Abstract
Background. In adults and children with respiratory syncytial virus (RSV) infection, a polymorphism in the interleukin 6 (IL-6) promoter at position −174 predicts illness magnitude. In addition, polymorphisms in the interleukin 10 (IL-10), tumor necrosis factor α (TNF-α), and interferon γ (IFN-γ) genes are associated with immune responsiveness and the frequency of complications. Here, the effect of these polymorphisms on illness and seroconversion during infection with rhinovirus type 39 (RV39) was evaluated.
Methods. Seventy-two adults were genotyped for the selected polymorphisms, experimentally exposed to RV39, and followed to track infection, seroconversion, and symptoms and signs of illness. Regression analysis was used to determine whether these polymorphisms predicted seroconversion and illness magnitude in 57 infected subjects.
Results. The low-production IL-6 −174 phenotype (C/C genotype) was associated with greater symptom magnitudes, and the IFN-γ phenotype +874 predicted the frequency of seroconversion. No relationship between the IL-10 or TNF-α polymorphisms and any measured outcome was documented. The concentration of IL-6 protein, as measured in nasal wash fluids from subjects, was positively correlated with symptom magnitude, but it was independent of the IL-6 −174 genotypes representing the high- and low-production phenotypes.
Conclusions. These results document statistically significant associations between the IL-6 −174 and IFN-gγpolymorphisms and specific responses to experimental RV39 infection. For the IL-6 −174 polymorphism, the results replicate those for experimental RSV infection.
Coldlike illness (CLI) is the most common disease affecting humans. The majority of CLIs are caused by a viral upper respiratory tract infection (vURTI), which could be due to any of a number of viruses, including rhinovirus, respiratory syncytial virus (RSV), influenza virus, coronavirus, and enterovirus, among others [1, 2]. Although CLIs are usually self-limited, they cause significant morbidity and are known precipitants of complications in the sinuses, lungs, and middle ears [3].
During a vURTI, a CLI is usually diagnosed on the basis of the perceived significance of the expressed signs and symptoms of illness. The viral symptom and sign complex (vSSC) is a summary measure of the degree of illness defined by the magnitudes and durations of those symptom and sign elements typically associated with a vURTI [4]. The vSSC elements are similar for the different causal viruses [1, 5] and are believed to represent the overt expression of the host immune and inflammatory responses to a vURTI [6]. This commonality suggests that these viruses provoke their vSSCs and cause CLIs via activation of similar immune and inflammatory pathways.
Cytokines are known to orchestrate and coordinate the host immune and inflammatory responses during a vURTI [7, 8]. For example, assays of serial nasal wash specimens from adult subjects experimentally infected with rhinovirus, RSV, influenza virus, and coronavirus document consistent patterns of interleukin 1 (IL-1),interleukin 6 (IL-6), interleukin 8 (IL-8), and interleukin 10 (IL-10) production in nasal secretion that temporally track the vSSC and respond appropriately to interventions and factors that modulate the vSSC [4, 9–16].
In the population of individuals with a vURTI, the vSSC is variable, such that not all vURTIs are associated with a CLI [17], and past studies have documented a variety of factors that modulate the vSSC and consequently CLI expression [18]. Regarding the latter, a study of natural RSV infection in infants [19] and a follow-up study of experimental RSV infection in adults [20] reported that the C/C genotype of a single-nucleotide polymorphism in the IL-6 promoter at position −174 was associated with a greater vSSC. The primary hypothesis tested in the present study was that this IL-6 genotype is associated with a greater vSSC and a more prolonged CLI during experimental infection with another virus, rhinovirus type 39 (RV39). Because the studies of RSV also documented the effects of polymorphisms in the IL-10, interferon γ (IFN-γ), and tumor necrosis factor α (TNF-α) genes on immune responses and complication expression, a secondary hypothesis tested in the present study was that these polymorphisms also affect the host responses to RV39 infection [19, 20].
Methods
Healthy adult subjects were recruited by advertisement. The protocol, study methods, and potential risks were explained to the recruited subjects; written informed consent for study participation and for human immunodeficiency virus (HIV) screening was obtained; and a general physical examination was performed and a health history taken for each recruit at screening. From the subjects who were not excluded for medical reasons, standard demographic information was collected; blood was drawn for total and differential blood cell counts, clinical chemical analysis, and assays of serum RV39 neutralizing antibody titer and HIV antibodies; urine was collected for clinical urinalysis; and buccal swabs were collected for DNA isolation.
Seventy-two healthy (according to history, physical examination, total and differential blood cell count, blood chemistry, urine profile, and negative test result for HIV antibodies), susceptible (RV39 serum antibody titer, <4), adult (18–55 years old) subjects were enrolled. Two days before admittance to cloister (day -2), blood was drawn from each subject for repeat assay of serum RV39 neutralizing antibody titer. Subjects were then admitted to the cloister site, where they were confined to an isolated floor of a hotel for 6 days (day 0, 24-h preexposure period; days 1–5, sequential 24-h postexposure periods). On day 0, female subjects with positive urine pregnancy test results were excluded, as were subjects presenting with a CLI. At the end of day 0, each subject was inoculated with RV39 by direct instillation of course drops to the nasal mucosa at an estimated dose of ∼100 times the median tissue culture infective dose [21]. On each day of cloister, all subjects underwent a general physical examination as well as an examination of the ears, nose, and throat and completed a symptom diary; in addition, nasal mucociliary clearance function was assessed, nasal secretion production was measured, and a nasal wash was performed, with samples aliquoted and frozen at −70°C for later assay. Throughout the study period, subjects were not permitted to take prescription or over-the-counter medications, with the exception of acetaminophen (which was dispensed by study personnel) and birth control drugs. At the end of day 5, the subjects were interviewed by a physician, provided with specific instructions for follow-up, and discharged from cloister. On or around study day 28, a convalescent blood sample was drawn from each subject. The study protocol and informed consent forms were approved by the institutional review boards at the University of Pittsburgh and Carnegie Mellon University.
VirologyThe challenge virus was a safety-tested strain of RV39. RV39 serum neutralizing antibodies were assayed at screening, on day -2, and on day 28 by a standard 2-fold dilution method, with titers reported as reciprocals of the final dilution [22]. Seroconversion was defined as a 4-fold increase in titer between days -2 and 28. A sample of nasal wash fluid from each subject on each study day was placed (3:1, vol/vol) in a cryovial containing 4× concentrated virus collecting broth and then frozen at −70°C. For RV39 detection, the sample was thawed, 0.2 mL of the mixture was inoculated into tubes of human embryonic lung fibroblast cells, and the cells were observed for rhinovirus cytopathic effect [22]. Infection was defined as detection of the challenge virus on any of days 1–5. A variable representing infection duration, “days shed,” was constructed as the sum of the days on which RV39 was isolated.
DNA isolation, genotyping, and phenotype assignment. Genomic DNA was isolated from the buccal cells by means of a QIAamp DNA Mini kit (Qiagen), amplified using a Repli-g Whole Genome Amplification kit (Qiagen), and quantified with a Quant-iT PicoGreen dsDNA Assay kit (Invitrogen) [23]. The primer and probe sequences for the cytokine genotype assays were based on those used in previous publications for IL-6 −174 [224]; TNF-α08 [25]; IL-10 −1082, −819, and −592 [25]; and IFN-γ+874 [26]. Genotyping was performed with a TaqMan Genotyping Master mix (Applied Biosystems) according to a published protocol [27]. The 7300 System SDS software (version 1.4; Applied Biosystems) was used for instrument control, automated data collection, and genotype assignment.
Phenotypes corresponding to in vitro cytokine production by stimulated lymphocytes [24, 28–33] were assigned to each genotype, using the convention published by Gentile and colleagues [19]. Specifically, for IL-6 −174, the G/G and G/C genotypes were assigned to the high-production phenotype and C/C to the low-production phenotype; for TNF-α308, the G/A and A/A genotypes were assigned to the high-production phenotype and G/G to the low-production phenotype; for IFNγ +874, the T/T genotype was assigned to the high-production phenotype, T/A to the intermediate-production phenotype, and A/A to the low-production phenotype; and for IL-10 −1082, −819, and −592, the GCC/GCC haplotype was assigned to the high-production phenotype, GCC/ACC and GCC/ATA to the intermediate-production phenotype, and ACC/ACC, ACC/ ATA, and ATA/ATA to the low-production phenotype. For IFNγ +874 and IL-10 −1082, −819, and −592, phenotypes were coded as 1 for low production, 2 for intermediate production, and 3 for high production; for TNF-α308 and IL-6 −174, phenotypes were coded as 1 for low production and 2 for high production.
IL-6 protein assay. One aliquot of the nasal wash fluid collected on each day from each of 65 subjects was thawed and assayed for IL-6 by means of a commercially available enzymelinked immunosorbent assay (BioSource) [34]. The lower detection limit of the assay was 0.1 pg/mL. For each subject, a summary variable for IL-6 production was constructed as the log of the baseline-adjusted IL-6 concentrations summed over the 5 postexposure days of cloister. Summed values that were <0 were assigned a value of log (0.1) = −1.
Symptom assessment. Thirteen vSSC elements were evaluated by means of a self-completed diary on each study day, using a 4-point scale (none, mild, moderate, or severe). These elements represented 4 symptom domains: nasal symptoms (rhinorrhea, nasal congestion, and sneezing), throat symptoms (sore throat and cough), general symptoms (headache, malaise, chills, sweats, and fever), and complication symptoms (earache, chest congestion, and sinus pain). For each element, the baseline score on day 0 was subtracted from the score on each of the 5 postchallenge days. Summary variables for each domain and for the total symptom score were calculated as the sum over all days of the baseline-adjusted scores for all contained elements (negative summed values were assigned a value of 0). The daily diary included a question as to the presence or absence of a “cold.” A continuous variable, “cold days,” was constructed by summing the yes responses over days 1–5.
Objective sign assessment. On each cloister day, subjects expelled their nasal secretions into preweighed tissues and sealed the tissues in plastic bags of known weight. At the end of each 24-h period, the bags with the expended tissues were weighed, and the secretion weight (ie, the nasal secretion production) was determined by subtraction [21]. Nasal mucociliary clearance function was measured by placing ∼20 µL of a dyed saccharin solution bilaterally onto the anterior aspect of the inferior turbinate using a calibrated pipette. The nasal mucociliary clearance time was calculated as the difference between the time when the solution was placed and the time when the subject reported a sweet taste [35]. Summary nasal secretion production and mucociliary clearance time variables were calculated as the baseline-adjusted sum of the values over days 1–5. Because these summary variables were not normally distributed, the data for both were log-transformed, and values that were <0 were assigned a value of log (0.1) = −1.
Data analysis. The 9 continuous outcome variables were the log of the summary nasal secretion weight, mucociliary clearance time, and IL-6 concentration; the summary nasal, throat, general, complication, and total symptom scores; and the number of cold days. The presence or absence of seroconversion was treated as a dichotomous outcome variable. The potential predictor variables consisted of the IL-6, IL-10, TNFα and IFN-γ phenotypes, day -2 RV39 titer, number of virusshedding days, age in years, race (white was coded as 1, and all other races were coded as 2), and sex (female was coded as 1, and male was coded as 2). To determine whether these variables were statistically significant predictors of each continuous outcome variable, multiple regression analysis was used. To determine whether these variables were statistically significant predictors of seroconversion, logistic regression was used. The relationships between the log of the baseline-adjusted IL-6 protein concentration and the continuous outcome variables were evaluated using simple linear regression. All statistical procedures were performed using NCSS 2000 Statistical Package software (Kaysville). Unless otherwise indicated, data are presented as mean ± standard deviation.
Results
Of the 72 enrolled subjects, 8 had a serum RV39 titer of >4 on study day -2 and were defined by the protocol as nonsusceptible. In addition, 5 subjects from whom RV39 was not recovered on any day were defined as uninfected (no “wild” viruses were isolated from any nasal secretion sample), and 2 subjects did not have buccal samples available for cytokine genotype analysis. The data for these 15 subjects were eliminated from the data set.
Of the remaining 57 subjects, 31 were male, 36 were white, and the mean age was 30.3±10.7 years. The data for demographics, symptoms and signs, virology assays, and genotype data for IFN-γ +874 and TNF-α −308 were complete for all subjects. The genotype assay for IL-6 −174 and the haplotype assay for IL-10 −1082, −819, and −592 failed for 1 and 3 subjects, respectively. IL-6 protein concentrations in nasal secretions collected on each study day were available for 51 subjects.
None of the cytokine genotype distributions deviated significantly from Hardy-Weinberg equilibrium. The distribution of cytokine phenotypes was as follows: for TNF-α, 12 for high and 45 for low production; for IL-6, 47 for high and 9 for low production; for IFN-γ 9 for high, 24 for intermediate, and 24 for low production; and for the IL-10 haplotype, 16 for high, 21 for intermediate, and 17 for low production. The number of subjects shedding virus on a total of 1, 2, 3, 4, and 5 days was 4, 6, 12, 24, and 11, respectively. The day -2 RV39 antibody titer was 1 (defined as a titer of <2), 2, and 4 in 40, 15, and 2 subjects, respectively. Thirty-four (60%) subjects seroconverted. The mean number of cold days was 1.68±1.79 days; the mean summary nasal, throat, general, complication, and total symptom scores were 9.39±7.32, 4.75±4.45, 3.67±4.84, 3.11±4.84, and 20.91±17.85, respectively; the mean summary log clearance time and mucus weight were 0.99±0.69 min and 0.61±75 g, respectively; and the mean summary log IL-6 concentration was 1.85±0.93 pg/mL.
Logistic regression identified the IFN-γ +874 phenotype as the only significant predictor of the frequency of seroconversion (β = −.42 [standard error, 0.63], where β is the odds ratio per unit change in the predictor; Z = 2.23; P = .024). Seroconversion rates for the low-, intermediate-, and high-production IFN-γ phenotypes were 0.79, 0.54, and 0.22, respectively.
The statistical results of the multiple regression analyses of the continuous outcome variables are presented in Table 1, which lists only those predictors for which P < .1. Age was not a significant predictor of any variable, but white race was a significant predictor of higher throat symptom score, and male sex was a significant predictor of more cold days, higher throat symptom score, and higher total symptom score and was a marginally significant predictor of greater log secretion weight. Higher baseline RV39 titer was a significant predictor of fewer cold days and of lower nasal, throat, and total symptom scores and a marginally significant predictor of lower complication symptom score. More virus-shedding days was a significant predictor of more cold days, greater log secretion weight, and higher summary nasal, throat, complication, and total scores. The low-production IL-6 −174 phenotype was a significant predictor of more cold days and higher general and total symptom scores and was a marginally significant predictor of higher nasal and complication symptom scores. Neither the TNF-α308 nor the IL-10 1082, 819, or 592 phenotype was significantly associated with any outcome variable.
Table 1.
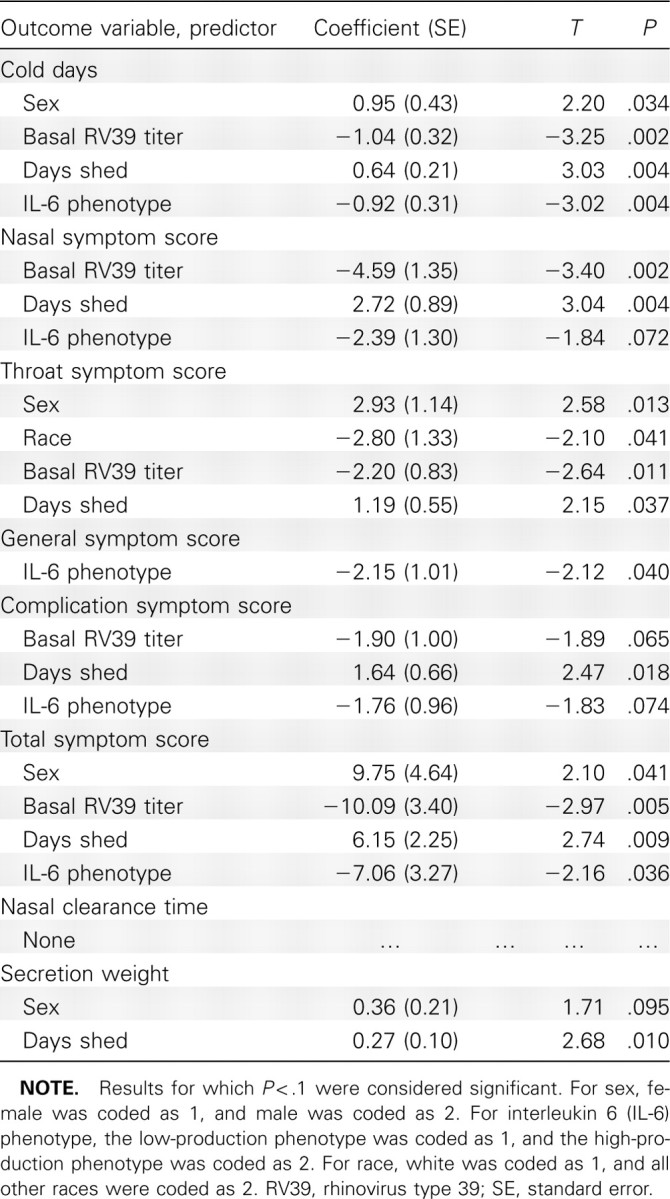
Summary Statistics for the Significant Predictor Variables of Each Continuous Outcome Variable Identified by multiple Regression
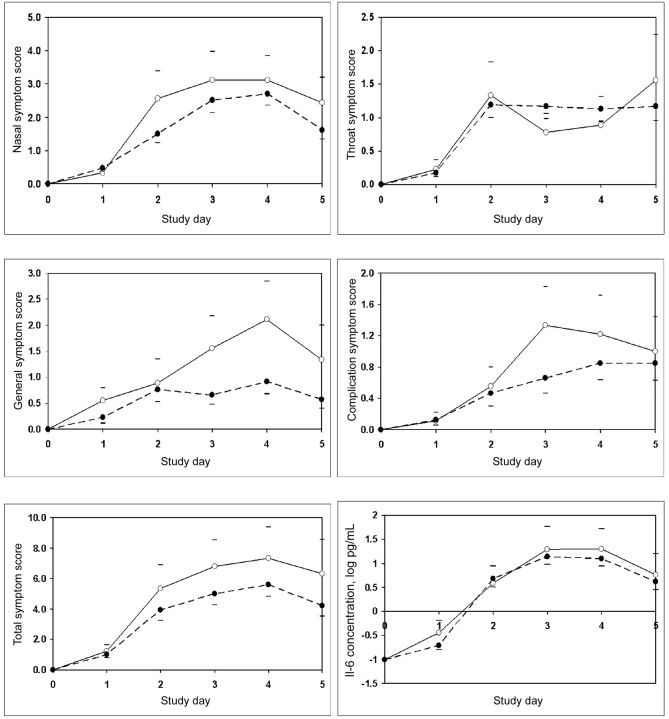
figure 1 shows the baseline-adjusted mean daily symptom scores in each of the 4 domains and total symptoms for groups defined by the low- and high-production IL-6 −174 phenotypes. With the exception of the throat symptom score, the mean scores for the other domains and for total symptoms were greater on most days among the subjects with the lowproduction phenotype (ie, C/C) compared with the subjects with the high-production phenotype (ie, C/G and G/G).
Figure 1.
Mean daily nasal, throat, general, complication, and total symptom scores and mean daily concentration of interleukin 6 (IL-6) for groups defined by the low-production (white circles) and high-production (black circles) IL-6 −174 phenotypes. The upper horizontal bars indicate the mean value of the low-production IL-6 phenotype plus its standard error, and the lower horizontal bars indicate the mean value of the high-production IL-6 phenotype minus its standard error.
The multiple regression analysis was overspecified for the contribution of all possible predictors to the log baseline-adjusted total IL-6 protein concentration. Limiting the possible predictors to the cytokine phenotypes, number of days shed, and baseline RV39 titer produced no significant effect of any variable on that outcome. The mean log values of that outcome variable for the low- and high-production IL-6 −174 phenotypes were 1.84±1.31 and 1.83±0.86 pg/mL, respectively; the between-group difference was not significant (P = .99, 2-tailed Student t test). figure 1 shows the mean log IL-6 concentration as a function of study day for the 2 groups defined by the IL- 6 −174 phenotype. The 2 curves were closely approximated, with no obvious differences on any study day. The log of the baseline-adjusted total IL-6 protein concentration was positively correlated with the number of cold days (r = .50; P < .001); the summary nasal (r = .54; P < .001), throat (r = .37; P = .008), general (r = .37; P = .007), complication (r = .36 P = .009 and total (r = .50; P<.001) symptom scores; and the log secretion weight (r = .26; P = .061).
Discussion
There is a vast literature relating cytokine polymorphisms to various inflammatory and infectious diseases, but few studies have examined the possible role played by these polymorphisms in modulating the various expressions of a vURTI. One study of RSV infection in hospitalized infants reported that the lowproduction IL-6 −174 phenotype was associated with greater illness severity, and a study of experimental RSV infection in adults reported that the low-production IL-6 −174 phenotype was associated with a greater vSSC. The results of the presentstudy of experimental RV39 infection in adults are consistent with those findings and support our primary hypothesis. Specifically, the low-production IL-6 −174 phenotype predicted a greater vSSC and greater symptom scores for most symptom domains in the RV39-infected subjects, with the effect realized across all postexposure days (figure 1). Although the IL-6 −174 phenotype had no effect on the 2 objective illness measures(nasal secretion production and mucociliary clearance time), these outcome variables were also not predicted by the IL-6 −174 phenotype in adults experimentally infected with RSV [20].
The results of 2 other studies support the hypothesis that the IL-6 −174 phenotype has an effect on vURTI expression as reflected in the frequency of a common vURTI complication, otitis media [36]. Specifically, in a cross-sectional study Patel and colleagues [37] reported that the low-production IL-6 −174 phenotype was significantly more frequent in children with a history of recurrent acute otitis media compared with children without a history of otitis media, and in a longitudinal study of young children Alper and colleagues [23] reported that the low-production IL-6 −174 phenotype was a significant predictor of the otitis media coincidence rate for rhinovirus infections.
A second objective of this study was to explore the effect of TNF-α −308, IFN-γ +874, and IL-10 −592, −819, and −1082 polymorphisms on the host responses to RV39 infection. These polymorphisms were shown to affect other host responses during RSV infection [20] and the rate of vURTI complications [19, 23, 37]. For the outcomes examined in this study, only the IFN-γ +874 polymorphism was shown to have a significant effect, with its phenotype being an inverse predictor of the rate of seroconversion during RV39 infection. In adults experimentally infected with RSV, the TNF-α −308 phenotype was directly related to the frequency of seroconversion, whereas the IFN-γ +874 phenotype was indirectly related to the frequency of subjects developing a 2-fold increase in nasal RSV-specific secretory immunoglobulin A antibody titers [20]. In the present study, the lack of significant effects of the other cytokine polymorphisms may be explained by their primary effects in previous studies on vURTI complications such as the frequencies of otitis media and pneumonia [19, 23, 37], events that were not observed in the present study.
From these results, there is sufficient evidence to advance the hypothesis that the IL-6 −174 C/C genotype (low-production phenotype) is associated with up-regulated inflammatory responses to a vURTI as reflected in the vSSC and the frequency of expressed complications. Although the mechanism(s) by which that polymorphism exerts these effects remains elusive, given the complex and poorly understood temporal interactions among the various cytokines and other inflammatory chemicals in provoking the vSSC and other expressions of a vURTI [4], the simplest hypothesis can be rejected. Specifically, as reported here and in past studies [12, 14, 16], higher nasal IL-6 concentrations are positively correlated in both phase and magnitude with the expressed vSSC, leading to the expectation that the high-production IL-6 −174 phenotype would predict higher nasal IL-6 concentrations and consequently greater illness magnitudes. However, neither of these expectations was supported by the experimental data; that is, no statistically significant relationship was demonstrated between the IL-6 −174 phenotype and local IL-6 nasal secretion concentration for either experimental RV39 or experimental RSV infection in adults [20], and for all studies reviewed the low-production (not the high-production) IL-6 −174 phenotype predicted greater illness and complications [19, 20, 23, 37].
In interpreting these results, it should be recognized that the cytokine phenotypes were assigned on the basis of in vitro results for stimulated cytokine release by leukocytes that may [28, 30, 38] or may not relate to in vivo cytokine production in the nasal mucosa, blood [39, 40], and local secretions [20] during a vURTI. Moreover, there are a large number of polymorphisms in the genes for IL-6 and other cytokines that were not assayed in the present study but that have the potential to interact with the assayed polymorphisms to influence cytokine production [33, 41–44], and past studies have shown that the local production of specific cytokines was influenced by polymorphisms in the genes for other cytokines (eg, the TNF-α -308 polymorphism predicted IL-6 and IL-8 protein concentrations in nasal secretions) [20]. These observations emphasize the difficulties of predicting cytokine production for any biological compartment in response to a vURTI using the genotype-phenotype associations for a given cytokine polymorphism on the basis of in vitro studies.
In conclusion, the present study documents statistically significant effects of certain cytokine polymorphisms on selected expressions of RV39 infection. Limitations of this study include the relatively few cytokine polymorphisms assayed, the assay of only IL-6 protein in nasal secretions, and the restriction of outcomes to seroconversion, provoked symptoms, and 2 objective measures of illness. In addition, because this study was limited to adults, in whom the frequency of vURTI complications is low, the effects of these cytokine polymorphisms on complication frequency could not be assessed. Nonetheless, it is highly likely that continued study of cytokine polymorphisms in the settings of natural and experimental vURTIs will yield important information regarding the genetic basis for CLIs, vURTIs, and vURTI complications.
Acknowledgments
We thank Dr Ellen Mandel, Dr Sancak Yuksel, James Seroky, Ellen Conser, Wesley Barnhart, Julianne Banks, Brendan Cullen-Doyle, and Amy Seroky for their assistance in screening and evaluating the study subjects before, during, and after the cloister phase of the study.
Footnotes
Potential conflicts of interest: none reported
Financial support: National Institute of Allergy and Infectious Disease (grant AI066367); Pennsylvania Department of Health (Commonwealth Research Enhancement grant); John D. and Catherine T. MacArthur Foundation's Network on Socioeconomic Status and Health; Eberly Research Endowment to the Department of Pediatric Otolaryngology, Children's Hospital of Pittsburgh.
References
- 1.Monto AS. Epidemiology of viral respiratory infections. Am J Med. 2002;112((suppl 6A)):4–12. doi: 10.1016/s0002-9343(01)01058-0. [DOI] [PubMed] [Google Scholar]
- 2.Heikkinen T, Jarvinen A. The common cold. Lancet. 2003;361((9351)):51–59. doi: 10.1016/S0140-6736(03)12162-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Proud D. Upper airway viral infections. Pulm Pharmacol Ther. 2008;21((3)):468–473. doi: 10.1016/j.pupt.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doyle WJ, Skoner DP, Gentile D. Nasal cytokines as mediators of illness during the common cold. Curr Allergy Asthma Rep. 2005;5((3)):173–181. doi: 10.1007/s11882-005-0034-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tyrrell DA, Cohen S, Schlarb JE. Signs and symptoms in common colds. Epidemiol Infect. 1993;111((1)):143–156. doi: 10.1017/s0950268800056764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eccles R. Understanding the symptoms of the common cold and influenza. Lancet Infect Dis. 2005;5((11)):718–725. doi: 10.1016/S1473-3099(05)70270-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melchjorsen J, Sørensen LN, Paludan SR. Expression and function of chemokines during viral infections: from molecular mechanisms to in vivo function. J Leukoc Biol. 2003;74((3)):331–343. doi: 10.1189/jlb.1102577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Message SD, Johnston SL. Host defense function of the airway epithelium in health and disease: clinical background. J Leukoc Biol. 2004;75((1)):5–17. doi: 10.1189/jlb.0703315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linden M, Greiff L, Andersson M, et al. Nasal cytokines in common cold and allergic rhinitis. Clin Exp Allergy. 1995;25((2)):166–172. doi: 10.1111/j.1365-2222.1995.tb01022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayden FG, Fritz R, Lobo MC, Alvord W, Strober W, Straus SE. Local and systemic cytokine responses during experimental human influenza A virus infection: relation to symptom formation and host defense. J Clin Invest. 1998;101((3)):643–649. doi: 10.1172/JCI1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner RB, Weingand KW, Yeh CH, Leedy DW. Association between interleukin-8 concentration in nasal secretions and severity of symptoms of experimental rhinovirus colds. Clin Infect Dis. 1998;26((4)):840–846. doi: 10.1086/513922. [DOI] [PubMed] [Google Scholar]
- 12.Cohen S, Doyle WJ, Skoner DP. Psychological stress, cytokine production, and severity of upper respiratory illness. Psychosom Med. 1999;61((2)):175–180. doi: 10.1097/00006842-199903000-00009. [DOI] [PubMed] [Google Scholar]
- 13.Fritz RS, Hayden FG, Calfee DP, et al. Nasal cytokine and chemokine responses in experimental influenza A virus infection: results of a placebo-controlled trial of intravenous zanamivir treatment. J Infect Dis. 1999;180((3)):586–593. doi: 10.1086/314938. [DOI] [PubMed] [Google Scholar]
- 14.Skoner DP, Gentile DA, Patel A, Doyle WJ. Evidence for cytokine mediation of disease expression in adults experimentally infected with influenza A virus. J Infect Dis. 1999;180((1)):10–14. doi: 10.1086/314823. [DOI] [PubMed] [Google Scholar]
- 15.Noah TL, Becker S. Chemokines in nasal secretions of normal adults experimentally infected with respiratory syncytial virus. Clin Immunol. 2000;97((1)):43–49. doi: 10.1006/clim.2000.4914. [DOI] [PubMed] [Google Scholar]
- 16.Doyle WJ, Gentile DA, Cohen S. Emotional style, nasal cytokines, and illness expression after experimental rhinovirus exposure. Brain Behav Immun. 2006;20((2)):175–181. doi: 10.1016/j.bbi.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Alper CM, Doyle WJ, Winther B, Hendley JO. Upper respiratory virus detection without parent-reported illness in children is virus-specific. J Clin Virol. 2008;43((1)):120–122. doi: 10.1016/j.jcv.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doyle WJ, Cohen S. In: Common cold. Eccles R, Weber O, editors. Basel, Switzerland: Birkhauser Verlag; 2009. pp. 149>–186. [Google Scholar]
- 19.Gentile DA, Doyle WJ, Zeevi A, et al. Cytokine gene polymorphisms moderate illness severity in infants with respiratory syncytial virus infection. Hum Immunol. 2003;64((1)):338–344. doi: 10.1016/s0198-8859(02)00827-3. [DOI] [PubMed] [Google Scholar]
- 20.Gentile DA, Doyle WJ, Zeevi A, Piltcher O, Skoner DP. Cytokine gene polymorphisms moderate responses to respiratory syncytial virus in adults. Hum Immunol. 2003;64((1)):93–98. doi: 10.1016/s0198-8859(02)00705-x. [DOI] [PubMed] [Google Scholar]
- 21.Doyle WJ, Skoner DP, Fireman P. Rhinovirus 39 infection in allergic and nonallergic subjects. J Allergy Clin Immunol. 1992;89((5)):968–978. doi: 10.1016/0091-6749(92)90219-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gwaltney JM, Jr, Colonno RJ, Hamparaian VV, Turner RB. Rhinovirus. In: Schmidt NJ, Emmons RW, editors. Diagnositic procedures for viral, rickettsial and chlamydial infections. 6th ed. Washington, DC: American Public Health Association; 2001. [Google Scholar]
- 23.Alper CM, Winther B, Hendley JO, Doyle WJ. Cytokine polymorphisms predict the frequency of otitis media as a complication of rhinovirus and RSV infections in children. Eur Arch Otorhinolaryngol. 2009;266((2)):199–205. doi: 10.1007/s00405-008-0729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hegedus CM, Skibola CF, Bracci P, Holly EA, Smith MT. Screening the human serum proteome for genotype-phenotype associations: an analysis of the IL6 −174G1C polymorphism. Proteomics. 2007;7((4)):548–557. doi: 10.1002/pmic.200600366. [DOI] [PubMed] [Google Scholar]
- 25.Koch W, Kastrati A, Bottiger C, Mehilli J, von Beckerath N, Schomig A. Interleukin-10 and tumor necrosis factor gene polymorphisms and risk of coronary artery disease and myocardial infarction. Atherosclerosis. 2001;159((1)):137–144. doi: 10.1016/s0021-9150(01)00467-1. [DOI] [PubMed] [Google Scholar]
- 26.Rad R, Prinz C, Neu B, et al. Synergistic effect of Helicobacter pylori virulence factors and interleukin-1 polymorphisms for the development of severe histological changes in the gastric mucosa. J Infect Dis. 2003;188((2)):272–281. doi: 10.1086/376458. [DOI] [PubMed] [Google Scholar]
- 27.Johnson VJ, Yucesoy B, Luster MI. Genotyping of single nucleotide polymorphisms in cytokine genes using real-time PCR allelic discrimination technology. Cytokine. 2004;27((6)):135–141. doi: 10.1016/j.cyto.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Fishman D, Faulds G, Jeffery R, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998;102((7)):1369–1376. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heesen M, Kunz D, Bachmann-Mennenga B, Merk HF, Bloemeke B. Linkage disequilibrium between tumor necrosis factor (TNF)-α-308 G/A promoter and TNF-β NcoI polymorphisms: association with TNF alpha response of granulocytes to endotoxin stimulation. Crit Care Med. 2003;31((1)):211–214. doi: 10.1097/00003246-200301000-00032. [DOI] [PubMed] [Google Scholar]
- 30.Hoffmann SC, Stanley EM, Darrin Cox E, et al. Association of cytokine polymorphic inheritance and in vitro cytokine production in anti-CD3/CD28-stimulated peripheral blood lymphocytes. Transplantation. 2001;72((8)):1444–1450. doi: 10.1097/00007890-200110270-00019. [DOI] [PubMed] [Google Scholar]
- 31.Karjalainen J, Hulkkonen J, Nieminen MM, et al. Interleukin-10 gene promoter region polymorphism is associated with eosinophil count and circulating immunoglobulin E in adult asthma. Clin Exp Allergy. 2003;33((1)):78–83. doi: 10.1046/j.1365-2222.2003.01577.x. [DOI] [PubMed] [Google Scholar]
- 32.Kilpinen S, Huhtala H, Hurme M. The combination of the interleukin-1α(IL-1α-889) genotype and the interleukin-10 (IL-10 ATA) haplotype is associated with increased interleukin-10 (IL-10) plasma levels in healthy individuals. Eur Cytokine Netw. 2002;13((1)):66–71. [PubMed] [Google Scholar]
- 33.Rivera-Chavez FA, Peters-Hybki DL, Barber RC, O'Keefe GE. Interleukin-6 promoter haplotypes and interleukin-6 cytokine responses. Shock. 2003;20((3)):218–223. doi: 10.1097/01.shk.0000079425.52617.db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chehimi J, Starr SE, Frank I, et al. Impaired interleukin 12 production in human immunodeficiency virus-infected patients. J Exp Med. 1994;179((4)):1361–1366. doi: 10.1084/jem.179.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doyle WJ, van Cauwenberge PB. Relationship between nasal patency and clearance. Rhinology. 1987;25((3)):167–179. [PubMed] [Google Scholar]
- 36.Winther B, Alper CM, Mandel EM, Doyle WJ, Hendley JO. Temporal relationships between colds upper respiratory viruses detected by polymerase chain reaction and otitis media in young children followed through a typical cold season. Pediatrics. 2007;119((6)):1069–1075. doi: 10.1542/peds.2006-3294. [DOI] [PubMed] [Google Scholar]
- 37.Patel JA, Nair S, Revai K. Association of proinflammatory cytokine gene polymorphisms with susceptibility to otitis media. Pediatrics. 2006;118((6)):2273–2279. doi: 10.1542/peds.2006-0764. [DOI] [PubMed] [Google Scholar]
- 38.Unal S, Gumruk F, Aytac S, Yalnzoglu D, Gurgey A. Interleukin-6 (IL-6), tumor necrosis factor-α (TNF-α) levels and IL-6, TNF-polymorphisms in children with thrombosis. J Pediatr Hematol Oncol. 2008;30((1)):26–31. doi: 10.1097/MPH.0b013e31815b1a89. [DOI] [PubMed] [Google Scholar]
- 39.Mysliwska J, Wieckiewicz J, Hak L, et al. Interleukin 6 polymorphism corresponds to the number of severely stenosed coronary arteries. Eur Cytokine Netw. 2006;17((3)):181–188. [PubMed] [Google Scholar]
- 40.Jones KG, Brull DJ, Brown LC, et al. Interleukin-6 (IL-6) and the prognosis of abdominal aortic aneurysms. Circulation. 2001;103((18)):2260–2265. doi: 10.1161/01.cir.103.18.2260. [DOI] [PubMed] [Google Scholar]
- 41.Nieters A, Brems S, Becker N. Cross-sectional study on cytokine polymorphisms cytokine production after T-cell stimulation and clinical parameters in a random sample of a German population. Hum Genet. 2001;108((3)):241–248. doi: 10.1007/s004390100464. [DOI] [PubMed] [Google Scholar]
- 42.Gu W, Du DY, Huang J, et al. Identification of interleukin-6 promoter polymorphisms in the Chinese Han population and their functional significance. Crit Care Med. 2008;36((5)):1437–1443. doi: 10.1097/CCM.0b013e31816a0adb. [DOI] [PubMed] [Google Scholar]
- 43.Hamid YH, Rose CS, Urhammer SA, et al. Variations of the interleukin-6 promoter are associated with features of the metabolic syndrome in Caucasian Danes. Diabetologia. 2005;48((2)):251–260. doi: 10.1007/s00125-004-1623-0. [DOI] [PubMed] [Google Scholar]
- 44.Acalovschi D, Wiest T, Hartmann M, et al. Multiple levels of regulation of the interleukin-6 system in stroke. Stroke. 2003;34((8)):1864–1869. doi: 10.1161/01.STR.0000079815.38626.44. [DOI] [PubMed] [Google Scholar]