Abstract
Recent advances in optical imaging and molecular manipulation techniques have made it possible to observe the activity of individual enzymes and study the dynamic properties of processes that are challenging to elucidate using ensemble-averaging techniques. The use of single-molecule approaches has proven to be particularly successful in the study of the dynamic interactions between the components at the replication fork. In this section, we describe the methods necessary for in vitro single-molecule studies of prokaryotic replication systems. Through these experiments, accurate information can be obtained on the rates and processivities of DNA unwinding and polymerization. The ability to monitor in real time the progress of a single replication fork allows for the detection of short-lived, intermediate states that would be difficult to visualize in bulk-phase assays.
Keywords: Single-molecule detection, E. coli replication, Bacteriophage T7 replication
1. Introduction
DNA replication involves the coordinated activity of a large number of proteins. The replisome, the molecular machinery of DNA replication, unwinds the double-stranded DNA, provides primers to initiate synthesis, and polymerizes nucleotides onto each of the two growing strands (1). Remarkable progress has been made in characterizing the structural and functional properties of the individual components; their coordination at the replication fork is less well understood.
The dynamic nature of the replisome makes it hard to probe its coordination with ensemble-averaging techniques. We describe here single-molecule techniques to observe, in real time, the replication of individual DNA molecules by replication complexes of E. coli and the bacteriophage T7. Replication reactions from both these systems can be reconstituted in vitro with a relatively small number of proteins and are compatible with the optical imaging and manipulation techniques developed to study DNA-protein interactions at the single-molecule level (2–4).
In this chapter, we describe how individual DNA molecules can be mechanically stretched and their lengths used as a probe for enzymatic activity at the replication fork. Linearized λ DNA is modified to have a biotin on one end and a digoxigenin moiety on the other. The biotinylated end is attached to a functionalized glass coverslip and the digoxigeninated end to a small bead (Figure 1). The assembly of these DNA-bead tethers on the surface of a flow cell allows a laminar flow to be applied and a drag force on the bead to be exerted. As a result, the DNA is stretched close to and parallel to the surface of the coverslip at a force that is determined by the flow rate. The length of the DNA is measured by monitoring the position of the bead. Length differences between single- and double-stranded DNA are utilized to obtain real-time information on the activity of the replication proteins at the fork (2–4) (Figure 2).
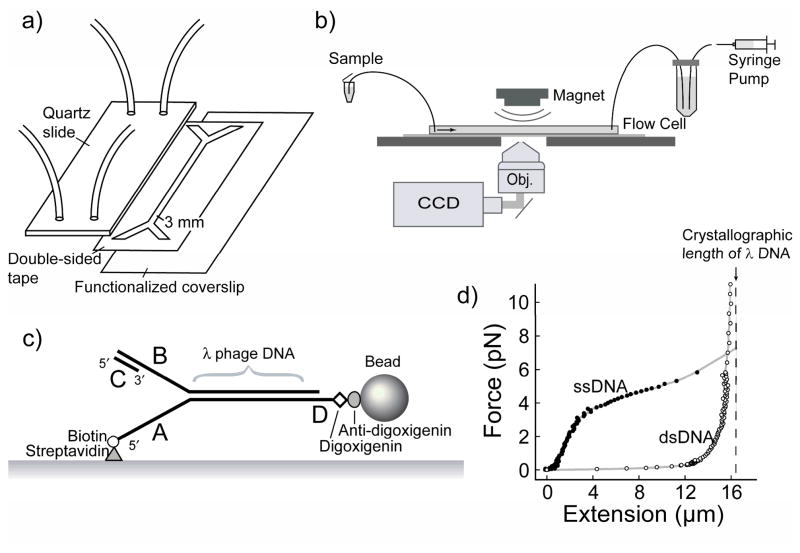
Figure 1.
Single-molecule experimental setup. (a) Flow cell is constructed by cutting a forked, 3 mm-wide channel out of a nonreactive double-sided adhesive ~100 μm thick. The channel is affixed to a quartz slide and to the functionalized coverslip. Tubing is inserted through holes drilled in the quartz and bonded with epoxy, creating a sealed chamber that is used for the single-molecule experiments (b) Bead-DNA assemblies are stretched using laminar flow of buffer and imaged using wide-field optical microscopy. By tracking the positions of the beads over time, while maintaining a constant stretching force, the lengths of the DNA constructs can be monitored. (c) Duplex λ DNA (48.5 kb) is attached to the surface of the flow cell via the 5′ end of the fork using a biotin-streptavidin interaction, and the 3′ end is attached to a bead using a digoxigenin anti-digoxigenin interaction. A primed replication fork is formed at the end opposite the bead to allow loading of the replication proteins. (d) Extension profile of ssDNA (filled circle) and dsDNA (open circle) under low forces. Dashed line shows crystallographic length of fully ds-λ DNA, 16.3 μm. The large difference in length between ssDNA and dsDNA at forces around 2 pN allows a direct observation of conversions between ss- and dsDNA by monitoring changes in the DNA length. The simultaneous visualization of large numbers of DNA-coupled beads allows for the study of many individual replisomes in one experiment. Figure adapted from Tanner et al(4).
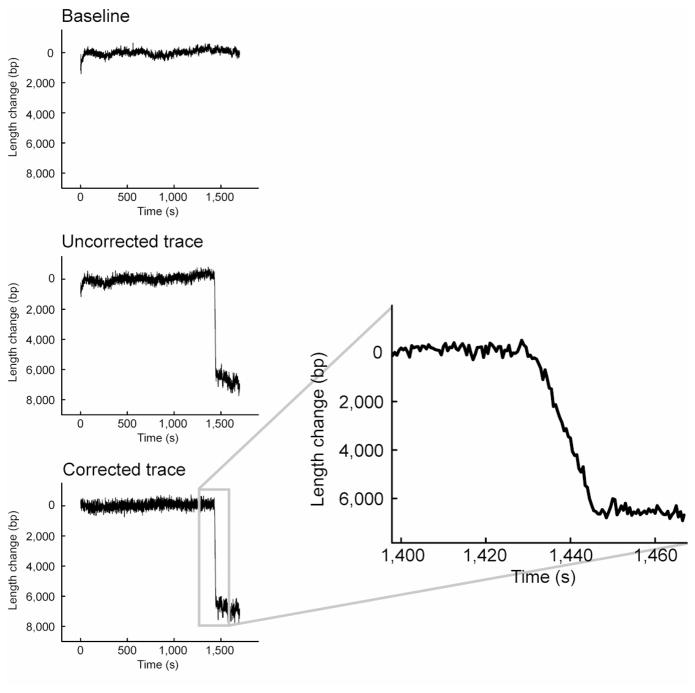
Figure 2.
Example of typical data from a single-molecule leading-strand synthesis experiment. DNA length baseline values are determined from a tethered DNA that is not enzymatically altered. This baseline trace is subtracted from altered substrates to remove global flow instabilities from the data. The inset shows a zoomed view of the length change, which appears instantaneous in the overall view (left) due to the compressed time axis, but in fact represents a shortening over many seconds. Figure from Tanner et al(4).
2. Materials
2.1 Surface Functionalization
Glass coverslips, staining jars.
Ethanol, absolute anhydrous.
1M potassium hydroxide.
Acetone.
3–aminopropyltriethoxysilane.
Functionalized PEGs, here succinimidyl propionate-PEG (M-SPA-5000) and Biotin-PEG-CO2NHS-5000 (Nektar™).
PEG coupling buffer: 100 mM NaHCO3, pH 8.2.
Oven (110 °C).
Bath sonicator.
Compressed N or Ar gas.
2.2 DNA Substrate Preparation
Bacteriophage λ DNA (14 nM stock; New England Biolabs; see Note 1).
Oligonucleotides (100 μM stocks, see Fig 1A): biotinylated fork arm (A: 5′-biotin-AAAAAAAAAAAAAAAAGAGTACTGTACGATCTAGCATCAATCA CAGGGTCAGGTTCGTTATTGTCCAACTTGCTGTCC-3′); λ-complementary fork arm (B: 5′-GGGCGGCGACCTGGACAGCAAGTTGGACAATCTCGTTC TATCACT AATTCACTAATGCAGGGAGGATTTCAGATATGGCA-3′); fork primer (C: 5′-TGCCATATCTGAAATCCTCCCTGC-3′); λ-complementary digoxigenin end (D: 5′-AGGTCGCCGCCCAAAAAAAAAAAA-digoxigenin-3′).
T4 DNA Ligase (New England Biolabs).
T4 Polynucleotide Kinase (New England Biolabs).
Ligase/Kinase Buffer (10X): 500 mM Tris-HCl pH 7.5, 100 mM DTT, 100 mM MgCl2, 10 mM ATP.
Heat block (up to 65 °C).
2.3 Bead Functionalization
Paramagnetic beads (described are Dynal 142.03, tosyl activated, 2.8 μm diameter, 2×109 beads/mL).
Antibody solution (here, α-digoxigenin Fab 1 mg/ml, Invitrogen).
Buffer A: 0.1 M H3BO3, pH 9.5.
Buffer B: 1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na-phosphate), pH 7.4, 0.1% w/v BSA.
Buffer C: 0.2 M Tris-HCl, pH 8.5, 0.1% w/v BSA.
Magnetic Separator (e.g. Dynal MPC).
Rotator.
2.4 Experimental Setup
Functionalized coverslip (see Subheading 3.1).
2 × 5 cm quartz slide with drilled holes for outlet/inlet tubes.
Double-sided tape (Secure-Seal, Grace BioLabs).
Tubing, Intramedic™ PE60 (polyethylene 0.76 mm inner diameter, 1.22 mm outer diameter).
Quick-dry epoxy.
Blocking buffer (5X): 20 mM Tris-HCl pH 7.5, 2 mM EDTA, 50 mM NaCl, 1 mg/mL BSA, 0.025% Tween-20.
Working buffer (1X): 20 mM Tris-HCl pH 7.5, 2 mM EDTA, 50 mM NaCl.
Streptavidin (Sigma S4762), 1 mg/mL in PBS pH 7.3.
Permanent rare-earth magnets (National Imports).
Inverted optical microscope (Olympus IX-51) with 10X objective.
Syringe pump (Harvard Apparatus 11 Plus).
Vacuum desiccator
5 mL syringe with 21-gauge needle
2.5 Experiment and Data Analysis
T7 Replication Buffer 1: 40 mM Tris-HCl pH 7.5, 50 mM K-glutamate, 2 mM EDTA, 100 μg/ml BSA, 5 mM DTT, 760 μM dNTPs
T7 Replication Buffer 2: 40 mM Tris-HCl pH 7.5, 50 mM K-glutamate, 10 mM MgCl2, 100 μg/ml BSA, 5 mM DTT, 760 μM dNTPs (add 200 μM rNTPs to support primase activity for lagging-strand synthesis)
Purified components of T7 replisome: helicase, gp4; polymerase, gp5-thioredoxin complex; single-stranded DNA binding protein, gp2.5 for coordinated replication.
E. coli Replication Buffer: 50 mM HEPES-KOH pH 7.9, 80 mM KCl, 12 mM Mg-Acetate, 100 μg/ml BSA, 2.5 mM DTT, 1 mM ATP, 760 μM dNTPs (add 200 μM UTP, CTP, GTP to support primase activity for lagging-strand synthesis)
Purified components of E. coli replisome: helicase, DnaB6; helicase loader, DnaC6; core polymerase, αεθ processivity clamp, β2; clamp loader, τ1γ2δδ′ (or other stoichiometry).
CCD camera (Q-Imaging Rolera Fast), computer and imaging software (MetaVue™; Molecular Devices)
Particle tracking software (DiaTrack, Semasopht)
3. Methods
3.1 Surface Functionalization
To minimize nonspecific interactions between the glass surface and proteins, we covalently couple high-molecular-weight polyethylene glycol (PEG) to the surface. First, the glass is coupled to the alkoxy group of an aminosilane, creating a surface with reactive amine groups that can subsequently be coated with a polymer of choice (5). Here, we describe how a mixed population of biotinylated and non-biotinylated succinimidyl propionate-PEG is coupled to the amine-functionalized glass, coating the coverslip in a layer of PEG displaying a mixture of biotin and nonreactive methyl groups. The biotin is used to tightly bind streptavidin, allowing for a subsequent coupling of biotinylated DNA to the surface. Any functionalized PEG can be used to allow a customizable surface based on choice of DNA modification.
Clean glass coverslips (in a staining jar or equivalent) by sonicating for 30 min in ethanol (EtOH), followed by 30 min sonication in 1M KOH. Repeat both steps, rinsing in Millipore water between each step. After cleaning, it is essential to remove all traces of water from the coverslips and their containers, as the silanization reagent in the next step will hydrolyze rapidly in the presence of water. First decant all water from the container and wash 2–3 times with acetone, sonicating for 10 min at the last wash. Dry the exterior of the containers thoroughly with a towel, rinse the coverslips in acetone again and decant for next step.
Prepare a 2% (or other amount based on desired surface group density) 3-aminopropyltriethoxysilane solution in acetone and pour into containers. Vigorously agitate the containers either manually or on a rocker for 2–3 min and quench the reaction by addition of a large excess of water (10–15 volumes) to the container, simply by pouring water directly into the container for rapid solvent exchange. Rinse coverslips 2–3 times in water. Place coverslips onto foil or baking sheet and bake at 110°C for 30 min to cure the silane and dry the coverslips for addition of PEG.
Remove functionalized PEG from freezer and allow to warm to room temperature. Mix the methylated (M-SPA-5000) and biotinylated (Biotin-PEG-CO2NHS-5000) PEG at a ratio of ~50–100:1 in PEG coupling buffer to a final concentration of 0.2% w/v biotinylated PEG (for example, 150 mg M-SPA-5000 and 2 mg Biotin-PEG-CO2NHS-5000 in 1 mL buffer). This ratio can be adjusted to achieve an optimal density of functional groups in the flow cell based on the number of DNA molecules desired. Be sure to pipette the mixed PEG solution quickly, as hydrolysis will occur on a timescale of min (see Note 2). Apply 100 μL of the solution to the silanized surface, using the silanized side facing up. Take another baked coverslip and place it silanized side down on top of the slip with the PEG solution, making a slip-solution-slip sandwich and incubate for three h at room temperature. Separating the slips is easier if a nonfunctionalized coverslip is placed along the edge between the two silanized coverslips, allowing for the two sandwiched slips to be more easily separated later. Once the incubation is complete, peel the slips apart, rinse at least 6 times in water and dry under a flow of dry nitrogen gas. The dry slips can be stored at room temperature under vacuum for at least one week without loss of quality.
3.2 DNA Substrate Preparation
Bacteriophage λ DNA is 48.5 kb of double-stranded DNA readily purchased from suppliers, providing an ideal scaffold for single-molecule DNA manipulation. The linearized DNA has 12-base single-stranded overhangs at each end, to which we attach modified and unmodified oligonucleotides using standard annealing and ligating techniques. The following steps describe in detail how to prepare a DNA substrate with a primed replication fork at the surface-attached end and a site for bead attachment at the other (Figure 1). The following protocol will result in 0.5 mL of DNA substrate at a concentration of 1.4 nM.
Phosphorylate the 5′ ends of oligos A, λ-complementary fork arm, and D, λ-complementary digoxigenin end. For each of the two oligos, add 2.0 μL of oligonucleotide (100 μM stock), 15.5 μL of H2O, 2 μL of 10X ligase/kinase buffer, and 0.5 μL of T4 Polynucleotide Kinase (PNK) and incubate at 37°C for 1 hour. This procedure yields a final concentration of 10 μM phosphorylated oligo for the next steps.
Next, the fork oligos (A, B, and C) are annealed to the λ DNA in one step. Fork arm (A) anneals directly to its complementary end of the λ DNA, fork arm (B) anneals to its complementary sequence on arm (A) and primer (C) anneals to the end of arm (A) (See Figure 1c). Mix 51 μL of 10× ligase/kinase buffer in 400 μL H2O, then add 56 μL of λ DNA (14 nM stock), 1.0 μL of oligo A (from 10 μM solution), 1.0 μL of oligo B (from 10 μM solution), and 2.0 μL of oligo C (from 10 μM solution). This provides a 10X excess of fork oligos (and 10:1 primer: fork) to ensure that all λ DNA molecules are annealed to a fork and all forks are primed. To anneal, incubate at 65°C for 5 min and allow to cool to room temperature slowly by simply turning the power of the heat block off. Allow at least 30 min for the solution to gradually cool down. The resultant nicks between the oligos and the λ DNA are ligated by addition of 2.0 μL of T4 DNA ligase and incubating at room temperature for at least 2 h.
Finally, the digoxigenin end oligo (D) is annealed to the end of the λ DNA opposite the fork by adding 10 μL of phosphorylated (D) from the first step (~100× excess with respect to λ DNA). Incubate at 45°C for 30 min and cool to room temperature slowly by turning off heat block. Ligate the digoxigeninated oligo to the λ DNA by adding 2 μL of T4 Ligase and incubating at room temperature for at least 2 h. The final DNA construct is now ready for use at a concentration of 1.4 nM.
3.3 Bead Functionalization
In the flow-stretching single-molecule experiment, we measure DNA length change by observation of the position of a small bead bound to the end of the λ DNA. To achieve this, the beads are functionalized with a Fab fragment with specificity for the digoxigenin. Activated beads can then be attached to tethered DNA and used to manipulate the DNA. The following protocol will result in 1.0 mL of 1–2×109 beads/mL functionalized beads.
Resuspend the stock beads and transfer 0.4 mL to a 1.7 mL tube. Place tube into slot of magnet until solution clears, then remove supernatant by pipetting. Add 1.0 mL buffer A, which will activate the tosyl groups for antibody coupling. Mix gently by pipetting, clear solution using magnet, and remove buffer.
Add 0.4 mL of buffer A and 240 μL of Fab solution (1.0 mg/mL, gives 20 μg Fab/mg beads). Resuspend thoroughly, and incubate 16–24 h at 37°C using rotator.
After incubation, pull down beads using magnet and remove buffer. Add 1.0 mL of buffer B and incubate at 4°C for 5 min. Remove the buffer again and repeat buffer B wash and removal.
Add 1.0 mL of buffer C and incubate at 37°C for 4 h to block free tosyl groups. Pull down beads using magnet, remove buffer, add 1.0 mL of buffer B and incubate at 4°C for 5 min.
Remove buffer, resuspend beads in 1.0 mL of buffer B and aliquot for use. Beads can be stored at 4°C for several months without loss of quality.
3.4 Experimental Setup
Once the DNA and functionalized beads have been prepared and microscope coverslips have been functionalized, a flow cell can be assembled and single-molecule experiments performed. Here we describe how a flow chamber is prepared with the functionalized coverslip and a quartz slide, and how the substrate is constructed in situ by flowing λ DNA fork substrates and functionalized beads.
Immediately before assembling the flow cell, incubate a functionalized coverslip with streptavidin solution, 25 μL (1 mg/mL) in 100 μL of PBS. Spread the solution across the surface and leave at room temperature for 30 min. During incubation, place the slip in a humid atmosphere to prevent drying out of the solution. A simple way to achieve this is to place the slip in a covered and empty pipette-tip box, with some water on the bottom of the box to keep the air saturated with moisture.
Dilute 5 mL of 5X blocking buffer into 20 mL of working buffer. Degas the solution by placing it, with loosely screwed cap, in a vacuum desiccator.
Create the flow cell by cutting the desired shape of the channel in a double-sided tape with external dimensions of 2 × 5 cm to match the quartz size. For experiments with beads, it is advisable to use two inlet and two outlet channels, as the possibility to switch inlets after flowing beads reduces the amount of washing needed to remove free beads from the tubing (see Figure 1a). The height of the chamber is set by the thickness of the tape, but the channel width is variable. Our experiments use a 3.0 mm wide channel with a height of 100 μm and are cut into a double Y shape, with two holes drilled 8 mm apart for both inlet and outlet pairs. The pattern of the channel should be aligned with the holes drilled in the quartz slide simply by marking the hole position on the tape with a pencil. The slide should be cleaned thoroughly prior to placement of the channel cutout.
Holes in the slide should be only slightly bigger than the outer diameter of the tubing, just loose enough to allow the tube to slide if pushed through the hole. Cut tubing to desired length based on the distance from the microscope to the pump. Also, cut the end of the tube that will be inserted through the quartz into the chamber at a ~30° angle. This angled cut prevents the face of the tube to sit too tightly against the flow-cell surface and block the flow. After cutting the chamber pattern and applying it to the quartz, dust should be removed with compressed air. Using plastic forceps, apply slight pressure to the tape after adhering in order to rub out any bubbles and fully seal the border of the chamber.
Wash the streptavidin-coated coverslip thoroughly in water and fully dry using compressed air. Once dry, the next steps should be done quickly to minimize air exposure and avoid surface degradation. Remove the backing from the tape/quartz and press onto the functionalized surface of the coverslip and apply slight pressure onto the thin cover slip by rubbing with plastic forceps to form a complete seal. Apply a quick-dry epoxy to the interface between the slide and coverslip, forming a seal around the outer edges of the chamber.
Insert the four cut tubes into the holes, supporting the tube length with a taller object (e.g. test tube rack) in order for the tubes to be placed vertically into the holes. Apply epoxy around each tube inserted and let dry to seal tube in hole.
Once the epoxy has dried, place the two outlet and one of the inlet tubes into degassed blocking buffer and insert a 21-gauge needle (or other size depending on tube inner diameter) attached to a 5 mL syringe into the second inlet tube. Slowly draw buffer into the syringe, checking that all tubes permit flow. Expel/draw buffer 2–3 times and leave flow cell full of buffer for 20 min to block surface.
After incubation, the flow cell is ready for use in experiments. Attach both outlet tubes to the inlet tubes of the airspring (see Note 3). Block end of one inlet tube by inserting a needle previously filled with epoxy. Clear air from tubes by manually pulling on syringe pump drive arm and flicking the outlet tubes. Switch inlet tubes and repeat. Before flowing DNA, block one outlet tube by kinking to create a flow path from left inlet to right outlet or right inlet to left outlet, facilitating a laminar flow profile.
Flow DNA into flow cell at desired rate and concentration (0.3–2 μL of DNA stock diluted in 1–1.5 mL of working buffer should be sufficient for several tens to hundreds of tethered beads per field of view). Slower flow rates allow for higher surface binding efficiency.
Dilute beads (2–3 μL stock into 1 mL of blocking buffer) and mix thoroughly by vortexing, then sonicate for 30 s to disrupt any bead aggregates. Flow beads into cell based at a flow rate of ~1 mL/hr. Since the beads are large enough to settle on the surface of the flow cell, periodically tap the microscope stage to ensure that the unbound beads will continue to slowly move through the flow cell.
Once beads are added, switch inlet tubes, being sure to close both outlet tubes. Any change to the tubes without the chamber being closed to pressure fluctuations (i.e. if an outlet is open) will exert a strong force on the beads and shear any tethered DNA. Begin to wash flow cell extensively, manually agitating the stage by tapping to remove any beads nonspecifically stuck to the surface.
Tethered DNA can be seen by gentle agitation of outlet tube or lifting of airspring to see bead movement based on flow direction. Once free beads are sufficiently removed, the enzymatic reaction can be performed.
3.5 Experiment and Data Analysis
Solutions containing replication proteins are introduced into the flow cell and any bead movement is observed in real time by imaging the tethered bead positions with a CCD camera. Movement of the beads is converted to length change of the DNA, allowing temporal and kinetic analysis of single replication events. Several methods to relate DNA length changes to replication can be employed. In the first method, we make use of the fact that at stretching forces lower than 6 pN, DNA in the single-stranded form is considerably shorter than DNA in the duplex form (Figure 1d). In the case of leading-strand synthesis, only one of the two unwound parental DNA strands will be converted to duplex DNA. In the absence of lagging-strand synthesis, the other strand will remain in the single-stranded form after helicase-mediated unwinding at the fork. By attaching the 5′ lagging strand of the DNA to the surface, we can observe leading-strand synthesis by the effective conversion of parental duplex DNA into single-stranded DNA, resulting in a shortening of the DNA. The second method is employed in those reactions with both leading- and lagging-strand synthesis. Here, no net conversion between single- and double-stranded DNA takes place, but the transient formation of a replication loop at the lagging strand can be observed as a brief and gradual shortening of the DNA, followed by a sudden lengthening. These two events correspond to the formation and release of a replication loop, respectively.
Enzyme solutions should be prepared in degassed buffers to avoid introduction of air bubbles to the flow cell. Once flow cell is ready for reaction, stop flow and allow chamber to reach equilibrium. Close outlet tube and switch the inlet reservoir to protein solution, slowly opening the outlet to avoid rapid pressure changes. Apply the magnet at measured distance above the flow cell before restarting flow (see Note 4).
For leading-strand synthesis experiments with the T7 replisome, we preassemble the helicase, gp4, and polymerase, gp5 with thioredoxin on the DNA in the presence of dNTPs but in the absence of Mg2+, a cofactor required for helicase activity but not assembly (2, 3). We prepare a 20 nM solution of the proteins in T7 replication buffer 1 at room temperature and wash free proteins with buffer 1 alone. After washing, we flow replication buffer 2 and begin data acquisition. Alternatively, as in our E. coli experiments, the proteins can be flowed continuously during data acquisition (4). This can lead to multiple replication events on a single substrate, but allows for data collection even with less robust or stable replication systems. We flow a solution of all leading-strand proteins (αεθ, DnaB6, DnaC6, τ1γ2δδ′ β2) at 30 nM in E. coli replication buffer.
During acquisition, use a flow rate corresponding to a low pN drag force (1–5 pN) on the DNA tethers to take advantage of the ssDNA-dsDNA length disparity. Based on the pump, tubing, bead and flow cell size described above, 3 pN drag force corresponds to a flow rate of 0.0125 mL/min, but will vary if parameters, such as syringe diameter, flow cell volume or bead size are changed.
View field with CCD camera and focus using tethered bead (see Note 5). Data is acquired for 20–30 min at a slow frame rate (typically 2–4 Hz) and transferred to tracking software of choice, either purchased or designed in house.
Trajectories are extracted by tracking bead position throughout course of experiment and exporting as text file. Several commercial packages exist to track particles (MetaVue™ from Molecular Devices and DiaTrack from Semasopht are two suitable packages).
Using graphing and analysis software (Origin®, OriginLab), visualize trajectories by plotting bead position versus time. For increased accuracy, subtract from the traces of interest a baseline trace of a bead tether that is not enzymatically altered (Figure 2). To determine enzymatic processivity, use the Δy from start to end of a shortening event. For rate calculation, fit the event itself with a linear regression and take the Δy/Δx (slope) of the line as a measure of base pairs/second.
Once the experiment is complete, the drilled quartz slide can be reused after soaking the intact flow cell in acetone, which will dissolve the tape and epoxy from the slide.
Acknowledgments
The authors would like to thank Charles Richardson and Nick Dixon for their generous gifts of T7 and E. coli replication proteins, respectively. The authors would also like to acknowledge contributions from Paul Blainey, Candice Etson, Jong-Bong Lee, and Joseph Loparo towards the development of the single-molecule replication assay.
Footnotes
When used in a single-molecule experiment, observation of tethered λ DNA serves as an internal control that the substrate is assembled properly. As the biotinylated oligo does not complement the λ itself but rather the other fork arm, if DNA is attached to the surface it immediately shows correct fork assembly. Any bead attached similarly confirms annealing of the digoxigenin oligo. As a caveat, the fork oligos are present at a high concentration and can anneal without the λ DNA to form small forks which can bind to the surface. This is typically not a matter of concern, but for troubleshooting or adapting the technique to higher resolution experiments or other protein systems this fact should be considered. Eliminating the excess forks is simply a matter of reducing the oligonucleotide ratios in substrate construction or purifying the free oligos away after preparation of the DNA constructs.
The PEG powders and silane solutions are extremely sensitive to hydrolysis, and care must be taken to prevent their degradation. Immediately after use, place containers in a desiccator and remove air. Replace with inert atmosphere (N2 or Ar) and seal lid with plastic wrap until next use. Batches of functionalized coverslips that display poor tethering capability are frequently due to degraded reagents.
Syringe pumps often exhibit small irregularities in flow, resulting in significant force fluctuations. A simple way to reduce these flow instabilities is to place an airspring between the flow cell and the syringe pump (see Figure 1b). A 50 ml plastic tube is sealed and the lid affixed using epoxy. Three holes are pierced in the lid, and three lengths of tubing (same tubing as flow cell) are inserted to ~1 cm from the bottom. Using epoxy, the tubes are sealed to the lid, preventing any air from entering or escaping. The tube is filled with 40–45 mL water and connected to the syringe pump with one of the three tubing pieces. The remaining two connect to the flow cell tubing using an adaptor piece of slightly larger tubing. Upon starting the syringe pump, the withdrawal of water from the air spring will result in a pressure drop in the closed air volume. This negative pressure will cause buffer to flow through the flow cell. Any irregularity in the syringe pump will not immediately change the negative pressure in the air spring and will be dampened out very effectively. The airspring provides two additional benefits: 1) a simple method of connecting the two outlet channels to a single syringe pump, and 2) an easy way of changing the flow direction. By lifting the airspring, gravity will force the flow to reverse direction and cause the bead-DNA tethers to flip back and forth with the flow.
A common problem in the experiment occurs when the large beads nonspecifically stick to the surface of the coverslip, preventing movement and measurement. As a solution, we apply a small magnetic force (~1.7 pN) perpendicular to the flow direction to lift the beads off the surface. Permanent rare-earth magnets are moved into place above the flow cell using a 2-axis translational stage (Thor Labs) immediately prior to data acquisition.
Dark-field illumination can be used to increase the contrast in the bead imaging. A fiber illuminator (Thor Labs OSL1) is positioned at an incidence angle between 10 degrees and parallel to the microscope stage ~0.5m away. The low numerical aperture of the 10X objective will not allow the illumination light to be transmitted, but will allow the light scattered by the beads to be imaged. As a result, the beads can now be seen as bright objects against a dark background.
References
- 1.Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu Rev Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 2.Lee JB, Hite RK, Hamdan SM, Xie XS, Richardson CC, van Oijen AM. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–4. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- 3.Hamdan SM, Johnson DE, Tanner NA, et al. Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. Mol Cell. 2007;27:539–49. doi: 10.1016/j.molcel.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 4.Tanner NA, Hamdan SM, Jergic S, Schaeffer PM, Dixon NE, van Oijen AM. Single-molecule studies of fork dynamics in Escherichia coli DNA replication. Nat Struct Mol Biol. 2008;15:170–6. doi: 10.1038/nsmb.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sofia SJ, Premnath VV, Merrill EW. Poly(ethylene oxide) Grafted to Silicon Surfaces: Grafting Density and Protein Adsorption. Macromolecules. 1998;31:5059–70. doi: 10.1021/ma971016l. [DOI] [PubMed] [Google Scholar]