, et al. ABO blood group and the risk of pancreatic cancer. J Natl Cancer Inst. 2009;101:424–31. doi: 10.1093/jnci/djp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41:986–90. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pare G, Chasman DI, Kellogg M, et al. Novel association of ABO histo-blood group antigen with soluble ICAM-1: results of a genome-wide association study of 6,578 women. PLoS Genet. 2008;4:e1000118. doi: 10.1371/journal.pgen.1000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kraft P, Cox DG, Paynter RA, Hunter D, De Vivo I. Accounting for haplotype uncertainty in matched association studies: a comparison of simple and flexible techniques. Genet Epidemiol. 2005;28:261–72. doi: 10.1002/gepi.20061. [DOI] [PubMed] [Google Scholar]
- 18.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 19.Rothman KJ, Greenland S, Lash TL. Modern Epidemiology. Philadelphia: Lippincott, Williams & Wilkins; 2008. [Google Scholar]
- 20.Gail MH. Models of absolute risk: interpretation, estimation, validation and application. In: Rebbeck TR, Ambrosone CB, Shields PG, editors. Molecular epidemiology: applications in cancer and other human diseases. New York: Informa Healthcare; 2008. [Google Scholar]
- 21.Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–29. [Google Scholar]
- 22.Blumenfeld OO, Patnaik SK. Allelic genes of blood group antigens: a source of human mutations and cSNPs documented in the Blood Group Antigen Gene Mutation Database. Hum Mutat. 2004;23:8–16. doi: 10.1002/humu.10296. [DOI] [PubMed] [Google Scholar]
- 23.Garratty G, Glynn SA, McEntire R. ABO and Rh(D) phenotype frequencies of different racial/ethnic groups in the United States. Transfusion. 2004;44:703–6. doi: 10.1111/j.1537-2995.2004.03338.x. [DOI] [PubMed] [Google Scholar]
- 24.Yip SP. Single-tube multiplex PCR-SSCP analysis distinguishes 7 common ABO alleles and readily identifies new alleles. Blood. 2000;95:1487–92. [PubMed] [Google Scholar]
- 25.Gassner C, Schmarda A, Nussbaumer W, Schonitzer D. ABO glycosyltransferase genotyping by polymerase chain reaction using sequence-specific primers. Blood. 1996;88:1852–6. [PubMed] [Google Scholar]
- 26.Marcus DM. The ABO and Lewis blood-group system. Immunochemistry, genetics and relation to human disease. N Engl J Med. 1969;280:994–1006. doi: 10.1056/NEJM196905012801806. [DOI] [PubMed] [Google Scholar]
- 27.Hakomori S. Tumor-associated carbohydrate antigens defining tumor malignancy: basis for development of anti-cancer vaccines. Adv Exp Med Biol. 2001;491:369–402. doi: 10.1007/978-1-4615-1267-7_24. [DOI] [PubMed] [Google Scholar]
- 28.Garcea G, Dennison AR, Steward WP, Berry DP. Role of inflammation in pancreatic carcinogenesis and the implications for future therapy. Pancreatology. 2005;5:514–29. doi: 10.1159/000087493. [DOI] [PubMed] [Google Scholar]
- 29.Guerra C, Schuhmacher AJ, Canamero M, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 30.Melzer D, Perry JR, Hernandez D, et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4:e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meigs JB, Hu FB, Rifai N, Manson JE. Biomarkers of endothelial dysfunction and risk of type 2 diabetes mellitus. JAMA. 2004;291:1978–86. doi: 10.1001/jama.291.16.1978. [DOI] [PubMed] [Google Scholar]
- 32.Zollner S, Pritchard JK. Overcoming the winner's curse: estimating penetrance parameters from case-control data. Am J Hum Genet. 2007;80:605–15. doi: 10.1086/512821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wacholder S, Rothman N, Caporaso N. Population stratification in epidemiologic studies of common genetic variants and cancer: quantification of bias. J Natl Cancer Inst. 2000;92:1151–8. doi: 10.1093/jnci/92.14.1151. [DOI] [PubMed] [Google Scholar]
- 34.Shibuya K, Mathers CD, Boschi-Pinto C, Lopez AD, Murray CJ. Global and regional estimates of cancer mortality and incidence by site: II. Results for the global burden of disease 2000. BMC Cancer. 2002;2:37. doi: 10.1186/1471-2407-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.