Abstract
Concordance of MLL-rearranged acute leukemia in infant monozygotic twins is thought to be 100% with a very short latency period, suggesting that either the MLL fusion itself is sufficient to cause leukemia or that it promotes the rapid acquisition of additional oncogenic events that result in overt disease. We report the first case of discordance in an infant monozygotic twin pair. Twin A presented at age 9 months with MLL-ENL+ acute lymphoblastic leukemia and twin B remains healthy 3 years later. The presence and eventual clearance of a clonal population of MLL-ENL+ cells was shown in the bone marrow and peripheral blood of twin B. Clearance of this clone was temporally associated with viral-induced cytopenias, suggesting an immune-mediated clearance of the clone before the development of leukemia. Thus, concordance of MLL-rearranged acute leukemia in infant monozygotic twins is not universal. The implications of this case for MLL-rearranged leukemogenesis are discussed.
Introduction
Reported concordance rates for acute leukemia in monozygotic twins range from less than 5% to 100%, depending on the age of twin A at diagnosis and the cytogenetics of the leukemia.1 Many reports of twin concordance have documented identical, clonal, nonconstitutional fusion sequences in both twins, suggesting that in these cases a preleukemic clone developed during the prenatal period in one twin and was transferred to the other by placental anastomoses.2,3
Up to 80% of infant leukemias possess rearrangement of the MLL gene at 11q23, most commonly fused with the AF4, ENL, or AF9 genes.4 These leukemias have a very short latency period, with diagnosis occurring at an average age of 6 months. Although the number of reported cases is small, the concordance rate for infant monozygotic twins with MLL-rearranged (MLL-R) leukemia is thought to be 100%.1 The rapid transition to leukemia (which is also seen in therapy-associated MLL-R leukemias) and high twin concordance have led some to speculate that MLL fusions alone are sufficient for the development of leukemia. This view is supported by recent studies showing a striking paucity of copy number alterations (CNAs) in cases of MLL-R acute lymphoblastic leukemia (ALL) compared with other leukemias.5 However, data from MLL-R transgenic murine models strongly suggest that additional events are required for the development of leukemia.6,7 Others have suggested that MLL fusions may promote genetic instability, rapid clonal expansion, epigenetic events, or defective DNA damage repair, leading to the rapid development of disease.1 It is possible that such events are undetectable at the resolution of the CNA studies.
The concordance rate for leukemia in monozygotic twins in childhood (index case older than 1 year) is much lower (approximately 10%).1 There is often a long latency period, as seen in a set of twins with ETV6/RUNX1 ALL diagnosed at 5 and 14 years of age. Retrospective analysis of bone marrow from the second twin showed the presence of clonal ETV6/RUNX1+ cells at the time of diagnosis of the sibling 9 years earlier.8
Many childhood leukemias are thought to initiate prenatally. Retrospective analyses of neonatal blood spots from children diagnosed with leukemia years later have shown the presence of cells with identical clonal fusion sequences. This phenomenon is especially common in younger children with a t(12;21) ETV6/RUNX1 translocation or hyperdiploidy and is distinctly uncommon in cases in older children or those with a t(1;19) E2A/PBX1 translocation.9–13 Further studies have established that the detection of leukemia-related translocations at birth does not inevitably result in leukemia. In a screen of unselected cord blood samples, 1% of the samples were positive for the ETV6/RUNX1 fusion gene, approximately 100 times the frequency of overt disease.14 The lower twin concordance rate, variable latency period, and excessive rate of ETV6/RUNX1 translocations in unaffected persons suggest that additional genetic alterations are required for the development of these leukemias. This is further supported by the recent discovery of more than 6 somatic CNAs per case in childhood ALL.5
We report a case of discordance in an infant monozygotic twin pair in which twin A presented with MLL-ENL+ ALL and twin B, who was transiently positive for MLL-ENL by reverse transcriptase–polymerase chain reaction (RT-PCR) in bone marrow (BM) and peripheral blood (PB), remains healthy more than 3 years later.
Study design
Patient chronology
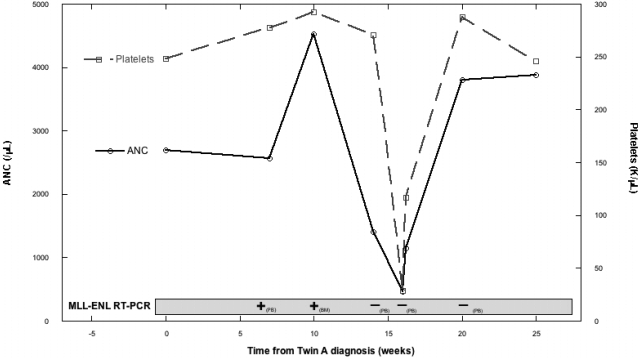
Twin A was diagnosed at age 9 months (T = 0) with CD10(-) B-precursor ALL, cytogenetics t(11;19)(q23;p13.3), fluorescence in situ hybridization (FISH) positive for MLL-R, and RT-PCR positive for MLL-ENL fusion transcript. Twin B was healthy with normal complete blood count (CBC) and physical examination (PE). At T +7weeks, twin B's CBC and PE remained normal, but RT-PCR on peripheral blood mononuclear cells (PBMNCs) was MLL-ENL positive. Three weeks later (T = +10 weeks), BM aspirate was also RT-PCR positive for MLL-ENL. Morphology, FISH, and cytogenetics were normal, as were CBC and PE. Two weeks later (T = +12 weeks), twin B developed neutropenia (white blood cell count = 2.88 × 109/L; absolute neutrophil count [ANC] = 0.46 × 109/L) and thrombocytopenia (platelets = 29 × 109/L) 1 week after a viral upper respiratory infection consisting of fever, rhinorrhea, cough, and a diffuse erythematous rash. Review of PB smear confirmed the cytopenias and showed atypical lymphocytosis and activated monocytes compatible with a viral process, but no lymphoblasts. Large platelets were noted. The constellation of clinical and laboratory findings led to a diagnosis of immune-mediated cytopenias. No treatment was given. Repeat CBC 2 days later showed recovery with white blood cell count of 6.39 × 109/L (ANC = 1.15 × 109/L) and platelet count of 117 × 109/L. RT-PCR of PBMNCs was negative for MLL-ENL. One week later, CBC had normalized (Figure 1). Over the next 4 months, CBC and PE were normal, and RT-PCR of PBMNCs was negative. Twin B remains healthy at T of +3 years.
Figure 1.
Twin B's ANC, platelet count, and MLL-ENL RT-PCR status over time from twin A's diagnosis. ANC indicates absolute neutrophil count; PB, peripheral blood; and BM, bone marrow.
Twin A was treated according to the Children's Oncology Group protocol P9407 followed by an additional year of maintenance therapy. End-induction BM showed morphologic remission, normal cytogenetics, and negative FISH, although RT-PCR remained positive for MLL-ENL. End-consolidation BM showed continued remission and was negative for MLL-ENL by cytogenetics, FISH, and RT-PCR. Two months after completion of therapy (26 months from diagnosis), twin A experienced an isolated testicular relapse of CD10(-) B-precursor MLL-ENL+ ALL. He received reinduction chemotherapy and testicular radiation and is currently receiving postinduction chemotherapy.
Laboratory methods
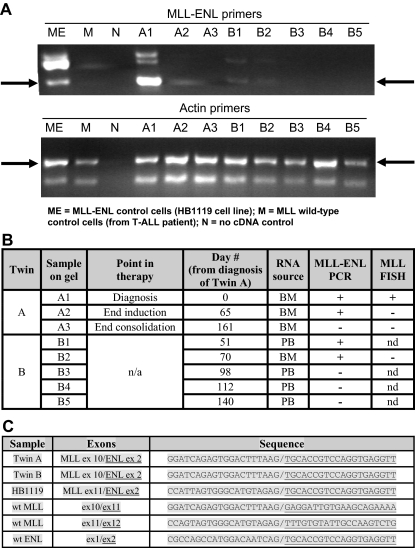
BM and PB samples were collected under Johns Hopkins University's Institutional Review Board–approved cell procurement protocols after informed consent was obtained in accordance with the Declaration of Helsinki. To avoid cross-contamination, all samples and intermediate products from the 2 twins were stored in separate locations and were manipulated at different times and in different areas of the laboratory. The only exception was after the addition of sample buffer, PCR products were loaded together on the same gel for the purpose of creating Figure 2A. Mononuclear cells were enriched by Ficoll–Hypaque centrifugation. Total RNA was extracted with QIAGEN RNeasy kits. RNA samples were reverse transcribed with random hexamer primers to generate cDNA. The cDNA corresponding to the MLL-ENL fusion and actin transcripts were amplified by PCR with the following primers: MLL-ENL(F), 5′-CGCCCAAGTATCCCTGTAAA; MLL-ENL(R), 5′-GCGTACCCCGACTCCTCTAC; ACTIN(F), 5′-CGCGAGAAGATGACCCAGAT; and ACTIN(R), 5′-ACTCCATCCCCAGGAAGG.
Figure 2.
Laboratory results. (A) MLL-ENL RT-PCR for twins A and B with actin control. (B) Summary of MLL-ENL RT-PCR and MLL FISH results for twins A and B; BM indicates bone marrow; PB, peripheral blood; n/a, not applicable; nd, not done. (C) Sequencing results (wt MLL and ENL sequences included as reference).
Samples were amplified with the following PCR conditions: 94°C for 3 minutes; 35 cycles of 94°C for 45 seconds, 52°C for 30 seconds, 72°C for 1 minute, and 72°C for 10 minutes. PCR products were loaded onto an agarose gel and electrophoresed. Resolved PCR products were stained with ethidium bromide and visualized with the use of a UV transilluminator. For all MLL-ENL reactions, purified PCR products were directly sequenced at the Genetic Resource Core Facility of Johns Hopkins University with the use of internal forward and reverse primers. Resultant sequences were analyzed with Sequencher 4.6 software (Gene Codes).
Results and discussion
MLL-ENL RT-PCR and MLL FISH results from twins A and B are shown in Figure 2A and 2B. Direct sequencing performed on the PCR products (Figure 2C) confirmed that the MLL-ENL transcript isolated from both twins had the identical sequence, fusing exon 10 of MLL to exon 2 of ENL. Simultaneous sequencing of PCR products from the cell line HB1119 confirmed the presence of a different transcript sequence, fusing exon 11 of MLL to exon 2 of ENL. Therefore, the MLL-ENL+ cells in both twins were derived from a single clone.
To our knowledge, this is the first report of discordant MLL-R leukemia in infant monozygotic twins. This is also the first report showing the presence and eventual clearance of a clonally derived, leukemia-associated fusion sequence–positive population of cells in a healthy twin B. There are 3 other reports of twins discordant for MLL leukemia, but placental status was either unknown or dichorionic, or the leukemia developed in the first twin at 5 years of age and was probably postnatally acquired.1 This case confirms prior reports that clonal MLL-R cells are shared between monozygotic twins; however, it presents evidence against the hypothesis that the presence of an MLL rearrangement (specifically an MLL-ENL fusion) in hematopoietic cells invariably leads to the development of leukemia.
It is thought that abnormal immune responses to common childhood infections may play a role in the development of childhood leukemia and that early infectious exposures might provide some protection from the development of disease.11,15–17 The temporal association of the disappearance of twin B's MLL-ENL+ clone with an episode of presumably viral-induced thrombocytopenia and neutropenia suggests the possibility that the immune process that caused the cytopenias may have been responsible for clearance of the MLL-ENL+ clone. There is also a report proposing that endogenous serum cortisol levels achieved during an infectious process can approximate the levels of glucocorticosteroids used in the treatment of ALL and could lead to the clearance of preleukemic clones in young children.18 It remains possible that twin B's MLL-ENL+ clone persists in a dormant state at a level beneath the sensitivity of our RT-PCR assay. There have been case reports of later onset childhood MLL-R leukemia and one report of a 6-year-old with MLL-R leukemia who had a positive blood spot at birth.19 What does seem clear is that short latency concordance of infant MLL-R leukemia in monozygotic twins is not universal.
Acknowledgments
This work was supported by grants from the NCI (K23 CA111728, P.B.), Damon Runyon-Lilly Clinical Investigator Award (New York, NY; 30-06, P.B.), the Leukemia & Lymphoma Society (White Plains, NY; SCOR 7372-07, P.B.) and the Children's Cancer Foundation (Owings Mills, MD; P.B.).
The publisher or recipient acknowledges the right of the US government to retain a nonexclusive, royalty-free license in and to any copyright covering the article. The views expressed in this paper do not necessarily reflect the views of the National Institutes of Health (NIH) or of the US government.
Footnotes
Presented in abstract/poster form at the 50th Annual Meeting of the American Society of Hematology, San Francisco, CA, December 7, 2008.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.K.C. wrote the paper; E.M. performed experiments; D.S. supervised experiments; and P.B. designed experiments, performed experiments, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Patrick Brown, Johns Hopkins Oncology, CRB1 2M49, 1650 Orleans St, Baltimore, MD 21231; e-mail: pbrown2@jhmi.edu.
References
- 1.Greaves MF, Maia AT, Wiemels JL, Ford AM. Leukemia in twins: lessons in natural history. Blood. 2003;102:2321–2333. doi: 10.1182/blood-2002-12-3817. [DOI] [PubMed] [Google Scholar]
- 2.Ford AM, Ridge SA, Cabrera ME, et al. In utero rearrangements in the trithorax-related oncogene in infant leukaemias. Nature. 1993;363:358–360. doi: 10.1038/363358a0. [DOI] [PubMed] [Google Scholar]
- 3.Gill Super HJ, Rothberg PG, Kobayashi H, Freeman AI, Diaz MO, Rowley JD. Clonal, nonconstitutional rearrangements of the MLL gene in infant twins with acute lymphoblastic leukemia: in utero chromosome rearrangement of 11q23. Blood. 1994;83:641–644. [PubMed] [Google Scholar]
- 4.Jansen MW, Corral L, van der Velden VH, et al. Immunobiological diversity in infant acute lymphoblastic leukemia is related to the occurrence and type of MLL gene rearrangement. Leukemia. 2007;21:633–641. doi: 10.1038/sj.leu.2404578. [DOI] [PubMed] [Google Scholar]
- 5.Mullighan CG, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 6.Lavau C, Luo RT, Du C, Thirman MJ. Retrovirus-mediated gene transfer of MLL-ELL transforms primary myeloid progenitors and causes acute myeloid leukemias in mice. Proc Natl Acad Sci U S A. 2000;97:10984–10989. doi: 10.1073/pnas.190167297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobson CL, Warren AJ, Pannell R, et al. The mll-AF9 gene fusion in mice controls myeloproliferation and specifies acute myeloid leukaemogenesis. EMBO J. 1999;18:3564–3574. doi: 10.1093/emboj/18.13.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiemels JL, Ford AM, Van Wering ER, Postma A, Greaves M. Protracted and variable latency of acute lymphoblastic leukemia after TEL-AML1 gene fusion in utero. Blood. 1999;94:1057–1062. [PubMed] [Google Scholar]
- 9.Gale KB, Ford AM, Repp R, et al. Backtracking leukemia to birth: identification of clonotypic gene fusion sequences in neonatal blood spots. Proc Natl Acad Sci U S A. 1997;94:13950–13954. doi: 10.1073/pnas.94.25.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiemels JL, Cazzaniga G, Daniotti M, et al. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet. 1999;354:1499–1503. doi: 10.1016/s0140-6736(99)09403-9. [DOI] [PubMed] [Google Scholar]
- 11.Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nat Rev Cancer. 2003;3:639–649. doi: 10.1038/nrc1164. [DOI] [PubMed] [Google Scholar]
- 12.Taub JW, Konrad MA, Ge Y, et al. High frequency of leukemic clones in newborn screening blood samples of children with B-precursor acute lymphoblastic leukemia. Blood. 2002;99:2992–2996. doi: 10.1182/blood.v99.8.2992. [DOI] [PubMed] [Google Scholar]
- 13.Wiemels JL, Leonard BC, Wang Y, et al. Site-specific translocation and evidence of postnatal origin of the t(1;19) E2A-PBX1 fusion in childhood acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2002;99:15101–15106. doi: 10.1073/pnas.222481199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mori H, Colman SM, Xiao Z, et al. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proc Natl Acad Sci U S A. 2002;99:8242–8247. doi: 10.1073/pnas.112218799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greaves MF. Speculations on the cause of childhood acute lymphoblastic leukemia. Leukemia. 1988;2:120–125. [PubMed] [Google Scholar]
- 16.Kinlen L. Evidence for an infective cause of childhood leukaemia: comparison of a Scottish new town with nuclear reprocessing sites in Britain. Lancet. 1988;2:1323–1327. doi: 10.1016/s0140-6736(88)90867-7. [DOI] [PubMed] [Google Scholar]
- 17.Greaves M. Infection, immune responses and the aetiology of childhood leukaemia. Nat Rev Cancer. 2006;6:193–203. doi: 10.1038/nrc1816. [DOI] [PubMed] [Google Scholar]
- 18.Schmiegelow K, Vestergaard T, Nielsen SM, Hjalgrim H. Etiology of common childhood acute lymphoblastic leukemia: the adrenal hypothesis. Leukemia. 2008;22:2137–2141. doi: 10.1038/leu.2008.212. [DOI] [PubMed] [Google Scholar]
- 19.Maia AT, Koechling J, Corbett R, Metzler M, Wiemels JL, Greaves M. Protracted post-natal natural histories in childhood leukemia. Genes Chromosomes Cancer. 2004;39:335–340. doi: 10.1002/gcc.20003. [DOI] [PubMed] [Google Scholar]