Abstract
We have evaluated the feasibility of using nanoparticle (NP)-based assays for improving detection sensitivity of HIV-1 p24 antigen. The first assay is a gold NP-based biobarcode amplification (BCA) assay which could detect HIV-1 p24 antigen at levels as low as 0.1 pg/ml. Compared with BCA, the lower limit of detection (LOD) for enzyme-linked immunosorbent assay (ELISA) was 10 ~ 15 pg/ml. These results demonstrate that the HIV-1 p24 BCA assay offers 100 ~ 150-fold enhancement in the detection limit over the traditional colorimetric ELISA. Furthermore, the BCA assay detected HIV-1 infection 3 days earlier than ELISA in seroconversion samples. A second assay is the europium (Eu+) NP-based immunoassay (ENIA), which uses Eu+ NPs to replace gold NPs in the BCA assay to further simplify the detection method and decrease the incubation time. For detection of HIV-1 p24, the lower LOD for ENIA was 0.5 pg/ml. These results indicate that the universal labeling technology based on NPs and its application may provide a rapid and sensitive testing platform for clinical diagnosis and laboratory research.
Introduction
Nanotechnology is research at the atomic, molecular, or macromolecular scale using nanomaterials that have a length scale of 1–100 nanometers (nm) although it may not be restricted to 100 nm.[1] Nanomaterials have some unique physical, chemical, and biological properties which are fundamentally different from those of the corresponding bulk material and could be widely used in medical testing.[2] For example, relative to small size of nanomaterials, they usually have large surface-to-volume ratio and could be easily labeled with large amounts of different molecules; their physical properties are chemically tailorable. In addition, nanomaterials also exhibit other unique physical properties, such as very sharp melting temperature, magnetic properties, unusual target binding properties, and different colors at different sizes of nanomaterials. In the past decade, nanotechnology, in particular nanoparticle (NP)-based assays have been adapted to improve the sensitivity and specificity of medical testing, and could provide new tools for clinical diagnosis due to their potential for high degrees of sensitivity, specificity, multiplexing capabilities and ability to operate without enzymes.[2]
Diagnosis of infection of human immunodeficiency virus type 1 (HIV-1), the cause of acquired immunodeficiency syndrome (AIDS), relies on the detection of HIV-1 RNA, capsid antigen (p24) and anti-HIV antibody.[3] HIV-1 p24 antigen levels are significantly high during the early, acute phase of infection and terminal stage of AIDS. It is also a useful marker for predicting CD4+ T cell decline and disease progression, for early detection of HIV-1 infection and improved patient management, as well as for testing the blood supply in regions of the world where HIV-1 RNA testing is not available or practical. A possible alternative to testing for HIV-1 RNA is HIV-1 p24 antigen which is usually detected by enzyme-linked immunosorbent assay (ELISA). However, the detection sensitivity of the conventional HIV-1 p24 ELISA is around 10–20 picograms per milliliter. It is an enzyme-based colorimetric assay that involves multiple steps of incubation. Due to its requirement for large sample volume, it is further restricted in the application of testing infant samples. During the past decade, HIV-1 p24 antigen assays have been significantly improved by implementing immune complex disruption methods, using more effective lysis buffer and incorporating a tyramide-mediated boosted assay. It has been reported that the boosted ELISA can decrease the lower limit of detection (LOD) of p24 antigen detection to ~1 pg/ml.[4–7] Another sensitive method called real-time immuno-polymerase chain reaction (IPCR) assay can detect 1000 HIV-1 RNA copies or 40 attograms of HIV-1 p24 antigen per reaction.[8] However, all these improvements of detection sensitivity increase the complexity of testing. Several years ago, we started to evaluate the feasibility to adapt nanotechnology and nanomaterials especially NPs to improve assay sensitivity for proteins including HIV-1 p24. In 2003, Mirkin’s group reported that by using gold NP-based biobarcode amplification (BCA) assay and adequate antibodies, they could detect 30 attomolar concentrations of prostate specific antigen (PSA) in the serum sample.[9] Dr. Bao from Nanosphere Inc. modified the system by using streptavidin (SA)-coated NPs. For detection of PSA, they found that the new format could improve the dose response over 10,000-fold and the detection sensitivity by 1000-fold compared to ELISA.[10] The enhanced sensitivity of the BCA assay is due to the amplification process that occurs in each antigen recognition and binding event,[10, 11] and a highly sensitive biobarcode detection process that involves NP-based silver enhancement and a microarray method.[12, 13] We then further modified the BCA assay for sensitive detection of HIV-1 p24 antigen and investigated other strategies that can simplify the detection method of BCA without having significant impact on detection sensitivity, and found that highly fluorescent europium (Eu+) NPs are suitable as replacement for gold NPs used in the original BCA assay. In our Eu+ NP–based immunoassay (ENIA), the antibody-antigen sandwich complex bound to Eu+ NPs can be directly measured using a fluorescence reader. Herein we report the progress of NP-based new testing methods (BCA and ENIA) for sensitive detection of HIV-1 p24.
Methods and Results
BCA assay
As shown in figure 1, our modified BCA assay adapts a sandwich immunoassay format in which HIV-1 p24 antigen is captured by a monoclonal anti-p24 antibody coated on the surface of microtiter plate wells and further complexed with a biotinylated polyclonal anti-p24 antibody. The monoclonal anti-p24 antibodies (#3537[14], #6521[15]) were obtained through the NIH AIDS Research and Reference Reagent Program (Germantown, MD); the biotinylated polyclonal anti-p24 antibody was purchased from ViroStat (Portland, MN). The immune sandwiched complex is then coupled to SA-coated 15 nm gold NPs (British Biocell Int., Cardiff, UK) through biotin-SA interaction followed by the binding of biotinylated biobarcode DNAs at the surface of the NPs. After extensive washing between steps with detergent solution to remove unbound or nonspecifically bound conjugates, the biobarcode DNA LT68 were released into the supernatant by incubation at 80°C for 5 min and detected by a scanometric assay[12] in which CodeLink Activated slides (Amersham Biosciences, Piscataway, NJ) printed with biobarcode capture oligonucleotides (complementary to the specific 25mer biobarcode LT68 sequence) were hybridized with the barcode DNA LT68, followed by the binding of barcode DNA to the dT20 gold NP chip-probe. The detection of the biobarcode signal was done by enhanced silver staining solution (Nanosphere, Inc., Northbrook, IL) and quantified on an image analyzer Verigene ID (Version 1.1.6, Nanosphere, Inc.).
Figure 1.
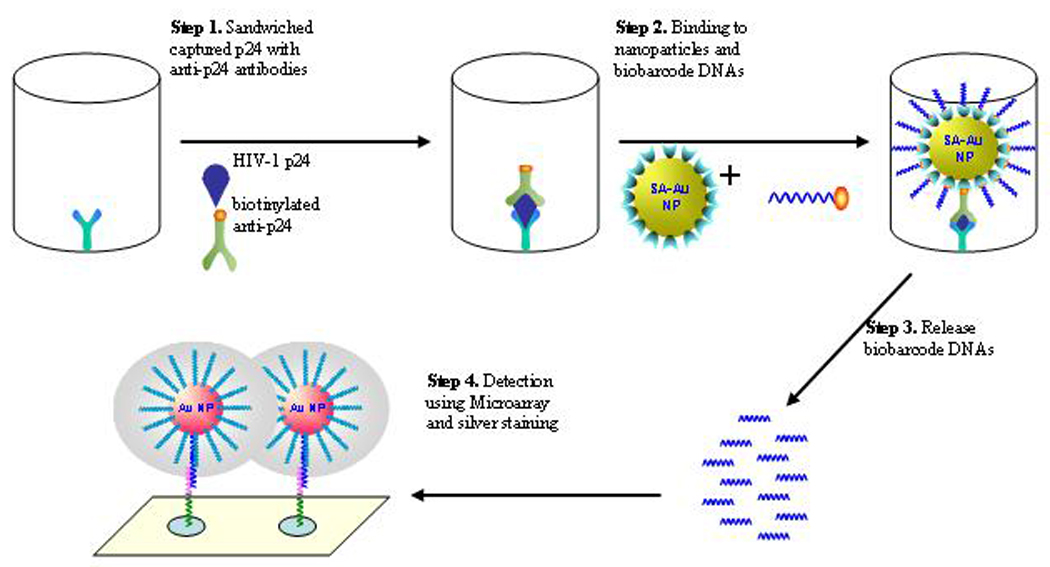
Scheme of Biobarcode amplification (BCA) assay for detection of HIV-1 p24 antigen. HIV-1 p24 is captured by anti-p24 coated microtiter wells and sandwiched with biotinylated secondary anti-p24 antibody. The immune complex is then coupled with 15nm gold NPs modified with SA that recognizes and binds biotinylated biobarcode DNA. After extensive washing and heating at 80°C for 5 min, biobarcode DNAs are eluted and detected by microarray method in which the released biobarcode DNAs are first hybridized with the capture oligos immobilized on the surface of glass slides. Gold NPs coated with T20 probe complementary to the other half of the biobarcode DNA are then hybridized to the captured biobarcode DNA. Finally, silver ions in the staining solution are reduced and grow around NPs to facilitate visualization of the hybridization event. The results are recorded and quantified with the Verigene ID system Nanosphere Inc., Northbrook, IL).
Before analyzing patient samples, we used BCA and conventional colorimetric in-house ELISA to establish calibration curves for purified HIV-1 p24 protein from PerkinElmer HIV-1 p24 kits (PerkinElmer, Waltham, MA) (Fig. 2). The BCA assay exhibited a linear detection range of 0.1 ~ 500 pg/ml, with a lower LOD of 0.1 pg/ml. However, at high target concentrations (>100 ng/ml), the assay responses becomes nonlinear, and eventually, the signal plateaus. Compared with the BCA assay, the detection range for ELISA was 10 ~ 1000 pg/ml while the lower LOD for ELISA was 10 ~ 15 pg/ml (Fig. 2). These results demonstrate that HIV-1 p24 BCA assay offers 100 ~ 150-fold improvements in the lower detection limit over traditional colorimetric and ELISA or at least 10-fold more sensitive than ultrasensitive ELISA under the current conditions. The BCA assay was further evaluated in HIV-1 regular negative and positive serum or plasma samples. No false positive results were observed in 30 HIV-1 negative normal healthy adult samples while HIV-1 p24 was detected in all 45 HIV-1 RNA positive samples including 23 seroconversion samples, 10 AIDS patient samples and 12 blood donor samples.[16] The average S/CO value for the normal healthy adult samples was 0.57 ± 0.2 while the S/CO values for samples from seroconversion panels, AIDS patients, blood donors and all HIV-1 positive were 4.3 ± 3.07, 9.34 ± 8.32, 5.33 ± 5.94 and 5.88 ± 6.01, respectively. Furthermore, for the seroconversion samples, the average time when HIV-1 p24 was first detected by BCA and ELISA were 12 and 15 days after the first HIV-1 RNA positive by PCR. The PCR results of these seroconversion samples were provided by Dr. Philip Norris. Therefore, the BCA assay detected HIV-1 infection 3 days earlier than ELISA in seroconversion samples.[16]
Figure 2.
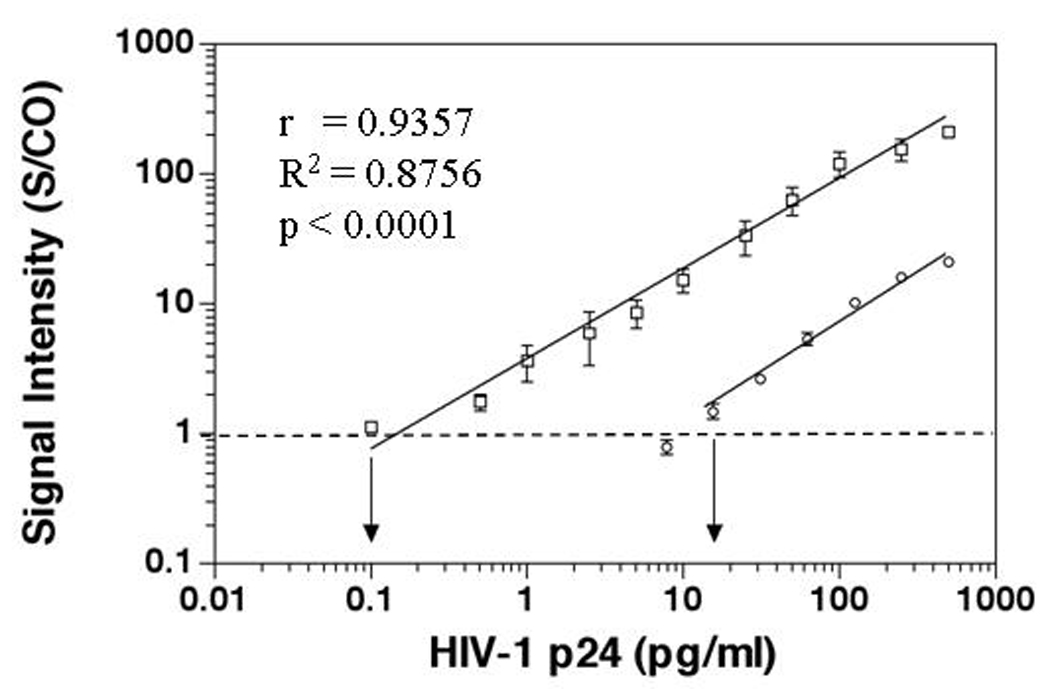
Increased sensitivity of BCA assay compared to in-house ELISA in detecting HIV-1 p24 antigen. HIV-1 p24 positive control antigen, ranging from 0.1 to 500 pg/ml in serial dilution in PBS, served as targets. The normalized relative signal intensities are represented as the ratios of samples over the cut-off value of the negative control (S/CO). BCA assay is represented by open square; ELISA by open circle. The error bar represents the standard deviation of at least three independent repeated experiments for each assay. The correlation between the HIV-1 p24 BCA assay and the concentrations of HIV-1 p24 was r = 0.9357 (R2 = 0.8756; p < 0.0001).
ENIA
Although the BCA assay successfully avoids the use of enzymes while achieving high sensitivity with significantI improvements in early detection of HIV-1 infection, it takes several hours to perform and needs microarray-based testing method for detection. The current BCA assay format is still not feasible for use in resource-limited areas. Therefore, we evaluated other NPs to simplify the assay format, and identified ENIA using Eu+ NP as suitable for rapid and sensitive detection of HIV-1 p24 (Fig 3). The difference between BCA and ENIA is that Eu+ NPs modified with SA were used to replace gold NPs to bind to the antigen-antibody sandwiched complex followed by the binding of biotinylated anti-SA antibody and SA-coated europium chelates (PerkinElmer). Since each Eu+ NP contains around 30,000 europium ions, the Eu+ NPs can produce intense long-lifetime fluorescence lights which are identical to those in the dissociation-enhanced lanthanide fluoroimmunoassay (DELFIA) method and can be measured directly in Victor Multilabel Counter (PerkinElmer).[17] A calibration curve for the purified HIV-1 p24 was established (Fig. 4). ENIA exhibits an analytical target concentration range of three orders of magnitude, with a lower LOD of 0.5 pg/ml. The preliminary evaluation showed that all 50 HIV-1 negative serum samples were HIV-1 p24 negative while 10 AIDS patient samples were positive for HIV-1 p24 by our ENIA assay. Furthermore, evaluation of 40 plasma samples infected with hepatitis B and C, and West Nile virus found no false positive. Thirty eight plasma samples composed of the duplicates of 19 HIV-1 seroconversion samples were tested. The results for the duplicate samples were consistent. Among the 19 HIV seroconversion samples, 8 were negative by both PCR and ENIA; 9 out of the 11 HIV RNA positive samples were positive for p24 by ENIA. One of the p24 antigen negative samples by ENIA was the first PCR positive bleed with viral load of <100 copies/ml; another p24 negative sample was the 7 day bleed after the first positive PCR and had a viral load of 2500 copies/ml. These results indicated the ENIA may miss samples of very early HIV infection and relative low viral load possibly due to its slightly lower sensitivity than BCA. Further evaluation of these samples is on-going with an enhanced ENIA. However, the ELISA missed 4 HIV-1 RNA positive samples. Among them, two samples which were bled 7 and 32 days after the positive PCR, respectively, were p24 positive by ENIA. These preliminary results indicate that the universal labeling technology based on Eu+ NPs and its application may provide a rapid and sensitive testing platform for clinical diagnosis and laboratory research.
Figure 3.
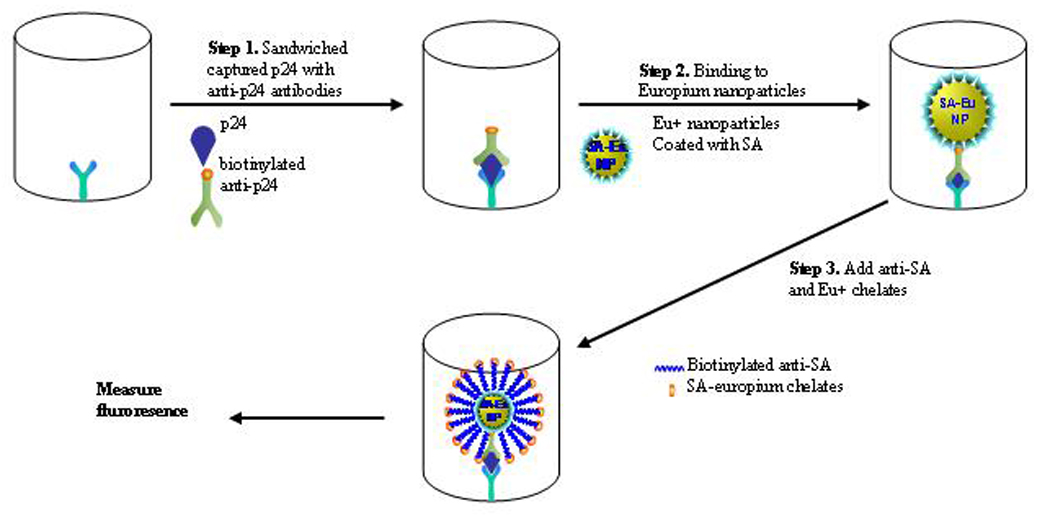
Scheme of Eu+ NP-based immunoassay (ENIA) for detection of HIV-1 p24 antigen. ENIA uses monoclonal anti-PA antibody coated microtiter wells to capture HIV-1 p24 that is then sandwiched with secondary biotinylated anti-p24 antibody. The Eu+ NPs modified with SA recognize and bind the above biotinylated antigen-antibody complex. The addition of biotinylated anti-SA antibody and SA-coated Eu+ chelates (PerkinElmer) facilitate the signal intensity of the binding event. After extensive washing, the fluorescence signal released from the sandwiched complex are then recorded and quantified with the Vector Mutlilabel Counter (PerkinElmer).
Figure 4.
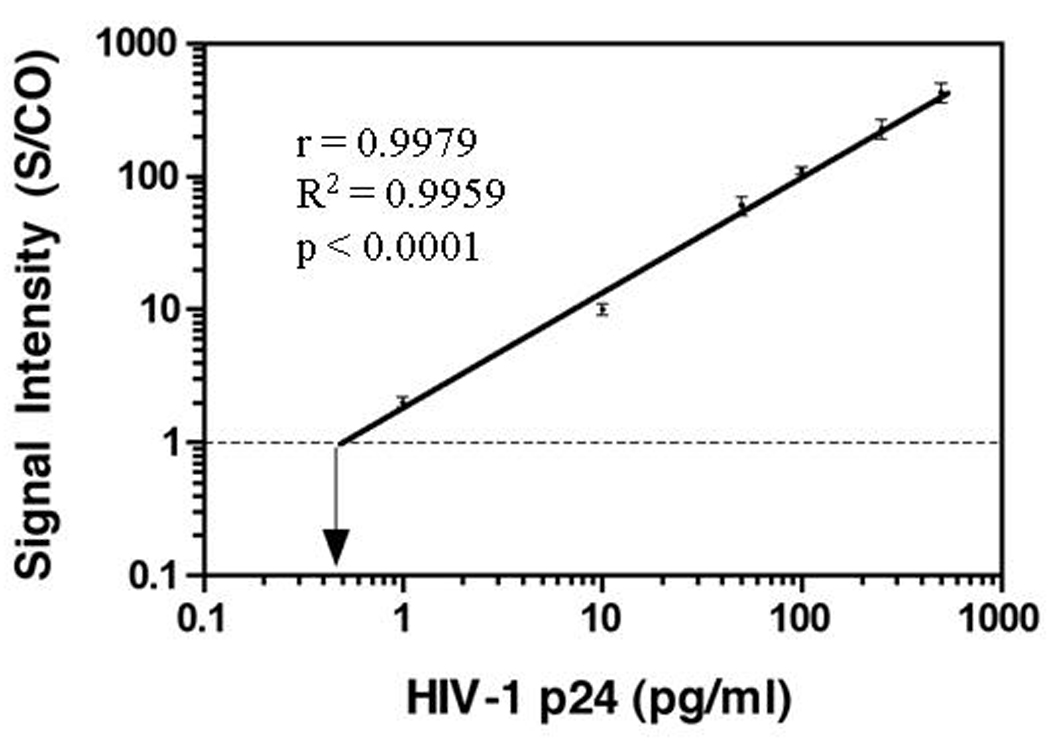
Increased detection sensitivity of ENIA in detecting HIV-1 p24 antigen. Purified HIV-1 p24 antigen, ranging from 0.5 pg/ml to 500 pg/ml in serial dilution in PBS, served as targets. The normalized relative signal intensities are represented as the ratios of samples over the cut-off value of the negative control (S/CO). ENIA assay is represented by closed cycle. The error bar represents the standard deviation of at least three independent repeated experiments for each assay. The correlation between the S/CO ratio by p24 ENIA assay and the concentrations of HIV-1 p24 was r = 0.9979; R2 = 0.9959; p < 0.0001.
Discussion
In this report, we have presented results using novel NP-based assays for sensitive and early detection of HIV-1 p24 antigen. Our preliminary data and other research results confirm that NP-based assays could dramatically increase the detection sensitivity of protein immunoassays, and have potential utility in improved diagnosis, point-of-care usage and even blood donor screening.[2, 16–18] This is an important advancement since unlike nucleic acid molecules, protein molecules per se cannot yet be duplicated in in vitro testing assays. There are several ways to improve the sensitivity of protein assays. One way to boost signal intensity for protein detection is amplification of surrogate markers. For example, immuno-PCR (IPCR) uses oligonucleotide as a surrogate marker and PCR for amplification.[19] However, PCR generally requires complex instrumentation for target amplification and detection, and is sensitive to contamination, ultimately limiting its utility in point-of-care venues, in particular in resource-limited countries. The BCA assay increases its detection sensitivity mainly through two steps: 1) to use the released biobarcode DNA as surrogate markers to amplify the target signal. Amplification occurs as a result of the large number of biobarcode DNA strands that bind to the NPs and are released from Ag-Ab-Ag-NPs complexes in each antigen recognition and binding event;[9, 11] 2) to use highly sensitive microarray method for detection.[12] The advantages of the BCA assay are its ultrasensitive detection capabilities in the absence of enzymatic reactions. However, specific instrumentation and long incubation times (~6 hours) are needed to achieve these limits of detection. Another way to improve detection sensitivity is to use novel labeling technology. Commonly used labeling technologies in immunoassays include enzyme activity, and chemicals (chemiluminescence and fluorescence). A major disadvantage of the enzyme-based colorimetric ELISA is its relatively low detection sensitivity. In the past decade, lanthanide chelates have been successfully used in immunoassay, such as the DELFIA technology. DELFIA immunoassay is a highly sensitive assay with stable signal, high signal-to-noise ratio, no enzymatic step, flexible platform and short incubation time. Recently, polystyrene nanoparticle containing lanthanide chelates have been reported to significantly improve detection sensitivity of PSA.[20, 21] The improved detection sensitivity is due to the huge content of Eu+ ions in a single NP, and in particular the unique characteristic of the time-resolved lanthanide chelates without self quenching even at high millimolar concentrations. Our study indicates that the Eu+ NPs, especially SA-coated NPs, may be more useful than Eu+ chelates in developing assays with high amplification ratios and extremely high detection sensitivity. Other advantages of our NP-based assays include 1) reagents are either biotinylated or labeled with SA, so the systems are easily adapted to detection of different targets; 2) the assays do not involve any enzymatic reactions for target amplification or detection, it does not need specific devices or equipments for reactions or storage of the reagents; 3) the assay format is similar to ELISA, which is widely used in laboratories and clinics, it does not need specific instruments and training; 4) like ENIA, it is more suitable for high-throughput screening. However, we need to further simplify the testing system to make it rapid and easy to use; and to further improve its detection sensitivity. We believe that with these modifications, NP-based assays could be useful for diagnostics and blood donor testing particularly in resource-limited settings.
Acknowledgment
We wish to acknowledge Office of Science and Health Coordination, FDA; National Heart, Lung and blood Institute, and National Institute of Allergy and Infectious Diseases, NIH for funding this work. We wish to acknowledge Dr. Philip J. Norris, Richard F. Little, Robert Yarchoan, Susan L. Stramer, Owen Wood, Ms. Kathleen M. Wyvill, Dr. Robert Gorelick, and Harri Harma for providing samples and reagents; Dr. Jiangqin Zhao, James J. Storhoff, Dr. C. Shad Thaxton, Dimitra Georganopoulou, Savka Stoeva and Mr. Jae-Seung Lee for providing technical support; Dr. Char A. Mirkin and Steven Wolinsky for helping to initiate the project. The findings and conclusions in this article have not been formally disseminated by the FDA and should not be construed to represent any Agency determination or policy.
Footnotes
Potential conflicts of interest: none reported
Reference
- 1.McNeil SE. Nanotechnology for the biologist. J Leukoc Biol. 2005;78:585–594. doi: 10.1189/jlb.0205074. [DOI] [PubMed] [Google Scholar]
- 2.Rosi NL, Mirkin CA. Nanostructures in biodiagnostics. Chem Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 3.Fiebig EW, Wright DJ, Rawal BD, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. Aids. 2003;17:1871–1879. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 4.Schupbach J, Boni J, Tomasik Z, Jendis J, Seger R, Kind C. Sensitive detection and early prognostic significance of p24 antigen in heat-denatured plasma of human immunodeficiency virus type 1-infected infants. Swiss Neonatal HIV Study Group. J Infect Dis. 1994;170:318–324. doi: 10.1093/infdis/170.2.318. [DOI] [PubMed] [Google Scholar]
- 5.Schupbach J, Flepp M, Pontelli D, Tomasik Z, Luthy R, Boni J. Heat-mediated immune complex dissociation and enzyme-linked immunosorbent assay signal amplification render p24 antigen detection in plasma as sensitive as HIV-1 RNA detection by polymerase chain reaction. Aids. 1996;10:1085–1090. [PubMed] [Google Scholar]
- 6.Sutthent R, Gaudart N, Chokpaibulkit K, Tanliang N, Kanoksinsombath C, Chaisilwatana P. p24 Antigen detection assay modified with a booster step for diagnosis and monitoring of human immunodeficiency virus type 1 infection. J Clin Microbiol. 2003;41:1016–1022. doi: 10.1128/JCM.41.3.1016-1022.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Layne SP, Merges MJ, Dembo M, et al. Factors underlying spontaneous inactivation and susceptibility to neutralization of human immunodeficiency virus. Virology. 1992;189:695–714. doi: 10.1016/0042-6822(92)90593-e. [DOI] [PubMed] [Google Scholar]
- 8.Barletta JM, Edelman DC, Constantine NT. Lowering the detection limits of HIV-1 viral load using real-time immuno-PCR for HIV-1 p24 antigen. Am J Clin Pathol. 2004;122:20–27. doi: 10.1309/529T-2WDN-EB6X-8VUN. [DOI] [PubMed] [Google Scholar]
- 9.Nam JM, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 10.Bao YP, Wei TF, Lefebvre PA, et al. Detection of protein analytes via nanoparticle-based bio bar code technology. Anal Chem. 2006;78:2055–2059. doi: 10.1021/ac051798d. [DOI] [PubMed] [Google Scholar]
- 11.Nam JM, Stoeva SI, Mirkin CA. Bio-bar-code-based DNA detection with PCR-like sensitivity. J Am Chem Soc. 2004;126:5932–5933. doi: 10.1021/ja049384+. [DOI] [PubMed] [Google Scholar]
- 12.Taton TA, Mirkin CA, Letsinger RL. Scanometric DNA array detection with nanoparticle probes. Science. 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 13.Storhoff JJ, Marla SS, Bao P, et al. Gold nanoparticle-based detection of genomic DNA targets on microarrays using a novel optical detection system. Biosens Bioelectron. 2004;19:875–883. doi: 10.1016/j.bios.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Wehrly K, Chesebro B. p24 antigen capture assay for quantification of human immunodeficiency virus using readily available inexpensive reagents. Methods. 1997;12:288–293. doi: 10.1006/meth.1997.0481. [DOI] [PubMed] [Google Scholar]
- 15.Simon JH, Fouchier RA, Southerling TE, Guerra CB, Grant CK, Malim MH. The Vif and Gag proteins of human immunodeficiency virus type 1 colocalize in infected human T cells. J Virol. 1997;71:5259–5267. doi: 10.1128/jvi.71.7.5259-5267.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang S, Zhao J, Storhoff JJ, et al. Nanoparticle-Based biobarcode amplification assay (BCA) for sensitive and early detection of human immunodeficiency type 1 capsid (p24) antigen. J Acquir Immune Defic Syndr. 2007;46:231–237. doi: 10.1097/QAI.0b013e31814a554b. [DOI] [PubMed] [Google Scholar]
- 17.Harma H, Soukka T, Lovgren T. Europium nanoparticles and time-resolved fluorescence for ultrasensitive detection of prostate-specific antigen. Clin Chem. 2001;47:561–568. [PubMed] [Google Scholar]
- 18.Tang S, Moayeri M, Chen Z, et al. Detection of anthrax toxin by an ultrasensitive immunoassay using europium nanoparticles. Clin Vaccine Immunol. 2009;16:408–413. doi: 10.1128/CVI.00412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Zhao Q, Li XF, Le XC. Ultrasensitive assays for proteins. Analyst. 2007;132:724–737. doi: 10.1039/b704256f. [DOI] [PubMed] [Google Scholar]
- 20.Harma H, Soukka T, Lonnberg S, Paukkunen J, Tarkkinen P, Lovgren T. Zeptomole detection sensitivity of prostate-specific antigen in a rapid microtitre plate assay using time-resolved fluorescence. Luminescence. 2000;15:351–355. doi: 10.1002/1522-7243(200011/12)15:6<351::AID-BIO624>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 21.Soukka T, Harma H, Paukkunen J, Lovgren T. Utilization of kinetically enhanced monovalent binding affinity by immunoassays based on multivalent nanoparticle-antibody bioconjugates. Anal Chem. 2001;73:2254–2260. doi: 10.1021/ac001287l. [DOI] [PubMed] [Google Scholar]


