Abstract
OTX2 is a developmentally regulated transcription factor that plays important roles in early morphogenesis of the central nervous system. We previously identified OTX2 amplification and overexpression in medulloblastoma cell lines. However, the nature and frequency of OTX2 genetic alterations have not been determined in large sample sizes of primary tumors. To refine our understanding of genomic events affecting OTX2, we analyzed genomic copy number and mRNA expression of OTX2 in a large cohort of primary medulloblastomas. We found that the most frequent focal copy number gain in these primary medulloblastomas (gained in 21% of the tumors, n=201) contained the single gene OTX2, indicating that copy number gain of OTX2 is a driver event in this tumor. In this cohort of tumors, we found that the event was restricted to tumor subtypes that do not express a molecular signature of either Wnt or Shh pathway activation. FISH analysis revealed that OTX2 copy number gain is present in a subset of cells within medulloblastoma samples, suggesting that it is a late event contributing to tumor progression. Gain of OTX2 copy number is associated with the presence of anaplastic histological features and shorter survival in medulloblastoma patients. To verify a functional role of the observed genomic events, we demonstrate that ectopic OTX2 expression enhances proliferation and tumorigenicity of immortalized primary cells and that OTX2 knockdown in medulloblastoma xenografts prolongs survival of recipient animals. Finally, we demonstrate that OTX2 transcriptionally upregulates the medulloblastoma oncogene MYC as a potential mechanism whereby OTX2 promotes tumor progression.
Keywords: OTX2, medulloblastoma, MYC
Background
Medulloblastoma is the most common malignant brain tumor in children, with a peak incidence at seven years of age (1–3). In recent years, medulloblastoma has been redefined by the development of treatment strategies, including surgical resection, radiation, and chemotherapy. Still, an unacceptably large percentage of children die of this disease, with five-year survival rates reaching only 50–60% (4–6). Moreover, long-term survivors frequently suffer life-long sequelae from aggressive treatment regimens (7–9). More refined tumor grading systems are therefore urgently needed to optimize efficacy and minimize treatment-related toxicity for each patient.
Histologically, medulloblastomas exhibit substantial heterogeneity both among and within tumors, and histological characteristics do not uniformly correlate with clinical prognosis or treatment response. However, molecular analysis of medulloblastomas has shown that incorporating gene expression and genetic alteration data along with clinical and histopathologic data into a patient’s profile may better stratify patients into particular treatment and prognostic groups (5, 10–13).
Recently, we and others identified OTX2 amplification and overexpression in medulloblastoma cell lines and primary tumors (14–17). Gene expression analyses revealed that OTX2 transcripts were present at high levels in 14 of 15 (93%) medulloblastomas with anaplastic histopathologic features. Additionally, we demonstrated that knockdown of OTX2 expression by siRNAs inhibited medulloblastoma cell growth in vitro (15).
OTX2 is a member of a well-conserved family of bicoid-like homeodomain (HD)-containing transcription factors that play important roles in embryo patterning, brain regionalization, and lineage specification (18). OTX2 is expressed in the prospective cerebellum (the midbrain/hindbrain boundary) of the early embryo as well as in the rapidly expanding population of cerebellar granule cell precursors (19, 20). OTX2-deficient mice fail to develop the mesencephalon and prosencephalon, and heterozygotes exhibit a lethal “headless” phenotype of variable penetrance (21, 22). Although the expression pattern and function of OTX2 in brain morphogenesis have been well-studied in progressive developmental stages of rodent models (20, 23), there is little understanding of how OTX2 relates to the clinical and histopathologic features of medulloblastoma or to the molecular pathways of medulloblastoma pathogenesis.
To further our understanding of the genetic mechanism of OTX2 activation and its clinical implications in medulloblastomas, we examined its copy number and expression status in a large cohort of tumors (201 and 103 primary medulloblastomas, respectively). Here we identify recurrent OTX2 gain (~21%) and, even more commonly, OTX2 overexpression (~74%) in primary medulloblastomas. OTX2 focal gains were limited to tumors not expressing signatures of Wnt or Shh pathway activation. FISH analysis demonstrated that OTX2 copy number gain was present in a subset of medulloblastoma cells and was associated with a more aggressive tumor phenotype and adverse clinical outcome. In order to understand its oncogenic functions, we overexpressed OTX2 in immortalized primary cells and demonstrated that OTX2 could promote cell proliferation and tumorigenicity. Furthermore, we determined that stable OTX2 knockdown prolonged survival in a medulloblastoma xenograft model. Finally, we demonstrated that OTX2 transcriptionally regulates the medulloblastoma oncogene MYC. These results suggest that OTX2 plays a role in progression and maintenance of a large subset of medulloblastoma tumors and thus represents a promising therapeutic target.
Material and methods
Patients
Medulloblastomas were obtained from the Duke University Medical Center Brain Tumor Biorepository and the Hospital for Sick Children, University of Toronto according to Internal Review Board (IRB) protocol. This biorepository contained specimens obtained from Duke and the (now defunct) Pediatric Oncology Group that had been originally studied for MYC amplification (24).
Pathology
Classic, desmoplastic/nodular, and anaplastic/large cell medulloblastomas used in FISH analysis were examined by a neuropathologist (RE McLendon) and classified using the World Health Organization guidelines (1). The classic, or diffuse, pattern of medulloblastoma presents a histologic appearance of densely packed cells with round-to-oval or carrot-shaped hyperchromatic nuclei surrounded by scanty cytoplasm growing in sheets. Neuroblastic, or Homer-Wright, rosettes are most commonly encountered in this group (25).
The anaplastic variant is composed of cells with elongated hyperchromatic nuclei that are densely crowded and exhibit characteristic nuclear wrapping against adjacent tumor cells, abundant individual cell necrosis frequently associated with geographic regions of necrosis, and numerous mitotic figures (26). Moderate anaplasia lacks the degree and extent of necrosis seen in severely anaplastic tumors and is more commonly recognized by a “starry sky” pattern of individual cell necrosis interrupting the sheets of tumor cells.
Single nucleotide polymorphism array and microarray analysis
Hybridization, detection, and analysis of Affymetrix single nucleotide polymorphism (SNP) and exon arrays were carried out as previously described (27, 28).
FISH
FISH was performed as described previously (29). BAC clones RP11-1085N6 (56.159277-56.402022 Mb, spanning the OTX2 gene) and CTD-2505P22 (103.186989-103.414534 Mb, located telomeric to OTX2 on chromosome 14q) were obtained (Invitrogen, Carlsbad, CA) and used to generate probes for the OTX2 gene and an internal control, respectively. One hundred cells were counted in each of 10 classic medulloblastomas, 11 desmoplastic/nodular medulloblastomas, 10 medulloblastomas with moderate anaplastic features, and 11 medulloblastomas with severe anaplastic features. OTX2 copy number was determined by the average ratio of green to red probes per cell, and copy number gain was designated as tumors with a ratio greater than 1.2.
Statistical analysis
The significance level for all tests was set a priori at 0.05 by using a 95% confidence interval. All statistical and survival analyses were performed with SAS E-guide software (SAS Institute, Cary, NC).
Plasmid constructs
A full-length cDNA of human OTX2 was derived from pCMV6-XL5-OTX2 (OriGene, Rockville, MD) and subcloned into the pEGFP-N1 vector.
Cell culture, proliferation, and apoptosis assays
Medulloblastoma and rat kidney epithelium (RK3E) cell lines were maintained as described previously (15, 30). Cell line D425MED was derived from a patient with a primary medulloblastoma, whereas D458MED was established independently from cerebrospinal fluid samples of the same patient at a later date. MTT and colony proliferation assays were performed as described (15, 30).
Establishing an RK3E line stably expressing OTX2
RK3E cells were transfected with pEGFP-N1 or pEGFP-OTX2. The transfected cells were cultured in DMEM media containing 500 µg/ml G418 (Invitrogen) for 3–4 weeks. Single cell dilution was used to select monoclonal cell lines that were resistant to G418 and expressing OTX2.
Tumorigenicity in athymic nude mice
Groups of seven or eight mice per cell line were used to investigate the tumorigenicity of RK3E cells stably transfected with pEGFP-N1 or pEGFP-OTX2, respectively. We implanted 106 cells stereotactically into the cerebral hemisphere of BALB/c athymic nu/nu mice. Brains of euthanized mice were collected, fixed in formalin, paraffin embedded, and sectioned. The presence of intracranial tumors, indicated by parenchymal and ventricular invasion, was assessed microscopically in multiple brain sections. The tumor proliferation activity was examined by Ki67 immunoreactivity using a Ki-67 monoclonal antibody (Lab Vision, Fremont, CA). OTX2 immunoreactivity was detected with biotinylated anti-human OTX2 antibody (BAF1979; R&D, Minneapolis, MN), followed by HRP-conjugated secondary antibody.
Immunoblotting
OTX2 was detected with biotinylated anti-human OTX2 antibody (BAF1979, R&D, Minneapolis, MN) and HRP-rabbit anti-goat IgG secondary antibody (Zymed, San Francisco, CA). MYC and GAPDH were detected with 9E10 (sc-40; Santa Cruz, CA) and FL-335 (sc-25778; Santa Cruz, CA), respectively, and enhanced chemiluminescence anti-rabbit IgG secondary antibody (Amersham Biosciences, Buckinghamshire, England).
siRNA and shRNA of OTX2 and MYC
Medulloblastoma cells were plated at 2 × 105 cells per well into a 6-well plate and transfected with siRNA or shRNA using Lipofectamine 2000 (Invitrogen). siRNA and shRNA sequences used are described in the Supplemental Methods section.
Animal survival studies
Approximately 107 D425MED cells were transfected with scrambled-shRNA or OTX2-shRNA, suspended in 10 µl of PBS, and stereotactically introduced into the right cerebral hemisphere of a BALB/c athymic nu/nu mouse. Mice were euthanized at the first sign of neurologic dysfunction. The date when each mouse became symptomatic was recorded to calculate the time-to-event. Standard Kaplan-Meier estimates were generated for each time-to-event, and groups were compared with log-rank tests.
Luciferase assays
For luciferase assays, we obtained the pBV-del-3-Luc plasmid, comprised of a 1.1-kb genomic fragment encompassing the MYC promoter inserted upstream of a minimal promoter and the firefly luciferase reporter gene in the pBV-Luc plasmid (Supplementary Fig 3B) (31). Medulloblastoma cells were seeded at 1 × 105 cells per well in 48-well plates for 24 h, and then each well was cotransfected with 0.8 µg of firefly luciferase reporter plasmid pBV-del-3-Luc, the internal control plasmid pRL-CMV, and 0.8 µg of pEGFP-OTX2 or the control vector pEGFP-N1. Dual-Luciferase Reporter assays were performed according to the manufacturer’s protocol (Promega, Madison, WI).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using a ChIP Assay Kit (Upstate Cell Signaling Solutions, Lake Placid, NY) according to the manufacturer’s protocol. Immunoprecipitation was performed overnight at 4°C with biotinylated anti-human OTX2 antibody (R&D, Minneapolis, MN). Immunoaffinity-enriched and control DNA fragments were then purified for DNA PCR amplification.
Results
Recurrent OTX2 focal gains and overexpression in medulloblastomas
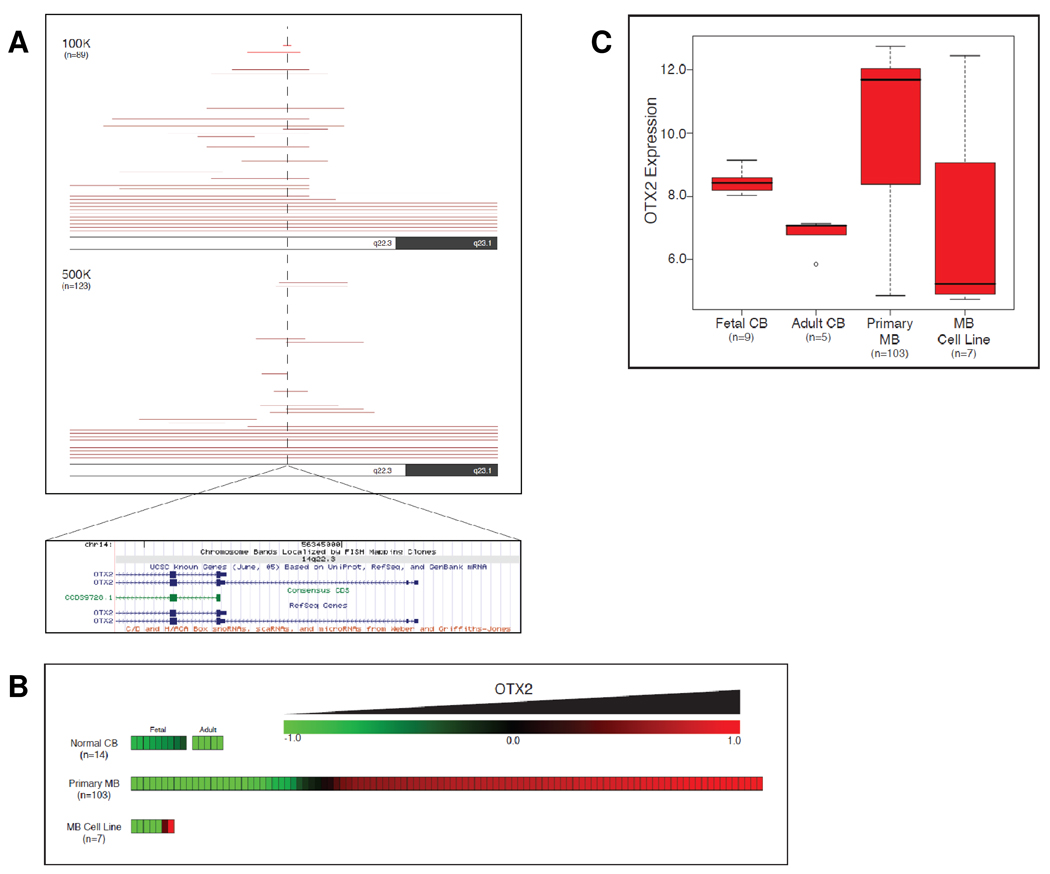
To determine the frequency of OTX2 genetic alterations in a large cohort of primary tumors, we utilized high-density SNP arrays to perform a genome-wide search for genetic changes in 11 medulloblastoma cell lines and 201 primary medulloblastomas (27). The most frequent target of focal gain among the 212 samples was a region mapping to chromosome 14q23, detected in ~10% (21/212) of the medulloblastomas (Fig 1A). This locus exhibited either focal (ie <1 Mb) or large (ie 14q gain) genomic gain in a total of ~21% (9.9% and 11.3%, respectively) of cases profiled. Examination of the minimal common region gained in these samples using the UCSC Genome Browser (NCBI Build 35, hg17; http://genome.ucsc.edu/) revealed that the single gene OTX2 was completely located within this minimal region of chromosomal gain (Fig 1A). OTX2 gain of copy number indicated by SNP arrays was verified by Real-Time PCR on genomic DNA of selected medulloblastoma samples (Supplementary Fig 1). The highly specific gain of OTX2 copy number in a large proportion of medulloblastomas implies that OTX2 gain is a driver event that confers a selective advantage during clonal expansion of this neoplasm.
Figure 1. Frequent gain and overexpression of OTX2 in medulloblastomas.
(A) OTX2 copy number status in 212 medulloblastomas (201 primary MBs and 11 MB cell lines) was inferred using Affymetrix SNP arrays (100K and 500K). Each red line shown in the ideogram represents copy number gain for a single sample in the region shown. (B) Affymetrix exon arrays were used for gene expression profiling of 9 fetal cerebella, 5 adult cerebella, 103 primary medulloblastomas, and 7 medulloblastoma cell lines. OTX2 expression levels are shown as a heatmap. (C) Box and whisker plot representing distribution of OTX2 expression level within normal and MB sample groups.
In addition to harboring focal gains and activating mutations, oncogenes are commonly overexpressed to activate tumorigenic pathways. To determine the prevalence of OTX2 mRNA overexpression, we utilized Affymetrix exon arrays to measure OTX2 mRNA level in 9 normal fetal cerebella, 5 normal adult cerebella, and 103 primary medulloblastomas. We found that fetal cerebellum expressed OTX2 at a modestly higher level than adult cerebellum; however, among the 103 medulloblastomas, 76 tumors (74%) expressed OTX2 at significantly (ie > two-fold) higher levels than fetal or adult cerebellum (P < 0.001, Student’s T-Test) (Fig 1B and C).
To understand OTX2 amplification at the cellular level, we performed dual-color FISH to validate OTX2 amplification in three medulloblastoma cell lines previously identified to bear OTX2 amplifications. We noted OTX2 genomic amplification as double-minute chromosomes in D425MED, D458MED, and D487MED cell lines (Supplementary Fig 2, panels a–c). Moreover, increased OTX2 copy number was also observed in the primary tumors from which the medulloblastoma cell lines D425MED and D487MED were derived (Supplementary Fig 2, panels d–e).
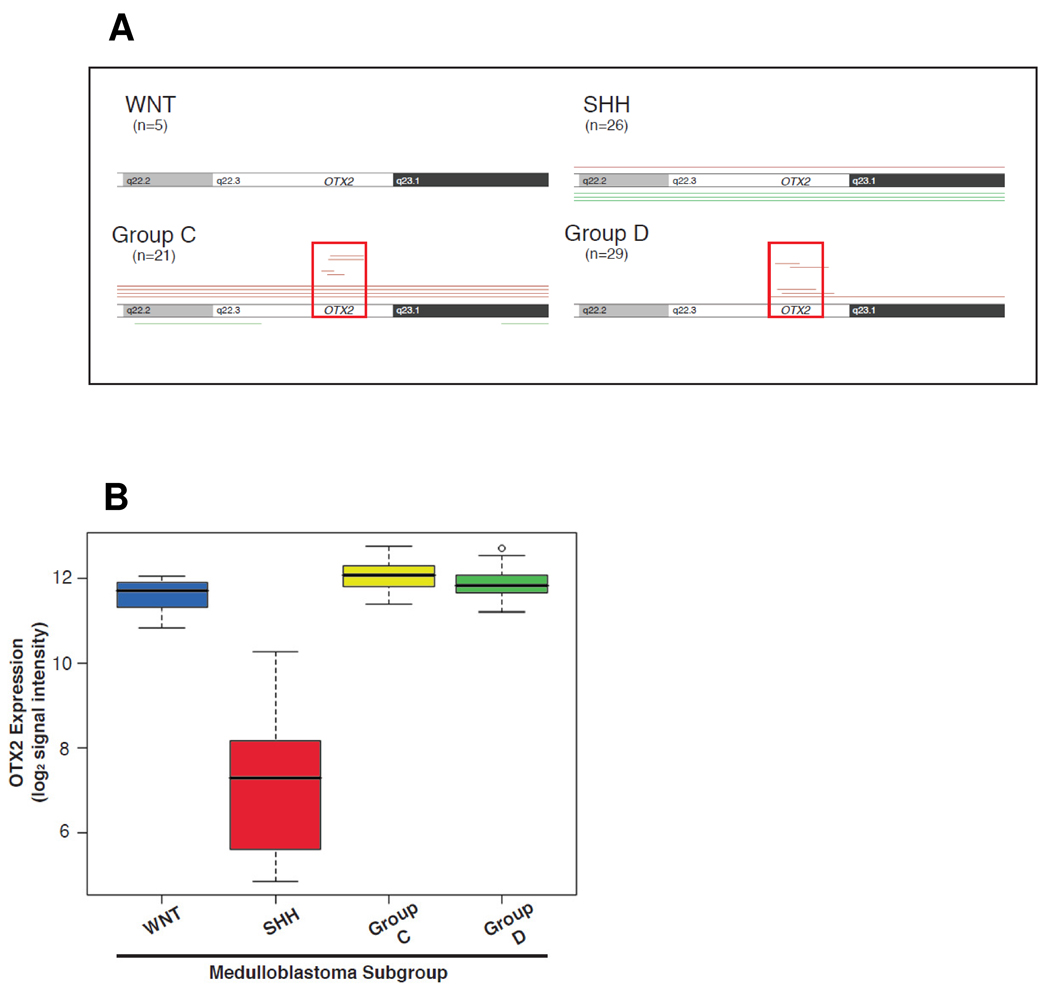
Medulloblastomas can be classified into distinct molecular subgroups based on gene expression signatures (28, 32, 33). Our clustering analysis of a large cohort (n=103) of tumors based upon Affymetrix exon arrays previously revealed four distinct medulloblastoma subtypes: tumors expressing a molecular signature of Wnt pathway activation, those exhibiting a signature of Shh pathway activation, and those bearing two additional, distinct molecular signatures designated Group C and D tumors (28). Group C and D tumors as well as those with Wnt pathway activation are typically of the classic histopathological subtype, whereas tumors with a Shh molecular signature are often of the nodular/desmoplastic histopathological subtype (32, 33). Examination of OTX2 focal gains among medulloblastoma subtypes reveals that this event is limited to Group C and D tumors (Figure 2A). Additionally, OTX2 is differentially expressed in the molecular subgroups of medulloblastoma (P < 0.001, One-way ANOVA), with the most aberrant expression observed in Group C and D tumors as well as tumors with a Wnt pathway signature (Figure 2B).
Figure 2. OTX2 gain and overexpression are specific to medulloblastoma subgroups.
A subset of the primary medulloblastomas (n=81) analyzed in Figure 1 were classified into four molecular subgroups based upon gene expression signatures obtained from exon arrays. (A) Focal gain of OTX2 (designated in red) is limited to Group C and D medulloblastomas. The minimal region of gain (~10.5 kb) is boxed. (B) OTX2 mRNA overexpression is highest in Group C and D tumors, as well as those expressing a signature of Wnt pathway activation.
OTX2 gain of copy number associates with anaplastic features and a worse patient outcome
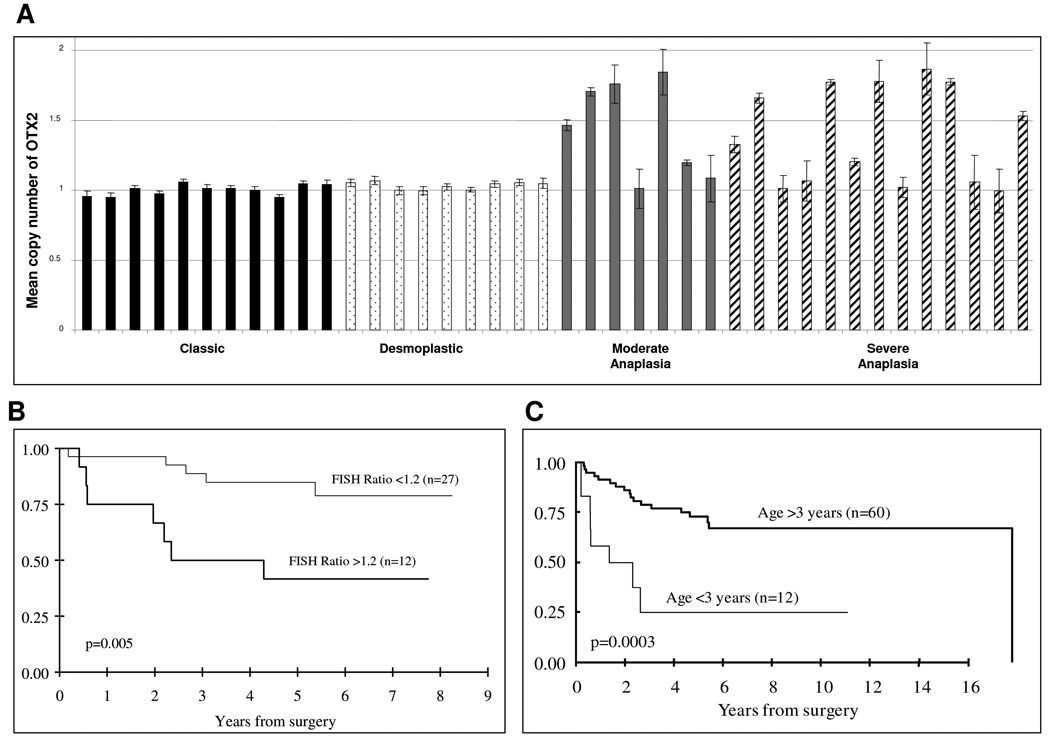
To understand the clinical implication of OTX2 activation in medulloblastomas, we collected medulloblastoma samples with linked clinical data and studied OTX2 genetic status in these cases using FISH. A total of 39 patients with complete clinical data were available for survival analysis on the basis of the FISH data, including 20 with tumors of classic or nodular histology and 19 with tumors of moderate to severe anaplasia. Classic and desmoplastic/nodular sections consistently demonstrated no evidence of increased OTX2 copy number (0/20). However, a significant number of tumors with moderate and severe anaplastic characteristics displayed increased OTX2 copy number (12/19, P < 0.05) (Fig 3A). Only a subpopulation of cells in those samples revealed increased copy number, with OTX2 copy number ranging from 3 to 24 per individual cell (mean = 2.3 ± 1.7). Among the 39 patients, 12 patients who had gain of OTX2 copy number in their tumors had a shorter overall survival compared to those with diploid OTX2 copy number (P = 0.005; Fig 3B). The 5-year survival estimate is 46% (95% CI: 24–87%) for the group with increased OTX2 copy numbers and was nearly doubled (86%; 95% CI: 74–99%) for the group with normal OTX2 copy numbers. Age stratification of this sample group revealed that survival of these patients mimics that of medulloblastoma patients in general, with age < 3 years indicating poor prognosis (P = 0.001) (Figure 3C).
Figure 3. Interphase FISH of OTX2 in primary medulloblastoma showing that OTX2 copy number gain is associated with tumor anaplasia and poor patient survival.
(A) Increased OTX2 copy number is seen in primary medulloblastoma tumors with anaplastic features. The Y axis is the ratio of the mean copy number of OTX2 to the control chromosome 14 probe in each cell of each sample. (B) Kaplan-Meier survival analysis of patients stratified by mean OTX2 copy number. (C) Kaplan-Meier survival analysis of patients stratified by age.
OTX2 promotes cell proliferation and tumorigenicity and is essential for medulloblastoma cell survival
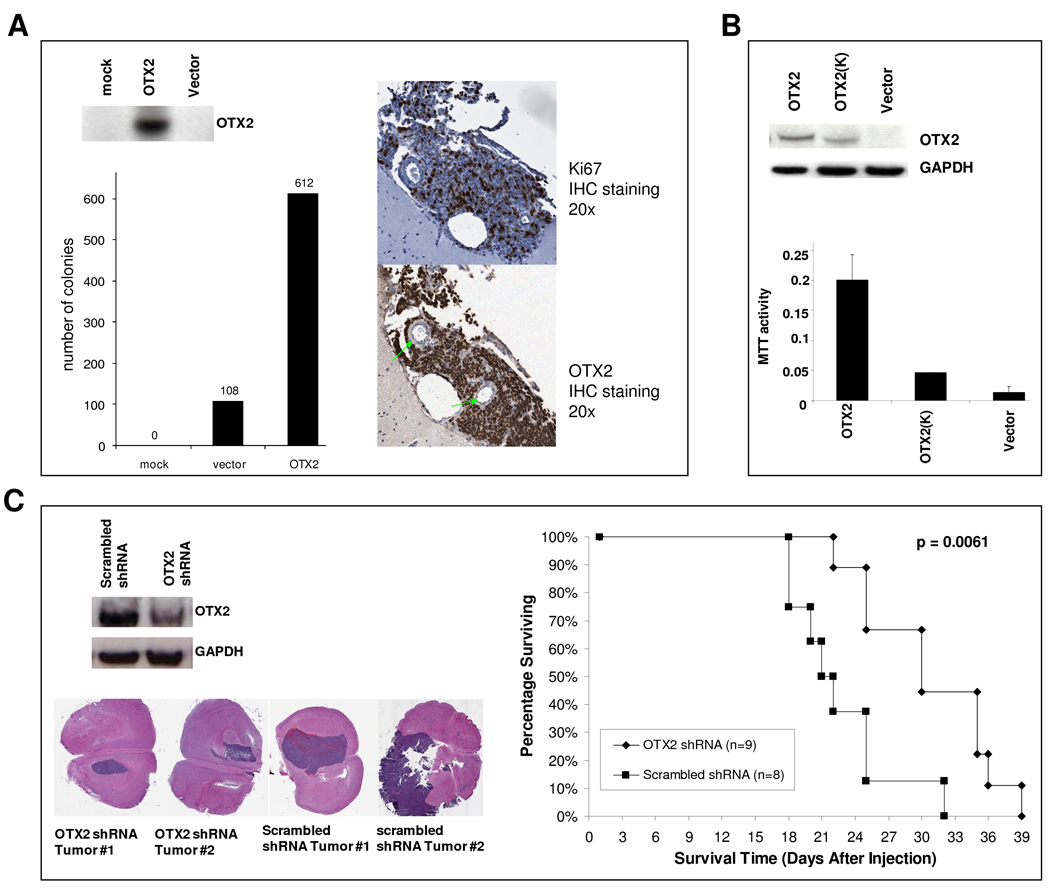
In order to examine the transforming potential of OTX2, we transfected the pEGFP-OTX2 plasmid into non-neoplastic RK3E cells, an adenovirus E1A-immortalized epithelial line that has been widely used for neoplastic transformation studies (30). This assay reveals oncogenic activity independent of cell origin effects, which is suitable considering the cell origin of medulloblastomas harboring OTX2 focal gains (Group C and D tumors) is unknown. OTX2 protein expression was detected by Western blotting (Fig 4A). Overexpression of OTX2 promoted a significant increase in colony formation (Fig 4A, P < 0.05). To assess the ability of OTX2 to promote in vivo tumor growth, we injected RK3E cells expressing either pEGFP-OTX2 or pEGFP-N1 into the cerebral hemispheres of nu/nu mice. We found that while all OTX2-expressing clones formed tumors and displayed high mitotic indices scored by Ki67 staining (Fig 4A), GFP-expressing clones formed tumors much less frequently and scored relatively lower for Ki67 staining (P < 0.001, Supplementary Table 1). To verify that the transformative ability of OTX2 relies upon its transcriptional activity rather than alternative functions, we introduced a loss of function mutation into the OTX2 homeodomain (HD) to abolish DNA binding (34) (Supplementary Fig 3A). We then transfected MHH-1 medulloblastoma cells with either OTX2 or its HD NAAIRS mutant [pEGFP-OTX2 (K)] and measured MTT activity. As expected, wild type OTX2 increased proliferation of MHH-1 cells, and the effect was greatly reduced in the NAAIRS mutant (Fig 4B).
Figure 4. OTX2 promotes cell proliferation in vitro and is required for tumor maintenance in vivo.
(A) RK3E cells were transfected with pEGFP-N1 or pEGFP-OTX2 and colony formation potential was determined. Tumors derived from intracranial xenografts of stably transfected RK3E cells were stained for Ki67 and OTX2, and representative samples are pictured. Green arrows identify intra-tumor blood vessels. (B) MHH-1 cells were transfected with pEGFP-N1, pEGFP-OTX2, or pEGFP-OTX2 (K). After three doubling times, proliferation was determined with an MTT assay. (C) D425MED cells derived from OTX2 shRNA knockdown or scrambled shRNA were injected into the right cerebral hemisphere of mice. Representative H & E-stained tumor sections from each group are pictured.
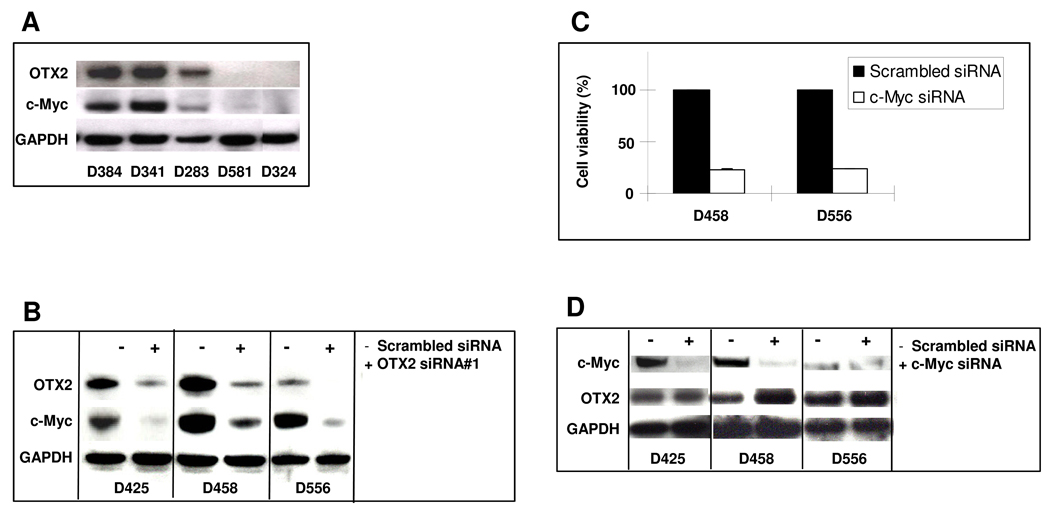
We have previously shown that OTX2 knockdown reduces viability of medulloblastoma cell lines (15). To determine if OTX2 expression is required for tumor maintenance in vivo as well as in vitro, we carried out RNA interference experiments in medulloblastoma xenografts. First, we utilized OTX2 siRNA or shRNA to confirm that OTX2 knockdown inhibited cell growth or survival in an expanded cohort of OTX2-expressing medulloblastoma cell lines (Supplementary Fig 4). We then applied the pSUPER vector system to stably suppress endogenous OTX2 expression in D425MED cells (Fig 4C) and examined whether downregulation of OTX2 expression affected survival in nude mice bearing intracranial xenografts. Although tumors were detected from both the OTX2-knockdown group and the control group, reducing OTX2 expression significantly increased the survival time of the animals from 22.6 ± 4.7 days to 30.8 ± 5.9 days (P = 0.0061, Fig 4C).
MYC expression is regulated by OTX2
Amplification and overexpression of the oncogene MYC are also common in medulloblastoma, particularly in those exhibiting anaplastic histopathological features (35). Therefore we performed biochemical studies to investigate the relationship between OTX2 and MYC.
We first examined the correlation between OTX2 and MYC expression by Western blot in medulloblastoma cell lines that do not have OTX2 or MYC amplification. We observed that detectable MYC expression level was limited to cell lines expressing OTX2 (Fig 5A). Using serial analysis of gene expression (SAGE) data from the Cancer Genome Anatomy Project, we found that presence of OTX2 mRNA correlated with the presence of MYC mRNA in medulloblastomas (P = 0.0140; Supplementary Fig 5).
Figure 5. MYC is a target gene of OTX2 in medulloblastoma cell lines.
(A) Expression of endogenous OTX2 and MYC in medulloblastoma cell lines demonstrated by Western blotting. (B) Medulloblastoma cell lines were treated with OTX2 siRNA (+) or scrambled siRNA (−), and protein expression of OTX2, MYC, and GAPDH was detected by Western blotting. (C) Cells were treated with MYC siRNA or scrambled siRNA and after three doubling times assessed for viability with the MTT assay. (D) Medulloblastoma cell lines were treated with MYC siRNA (+) or scrambled siRNA (−), and protein expression of OTX2, MYC, and GAPDH was detected by Western blotting.
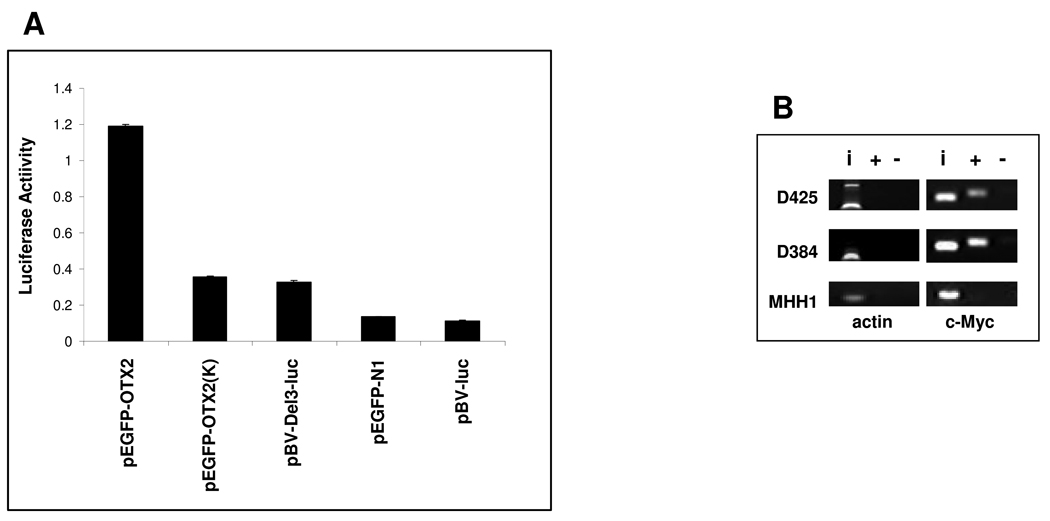
To determine whether OTX2 is required for MYC expression in medulloblastoma, we applied OTX2-specific siRNA to knock down OTX2 expression in medulloblastoma cell lines and found that MYC expression was also downregulated by the OTX2 siRNA (Fig 5B). The complementary experiment in which MYC is knocked down by siRNA resulted in cell growth inhibition (Fig 5C) but not downregulation of OTX2 (Fig 5D). We then utilized a luciferase assay to demonstrate that OTX2 upregulates MYC via cis-acting elements in its promoter (Fig 6A), which contains multiple putative OTX2 binding sites (Supplementary Fig 3B). Finally, chromatin immunoprecipitation of fragments enriched with an anti-OTX2 antibody in cell lines expressing both MYC and OTX2 revealed direct binding of endogenous OTX2 to the MYC promoter in the context of native chromatin (Fig 6B).
Figure 6. The OTX2 homeodomain is required for OTX2-mediated MYC transcription.
(A) MHH-1 cells were cotransfected with the MYC promoter reporter construct (pBV-Del-3-Luc) and either pEGFP-N1, pEGFP-OTX2 or pEGFP-OTX2 (K), and then the relative luciferase reporter activity was measured. (B) OTX2 ChIP and PCR analyses were used to validate binding of endogenous OTX2 to the promoter region of MYC (primers were designed to span the region 375 bp directly upstream of the MYC TSS) in OTX2-expressing cell lines. D425MED and D384MED are OTX2-expressing medulloblastoma cell lines, whereas MHH1 does not express OTX2. i, input of total DNA from cell lysates; +, OTX2 antibody immunoaffinity-enriched DNA; −, IgG control antibody immunoaffinity-enriched DNA.
Discussion
Medulloblastoma patients have widely disparate and often unpredictable clinical outcomes. It is therefore important to develop better predictive molecular diagnostic tools that could be used in clinical practice to better define prognosis and guide therapy. In the current report, the investigation of OTX2 genetic status and its pathogenic roles in medulloblastoma oncogenesis have revealed that OTX2 genetic alteration is associated with tumor progression and poor survival and thus may have diagnostic value. Furthermore, we have demonstrated a requirement for OTX2 in medulloblastoma viability, potentially revealing a novel therapeutic target in this tumor. We have also identified MYC as a downstream target gene of OTX2. This study advances our knowledge of the pathogenesis of medulloblastoma and has great potential to lead to significant contributions to the therapeutic intervention of this frequently lethal cancer.
The specific and recurrent focal gain of OTX2 represents a driver event in medulloblastoma tumorigenesis. OTX2 copy number gain is limited to tumors not bearing a molecular signature of Shh or Wnt pathway activation and thus represents an oncogenic event in tumor subtypes whose molecular progressions are currently unclear. The Group C and D tumors that distinctly harbor OTX2 copy number gains are typically of the classic histopathological subtype (22, 23). Considering that OTX2 gain is also associated with the presence of anaplastic features, it would be expected that there would be substantial overlap between Group C or D molecular signatures and the presence of anaplasia. It has been described previously that anaplastic features can be present in variable degrees of severity and extent among the medulloblastoma histopathological subtypes (36).
Using FISH, we found that OTX2 copy number varies widely among tumor cells, suggesting that OTX2 gain of copy number is a late event that is selected for within the tumor as a step of tumor progression. This model is consistent with the association of OTX2 copy number gain with worse patient survival as well as the presence of anaplastic features, which has been proposed to result from the progression of less aggressive tumors (37).
It remains a challenge to identify the patients with unfavorable prognoses who may benefit from the most aggressive treatment. Patient stratification is particularly important to medulloblastoma due to the deleterious effects of aggressive treatment in young patients. The Kaplan-Meier analysis revealed a significant association between increased OTX2 copy number and worse survival. Similar to other study populations, this group of patients also demonstrated a clear association between poor survival and age <3 years, a well-known negative predictive factor for medulloblastoma. These findings support the possible use of OTX2 genetic status as an important marker for prognosis. Also, OTX2 status may improve stratification schemes for current treatment protocols. de Haas et al. did not observe a similar correlation between OTX2 expression and outcome, which suggests that copy number gain is a more faithful indicator of poor prognosis than overexpression alone (38).
Although mutation and copy number analysis of tumor specimens has revolutionized our understanding of recurrent molecular events in cancer, it is important to confirm oncogenic activity by functional studies. Here we have demonstrated in vitro and in vivo that OTX2 promotes cell growth and enhances tumorigenicity, and that medulloblastoma cells rely on OTX2 transcriptional function for their aggressive tumorigenic phenotype. Therefore, we have demonstrated that the highly prevalent overexpression of OTX2 in medulloblastoma has functional significance.
Recent advances in cancer genetics and the development of mouse models have shown a relationship between developmentally associated molecular pathways and medulloblastoma pathogenesis (39). The current study supports previous work implicating OTX2 as one such developmental gene involved in tumorigenesis (16, 17). While medulloblastomas arising from oncogenic Shh signaling arise from committed granule cell precursors (40, 41), the cell origins of Group C and D medulloblastomas (which distinctly harbor OTX2 focal gains) are currently unknown. During normal cerebellum development, OTX2 is expressed in the prospective cerebellum as well as emergent and migrating granule cell precursors (20, 38). Mouse models have demonstrated that OTX2 coordinates lineage differentiation in the central nervous system, promoting generation of dopaminergic neurons and, in some domains, repressing lineage commitment to granule cell and serotonergic neural lineages (42, 43). Understanding the potential of OTX2 to facilitate tumorigenesis of particular cerebellum lineages will yield considerable insight into the cell origin of Group C and D medulloblastomas.
The molecular classification of medulloblastoma using gene expression signatures and select genomic features has been reported by three independent laboratories to date: Gilbertson (33), Kool (32), and ours (28). In all of these studies, very similar bioinformatics was employed. In each case, class discovery was achieved by performing unsupervised hierarchical clustering (Pearson correlation) using the most differentially expressed genes in the dataset. Thompson et al and Kool et al each identified 5 molecular subgroups (based upon 46 and 62 primary tumors, respectively), whereas we used 90 tumors and identified 4 molecular subgroups. The disparity between the 5 versus 4 subgroups pertains to what we have designated Group C and Group D, which are equivalent to what Kool et al referred to as C, D, and E. We have since analyzed a larger cohort of our own 103 cases, and again generated the same 4 molecular subgroups with extremely high confidence (≥97% reproducibility; Northcott et al, in preparation). The larger sample cohort and more robust array platform (ie Affymetrix exon arrays vs U133 arrays) utilized in the current study provides increased power and statistical confidence in class discovery. Further application of the proposed molecular clustering strategies will reveal the most faithful method of tumor classification.
This study provides important insights into the mechanisms by which medulloblastomas develop through the dysregulation of OTX2, a gene normally silenced in adult cerebellum. Furthermore, these results establish OTX2 as a potential therapeutic target, the inhibition of which could result in tumor cell growth inhibition. For the future development of targeted therapies, it will be important to perform detailed molecular studies of the oncogenic pathways driven by OTX2 as well as the pathways maintaining its expression in tumors. These studies will aide the development of molecular diagnostic and therapeutic strategies for patients whose tumors present with this molecular hallmark. Once such strategies become available, there is promise that in tumors with OTX2 expression, the dysregulated signaling pathway can be corrected by specifically reducing OTX2 expression or inhibiting other components of this pathway.
Supplementary Material
Acknowledgements
Grant support: this project is supported by The Pediatric Brain Tumor Foundation Institute at Duke; a Damon Runyon Foundation Scholar Award; a Southeastern Brain Tumor Foundation Research Grant; an Alex’s Lemonade Stand Foundation Innovation Award; a V Foundation Cancer Research Grant and NIH Grant R01CA118822. MDT was supported by grants from the Canadian Cancer Society and the Pediatric Brain Tumour Foundation as well as a Clinician-Scientist award from the Canadian Institutes of Health Research.
References
- 1.Kleihues P, Louis DN, Scheithauer BW, et al. The WHO classification of tumors of the nervous system. J Neuropathol Exp Neurol. 2002;61:215–225. doi: 10.1093/jnen/61.3.215. discussion 26–9. [DOI] [PubMed] [Google Scholar]
- 2.Pietsch T, Taylor MD, Rutka JT. Molecular pathogenesis of childhood brain tumors. J Neurooncol. 2004;70:203–215. doi: 10.1007/s11060-004-2750-7. [DOI] [PubMed] [Google Scholar]
- 3.Zakhary R, Keles GE, Aldape K, Berger MS. Medulloblastoma and primitive neuroectodermal tumors. London: Churchill Livingstone; 2001. [Google Scholar]
- 4.Packer RJ, Reddy A. New Treatments in Pediatric Brain Tumors. Curr Treat Options Neurol. 2004;6:377–389. doi: 10.1007/s11940-996-0029-3. [DOI] [PubMed] [Google Scholar]
- 5.Gajjar A, Hernan R, Kocak M, et al. Clinical, histopathologic, and molecular markers of prognosis: toward a new disease risk stratification system for medulloblastoma. J Clin Oncol. 2004;22:984–993. doi: 10.1200/JCO.2004.06.032. [DOI] [PubMed] [Google Scholar]
- 6.Ray A, Ho M, Ma J, et al. A clinicobiological model predicting survival in medulloblastoma. Clin Cancer Res. 2004;10:7613–7620. doi: 10.1158/1078-0432.CCR-04-0499. [DOI] [PubMed] [Google Scholar]
- 7.Ris MD, Packer R, Goldwein J, Jones-Wallace D, Boyett JM. Intellectual outcome after reduced-dose radiation therapy plus adjuvant chemotherapy for medulloblastoma: a Children's Cancer Group study. J Clin Oncol. 2001;19:3470–3476. doi: 10.1200/JCO.2001.19.15.3470. [DOI] [PubMed] [Google Scholar]
- 8.Xu W, Janss A, Packer RJ, Phillips P, Goldwein J, Moshang T., Jr Endocrine outcome in children with medulloblastoma treated with 18 Gy of craniospinal radiation therapy. Neuro Oncol. 2004;6:113–118. doi: 10.1215/S1152851703000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoppe-Hirsch E, Renier D, Lellouch-Tubiana A, Sainte-Rose C, Pierre-Kahn A, Hirsch JF. Medulloblastoma in childhood: progressive intellectual deterioration. Childs Nerv Syst. 1990;6:60–65. doi: 10.1007/BF00307922. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez-Teijeiro A, Betensky RA, Sturla LM, Kim JY, Tamayo P, Pomeroy SL. Combining gene expression profiles and clinical parameters for risk stratification in medulloblastomas. J Clin Oncol. 2004;22:994–998. doi: 10.1200/JCO.2004.03.036. [DOI] [PubMed] [Google Scholar]
- 11.Lee Y, Miller HL, Jensen P, et al. A molecular fingerprint for medulloblastoma. Cancer Res. 2003;63:5428–5437. [PubMed] [Google Scholar]
- 12.MacDonald TJ, Brown KM, LaFleur B, et al. Expression profiling of medulloblastoma: PDGFRA and the RAS/MAPK pathway as therapeutic targets for metastatic disease. Nat Genet. 2001;29:143–152. doi: 10.1038/ng731. [DOI] [PubMed] [Google Scholar]
- 13.Pomeroy SL, Tamayo P, Gaasenbeek M, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 14.Boon K, Eberhart CG, Riggins G. Genomic amplification of Orthodenticle Homolog 2 (OTX2) in Medulloblastomas. Cancer Research. 2005;65 [PubMed] [Google Scholar]
- 15.Di CH, Liao SH, Adamson DC, Parrett TJ, Broderick DK, Shi Q, Lengauer C, Cummins JM, Velculescu VE, Fults DW, McLendon RE, Bigner DD, Yan H. Identification of OTX2 as a medulloblastoma oncogene whose product can be targeted by all-trans-retinoic acid. Cancer Res. 2005;65:919–924. [PubMed] [Google Scholar]
- 16.Yokota N, Mainprize TG, Taylor MD, et al. Identification of differentially expressed and developmentally regulated genes in medulloblastoma using suppression subtraction hybridization. Oncogene. 2004;23:3444–3453. doi: 10.1038/sj.onc.1207475. [DOI] [PubMed] [Google Scholar]
- 17.Michiels EM, Oussoren E, Van Groenigen M, et al. Genes differentially expressed in medulloblastoma and fetal brain. Physiol Genomics. 1999;1:83–91. doi: 10.1152/physiolgenomics.1999.1.2.83. [DOI] [PubMed] [Google Scholar]
- 18.Simeone A, Puelles E, Acampora D. The Otx family. Curr Opin Genet Dev. 2002;12:409–415. doi: 10.1016/s0959-437x(02)00318-0. [DOI] [PubMed] [Google Scholar]
- 19.Broccoli V, Boncinelli E, Wurst W. The caudal limit of Otx2 expression positions the isthmic organizer. Nature. 1999;401:164–168. doi: 10.1038/43670. [DOI] [PubMed] [Google Scholar]
- 20.Frantz GD, Weimann JM, Levin ME, McConnell SK. Otx1 and Otx2 define layers and regions in developing cerebral cortex and cerebellum. J Neurosci. 1994;14:5725–5740. doi: 10.1523/JNEUROSCI.14-10-05725.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S. Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev. 1995;9:2646–2658. doi: 10.1101/gad.9.21.2646. [DOI] [PubMed] [Google Scholar]
- 22.Ang SL, Jin O, Rhinn M, Daigle N, Stevenson L, Rossant J. A targeted mouse Otx2 mutation leads to severe defects in gastrulation and formation of axial mesoderm and to deletion of rostral brain. Development. 1996;122:243–252. doi: 10.1242/dev.122.1.243. [DOI] [PubMed] [Google Scholar]
- 23.Simeone A, Acampora D, Mallamaci A, et al. A vertebrate gene related to orthodenticle contains a homeodomain of the bicoid class and demarcates anterior neuroectoderm in the gastrulating mouse embryo. Embo J. 1993;12:2735–2747. doi: 10.1002/j.1460-2075.1993.tb05935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aldosari N, Bigner SH, Burger PC, et al. MYCC and MYCN oncogene amplification in medulloblastoma. A fluorescence in situ hybridization study on paraffin sections from the Children’s Oncology Group. Arch Pathol Lab Med. 2002;126:540–544. doi: 10.5858/2002-126-0540-MAMOAI. [DOI] [PubMed] [Google Scholar]
- 25.McLendon RE, Rosenblum MK, Bigner DD, Russell DS, Rubinstein LJ. Russell and Rubinstein's pathology of tumors of the nervous system. 7th ed. London: Hodder Arnold; 2006. [Google Scholar]
- 26.Eberhart CG, Burger PC. Anaplasia and grading in medulloblastomas. Brain Pathol. 2003;13:376–85. doi: 10.1111/j.1750-3639.2003.tb00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Northcott PA, Nakahara Y, Wu X, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Northcott PA, Fernandez LA, Hagan JP, et al. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009;69:3249–3255. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang TL, Diaz LA, Jr, Romans K, et al. Digital karyotyping identifies thymidylate synthase amplification as a mechanism of resistance to 5-fluorouracil in metastatic colorectal cancer patients. Proc Natl Acad Sci U S A. 2004;101:3089–3094. doi: 10.1073/pnas.0308716101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolligs FT, Hu G, Dang CV, Fearon ER. Neoplastic transformation of RK3E by mutant beta-catenin requires deregulation of Tcf/Lef transcription but not activation of c-myc expression. Mol Cell Biol. 1999;19:5696–5706. doi: 10.1128/mcb.19.8.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He TC, Sparks AB, Rago C, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 32.Kool M, Koster J, Bunt J, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One. 2008;3:e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson MC, Fuller C, Hogg TL, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 34.Briata P, Ilengo C, Bobola N, Corte G. Binding properties of the human homeodomain protein OTX2 to a DNA target sequence. FEBS Lett. 1999;445:160–164. doi: 10.1016/s0014-5793(99)00113-1. [DOI] [PubMed] [Google Scholar]
- 35.Eberhart CG, Kratz J, Wang Y, et al. Histopathological and molecular prognostic markers in medulloblastoma: c-myc, N-myc, TrkC, and anaplasia. J Neuropathol Exp Neurol. 2004;63:441–449. doi: 10.1093/jnen/63.5.441. [DOI] [PubMed] [Google Scholar]
- 36.Eberhart CG, Kepner JL, Goldthwaite PT, et al. Histopathologic grading of medulloblastomas: a Pediatric Oncology Group study. Cancer. 2002;94:552–560. doi: 10.1002/cncr.10189. [DOI] [PubMed] [Google Scholar]
- 37.Stearns D, Chaudhry A, Abel TW, Burger PC, Dang CV, Eberhart CG. c-myc overexpression causes anaplasia in medulloblastoma. Cancer Res. 2006;66:673–681. doi: 10.1158/0008-5472.CAN-05-1580. [DOI] [PubMed] [Google Scholar]
- 38.de Haas T, Oussoren E, Grajkowska W, et al. OTX1 and OTX2 expression correlates with the clinicopathologic classification of medulloblastomas. J Neuropathol Exp Neurol. 2006;65:176–186. doi: 10.1097/01.jnen.0000199576.70923.8a. [DOI] [PubMed] [Google Scholar]
- 39.Wechsler-Reya R, Scott MP. The developmental biology of brain tumors. Annu Rev Neurosci. 2001;24:385–428. doi: 10.1146/annurev.neuro.24.1.385. [DOI] [PubMed] [Google Scholar]
- 40.Yang ZJ, Ellis T, Markant SL, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14:135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.vSchuller U, Heine VM, Mao J, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14:123–134. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vernay B, Koch M, Vaccarino F, et al. Otx2 regulates subtype specification and neurogenesis in the midbrain. J Neurosci. 2005;25:4856–4867. doi: 10.1523/JNEUROSCI.5158-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Omodei D, Acampora D, Mancuso P, et al. Anterior-posterior graded response to Otx2 controls proliferation and differentiation of dopaminergic progenitors in the ventral mesencephalon. Development. 2008;135:3459–3470. doi: 10.1242/dev.027003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.