Abstract
The modification of proteins with SUMO (small ubiquitin-related modifier) plays an important role in determining their functional properties. Importantly though, SUMOylation is a highly dynamic process enabling transient responses to be elicited. This dynamism is controlled by two competing conjugating and deconjugating activities. The latter activity is mediated by the SENP [SUMO1/sentrin/SMT3 (suppressor of mif two 3 homologue 1)-specific peptidase] family of SUMO-specific proteases. The transcription factor Elk-1 [ETS (E twenty-six)-like 1] undergoes rapid de-SUMOylation following cellular stimulation with growth factors, and this contributes to its conversion from a SUMO-dependent repressor into a potent transcriptional activator. In the present study we demonstrate an important role for SENP1 in the de-SUMOylation of Elk-1, and therefore an integral role in determining the Elk-1-dependent transcriptional programme. Among the SENPs, Elk-1 preferentially forms a complex with SENP1. This preferential binding is reflected by the higher efficiency of SENP1 in promoting Elk-1 transactivation. Moreover, depletion of SENP1 causes a reciprocal effect and reduces the transactivation properties of Elk-1. Partial redundancy of function with SENP2 is revealed by combinatorial knockdown studies. Importantly, depletion of SENP1 also reduces the activation of the Elk-1 target gene c-FOS. Taken together, these results therefore reveal an important role for SENP1 in the regulation of Elk-1-mediated gene expression in response to mitogenic signalling cues.
Keywords: ETS (E twenty-six)-like 1 (Elk-1), SUMO1/sentrin/SMT3 (suppressor of mif two 3 homologue 1)-specific peptidase (SENP), small ubiquitin-related modifier (SUMO), transcription
Abbreviations: Elk-1, ETS (E twenty-six)-like 1; ERK, extracellular-signal-regulated kinase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GST, glutathione transferase; HA, haemagglutinin; HDAC, histone deacetylase; HEK, human embryonic kidney; MAPK, mitogen-activated protein kinase; RT, reverse transcription; SENP, SUMO1/sentrin/SMT3 (suppressor of mif two 3 homologue 1)-specific peptidase; siRNA, small interfering RNA; SUMO, small ubiquitin-related modifier; WT, wild-type
INTRODUCTION
Protein SUMOylation is being increasingly recognized as an important post-translational modification. Both cytoplasmic and nuclear proteins have been shown to be SUMO (small ubiquitin-related modifier) substrates, but the majority of substrates fall into the latter class, with one major function of SUMO being in transcriptional control (reviewed in [1–3]). Importantly, protein SUMOylation is a dynamic process. SUMO is added to proteins through the action of an enzymatic cascade involving an E1 SUMO-activating enzyme, and the E2 SUMO-conjugating enzyme Ubc9 which transfers the SUMO to substrates. In several cases, E3 ligases can act as molecular bridges to facilitate the action of Ubc9 and promote substrate SUMOylation (reviewed in [2,4]). Conversely, SUMOylation can be reversed through the activity of SENPs [SUMO1/sentrin/SMT3 (suppressor of mif two 3 homologue 1)-specific peptidases] which cleave SUMO from substrates (reviewed in [5,6]). There are currently six known SENPs in humans, which are thought to act, in part, in distinct subnuclear structures with SENP1 and 2 localized to the nuclear pore complex and nucleoplasm, whereas SENP3 and SENP5 are localized to the nucleolus, at least under normal conditions (reviewed in [5,6]).
Protein SUMOylation can be controlled through the activity of protein kinase cascades in response to extracellular signals (reviewed in [7]). For example, HSF-1 (heat-shock factor-1) SUMOylation status is enhanced following heat-shock-mediated phosphorylation [8]. Similarly, SUMOylation is promoted in several other proteins following signal-dependent phosphorylation (reviewed in [9,10]). In contrast, following mitogen-dependent activation of the ERK (extracellular-signal-regulated kinase) MAPK (mitogen-activated protein kinase) pathway, reduced SUMOylation of the transcription factor Elk-1 [ETS (E twenty-six)-like 1] occurs [11]. De-SUMOylation of Elk-1 is a component of the transcriptional activation process that is orchestrated by this transcription factor in response to ERK pathway signalling. In contrast, anisomycin-dependent activation of the p38 MAPK cascade does not result in Elk-1 de-SUMOylation, although the activation component of the signalling pathway still occurs [12]. Thus Elk-1 de-SUMOylation is an important mechanism that dictates the outcome of MAPK signalling through Elk-1 and its target genes, and helps set the amplitude of the resulting transcriptional response.
In the present study, we further investigated how mitogenic signalling promotes Elk-1 de-SUMOylation and hence the functional outcome in terms of target gene expression. We sought to identify which SENP is involved in the de-SUMOylation process. Through a combination of overexpression and loss-of-function approaches, we have identified SENP1 as the predominant SENP acting on Elk-1, and hence placing SENP1 as a key player in determining the transcriptional outcomes to mitogenic signalling.
MATERIALS AND METHODS
Plasmid constructs
The following plasmids were used in mammalian cell transfections. pRL-TK-Renilla (Promega; TK is thymidine kinase), pG5-E1B-Luc, pAS1561 [encoding GAL–Elk(1–428)WT; WT is wild-type], pAS2058 [encoding GAL–Elk(1–428)K2R], pAS383 [encoding Flag-His-tagged Elk(1–428)WT] [11] and pCDNA3-HA-SUMO-2 (HA is haemagglutinin) [13] have been described previously.
pAS1138 (pCDNA3-FlagB-SENP1WT), pAS1140 (pCDNA3-FlagB-SENP2WT), pAS1139 (pCDNA3-FlagB-SENP1C602S) and pAS1141 (pCDNA3-FlagB-SENP2C547S) were constructed by inserting EcoRI-NotI fragments from pAS1134, pAS1136, pAS1135 and pAS1137 respectively, into pCDNA3-FlagB (pAS2236). pAS1134, pAS1135, pAS1136 and pAS1137 were constructed by inserting EcoRI-SalI fragments from pCMV-Flag-SENP1WT, pCMV-Flag-SENP1C602S, pCMV-Flag-SENP2WT and pCMV-Flag-SENP2C547S respectively (WT versions were kindly provided by Professor Edward Yeh, Department of Cardiology, The University of Texas, M.D. Anderson Cancer Center, Houston, TX, U.S.A.; [14]) into the same sites in pGEX4T3. pCMV-Flag-SENP1C602S and pCMV-Flag-SENP2C547S were constructed using QuikChange® mutagensis (Stratagene) using the template and the primer pair combinations: pCMV-Flag-SENP1WT and ADS2518/2519 and pCMV-Flag-SENP2WT and ADS2520/2521 respectively. pAS1142 (pCDNA3-FlagC-SENP3WT) and pAS1143 (pCDNA3-FlagC-SENP3C532S) were constructed by inserting BamHI-XhoI fragments from pCDNA3-RGS-SENP3WT and pCDNA3-RGS-SENP3C532S respectively (kindly provided by Professor Edward Yeh; [15]), into pCDNA3-FlagC (pAS2237). For bacterial expression, GST (glutathione transferase; pAS2751) and GST–Elk-1(205–428) (pAS407) were used [16].
Tissue culture, cell transfections, siRNA (small interfering RNA), reporter gene assays and RT (reverse transcription)–PCR
HEK (human embryonic kidney)-293T and HeLa cells were grown in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% FBS (fetal bovine serum). Transfections were performed using Polyfect™ (Qiagen) according to the manufacturer's instructions. Where indicated, cells were serum-starved for 24 h, then treated with PMA (10 nM) or anisomycin (250 ng/ml) prior to luciferase assays (6 h) or RT–PCR analysis (40 min).
Transfections of siRNAs were achieved using Lipofectamine™ siRNAMax (Invitrogen) according to the manufacturer's instructions. ON-TARGETplus SMARTpool siRNAs (Dharmacon) against SENP1, SENP2, SENP3 and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were used.
For reporter gene assays, typically 0.25 μg of reporter plasmid and 50 ng of pRL-TK-Renilla were co-transfected with 0.1–1 μg of expression plasmids. Cell extracts were prepared and equal amounts of protein were used in luciferase assays using the dual-luciferase kit (Promega). Data were normalized against the expression of the Renilla luciferase.
Real-time RT–PCR was carried out using the QuantiTect SYBR Green RT–PCR mix (Qiagen). All data were normalized to the levels of 18S rRNA. The following primer pairs were used for RT–PCR experiments. 18S RNA, ADS4005 (5′-TCAAGAACGAAAGTCGGAGGTT-3′) and ADS4006 (5′-GGACATCTAAGGGCATCACAG-3′); c-FOS, ADS1690 (5′-AGAATCCGAAGGGAAAGGAA-3′) and ADS1691 (5′-CTTCTCCTTCAGCAGGTTGG-3′); SENP1, ADS2502 (5′-TTAACTAACCAGGAACAGCTG-3′) and ADS2503 (5′-GAGTCTGATCCTTCAGATTGTG-3′); SENP2, ADS2506 (5′-GAACTTACAGAGGACATGGA-3′) and ADS2507 (5′-CTGAATACATGAAGTGCTGG-3′); SENP3, ADS2512 (5′-GAGCATCTTGGACGAATTCC-3′) and ADS2513 (5′-GTTCATCACCTGGTCATTGAG-3′). Three independent RT–PCRs were run on the same RNA samples to reduce variability and produce a mean value for each data point.
In vivo SUMOylation and in vitro de-SUMOylation assays
In vivo SUMOylation of overexpressed Elk-1 was detected by co-transfection of His-tagged Elk-1 and HA-tagged SUMO-2 proteins, followed by purification of the conjugates under denaturing conditions as described previously [11].
In vitro de-SUMOylation assays were performed using SUMO-modified recombinant GST–Elk-1 fusion proteins and immunoprecipitated FLAG-tagged SENP proteins. To prepare SUMOylated recombinant Elk-1, a reconstituted SUMOylation system in Escherichia coli was used [18]. To prepare SENPs, HEK-293T cells were transfected with 30 μg of FLAG–SENP1 DNA. At 48 h post-transfection, cells were resuspended in Buffer II [25 mM Tris/HCl (pH 8), 150 mM NaCl, 0.1% Tween 20, 2 mM DTT (dithiothreitol) and Complete™ protease inhibitors (Roche), diluted 1:100] and lysed by passing them trough a 25-gauge needle. The SENP protein was purified using an anti-FLAG antibody conjugated to agarose beads. Beads containing the protein were washed twice in Buffer II containing 1 M NaCl and twice in Buffer II. Protein was eluted by adding FLAG peptide. Assays were carried out as described previously [18]. SUMOylated Elk-1 from 50 ml cultures was purified and kept bound to the GSH–agarose. Reactions (50 μl volume) in Buffer II were incubated at 37 °C, and shaken at 700 rev./min for the times indicated. The samples were spun down, the supernatant was removed and proteins were eluted by adding 1×SDS/PAGE loading buffer. Gels were stained overnight using SYPRO-Ruby and visualized using a Bio-Rad gel doc.
GST-pulldown assays
GST-pulldown assays were carried put using recombinant GST–Elk-1 and cell lysates of HEK-293T cells transfected with constructs encoding each of the catalytically dead FLAG-tagged SENP1–3 enzymes, essentially as described previously [19].
Immunoprecipitation and Western blot analysis
Western blotting and immunoprecipitations were carried out using SuperSignal™ West Pico or Dura (Pierce) chemiluminescent substrates with the primary antibodies anti-FLAG (Sigma), anti-Elk-1 (Santa Cruz Biotechnology) or anti-HA (Roche) according to the manufacturer's instructions. Data were analysed using Quantity-One software (Bio-Rad). Co-immunoprecipitation assays were performed in 50 mM potassium phosphate (pH 7.8), 0.1% Triton X-100, 150 mM NaCl, 10 nM N-ethylmaleimide, 10 nM E64 (Sigma) and 1:100 protease inhibitor cocktail (Roche), followed by three washes in the same solution.
RESULTS
Elk-1 is preferentially activated by SENP1
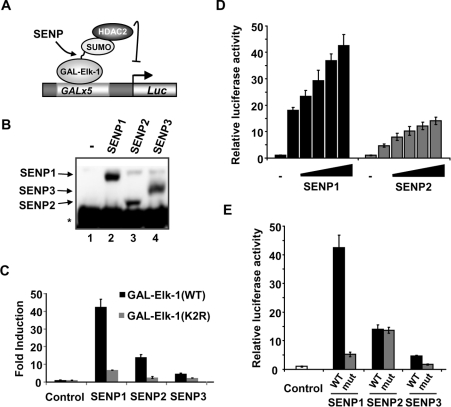
Elk-1 is SUMO-modified and this dampens down its transactivation activity and promotes active transcriptional repression. SUMOylation is lost during the transition to a transcriptional-activating state [11]. To begin to probe which SUMO protease(s) is involved in reversing Elk-1 SUMOylation we compared the abilities of different SUMO proteases (SENPs) to activate Elk-1 through de-SUMOylation. We first compared the expression levels of the SENPs. SENP1 and 3 were expressed at similar levels, with SENP2 being expressed to a slightly lesser extent (Figure 1B). Next, we compared the abilities of the SENPs to activate a GAL–Elk-1(1–428) fusion protein in a luciferase reporter assay (Figure 1A). This fusion protein is regulated by the SUMO pathway in an analogous manner to full-length Elk-1 [11,20]. SENP1 had the biggest effect on Elk-1 activation, whereas SENP3 barely affected Elk-1 activity; SENP2 had an intermediary effect (Figure 1C). Owing to the reduced expression levels of SENP2, we performed a dose–response experiment to compare the ability of SENP1 and SENP2 to activate Elk-1. However, even at 10-fold higher levels, SENP2 still activated Elk-1 to a lesser extent than SENP1 (Figure 1D, compare column 2, black bars with column 6, grey bars).
Figure 1. SENP1 preferentially activates Elk-1.
(A) Schematic diagram of the GAL-driven reporter system used to assay Elk-1-mediated transactivation. (B) Immunoprecipitation/Western blot analysis of the expression of the indicated FLAG-tagged SENP constructs. HEK-293T cells were transfected with different SENP expression vectors followed by immunoprecipitation with an anti-FLAG antibody and detection by Western blot analysis with the same antibody. The * represents a band corresponding to the antibody heavy chain. (C and D) Luciferase reporter analysis of the activity of GAL–Elk-1(1–428) constructs in the presence of co-transfected SENP expression constructs. (C) Wild-type (black bars) and K230R/K249R(K2R) mutant (grey bars) Elk-1 constructs were transfected in the absence and presence of the indicated SENP constructs. (D) Wild-type GAL–Elk-1 constructs were transfected in the presence of increasing amounts (0 ng, 125 ng, 250 ng, 500 ng and 1 μg) of the indicated SENP constructs. (E) Wild-type GAL–Elk-1 constructs were transfected in the presence of 1 μg of the indicated WT (black bars) or catalytically dead (mut; grey bars) SENP constructs. In all cases data are presented relative to the activity of either WT or K2R versions of GAL–Elk-1(1–428) in the presence of the control empty vector (taken as 1). Data are the average of two independent experiments, each with triplicate samples.
The effects of the SENPs could be indirect and/or independent of their SUMO-deconjugating activities. We therefore first established whether the Elk-1 SUMOylation sites are required for SENP-mediated activation. Again, we used a luciferase reporter assay but, instead of the WT protein, we tested GAL–Elk-1(1–428)(K2R), which lacks the two major SUMOylation sites at Lys230 and Lys249 [11]. In comparison with WT Elk-1, Elk-1(K2R) showed much reduced activation by the SENPs (Figure 1C), consistent with a role for SENPs in de-SUMOylating Elk-1. Conversely, we tested the activity of catalytically inactive mutant versions of the SENPs which had their active-site cysteine residues mutated. The ability of SENP1 to activate Elk-1 was much reduced, and a similar effect was seen on the smaller activation caused by SENP3 (Figure 1E). In contrast, the mutant version of SENP2 was able to activate Elk-1 to a similar extent as the WT version (Figure 1E), demonstrating that the activating effect of SENP2 on Elk-1 occurs by a mechanism that is independent from its SUMO-deconjugating activity.
Taken together, these results demonstrate that SENP1 is the most efficient SUMO protease acting on Elk-1, and that SENP3 has little effect on Elk-1. SENP2 has an intermediate effect, but its ability to activate Elk-1 is independent from its SUMO-deconjugating activity.
SENP1 binds to Elk-1
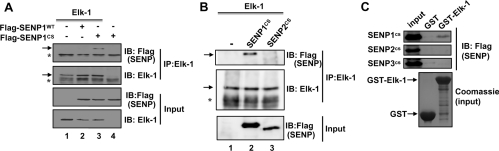
As SENP1 was established as the most efficient SUMO protease acting on Elk-1, we investigated whether we could detect complexes between these two proteins. Initial attempts at detecting interactions between Elk-1 and WT SENP1 were unsuccessful, either at endogenous or overexpressed levels. We reasoned that this might be due to the transient interactions between the enzymatic SENPs and their substrate Elk-1. Therefore, to potentially trap these interactions, we used a catalytically dead version of SENP1 (SENP1CS) in co-immunoprecipitation assays. In contrast with WT SENP1 where no complex could be detected, a complex detected between Elk-1 and the mutant SENP1CS (Figure 2A, lanes 2 and 3). Next, we compared the ability of SENP1CS and the equivalent catalytically dead mutant version of SENP2 (SENP2CS) to bind Elk-1. Preferential binding of SENP1 was observed in a co-immunoprecipitation assay (Figure 2B). To further probe the specificity of these interactions we performed GST-pulldown assays with recombinant GST–Elk-1 and HEK-293T cell lysates containing each of the catalytically dead SENP1–3 enzymes. Preferential interactions were observed with SENP1 (Figure 2C).
Figure 2. Elk-1 interacts with SENP1.
(A and B) Co-immunopreciptation analysis of Elk-1(1–428) and SENPs. (A) Elk-1 was transfected in the absence and presence of the indicated WT and catalytically inactive (CS) versions of FLAG-tagged SENP1. Following immunoprecipitation of Elk-1 (IP), bound SENPs were detected by immunoblotting (IB) with an anti-FLAG antibody. Immunoprecipitated proteins are shown in the top two panels and input proteins are shown in the bottom two panels. (B) Elk-1 was transfected in the absence and presence of the indicated catalytically inactive (CS) versions of FLAG-tagged SENPs. Following immunoprecipitation of Elk-1 (IP), bound SENPs were detected by immunoblotting (IB) with an anti-FLAG antibody. Immunoprecipitated proteins are shown in the top two panels and input proteins in the bottom panel. * Indicates non-specific cross-reacting bands in the IP samples. (C) GST-pulldown analysis of GST–Elk-1(205–428) binding to SENPs. The indicated catalytically inactive SENPs were expressed in HEK-293T cells, the lysates bound to recombinant GST or GST–Elk-1 and were detected by IB with an anti-FLAG antibody (top panel). A Coomassie-Blue-stained gel of the input bait proteins is shown in the bottom panel.
Collectively, these results demonstrate that SENP1 can form complexes with Elk-1 and this interaction appears to be specific among the SENPs.
SENP1 de-SUMOylates Elk-1
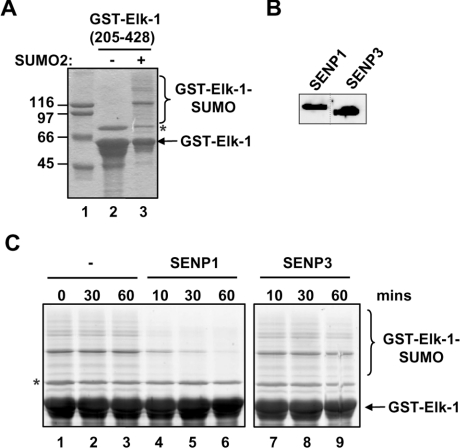
The ability of SENP1 to activate Elk-1 in a SUMO-dependent manner suggests that SENP1 can de-SUMOylate Elk-1. To prove that this is the case, we performed in vitro de-SUMOylation assays using recombinant SUMO-modified Elk-1 and FLAG-tagged SENP1 or SENP3 immunoprecipitated from HEK-293T cells. Elk-1 was modified by SUMO using an E. coli system containing a reconstituted SUMO pathway (Figure 3A; [18]), and equal amounts of immunoprecipitated SENP1 and SENP3 (Figure 3B) were incubated with this recombinant protein. The addition of SENP1 resulted in loss of SUMO conjugation to Elk-1 (Figure 3C, lanes 4–6), demonstrating that Elk-1 is a direct target of the SUMO proteolytic activity of SENP1. In contrast, little de-SUMOylation was seen upon incubation with SENP3 (Figure 3C, lanes 7–9). Thus, in agreement with the reporter and binding assays, SENP1 and SENP3 show different activities towards Elk-1, with SENP1 more efficiently de-SUMOylating Elk-1 in vitro.
Figure 3. SENP1 de-SUMOylates Elk-1 in vitro.
(A) A Coomassie-Blue-stained gel showing the non-SUMOylated and SUMOylated recombinant GST–Elk-1 fusion proteins. (B) Western blot analysis of FLAG-tagged SENP1 and SENP3 purified by immunoprecipitation from transfected HEK-293T cells. The broken line indicates where irrelevant lanes were removed. (C) SUMOylated recombinant GST–Elk-1(205–428) was incubated in the presence or absence of FLAG-tagged SENP1 or SENP3 for the times indicated and samples were detected by SYPRO-Ruby staining after SDS/PAGE. The positions of the recombinant Elk-1 protein and its SUMOylated forms are indicated. * Indicates the position of a co-purifying contaminant.
Endogenous SENP1 is important for Elk-1 activation
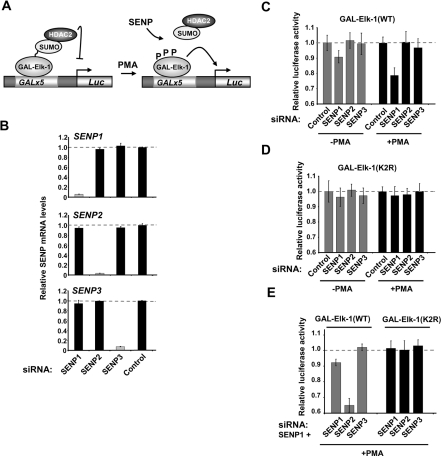
We have established that, among SENPs, Elk-1 is preferentially activated by overexpressed SENP1 and binds specifically to SENP1. However, to determine whether endogenous SENP1 is important for Elk-1 activation, we compared the effect of siRNA-mediated depletion of SENP1 with depletion of other SENPs on Elk-1 activity. First we determined the efficacy and specificity of each siRNA in depleting SENP mRNA levels. Each of the siRNA pools specifically depleted the corresponding SENP mRNA with high efficiency, with negligible effects on the other SENPs (Figure 4B), thereby demonstrating their utility for subsequent functional assays. Next, we compared the activity of Elk-1 in a luciferase reporter assay under serum-starved conditions or upon PMA treatment. PMA treatment leads to activation of the ERK pathway and loss of SUMO on Elk-1 and hence loss of its repressive properties (Figure 4A; [11]). In the absence of PMA, a 10% reduction in Elk-1 activity was observed following SENP1 depletion, but little effect was seen upon depleting the other SENPs (Figure 4C, grey bars). This effect was even more pronounced in the presence of PMA, with >20% reduction in Elk-1 activity seen following SENP1 depletion. Again, depletion of the other SENPs had a negligible effect (Figure 4C, black bars). We also probed the effect of SENP depletion on the activity of the Elk-1(K2R) mutant protein that could not be SUMO-modified. In contrast with the effects seen with the WT protein, depletion of any one of the SENPs had no effect on the activity of Elk-1(K2R), either in the presence or absence of PMA treatment (Figure 4D). This is consistent with a role for SENP1 in directly acting to de-SUMOylate Elk-1.
Figure 4. Depletion of SENP1 decreases the transactivation capacity of Elk-1.
(A) Schematic diagram of the GAL-driven reporter system used to assay Elk-1-mediated transactivation following ERK pathway activation. (B) RT–PCR analysis of the expression of the indicated SENPs in HeLa cells in the presence of control siRNAs or siRNA duplexes directed against the SENPs indicated. Data are representative of two independent experiments and are the average of two samples, presented relative to the transcript levels in the presence of control siRNA duplexes (taken as 1). (C–E) Luciferase reporter analysis of the activity of GAL–Elk-1(1–428) constructs in HEK-293T cells in the presence of co-transfected siRNA duplexes against SENPs. All data are the average of two experiments carried out in duplicate. Note that the axis does not begin at zero to emphasize the changes we observe. (C and D) Individual siRNA duplexes (25 pmol) were transfected in the presence of either WT (C) or mutant K230R/K249R(K2R) (D) forms of Elk-1. Cells were either untreated (grey bars) or treated with PMA for 6 h (black bars). (E) Cells were treated with PMA after transfection with vectors encoding WT (grey bars) or mutant (K2R) (black bars) versions of GAL–Elk-1(1–428) and with siRNA targeting SENP1 (12.5 pmol), and the additional siRNA constructs (12.5 pmol) against different indicated SENPs.
Although significant, the depletion of SENP1 caused relatively small changes in the activity of Elk-1. This could be due to functional redundancy whereby, in the absence of SENP1, another SENP can at least partially substitute for its activity. To test such a possibility, a combinatorial knockdown approach was undertaken. SENP1 siRNAs were used in combination with either SENP2 or SENP3 siRNAs. Again, SENP1 depletion caused a reduction in the activity of Elk-1 following PMA treatment, but this effect was amplified by the simultaneous depletion of SENP2, with Elk-1 activity reduced by nearly 40% (Figure 4E, grey bars). In contrast, co-depletion of SENP3 did not further enhance the activity of Elk-1, suggesting that, at the level of functional redundancy, there is still specificity of action. Again, neither the depletion of SENP1 alone nor in combination with other SENPs led to a reduction in the activity of the Elk-1(K2R) mutant (Figure 4E, black bars).
Taken together these results therefore demonstrate that SENP1 is the predominant SENP acting to promote Elk-1 activation in a SUMO-dependent manner, although there is a degree of functional redundancy with SENP2.
SENP1 plays a role in Elk-1 target gene activation in response to mitogenic signalling
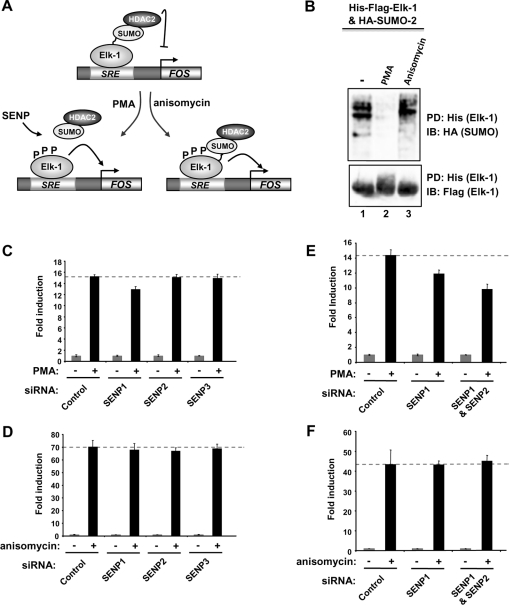
One of the best characterized Elk-1 target genes is c-FOS. Elk-1 orchestrates the activation of c-FOS in response to both mitogenic and stress signals, such as PMA and anisomycin. However, while PMA treatment leads to loss of SUMOylation, anisomycin treatment does not (Figure 5A; [12]), although both treatments lead to Elk-1 phosphorylation and c-FOS activation. We therefore probed the roles of SENPs in signal-dependent c-FOS activation. First we demonstrated that PMA and anisomycin had the predicted effects on Elk-1 SUMOylation, with only PMA promoting SUMO loss (Figure 5B). Next, we depleted SENP1, SENP2 or SENP3, and investigated c-FOS activation. The loss of SENP2 or SENP3 had little effect, but SENP1 loss caused a reduction in c-FOS activation following PMA treatment (Figure 5D). In contrast, little reduction was seen in c-FOS activation by anisomycin upon depletion of individual SENPs, including SENP1 (Figure 5D).
Figure 5. Depletion of SENP1 decreases the activation of the Elk-1 target gene c-FOS.
(A) Schematic diagram showing the different molecular events occurring on the c-FOS promoter after PMA (SUMO loss) or anisomycin (SUMO retention) stimulation. In both cases, c-FOS promoter activation is still observed. SRE, serum-response element. (B) Western blot analysis of Elk-1 SUMOylation levels. HEK-293T cells were transfected with expression vectors for His-FLAG-tagged Elk-1 and HA-tagged SUMO-2, and His-tagged Elk-1 was pulled down (PD) from lysates from HEK-293T cells treated with PMA or anisomycin for 40 min. Total and SUMOylated Elk-1 were detected by immunoblotting (IB) with anti-FLAG and anti-HA antibodies respectively. (C–F) RT–PCR analysis of the expression of c-FOS in HeLa cells in the presence of control GAPDH siRNAs or siRNA duplexes directed against the SENPs indicated. Either an siRNA duplex against a single SENP (25 pmol) or two siRNA duplexes against distinct SENPs (12.5 pmol each) were transfected and cells were either serum-starved (grey bars) or treated with PMA (C and E) or anisomycin (D and F) for 40 min (black bars). Data are the average of duplicate samples and are presented relative to the transcript levels in serum-starved cells in the presence of control GAPDH siRNA duplexes (taken as 1).
Studies using reporter gene analysis suggested that a degree of functional redundancy exists between SENP1 and SENP2 which might mask the effect of SENP1 depletion (Figure 4). We therefore compared the effect of depleting both SENP1 and SENP2 with depletion of SENP1 alone. Again, SENP1 depletion caused a reduction in c-FOS activation following PMA stimulation, but this effect was enhanced upon co-depletion of SENP2 (Figure 5E). In contrast, anisomycin-dependent c-FOS activation was unaffected by SENP1 depletion, either alone or in combination with SENP2 (Figure 5F). The lack of involvement of SENPs in c-FOS activation by anisomycin is consistent with the lack of requirement for SUMO loss in the activation of Elk-1 by this treatment.
These data therefore demonstrate that SENP1 is the predominant SENP acting to promote activation of the Elk-1 target gene c-FOS following mitogenic stimulation and, consistent with the reporter gene assays, there is a degree of functional redundancy with SENP2. In contrast, under signalling conditions where Elk-1 SUMOylation is not required (i.e. anisomycin treatment), SENP1 is not required for c-FOS induction.
DISCUSSION
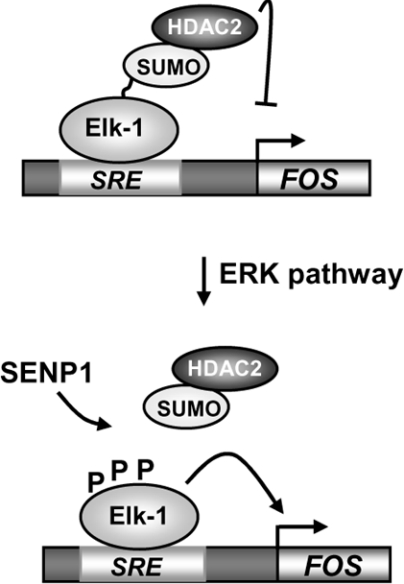
SUMOylation plays an important physiological role in determining the outcome of numerous processes in the cell. Importantly, SUMOylation is a dynamic process, enabling transient responses to be elicited, to processes such as cellular signalling events (reviewed in [7,21]). However, in most cases, it is unclear how SUMO conjugation is either promoted or subsequently lost in response to signalling cues. SUMO loss is promoted by the SENPs (reviewed in [5,6]). In the present study, we have focussed on Elk-1 and investigated the roles of SENPs in the de-SUMOylation process. Elk-1 makes an attractive model as SUMOylation promotes its transcriptional-repressive properties and, depending on the stimulus received, SUMOylation is either retained (upon stress signalling) or lost (upon mitogenic signalling). In the present study, we identified SENP1 as a key player in the loss of SUMO from Elk-1 in response to ERK pathway signalling (Figure 6).
Figure 6. Model for the role of SENP1 in Elk-1 regulation.
In the absence of ERK pathway signalling, Elk-1 is kept in a repressive state by SUMO-mediated HDAC-2 recruitment. Following ERK activation, Elk-1 is de-SUMOylated, leading to enhanced transactivation capacity, and SENP1 plays a pivotal role in this process. SRE, serum-response element.
Several lines of evidence establish SENP1 as the predominant SENP involved in Elk-1 de-SUMOylation: (i) SENP1 is the most efficient activator of Elk-1 in overexpression studies, (ii) Elk-1 binds preferentially to SENP1, (iii) SUMO-modified Elk-1 is a substrate for SENP1 in vitro, (iv) SENP1 depletion reduces the activation of Elk-1 by PMA, and (v) SENP1 depletion reduces the activation of the Elk-1 target gene c-FOS following mitogenic stimulation. Importantly, we compared the effect of several SENPs in each of the assays to demonstrate the specificity of SENP1 action. Moreover, we also verified that the effect of SENP1 was direct by determining the SUMO-dependence of its effects. SENP1-dependent activation of Elk-1 depended on its catalytic activity and the presence of SUMOylation sites in Elk-1 (Figure 1). Similarly, depletion of SENP1 only had an effect on Elk-1 activity when its SUMOylation sites were intact (Figure 4). Finally, SENP1 depletion only affected c-FOS activation under signalling conditions where SUMO loss is known to occur (Figure 5).
There are six known SENPs in humans and it is possible that some degree of functional redundancy occurs. In the present study, we only tested the effects of three different SENPs, although several assays were performed to test the role of SENP5, but this had little effect on the activity of Elk-1 or its target gene c-FOS (results not shown). It therefore remains possible that the additional SENPs SENP6 and SENP7 play a role in Elk-1 activation, maybe in a redundant manner. Nevertheless, it is clear that SENP1 plays a predominant role in Elk-1 de-SUMOylation. We do however observe functional redundancy in SENP-depletion assays where SENP2 specifically substitutes for SENP1 upon SENP loss (Figures 4 and 5). In this scenario though, the primary role of SENP1 is probably dictated due to its stronger binding to Elk-1 than SENP2 (Figure 2), thereby occluding SENP2 access to the SUMOylation sites. Only when SENP1 is lost will SENP2 be able to work on Elk-1.
Binding experiments demonstrated a specific interaction between SENP1 and Elk-1 (Figure 2). However, to perform such experiments in vivo, we had to use catalytically inactive versions of SENPs. The rationale behind this was that such mutants would stay associated for longer rather than merely transiently binding and dissociating following SUMO cleavage. Precedents for such behaviour exists in other enzyme–substrate systems where, for example, p38 MAPK binding to MEF2 (myocyte enhancer factor-2) could only be strongly detected using catalytically dead p38 MAPK [22]. Unfortunately, this observation precluded the possibility of detecting interactions between endogenous SENP1 and Elk-1, and probably will make any such substrate–SENP pairs difficult to detect at endogenous levels.
While SENP1 is the predominant SENP involved, overexpression of SENPs has various effects on Elk-1 activity (Figure 1). For example, SENP2 and SENP3 caused smaller, yet significant, increases in Elk-1 activity. This might merely be due to overexpression and displacement of endogenous SENP1, but might also have an important role. For example, SENP3 activates Elk-1 to a small extent, and this activation might be through p300 as this was recently shown to be a physiologically relevant SENP–substrate pairing [23]. In contrast, SENP2 can activate Elk-1, but this activation appears to be largely independent from its de-SUMOylation function as the catalytically dead version activates Elk-1 as efficiently as the WT version (Figure 2E). It is not clear what this de-SUMOylation-independent activity of SENP2 might be, although a similar phenomenon has previously been described for the SENP2-dependent activation of c-Jun [24]. The observation that SENP2 functions in a manner independent from its de-SUMOylation activity apparently contradicts its ability to substitute for SENP1 in regulating Elk-1 in a SUMO-site-dependent manner. However, it is possible that, in the case of overexpression, additional effects are triggered which would not been seen at endogenous levels, therefore interpretation of the results of these type of overexpression assays need to be treated with caution. Nevertheless, the assays do demonstrate a more potent activity of SENP1 towards Elk-1 as a SUMO protease, especially considering the alternative mode of action of SENP2 in these overexpression assays. Knockdown studies further support the role of SENP1 and more realistically delineate the potential functional redundancy of SENP2 at endogenous levels where its effects are, at least in part, elicited through its SUMO-protease activity.
Initially, it was thought that de-SUMOylation would be a fairly non-specific process. However, our demonstration that Elk-1 is preferentially targeted by SENP1 provides another member of a growing list of specific SENP–substrate interactions. Notable examples include SENP3 and p300 in response to mild oxidative stress [23], and the specific role of SENP1 in targeting HIF-1α (hypoxia-inducible factor-1α) [25] and HDAC (histone deacetylase)-1 [14]. At least part of the functional specificity of SENPs appears to result in their subnuclear localization, with SENP3 being localized to the nucleolus and thereby de-SUMOylating nucleolar substrates, such as nucleophosmin [26], although recent results demonstrate that nucleolar SENPs, such as SENP3, can be released into the nucleoplasm following mild oxidative stress [23]. In contrast, SENP1 and SENP2 are thought to be the major SENPs in the nucleoplasm, which is consistent with a role for SENP1 in controlling Elk-1 activity. It appears likely therefore that each SENP will have a specific spectrum of substrates, although some degree of functional redundancy is probably built in as a potential fail safe mechanism and subcellular localization will, to some degree, dictate the availability of substrates. It is, however, clear that the SENPs play a positive role in determining the dynamics and functional outcome of protein SUMOylation.
AUTHOR CONTRIBUTION
James Witty generated the data for Figures 1, 2(A), 2(B), 4 and 5; Elisa Aguilar-Martinez generated the data for Figures 2(C) and 3. James Witty and Elisa Aguilar-Martinez contributed to writing the paper. Andrew Sharrocks helped with experimental design and wrote the paper.
ACKNOWLEDGEMENTS
We thank Anne Clancy and Karren Palmer for excellent technical assistance, and Shen-Hsi Yang and members of our laboratory for comments on the manuscript and stimulating discussions. We thank Professor Ronald Hay (School of Life Sciences, University of Dundee, Dundee, Scotland, U.K.), Professor Víctor de Lorenzo (Centro Nacional de Biotecnologia CSIC, Madrid, Spain) and Edward Yeh for reagents.
FUNDING
This work was supported by the Biotechnology and Biological Sciences Research Council (a studentship to J.W.); the Wellcome Trust [grant number 078085]; and a Royal Society–Wolfson Award (to A.D.S.).
References
- 1.Garcia-Dominguez M., Reyes J. C. SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim. Biophys. Acta. 2009;1789:451–459. doi: 10.1016/j.bbagrm.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Geiss-Friedlander R., Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007;8:947–956.. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 3.Lyst M. J., Stancheva I. A role for SUMO modification in transcriptional repression and activation. Biochem. Soc. Trans. 2007;35:1389–1392. doi: 10.1042/BST0351389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rytinki M. M., Kaikkonen S., Pehkonen P., Jääskeläinen T., Palvimo J. J. PIAS proteins: pleiotropic interactors associated with SUMO. Cell Mol. Life Sci. 2009;66:3029–3041. doi: 10.1007/s00018-009-0061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J. H., Baek S. H. Emerging roles of desumoylating enzymes. Biochim. Biophys. Acta. 2009;1792:155–162. doi: 10.1016/j.bbadis.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Mukhopadhyay D., Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 2007;32:286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Guo B., Yang S. H., Witty J., Sharrocks A. D. Signalling pathways and the regulation of SUMO modification. Biochem. Soc. Trans. 2007;35:1414–1418. doi: 10.1042/BST0351414. [DOI] [PubMed] [Google Scholar]
- 8.Hietakangas V., Ahlskog J. K., Jakobsson A. M., Hellesuo M., Sahlberg N. M., Holmberg C. I., Mikhailov A., Palvimo J. J., Pirkkala L., Sistonen L. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol. Cell. Biol. 2003;23:2953–2968. doi: 10.1128/MCB.23.8.2953-2968.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X. J., Grégoire S. A recurrent phospho-sumoyl switch in transcriptional repression and beyond. Mol. Cell. 2006;23:779–786. doi: 10.1016/j.molcel.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Anckar J., Sistonen L. SUMO: getting it on. Biochem. Soc. Trans. 2007;35:1409–1413. doi: 10.1042/BST0351409. [DOI] [PubMed] [Google Scholar]
- 11.Yang S. H., Jaffray E., Hay R. T., Sharrocks A. D. Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol. Cell. 2003;12:63–74. doi: 10.1016/s1097-2765(03)00265-x. [DOI] [PubMed] [Google Scholar]
- 12.Yang S. H., Sharrocks A. D. PIASxα differentially regulates the amplitudes of transcriptional responses following activation of the ERK and p38 MAPK pathways. Mol. Cell. 2006;22:477–487. doi: 10.1016/j.molcel.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 13.Tatham M. H., Jaffray E., Vaughan O. A., Desterro J. M., Botting C. H., Naismith J. H., Hay R. T. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 14.Cheng J., Wang D., Wang Z., Yeh E. T. SENP1 enhances androgen receptor-dependent transcription through desumoylation of histone deacetylase 1. Mol. Cell. Biol. 2004;24:6021–6028. doi: 10.1128/MCB.24.13.6021-6028.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong L., Yeh E. T. Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J. Biol. Chem. 2006;281:15869–15877. doi: 10.1074/jbc.M511658200. [DOI] [PubMed] [Google Scholar]
- 16.Yang S. H., Yates P. R., Whitmarsh A. J., Davis R. J., Sharrocks A. D. The Elk-1 ETS-domain transcription factor contains a mitogen-activated protein kinase targeting motif. Mol. Cell. Biol. 1998;18:710–720. doi: 10.1128/mcb.18.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mencía M., de Lorenzo V. Functional transplantation of the sumoylation machinery into Escherichia coli. Protein Expr. Purif. 2004;37:409–418. doi: 10.1016/j.pep.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Reverter D., Lima D. C. Preparation of SUMO proteases and kinetic analysis using endogenous substrates. Methods Mol. Biol. 2009;497:225–239. doi: 10.1007/978-1-59745-566-4_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tatham M. H., Geoffroy M. C., Shen L., Plechanovova A., Hattersley N., Jaffray E. G., Palvino J. J., Hay R. T. RNF is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 20.Yang S. H., Sharrocks A. D. SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell. 2004;13:611–617. doi: 10.1016/s1097-2765(04)00060-7. [DOI] [PubMed] [Google Scholar]
- 21.Liu B., Shuai K. Regulation of the sumoylation system in gene expression. Curr. Opin. Cell Biol. 2008;20:288–293. doi: 10.1016/j.ceb.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han J., Jiang Y., Li Z., Kravchenko V. V., Ulevitch R. J. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 23.Huang C., Han Y., Wang Y., Sun X., Yan S., Yeh E. T., Chen Y., Cang H., Li H., Shi G., et al. SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation. EMBO J. 2009;28:2748–2762. doi: 10.1038/emboj.2009.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Best J., Ganiatsas S., Agarwal S., Changou A., Salomoni P., Shirihai O., Meluh P., Pandolfi P., Zon L. SUMO-1 protease-1 regulates gene transcription through PML. Mol. Cell. 2002;10:843–855. doi: 10.1016/s1097-2765(02)00699-8. [DOI] [PubMed] [Google Scholar]
- 25.Cheng J., Kang X., Zhang S., Yeh E. T. SUMO-specific protease 1 is essential for stabilization of HIF1α during hypoxia. Cell. 2007;131:584–595. doi: 10.1016/j.cell.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haindl M., Harasim T., Eick D., Muller S. The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep. 2008;9:273–279. doi: 10.1038/embor.2008.3. [DOI] [PMC free article] [PubMed] [Google Scholar]